Abstract
Background
NLPR3 is an important gene that belongs to the family of NOD-like receptors and is thought to play an important role in psoriasis. The aim of the present study was to investigate the expression of NLRP3 in psoriasis biopsy samples and to assess the possible correlation of its expression with that of interleukin IL-1β and Caspase-1.
Material/Methods
The mRNA expression was checked by qRT-PCR. The expression of the proteins was checked by Western blotting. The distribution of the proteins was determined by immunohistochemistry.
Results
The results of our study indicate that the expression of NLRP3 was significantly upregulated in all the psoriatic biopsy samples as indicated by qRT-PCR and Western blotting. The expression of NLRP3 in psoriatic samples was 3.5 to 4.3 times higher than the expression of NLRP3 in normal skin biopsy samples. Moreover, our results indicated that the expression levels of IL-1β were higher as compared to the normal skin biopsy samples. Relative to the expression of IL-1β in normal skin biopsy samples, the expression of IL-1β was about 2.7–4.6 times higher. Additionally, the expression of caspase-1 was considerably upregulated in the psoriatic samples. Caspase-1 gene expression was 2.2–3.4 times higher than in normal skin biopsy samples.
Conclusions
NLPR3 may prove to be an important therapeutic target for psoriasis.
MeSH Keywords: Arthritis, Psoriatic; Blotting, Far-Western; Gene Expression Profiling
Background
Psoriasis is one of the most prevalent skin diseases and affects about 3% of the world population. It is characterized by clearly delineated erythematous plaques with associated silvery scales. It is also associated with extreme keratinocyte proliferation, which leads to thickening of the epidermis [1]. It has been reported that psoriasis can be prompted by the impairment of T cell-arbitrated immune response, ultimately causing epidermal hyperplasia, penetration of leukocytes into dermal tissues, and proliferation of blood vessels [2]. The exact cause of psoriasis is not fully known. However, it is believed that genetic history of the individual and environmental factors have important roles in the onset of this chronic skin disease [3]. Normally, psoriasis is less prevalent in children and more often affects adults [4]. The treatment of psoriasis has been challenging for clinicians because it is a very a complex disease and the available treatment options are limited. The selection of a suitable treatment for psoriasis is difficult and must always take into consideration the appearances and the location of the psoriatic lesions and the extent to which the tissue is involved [5]. Previous studies have reported that about 75% of psoriasis patients exhibit slight severity, generally with about less than 20% of the body affected. Patients with mild psoriasis can be treated with topical therapy with satisfactory results [6]. However, complete disappearance of the lesions is still not possible with topical therapy. Medications like combination tazarotene gel, calcipotriol, and betamethasone achieve only 2–45% clearance of psoriatic lesions. Moreover, these therapies are associated with adverse effects that decrease quality of life [7]. The heterogeneity of the adverse effects of the therapies among patients poses further challenges in the selection of treatments for psoriasis [8].
Therefore, knowledge of factors responsible and the genetic basis for the onset of the disease could prove crucial in the development of treatment strategies for psoriasis. NLRP3 (NACHT, LRR, and PYD domains-containing protein 3) is an important protein, often referred to as cryopyrin [9]. NLPR3 belongs to the family of NOD-like receptors and is activated by microbial toxins, their cell wall components, ultraviolet radiation (UV), and reactive oxygen species. It is the basic component of the innate immune system [10]. Previous studies have showed the role of NLPR3, caspase 1, and interleukin IL-1β (IL-1β) in psoriasis [11]. The present study was designed to investigate the expression of NLPR3 in psoriasis biopsy samples and to assess the possible correlation of its expression with that of interleukin IL-1β and caspase 1. Our results revealed that the expression of NLRP3 was positively associated with the expression of both IL-1β and caspase 1. Therefore, we believe that NLPR3 may prove to be good therapeutic target for the management of psoriasis.
Material and Methods
Collection of biopsy samples
For the collection of skin biopsy samples, biopsies were obtained from untreated psoriasis patients and as well as the normal skin tissues of healthy volunteers. Informed consent was obtained from all patients under protocols approved by the Institutional review board of Wuhan No. 1 Hospital, Wuhan, Hubei Province, P. R. China under approval number VCXBB21458/2017. For carrying out the immunohistochemistry analyses, both the normal and psoriatic biopsy samples were administrated with formalin and embedded in paraffin. The samples were frozen in liquid nitrogen and stored at −80°C for further experimentation.
