Abstract
Background
Drylands cover nearly 41% of Earth’s land surface and face a high risk of degradation worldwide. However, the actual timeframe during which dryland floras rose on a global scale remains unknown. Zygophyllaceae, an important characteristic component of dryland floras worldwide, offers an ideal model group to investigate the diversification of dryland floras. Here, we used an integration of the phylogenetic, molecular dating, biogeographic, and diversification methods to investigate the timing and patterns of lineage accumulation for Zygophyllaceae overall and regionally. We then incorporated the data from other dominant components of dryland floras in different continents to investigate the historical construction of dryland floras on a global scale.
Results
We provide the most comprehensive phylogenetic tree for Zygophyllaceae so far based on four plastid and nuclear markers. Detailed analyses indicate that Zygophyllaceae colonized Africa, Asia, Australia, and the New World at different periods, sometimes multiple times, but Zygophyllaceae lineages in the four regions all experienced a rapid accumulation beginning at the mid-late Miocene (~ 15–10 Ma). Other eleven essential elements of dryland floras become differentiated at the same time.
Conclusions
Our results suggest that the rise of global dryland floras is near-synchronous and began at the mid-late Miocene, possibly resulting from the mid-Miocene global cooling and regional orogenetic and climate changes. The mid-late Miocene is an essential period for the assembly and evolution of global dryland floras.
Electronic supplementary material
The online version of this article (10.1186/s12862-018-1277-z) contains supplementary material, which is available to authorized users.
Keywords: Drylands, Climate change, Divergence time, Diversification rate, Miocene, Phylogeny
Background
Drylands form nearly 41% of Earth’s land surface [1] and are by definition areas where precipitation is scarce (≤ 250 mm/y) and typically of high variability [2, 3]. They are mainly distributed in tropical and subtropical dryland zones (0–30° latitude), occurring in Africa, Australia, and the New World, and in temperate dryland zones (> 30° latitude), largely limited to the Asian interior and western North America [4]. These dryland regions are home to more than 38% of the world’s population [5] and harbor rich biodiversity [4, 6]. Due to climate change and human activity, drylands worldwide face a high risk of degradation [3, 7]. The remarkable characteristic of dryland floras is a tight coupling of water availability and vegetation dynamics [8]. Compared with our knowledge regarding tropical/temperate forest and arctic regions [9–11], information about the main evolutionary processes underlying species richness in dryland floras is limited.
Geologic sediments indicate that the first onset of arid conditions in Africa and the New World occurred during the early Tertiary period [12, 13]. The aridification in the Asian interior first occurred near the Eocene-Oligocene boundary ~ 34 million years ago (Ma) [14]. Aridification in Australia began in the mid-Miocene, ~ 15 Ma [15]. Thus, the initial aridity in different continents was non-synchronous, which could suggest that the establishment of modern dryland floras in different continents was non-contemporaneous. The global cooling process between the Miocene and Pliocene also enhanced aridity on a global scale [13, 16]. The relation between the evolution of dryland floras and the cooling was not well understood.
In recent years, increasing numbers of studies have focused on the evolutionary history of arid-adapted plants, contributing greatly to our understanding of the assembly and evolution of dryland floras. Southern African ice plants (Aizoaceae), an important and characteristic component in the succulent biome of Africa, are dated to ~ 26 Ma and have diversified rapidly between 8.7 to 3.8 Ma [17]. Agave (Agavaceae), keystone plants of semiarid to arid regions in North American deserts, originated ~ 10 Ma and experienced two pulses of increased diversification: 8–6 Ma and 3–2.5 Ma [18]. The seven major lineages of Tiquilia (Boraginaceae), another diverse North American desert plant group, became differentiated ~ 23–13 Ma and experienced a marked increase in diversification beginning ~ 7 Ma [19]. Cacti (Cactaceae), the most spectacular New World succulent plants, originated ~ 35 Ma, but their species-rich clades originated in the late Miocene (~ 10–5 Ma) [20]. Central Asian Reaumuria (Tamaricaceae) began to diverge at the Eocene-Oligocene boundary and its two sections diversified in the early Miocene (22.5–19.8 Ma) [21]. In Australia, arid-adapted Camphorosmeae (Chenopodiaceae) became differentiated in the late Miocene (~ 7 Ma) [22]. These studies indicate that the timing of diversification of arid-adapted plant groups in different continents might have been inconsistent, but the period between the Miocene to the Pliocene was vital for the establishment of dryland floras globally. The majority of current studies focused on the evolutionary history of local arid-adapted plant groups, and did not investigate the evolution of dryland floras from a global view. Moreover, organismal capacities for adapting to novel ecological conditions can differ markedly [23]. To better understand the evolutionary processes that contributed to species richness in the different dryland floras, the diversification patterns of arid-adapted plant groups must be investigated in a broader phylogenetic context.
As an important representative in drylands [24, 25], Zygophyllaceae offers an opportunity for studying the diversification of dryland floras on a worldwide basis. This family consists of 22 genera and ~ 285 species, which are distributed throughout drylands of the world with a few extending to neighboring regions [25]. Zygophyllaceae plants are among the most important and characteristic components on the basis of their contribution to the flora and impact on the environment [25, 26]. For example, Zygophyllum and Fagonia are distributed widely in African dryland and are regarded as major components of the flora [27]; Creosotebush (Larrea tridentata) is one of the most dominant species in the New World dryland [24]. Morphological and anatomical features indicate that Zygophyllaceae plants can use water efficiently and are well adapted to dryland habitats [28, 29]. Additionally, Zygophyllaceae is one of 19 angiosperm families that use the C4 photosynthetic pathway [30], which is advantageous under the threat of extreme conditions (e.g. drought, sun, and high temperature) [31]. According to a large-scale molecular dating analysis in angiosperms, Zygophyllaceae originated in the early Paleocene [32].
In this study, we reconstruct a large-scale phylogenetic tree for Zygophyllaceae using four DNA markers with the most extensive taxon sampling to date. Within the large-scale phylogenetic framework, we then estimate the timing and patterns of lineage accumulation of Zygophyllaceae on global and regional scales. Finally, we investigate whether the rise of dryland floras in different continents were synchronous.
Methods
Molecular sequence data
One protein-coding gene (rbcL) and two noncoding regions (trnL intron and trnL-F intergenic spacer) of plastid DNA and one noncoding region, the internal transcribed spacers (ITS) of nuclear DNA were used for phylogenetic analysis. DNA sequences for all taxa were obtained from GenBank (www.ncbi.nlm.nih.gov). Accession numbers are listed in Additional file 1: Table S1. A total of 157 species from 21 of the 22 genera of Zygophyllaceae were sampled, with an additional 7 outgroup species from other rosids. Only the monotypic genus Metharme was not included because material was not available. At the species level, 55% of the currently 285 recognized species were included. The sequences of each marker were aligned and manually adjusted in BioEdit v.7.0.1 [33].
Phylogenetic analysis
Phylogenetic analyses were initially performed for each marker using maximum likelihood (ML) method in RAxML [34]. The bootstrap support for conflicting nodes showed no significance among the individual markers (considered to exceed 70%), and the four individual datasets were therefore combined. Detailed analyses were carried out using ML and Bayesian inference (BI) methods. The best-fit model of nucleotide substitutions for each DNA region was determined by the Akaike information criterion in jModelTest v.2.1.4 [35], as follows: GTR + Γ for ITS and GTR + I + Γ for rbcL, trnL, and trnL-F. The parameters of the best-fit model for each DNA region are listed in Additional file 1: Table S2. RAxML was conducted using 1000 replicates with the fast bootstrap option. Bayesian analyses were carried out in MrBayes v.3.2.2 [36]. Two independent runs with each comprised four Markov Chain Monte Carlo (MCMC) chains (one cold and three heated) were conducted, starting from random trees. Chains were run for 30 million generations, sampling one tree every 1000 generations. The stationarity of the runs was assessed using Tracer v.1.6 [37]. A majority rule (> 50%) consensus tree was constructed after removing a burn-in of the initial 25% of the sampled trees, and the posterior probability was used to evaluate nodal robustness.
In order to assess the impact of missing data on phylogenetic reconstruction, we generated three additional matrixes of Zygophyllaceae: matrix 1 including 22 taxa, each taxon with four loci (22 taxa, 4 markers), matrix 2 (76 taxa, 3–4 markers), and matrix 3 (142, 2–4 markers). These three matrices were analysed in RAxML. The results indicated that the backbone of Zygophyllaceae generated from the dataset with 164 taxa (Additional file 2: Figure S1) is highly congruent with those obtained from the above three matrices (Additional file 2: Figure S2) except for nodes with poor support. The dataset with 164 taxa (each taxon with 1–4 markers) was therefore used in followed analyses.
Divergence time estimates
Divergence times were estimated in BEAST v.1.8.1 [38], which employs a Bayesian MCMC approach to co-estimate topology and node ages. Dating analyses were conducted under the best-fit model for each marker partition, and a birth-death tree prior, with rate variation across branches uncorrelated and lognormal distributed. The MCMC chains were run for 50 million generations with parameters sampled every 1,000 generations. Tracer v.1.6 [37] was used to assess the adequate effective sample size (ESS) values (> 200) and the appropriate burn-in. The maximum clade credibility (MCC) tree with mean ages and 95% highest posterior density (HPD) intervals on nodes was generated using TreeAnnotator v1.8.0.
Phylogenetic position of Zygophyllales is poorly resolved within rosids [39]. The inclusion of sparse outgroups may violate a basic assumption of Bayesian dating approaches, namely even taxon sampling across lineages [40]. Therefore, rather than including rosid outgroups, we rooted the timetree using the mono-generic Krameriaceae (represented by Krameria ixine and Krameria lanceolata), sister to Zygophyllaceae [39]. The Zygophyllaceae fossil record is limited and cannot be placed confidently in the tree of extant taxa [41]. Moreover, no fossil has been reported for Krameriaceae. We used a secondary calibration point with normal prior distributions, 70 Ma (95% HPD: 49–88 Ma) for the split between Krameriaceae and Zygophyllaceae, obtained from a dense fossil-calibrated family-level phylogeny of angiosperms [32]. For comparison, we also used 60.9 Ma (34–90 Ma) recently estimated by Magallón et al. [42] to constrain the same node. Between these analyses, divergence times are highly congruent except that deep nodes vary less than c. 4%. The divergence times obtained using the age of the family-level timetree [32] as calibration was therefore reported and used for biogeographic and diversification analyses.
Ancestral area reconstructions
Ancestral area states were reconstructed using Statistical Dispersal-Vicariance Analysis (S-DIVA) in the RASP software package [43]. We scored four main regions: A, Africa; B, Asia; C, Australia; and D, New World. In this analysis, the New World was not divided into North America and South America, because North American Zygophyllaceae species are clustered with South American species in our phylogeny, and the biotic interchange across the Americas occurred continually at ~ 20–6 Ma [44]. The S-DIVA analysis was performed using 1000 trees sampled randomly from the BEAST output as a “trees file”, and the MCC tree was used as a final representative tree.