RNA isolation and quantitative RT-PCR
RNA was isolated by use of the RNeasy RNA isolation kit (Qiagen) as per the manufacturer’s instructions. Absorbance of RNA samples was determined using a NanoDrop®ND-1000 spectrophotometer (NanoDrop Technologies). RNA quality was assessed by separation on 1% agarose gel and by the optical density (OD) ratios at 260 nm/280 nm. This was followed by treatment with DNase I (Fermentas) to eliminate genomic DNA contamination. cDNA was synthesized using the RevertAid cDNA synthesis kit (Fermentas) following the manufacturer’s instructions. For real-time PCR, the cDNA was diluted 10 times and qRT-PCR was performed in triplicates using ABI StepOne Real-Time (Applied Biosystems), SYBR Green Master Mix (Fermentas), and gene specific primers. The relative quantification method (ΔΔ-CT) was used to evaluate quantitative variation between the replicates examined. The amplification of actin was used as an endogenous control to normalize all data.
Analysis of protein expression by Western blotting
For analysis of protein expression, the skin biopsy samples were lysed and the total protein extracts were collected. Equal protein extracts from each group were run on SDS PAGE and then transferred to a polyvinylidene fluoride membrane. This was followed by blocking with 5% non-fat milk and incubation at 25°C for 1 h. Thereafter, the membranes were incubated with a specific primary antibody (NLPR3, caspase 1, and IL-1β) at 4°C overnight. This was followed by washing in washing buffer and incubation for 1 h with the suitable secondary antibody. The protein bands of interest were visualized using the ECL Advanced Western Blot Detection Kit.
Statistical analysis
The experiments were carried out in triplicates and the data are presented as mean ±SD.
Statistical analysis was done by using the t test (for comparisons between 2 samples) and one-way ANOVA followed by Tukey’s test (for comparison between more than 2 samples) using GraphPad Prism 7 software. The values were considered significant at * p<0.01.
Results
NLRP3 was enhanced in psoriatic biopsy samples
NLPR3 is an important component and its expression is often upregulated by several biotic and abiotic stresses. To examine the status of NLRP3 in psoriasis, the cDNA synthesized from RNA isolated from psoriatic and normal skin biopsy samples was subjected to quantitative PCR. The results of our study indicated that the expression of NLRP3 was significantly upregulated in all the psoriatic biopsy samples. The increase in the expression of NLRP3 in psoriatic samples was 3.5–4.3 times higher than the expression of NLRP3 in normal skin biopsy samples (Figure 1). To further validate the expression of NLPR3, we examined its expression by Western blotting (Figure 2). The results showed that the protein expression of NLRP3 was also upregulated and followed the same trend as that of the quantitative RT-PCR expression.
Figure 1.
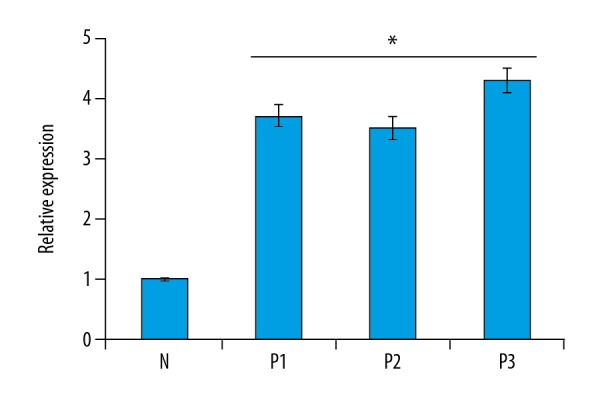
Expression of NLRP3 gene in normal (N) and psoriatic biopsy samples (P1, P2, and P3) as determined by quantitative RT-PCR analysis. The experiments were carried out in triplicates and the data are presented as mean ±SD. The values were considered significant at * p<0.01.
Figure 2.
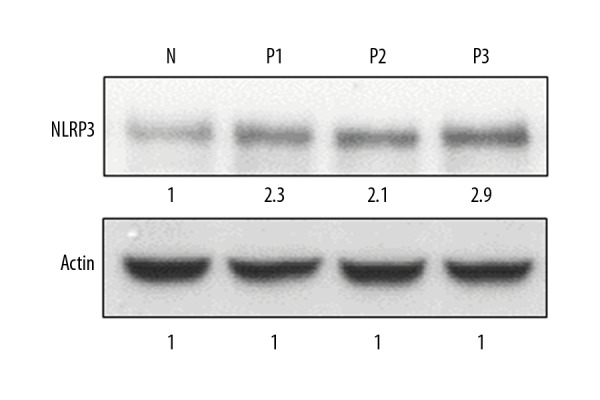
Expression of NLRP3 protein in normal (N) and psoriatic biopsy samples (P1, P2, and P3) as determined Western blotting. The experiments were carried out in triplicates.