Diversification analyses
We implemented four methods to shed light on the temporal diversification patterns of Zygophyllaceae on global and regional scales. We employed Bayesian analysis of macro-evolutionary mixtures (BAMM), implemented in BAMM v.2.5 [45] to assess diversification rate heterogeneity along the branches. Incomplete taxon sampling was accounted for by assigning the percentage of species sampled (55%). We performed two independent runs on the MCC tree with a reversible jump MCMC run of 10 million generations, sampling parameters every 1000 generations. ESS values were computed in the R package CODA (> 200). The configuration of the diversification rate shifts was estimated using the posterior distribution, and Bayes factors were used to compare alternative diversification models. Results were analyzed and plotted using the R package BAMMtools v.2.0.2 [46]. Moore et al. [47] criticized this method and considered that the posterior on the number of shifts was overly sensitive to Poisson prior values. However, Rabosky et al. [48] demonstrated that the conclusion of Moore et al. [47] resulted from an invalid likelihood function, and suggest that in the most recent version of BAMM (v.2.5), the extreme sensitivity to the prior cannot be replicated. Considering this ongoing debate, we performed multiple BAMM analyses, similar to previous studies of Campanula [49] and Theaceae [11], by using various prior values (0.5, 1, 2, 5, and 10) to determine whether our results were affected by different prior settings.
We also used TreePar [50] to identify the locations of temporal shifts in global diversification rates of Zygophyllaceae. Similar to the way Couvreur et al. [51] examined the diversification rates of Arecaceae, we first determined the time point at which incomplete taxon sampling might begin to have a significant effect on TreePar analysis. Here, we found a dramatic increase of missing taxa occurred at 8.7 Ma (21% rising to 30%). TreePar analyses were run under the BD model with the following settings: start = 8.7, end = crown age of Zygophyllaceae, grid = 0.1 Ma. Rate shifts were determined using the likelihood ratio test (p < 0.05).
To visualize the accumulation of lineages within different regions, we fit exponential curves to the plots of Africa, Asia, Australia, and New World divergences drawn from the MCC tree and 100 trees sampled randomly from the post-burnin posterior distribution of the BEAST analysis as implemented by in-house R script (available upon request from the corresponding authors). In addition, we examined regional diversification of Zygophyllaceae using nodal “lineage density” method [52], which can reflect diversity due to both situ diversification and independent immigration events [53]. Geographic distribution at each node was obtained from S-DIVA analysis. Six species have two or multiple distribution states and were excluded from these two analyses.
Results and discussion
Phylogeny of Zygophyllaceae
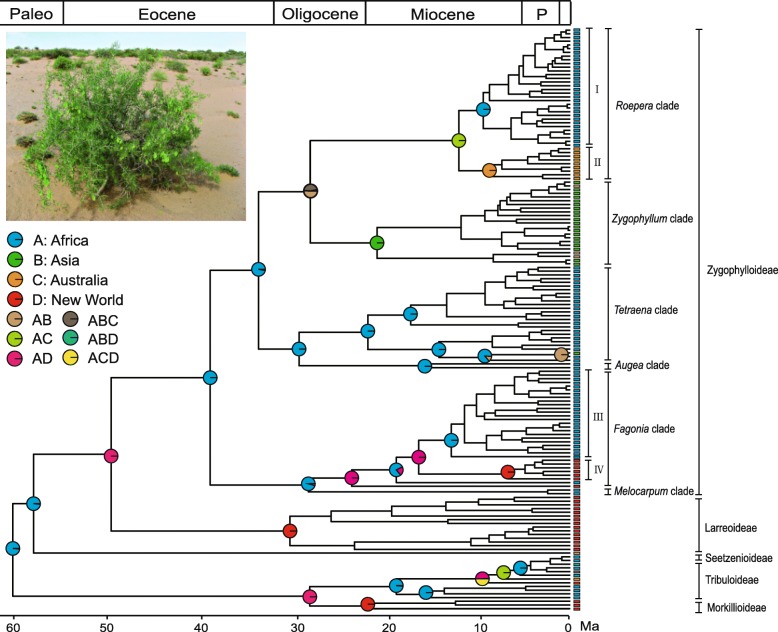
Phylogenetic analyses of the combined four-marker datasets with different taxon and character sampling strategies yielded topologies (Additional file 2: Figures S1 and S2) that are highly congruent with those of previous studies [41, 54]. Zygophyllaceae and its five subfamilies (Zygophylloideae, Larreoideae, Seetzenioideae, Tribuloideae, and Morkillioideae) are all strongly supported as monophyletic. Within Zygophylloideae, six major clades are identified, which correspond to Augea, Fagonia, Melocarpum, Roepera, Tetraena, and Zygophyllum clades [41, 55]. Based on the geographic distribution, the Roepera clade can be divided further into two subclades: African subclade (I) and Australian subclade (II), nevertheless the supports for these two subclades are weak. Similarly, two major subclades (III and IV) are recognized in the Fagonia clade, distributed in Africa and the New World, respectively.