NLRP3 expression was associated upregulation of with IL-1β
We further examined the expression of IL-1β in both the psoriatic and normal skin biopsy samples by quantitative RT-PCR. Our results indicated that the expression levels of IL-1β were higher as compared to the normal skin biopsy samples (Figure 3). Relative to the expression of IL-1β in normal skin biopsy samples, the expression of IL-1β was about 2.7–4.6 times higher. The analysis of expression of IL-1β by Western blotting also showed higher expression of IL-1β in psoriatic biopsy samples as compared to the normal skin biopsy samples (Figure 4). Taken together, these results show that the expression of IL-1β exhibited a similar trend as that of NLRP3.
Figure 3.
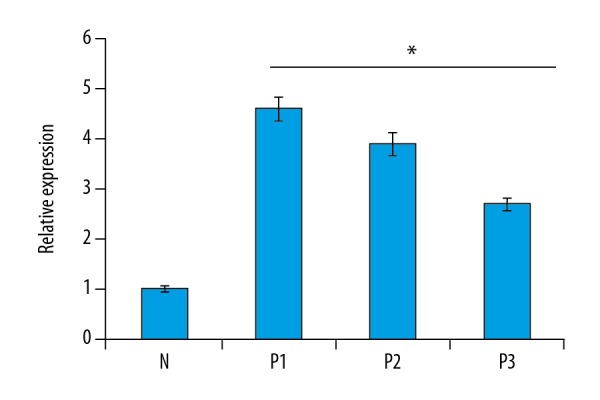
Expression of IL-1β gene in normal (N) and psoriatic biopsy samples (P1, P2, and P3) as determined by quantitative RT-PCR analysis. The experiments were carried out in triplicates and the data are presented as mean ±SD. The values were considered significant at * p<0.01.
Figure 4.
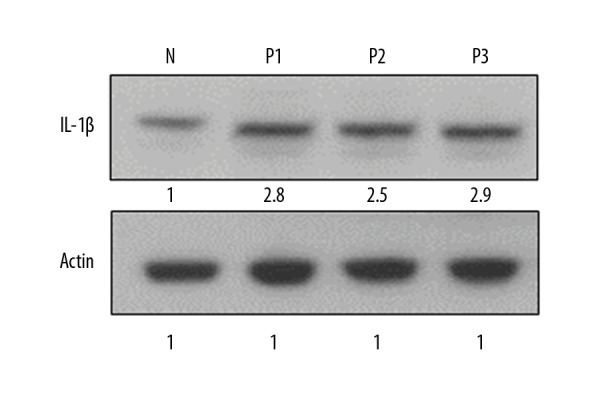
Expression of IL-1β protein in normal (N) and psoriatic biopsy samples (P1, P2, and P3) as determined by Western blotting. The experiments were carried out in triplicates and the data are presented as mean ±SD. The values were considered significant at * p<0.01.
NLRP3 expression was positively associated with the caspase-1 expression
Next, we examined the expression of caspase-1 gene in psoriatic and in normal skin biopsy samples by quantitative RT-PCR. The results again revealed that the expression of caspase-1 was considerably upregulated in the psoriatic samples. The increase of caspase-1 gene expression was 2.2–3.4 times higher than in normal skin biopsy samples (Figure 5). To confirm if mRNA of caspase-1 gene detected by RT-PCR is really converted into protein, we carried out Western blotting analysis. The results showed that the expression of caspase-1 was also upregulated at the protein level and followed the same trend as that of NLRP3 and IL-1β (Figure 6).
Figure 5.