Time and mode of diversification in Zygophyllaceae
Divergence time estimates for Zygophyllaceae are shown in Additional file 2: Figure S3. Ancestral area reconstruction using the S-DIVA analysis is shown in Fig. 1. The crown group age of Zygophyllaceae is here estimated to be 59.89 Ma (95% HPD: 38.14–80.95 Ma), overlapping with the ages inferred by using rbcL data with two secondary calibrations and using ITS data with one secondary calibration [55]. The most recent common ancestor of the family occupied Africa, and multiple independent migrations from ancestral region to other dryland regions took place since the Eocene. Five dispersals into the New World occurred at ca. 49 Ma, 28 Ma, 23 Ma, 16 Ma, and 9 Ma, correspondingly generating Larreoideae, Morkilloideae, Fagonia scoparia, Fagonia subclade IV, and Kallstroemia maxima, respectively. Two dispersals into Asia generated Zygophyllum clade and Tetraena mongolica at ca. 28 Ma and 10 Ma, respectively. Two dispersals into Australia resulted in Roepera subclade II and Tribulopis at ca. 12 Ma and 7 Ma, respectively.
Fig. 1.
Chronogram of Zygophyllaceae with ancestral area reconstructions. Color-coded bars at the tips of the tree indicate the contemporary distribution of the corresponding species. Color-coded pie diagrams at each node show the relative probabilities of alternative ancestral distributions obtained by Statistical Dispersal-Vicariance Analysis (S-DIVA) optimizations over the 1000 trees from the BEAST analysis. An example of dryland flora dominated by Zygophyllum xanthoxylum is presented in the upper left. Photograph by S-X Yu
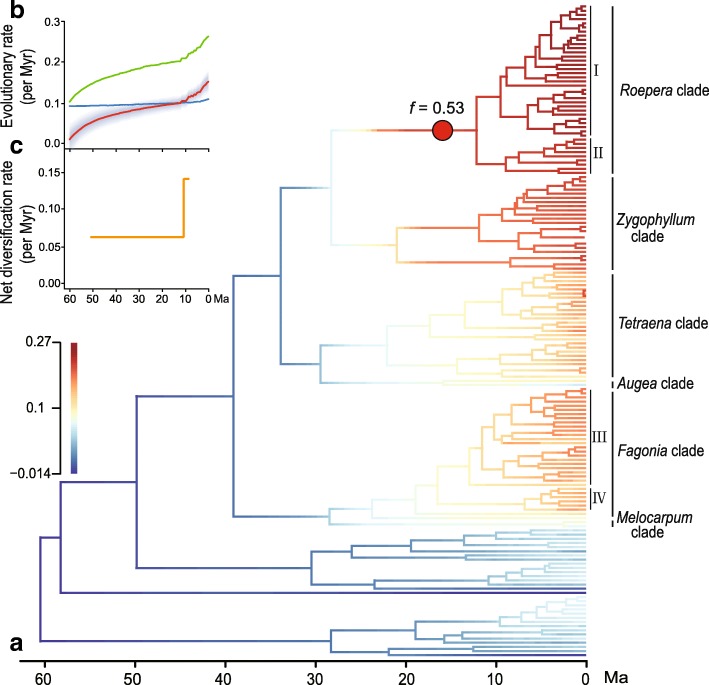
Different tested Poisson priors (0.5, 1, 2, 5, and 10) from the BAMM analyses generated highly similar results. Following the suggestion of Rabosky et al. [45] for small trees (< 500 tips), we here described the results based on the analysis of prior 1 (Fig. 2). One acceleration of diversification rates was detected in the Roepera clade by sampling the maximum a posteriori configuration with the highest frequency (f = 0.53), ~ 15 Ma (Fig. 2a), and its net diversification rates are 0.23 species per million years (myr). BAMM analyses also indicate that the branch colors of six monophyletic groups, African Roepera (clade I), Australian Roepera (clade II), African Tetraena and Fagonia (clade III), and Asian Zygophyllum, and New World Fagonia (clade IV), became increasingly warm since the mid to late Miocene (Fig. 2a), displaying elevated rates of net diversification for each of these regional clades compared with the rest of the family (see inset scale in Fig. 2).
Fig. 2.
Diversification rates through time and among lineages during the evolutionary history of Zygophyllaceae. a Phylorate inferred from BAMM analysis under the prior of ‘ExpectedNumberofShifts’ as 1. Colors of branches denote directionality and strength of rate change, cooler and warmer colors designate slower and faster rates, respectively (see inset scale). The red circle indicates the position of shift in the maximum sampled posterior configuration. b Rate-through-time plots for rates of speciation (green), extinction (blue) and net species diversification (red, with grey probability distribution). c Maximum likelihood diversification rate estimates according to TreePar
The rate-through-time plots suggest that the global speciation and net diversification rates of Zygophyllaceae significantly accelerated from about 10 Ma towards the present (Fig. 2b). TreePar analyses also rejected the null hypothesis of the constant diversification rate of the family (χ2 = 15.91, p = 0.0004), and found the one-shift model as the best. Net diversification rates of Zygophyllaceae on the whole increased from 0.053 species/myr to 0.142 species/myr at 10.4 Ma (Fig. 2c).
The extant Zygophyllaceae dates to the Paleocene of Africa, ~ 60 Ma, but based on our region-specific divergence plots, the rapid accumulation of African lineages did not occur until the mid-Miocene (Figs. 3 and 4a). At the same time, lineages of Zygophyllaceae in Asia, Australia and the New World all experienced a dramatic increase after the mid-Miocene Climatic Optimum (MMCO, ~ 15 Ma; Figs. 3 and 4a).
Fig. 3.