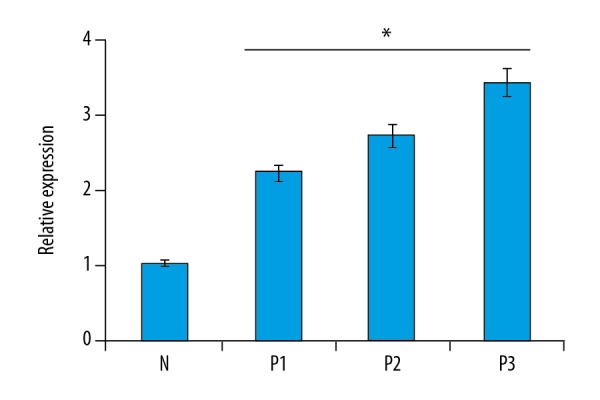
Expression of caspase-1 gene in normal (N) and psoriatic biopsy samples (P1, P2, and P3) as determined by quantitative RT-PCR analysis. The experiments were carried out in triplicates and the data are presented as mean ±SD. The values were considered significant at * p<0.01.
Figure 6.
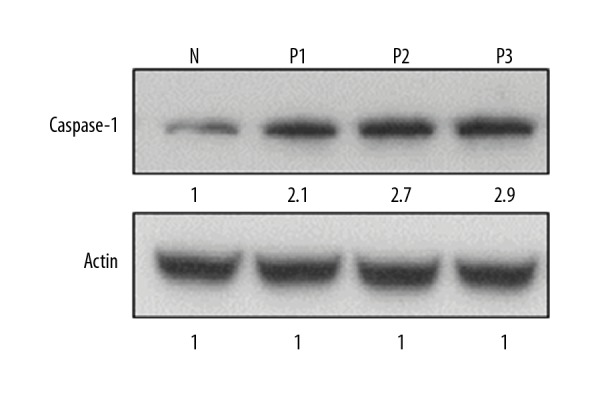
Expression of caspase-1 protein in normal (N) and psoriatic biopsy samples (P1, P2, and P3) as determined by Western blotting. The experiments were carried out in triplicates and the data are presented as mean ±SD. The values were considered significant at * p<0.01.
Discussion
Psoriasis is a chronic inflammatory skin disorder affecting about the 3% of the human population across the world [1]. However, studies have reported that the incidence of psoriasis may vary with race, geographic locality, and environmental conditions [1,2]. Psoriasis has a broad clinical spectrum, ranging from the involvement of skin to erythrodermia [12]. The cause of the onset of psoriasis has not been defined. Moreover, the treatment options are limited and have a number of adverse effects. It is also associated with extreme keratinocyte proliferation, which leads to thickening of the epidermis [1]. It has been reported that psoriasis can be prompted by the impairment of T cell-arbitrated immune response, ultimately causing epidermal hyperplasia, penetration of leukocytes into dermal tissues, and proliferation of blood vessels [2]. The exact cause of psoriasis is not fully known. However, it is believed that genetic history of the individual and environmental factors have important roles in the onset of this chronic skin disease [3]. NLRP3 is an important gene that forms an important component of the innate immune system. It belongs to the NOD-like receptor family. Several studies have reported that the expression of NLPR3 is upregulated in response to stress stimuli [13]. In the present study we examined the expression of NLPR3 in psoriatic and normal skin biopsy samples. Our results clearly showed that the expression of NLPR3 was significantly (p<0.01) upregulated in psoriatic skin biopsy samples as compared to the normal skin biopsy samples. Our results agree with several other report that NLPR3 exhibits enhanced expression in psoriatic skin samples and in murine models of psoriasis [14,15]. Further, we examined the expression of interleukin IL-1β in both the normal and the psoriatic samples, finding that IL-β showed the same expression as that of NLPR3. These results confirm the reports that the inflammasome plays an important role in auto-inflammatory disorder pathogenesis.
Our results are also supported by previous investigations wherein in IL-1β has been reported to be upregulated in psoriatic lesions [16]. Since there is concrete evidence that IL-1β is activated by caspase-1, we therefore examined the expression of caspase-1 in both the psoriatic and normal skin samples. We found that the expression of caspase-1 exhibited similar expression as that of IL-1β and NLPR3, both at the mRNA level and the protein level. As IL-1β is secreted from epithelial cells, it prompts T cell-dependent skin inflammation [17]. Additionally, caspase 1 and the subsequent enhanced IL-1 signaling cause upregulation of the Th17 cytokine profile and a psoriasis-like phenotype in mice [18]. The results of the present study reveal that NLPR3 is significantly upregulated in psoriatic tissues and its expression is associated with upregulation of caspase-1. Caspase-1 subsequently causes upregulation of IL-1β. The altered expression of these proteins indicates their potential role in psoriasis. Further research is needed to determine whether these proteins can be targeted for the treatment of psoriasis.