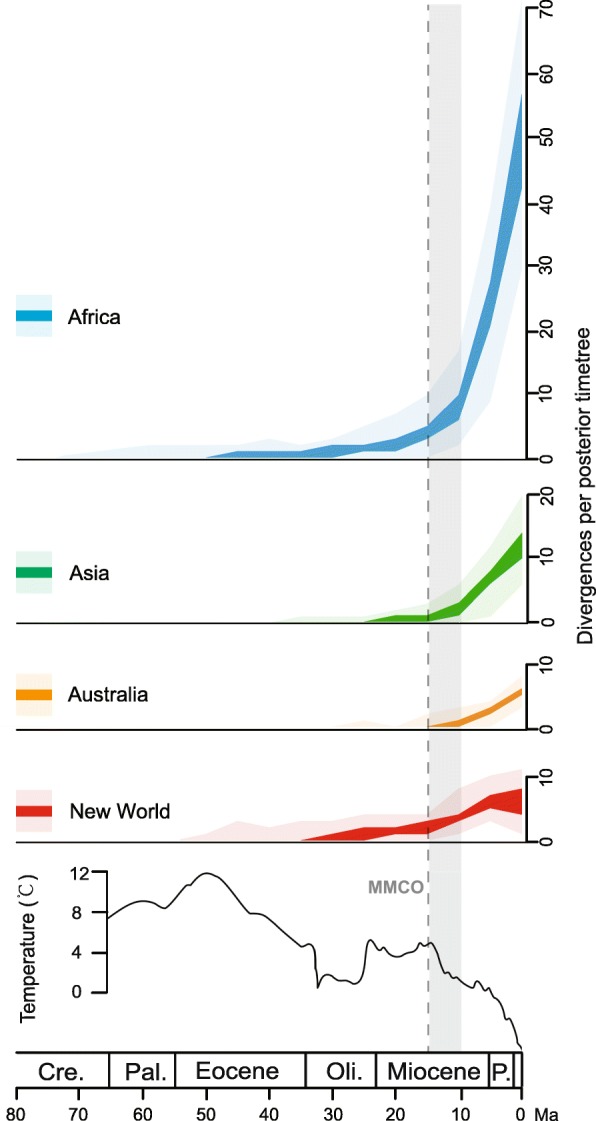
Zygophyllaceae divergences through time, according to geographic areas. Plots summarize the results of BEAST analyses of, and ancestral area reconstructions across, 101 posterior trees (see Materials and Methods). For each 5-million-year interval, the interquartile range (dark colors) and the complete span (light colors) of observed divergences are provided. The depiction of sea-surface temperature changes is modified from Zachos et al. [16]. Abbreviations: Cre. = Cretaceous; Pal. = Paleocene; Oli. = Oligocene; P. = Pliocene. Our results indicate that the initiation of diversification of Zygophyllaceae in different regions (light-grey shade) occurred after the MMCO (dashed line)
Fig. 4.
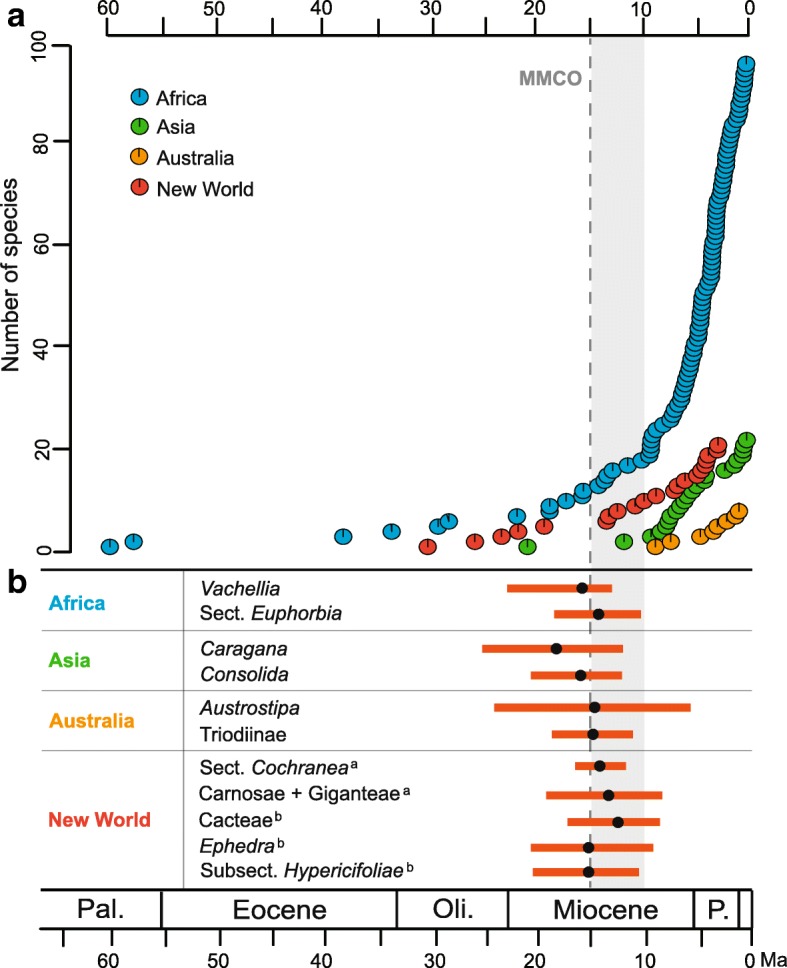
Diversification of plant groups in drylands. a Zygophyllaceae species accumulation estimates over time. Pie diagrams are color coded to reflect the proportion of alternative ancestral distributions obtained by the S-DIVA. b The timing of diversification for eleven representative plant groups in different dryland regions. The dark spots and their corresponding scale bars represent the mean ages and 95% HPD intervals (see Discussion for details). a, bTaxa are from tropical America and western North America, respectively. Abbreviations: Pal. = Paleocene; Oli. = Oligocene; P. = Pliocene
Diversification burst of the global dryland floras
Our analyses show that extant Zygophyllaceae date to the Paleocene of Africa, and migrations from ancestral areas to other regions did not take place synchronously. However, rapid accumulations of Zygophyllaceae lineages in four dryland regions (Africa, Asia, Australia, and the New World) occurred relatively recently, beginning at the mid-late Miocene (~ 15–10 Ma). The timing of these major diversification events within Zygophyllaceae is similar to those of other representative plant groups in different dryland regions, such as African Vachellia (Fabaceae) and Sect. Euphorbia (Euphorbiaceae), Asian Caragana (Fabaceae) and Consolida (Ranunculaceae), Australian Austrostipa (Poaceae) and Triodiinae (Poaceae), tropical American Sect. Cochranea (Heliotropiaceae) and Carnosae + Giganteae (Oxalidaceae), and western North American Cacteae (Cactaceae), Ephedra (Ephedraceae) and Subsect. Hypericifoliae (Euphorbiaceae) (Fig. 4b; Additional file 1: Table S3). Moreover, a rapid accumulation of spiny plant lineages, as well as mammalian herbivores in African savanna, occurred since the mid-Miocene [56]. Australia Acacia (Fabaceae) underwent an accelerated diversification at ~ 15 Ma [57]. The New World Cactaceae had a significant shift in diversification rates at 16.0–14.8 Ma [20]. In western North America, the ivesioids clade of Potentilla (Rosaceae) experienced a westward range expansion to the Sierra Nevada and the coast of California 12 Ma [58]. Thus, we suggest that the establishment of modern dryland floras in different continents was nearly synchronous, beginning in the mid-late Miocene.
The near-synchronous initiation of rise of dryland floras across the multiple continents indicates that a global trigger may have been responsible. After the MMCO, a steep and steady decline in global temperatures occurred (Fig. 3; [16, 59]), which possibly resulted in a decrease of global precipitation on account of a slowdown of the hydrological cycle [20]. Lines of evidence from paleoecology [60] and stable isotopes [61, 62] from multiple continents robustly support a general trend toward increasing aridity from the MMCO onwards. Several studies have suggested that global climate cooling was the main driver contributing to the long-term drying trend since the mid-Miocene and shaped the modern flora [63–66]. Thus, it is possible that the global rise of dryland floras was largely driven by the global climate change that reduced global precipitation and thereby led to a global expansion of arid environments.
The rise of dryland floras in different continents might have also been linked to the enhanced aridification and expansion of drylands due to regional orogenetic and climate changes. Uplift of the East African Plateau occurred during the early Miocene ~ 17 Ma, which resulted in the formation of aridity in east Africa [67, 68]. With the shrinkage of the Paratethys Sea during the late Miocene, the African monsoon was enhanced drastically, generating arid conditions across North Africa [69]. Broad-scale uplift of plateaus and mountains occurred in the northern and eastern Tibetan Plateau, the east-central Andes and Altiplano, the east African rift valley, and the northern Canadian Rockies in the last 15 Ma [6, 70–73]. Atmosphere and biosphere simulations indicate that the uplift itself might have led to a drastic reorganization of atmospheric circulation, engendering strong aridification and climate changes [74, 75]. A significant advance of Antarctic ice occurred at ~ 14 Ma [76], and at the same time, sea levels began to fall across southeastern Australia, eventually retreating towards the current coastline at ~ 10–9 Ma, heralding the onset of different conditions across continental Australia [77].
Conclusions
In summary, the present study reveals that the rise of dryland floras across multiple continents is near-synchronous, beginning at the mid-late Miocene. The causative factors for the global near-synchronous rise were suggested to be a band of a global climate cooling; however, regional orogenetic and climate changes may have played additional important roles. Our findings highlight the mid-late Miocene as an essential period for the assembly and evolution of modern dryland floras, although the original timings of dryland floras in different continents might have been asynchronous. Yet, Zygophyllaceae is only one of many components of dryland floras. This hypothesis needs to be further tested by studying other plant groups of dryland floras in a broad phylogenetic context.
Additional files
Table S1. Species, voucher information and GenBank accession numbers for the dataset of four markers. Table S2. The parameters of the best-fit model for each DNA region. Table S3. The crown group ages and ancestral habitat types of eleven representative plant groups in different drylands. (PDF 143 kb)
Figure S1. Phylogenetic relationships of Zygophyllaceae based on the 4-marker dataset with 164 taxa. Figure S2. Phylogenetic relationships of Zygophyllaceae based on three matrices (see Methods for details). Figure S3. Chronogram of Zygophyllaceae obtained from the BEAST analysis of the four-marker dataset. Figure S4. Ancestral habitat reconstructions for five representative plant groups inhabiting in both dryland and non-dryland floras. (PDF 2101 kb)
Acknowledgements
We sincerely thank Dinah Parker for carefully reading an early draft of the manuscript and Fernando T. Maestre for valuable suggestions. We also thank Alison Cuff and one anonymous reviewer for their comments and suggestions that greatly improved our manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (41571499, 31770231, 31770233, and 31470315), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB31030000), CAS International Research & Education Development Program (SAJC201315), the Ministry of Education (ZS12014), and Shanxi Scholarship Council of China [2010]14–62. The funding body had no role in the design, collection and conclusion of this study.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding authors on reasonable request.