Conclusions
We conclude that NLRP3 expression is upregulated in psoriasis and is associated with concomitant accumulation of IL-1β and caspase 1 and may therefore prove to be an important therapeutic target for psoriasis.
Footnotes
Source of support: Scientific Research Project Foundation of Health and Family Planning Commission of Wuhan Municipality (NO. WX16B03)
References
- 1.Lowes MA, Suárez-Fariñas M, Krueger JG. Immunology of psoriasis. Ann Rev Immunol. 2014;32:227–55. doi: 10.1146/annurev-immunol-032713-120225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harden JL, Krueger JG, Bowcock AM. The immunogenetics of psoriasis: A comprehensive review. J Autoimmun. 2015;64:66–73. doi: 10.1016/j.jaut.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim J, Krueger JG. The immunopathogenesis of psoriasis. Dermatol Clin. 2015;33(1):13–23. doi: 10.1016/j.det.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Strange A, Capon F, Spencer CC. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 2010;42:985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellinghaus E, Ellinghaus D, Stuart PE, et al. Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2. Nat Genet. 2010;42:991–95. doi: 10.1038/ng.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stuart PE, Nair RP, Ellinghaus E. Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat Genet. 2010;42:1000–4. doi: 10.1038/ng.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huffmeier U, Uebe S, Ekici AB, et al. Common variants at TRAF3IP2 are associated with susceptibility to psoriatic arthritis and psoriasis. Nat Genet. 2010;42:996–99. doi: 10.1038/ng.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng Y, Chang C, Lu Q. The inflammatory response in psoriasis: A comprehensive review. Clin Rev Allergy Immunol. 2016;50(3):377–89. doi: 10.1007/s12016-016-8535-x. [DOI] [PubMed] [Google Scholar]
- 9.Diani M, Altomare G, Reali E. T cell responses in psoriasis and psoriatic arthritis. Autoimmun Rev. 2015;14(4):286–92. doi: 10.1016/j.autrev.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Lee HJ, Hong YS, Yang SJ. Interaction between NLRP3 inflammasome and Sirt1/6: Metabolomics approach. FASEB J. 2015;29 Abstract: 913.12. [Google Scholar]
- 11.Gurung P, Kanneganti TD. Novel roles for caspase-8 in IL-1β and inflammasome regulation. Am J Pathol. 2015;185(1):17–25. doi: 10.1016/j.ajpath.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bronckers IM, Paller AS, Van Geel MJ, et al. Psoriasis in children and adolescents: Diagnosis, management and comorbidities. Pediatric Drugs. 2015;17(5):373–84. doi: 10.1007/s40272-015-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank MG, Hershman SA, Weber MD, et al. Chronic exposure to exogenous glucocorticoids primes microglia to pro-inflammatory stimuli and induces NLRP3 mRNA in the hippocampus. Psychoneuroendocrinology. 2014;40:191–200. doi: 10.1016/j.psyneuen.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaipiainen-Seppanen O, Punnonen K, van Gijn ME, Mononen T. Two pathogenic CIAS1 mutations and plasma cytokine profile in a Finnish patient with familial cold autoinflammatory syndrome responsive to anakinra. Scand J Rheumatol. 2008;37:75–76. doi: 10.1080/03009740701691491. [DOI] [PubMed] [Google Scholar]
- 15.Ślusarczyk J, Trojan E, Wydra K, et al. Beneficial impact of intracerebroventricular fractalkine administration on behavioral and biochemical changes induced by prenatal stress in adult rats: Possible role of NLRP3 inflammasome pathway. Biochem Pharmaco. 2016;113:45–56. doi: 10.1016/j.bcp.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Hanus JW, Abu-Asab MS, et al. NLRP3 upregulation in retinal pigment epithelium in age-related macular degeneration. Int J Mol Sci. 2016;17(1) doi: 10.3390/ijms17010073. pii: E73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villani AC, Lemire M, Fortin G, et al. Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat Genet. 2009;41:71–76. doi: 10.1038/ng285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts RL, Topless RK, Phipps-Green AJ, et al. Evidence of interaction of CARD8 rs2043211 with NALP3 rs35829419 in Crohn’s disease. Genes Immunol. 2010;11:351–56. doi: 10.1038/gene.2010.11. [DOI] [PubMed] [Google Scholar]