Abbreviations
- BAMM
Bayesian analysis of macro-evolutionary mixtures
- BI
Bayesian inference
- HPD
Highest posterior density
- ITS
Internal transcribed spacers
- Ma
Million years ago
- MCC
Maximum clade credibility
- MCMC
Markov Chain Monte Carlo
- ML
Maximum likelihood
- MMCO
Mid-Miocene Climatic Optimum
- myr
Million years
- S-DIVA
Statistical Dispersal-Vicariance Analysis
Authors’ contributions
WW conceived the research. SDW, LJZ, ZDC and WW contributed analysis tools. SDW, LL, SXY and WW performed the research and analyzed data. SDW and WW drafted the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sheng-Dan Wu, Email: shengdanwu@ibcas.ac.cn.
Lin-Jing Zhang, Email: linjingzh@aliyun.com.
Li Lin, Email: 474376879@qq.com.
Sheng-Xiang Yu, Email: yushengxiang@ibcas.ac.cn.
Zhi-Duan Chen, Email: zhiduan@ibcas.ac.cn.
Wei Wang, Email: wangwei1127@ibcas.ac.cn.
References
- 1.GLP, Global land project—science plan and implementation strategy” [IGBP (international geosphere biosphere program) report no. 53/international human dimensions Programme report No. 19, IGBP Secretariat, Stockholm, 2005]; www.globallandproject.org/documents.shtml. Accessed 15 Oct 2017.
- 2.Middleton NJ, Thomas DSG. World atlas of desertification. 2. New York: Edward Arnold; 1997. [Google Scholar]
- 3.Reynolds JF, Stafford Smith M, Lambin EF, Turner IIBL, Mortimore M, Batterbury SP, et al. Global desertification: building a science for dryland development. Science. 2007;316:847–851. doi: 10.1126/science.1131634. [DOI] [PubMed] [Google Scholar]
- 4.Walter H. Vegetation of the earth—ecological systems of the geobiosphere. 2. New York: Springer-Verlag; 1979. [Google Scholar]
- 5.MEA, Millennium Ecosystem Assessment . Ecosystems and Human Well-Being: Desertification synthesis. Washington, DC: World Resources Institute; 2005. [Google Scholar]
- 6.Manafzadeh S, Staedler YM, Conti E. Visions of the past and dreams of the future in the orient: the Irano-Turanian region from classical botany to evolutionary studies. Biol Rev. 2017;92:1365–1388. doi: 10.1111/brv.12287. [DOI] [PubMed] [Google Scholar]
- 7.Tietjen B, Jeltsch F, Zehe E, Classen N, Groengroeft A, Schiffers K, et al. Effects of climate change on the coupled dynamics of water and flora in drylands. Ecohydrology. 2010;3:226–237. [Google Scholar]
- 8.Noy-Meir I. Desert Ecosystems: Environment and producers. Annu Rev Ecol Syst. 1973;4:25–51. doi: 10.1146/annurev.es.04.110173.000325. [DOI] [Google Scholar]
- 9.Wang Q, Liu JQ, Allen GA, Ma YZ, Yue W, Marr KL, et al. Arctic plant origins and early formation of circumarctic distributions: a case study of the mountain sorrel, Oxyria digyna. New Phytol. 2016;209:343–353. doi: 10.1111/nph.13568. [DOI] [PubMed] [Google Scholar]
- 10.Eiserhardt WL, Couvreur TLP, Baker WJ. Plant phylogeny as a window on the evolution of hyperdiversity in the tropical rainforest. New Phytol. 2017;214:1408–1422. doi: 10.1111/nph.14516. [DOI] [PubMed] [Google Scholar]
- 11.Yu XQ, Gao LM, Soltis DE, Soltis PS, Yang JB, Fang L, et al. Insights into the historical assembly of east Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol. 2017;215:1235–1248. doi: 10.1111/nph.14683. [DOI] [PubMed] [Google Scholar]
- 12.Partridge TC. The evidence for Cainozoic aridification in southern Africa. Quatern Int. 1993;17:105–110. doi: 10.1016/1040-6182(93)90087-V. [DOI] [Google Scholar]
- 13.Graham A. Late cretaceous and Cenozoic history of Latin American vegetation and terrestrial environments. St Louis: Missouri Botanical Garden Press; 2010. [Google Scholar]
- 14.Sun JM, Windley BF. Onset of aridification by 34 ma across the Eocene-Oligocene transition in Central Asia. Geology. 2015;43:1015–1018. doi: 10.1130/G37165.1. [DOI] [Google Scholar]
- 15.Byrne M, Yeates DK, Joseph L, Kearney M, Bowler J, Williams MA, et al. Birth of a biome: insights into the assembly and maintenance of the Australian arid zone biota. Mol Ecol. 2008;17:4398–4417. doi: 10.1111/j.1365-294X.2008.03899.x. [DOI] [PubMed] [Google Scholar]
- 16.Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 17.Klak C, Reeves G, Hedderson T. Unmatched tempo of evolution in southern African semi-desert ice plants. Nature. 2004;427:63–65. doi: 10.1038/nature02243. [DOI] [PubMed] [Google Scholar]
- 18.Good-Avila SV, Souza V, Gaut BS, Eguiarte LE. Timing and rate of speciation in Agave (Agavaceae) Proc Natl Acad Sci U S A. 2006;103:9124–9129. doi: 10.1073/pnas.0603312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore MJ, Jansen RK. Molecular evidence for the age, origin and evolutionary history of the American desert plant genus Tiquilia (Boraginaceae) Mol Phylogenet Evol. 2006;39:668–687. doi: 10.1016/j.ympev.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Arakaki M, Christin P-A, Nyffeler R, Lendel A, Eggli U, Ogburn RM, et al. Contemporaneous and recent radiations of the world’s major succulent plant lineages. Proc Natl Acad Sci U S A. 2011;108:8379–8384. doi: 10.1073/pnas.1100628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang ML, Hao XL, Sanderson SC. Spatiotemporal evolution of Reaumuria (Tamaricaceae) in Central Asia: insights from molecular biogeography. Phytotaxa. 2014;167:89–103. doi: 10.11646/phytotaxa.167.1.5. [DOI] [Google Scholar]
- 22.Cabrera J, Jacobs SWL, Kadereit G. Biogeography of Camphorosmeae (Chenopodiaceae): tracking the tertiary history of Australian aridification. Telopea. 2011;13:313–326. doi: 10.7751/telopea20116023. [DOI] [Google Scholar]
- 23.Guerrero P, Rosas M, Arroyoa MTK, Wiens JJ. Evolutionary lag times and recent origin of the biota of an ancient desert (Atacama–Sechura) Proc Natl Acad Sci U S A. 2013;110:11469–11474. doi: 10.1073/pnas.1308721110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitford WG. Ecology of desert systems. New York: Academic Press; 2002. [Google Scholar]
- 25.Sheahan MC. Zygophyllaceae. In: Kubitzki K, editor. The families and genera of vascular plants. Hamburg: Springer; 2007. pp. 488–500. [Google Scholar]
- 26.Zhang XS, Wu ZY, Wang XP, Liu FX, Zhu YC, Li SY, Li B, et al. Chinese Vegetation. Beijing: Science Press; 1980. Desert; pp. 583–595. [Google Scholar]
- 27.Beier BA. A revision of the desert shrub Fagonia (Zygophyllaceae) Syst Biodivers. 2005;3:221–263. doi: 10.1017/S1477200005001684. [DOI] [Google Scholar]
- 28.Yang SM, Furukawa I. Anatomical adaptations of three species of Chinese xerophytes (Zygophyllaceae) J Forest Res. 2006;17:247–251. doi: 10.1007/s11676-006-0056-7. [DOI] [Google Scholar]
- 29.Lauterbach M, van der Merwe PW, Keßler L, Pirie MD, Bellstedt DU, Kadereit G. Evolution of leaf anatomy in arid environments – a case study in southern African Tetraena and Roepera (Zygophyllaceae) Mol Phylogenet Evol. 2016;97:129–144. doi: 10.1016/j.ympev.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Sage RF. A portrait of the C4 photosynthetic family on the 50th anniversary of its discovery: species number, evolutionary lineages, and hall of fame. J Exp Bot. 2016;67:4039–4056. doi: 10.1093/jxb/erw156. [DOI] [PubMed] [Google Scholar]
- 31.Christin PA, Osborne CP, Sage RF, Arakaki M, Edwards EJ. C4 eudicots are not younger than C4 monocots. J Exp Bot. 2011;62:3171–3181. doi: 10.1093/jxb/err041. [DOI] [PubMed] [Google Scholar]
- 32.Bell CD, Soltis DE, Soltis PS. The age and diversification of the angiosperms re-revisited. Am J Bot. 2010;97:1296–1303. doi: 10.3732/ajb.0900346. [DOI] [PubMed] [Google Scholar]
- 33.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 34.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 35.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 36.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 37.Rambaut A, Drummond AJ. Tracer v1.6. 2013. [Google Scholar]
- 38.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevens PF. Angiosperm Phylogeny Website. Version 12, July 2012 [and more or less continuously updated since]. 2001 onwards. Accessed 25 Sept 2017.
- 40.Drummond AJ, Bouckaert RR. Bayesian evolutionary analysis with BEAST 2. Cambridge: Cambridge University Press; 2015. [Google Scholar]
- 41.Bellstedt DU, Galley C, Pirie MD, Linder HP. The migration of paleotropical arid flora: Zygophylloideae as an example. Syst Bot. 2012;37:951–959. doi: 10.1600/036364412X656608. [DOI] [Google Scholar]
- 42.Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL. Hernández-Hernández T. a metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 2015;207:437–445. doi: 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- 43.Yu Y, Harris AJ, He XJ. S-DIVA (statistical dispersal-Vicariance analysis): a tool for inferring biogeographic histories. Mol Phylogenet Evol. 2010;56:848–850. doi: 10.1016/j.ympev.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 44.Bacon CD, Silvestro D, Jaramillo C, Smith BT, Chakrabarty P, Antonelli A. Biological evidence supports an early and complex emergence of the isthmus of Panama. Proc Natl Acad Sci U S A. 2015;112:6110–6115. doi: 10.1073/pnas.1423853112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rabosky DL. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One. 2014;9:e89543. doi: 10.1371/journal.pone.0089543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabosky DL, Grundler M, Anderson C, Title P, Shi JJ, Brown JW, et al. BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol Evol. 2014;5:701–707. doi: 10.1111/2041-210X.12199. [DOI] [Google Scholar]
- 47.Moore BR, Höhna S, May MR, Rannala B, Huelsenbeck JP. Critically evaluating the theory and performance of Bayesian analysis of macroevolutionary mixtures. Proc Natl Acad Sci U S A. 2016;113:9569–9574. doi: 10.1073/pnas.1518659113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabosky DL, Mitchell JS, Chang J. Is BAMM flawed? Theoretical and practical concerns in the analysis of multi-rate diversification models. Syst Biol. 2017;66:477–498. doi: 10.1093/sysbio/syx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones KE, Korotkova N, Petersen J, Henning T, Borsch T, Kilian N. Dynamic diversification history with rate upshifts in Holarctic bell-flowers (Campanula and allies) Cladistics. 2017;33:637–666. doi: 10.1111/cla.12187. [DOI] [PubMed] [Google Scholar]
- 50.Stadler T. Mammalian phylogeny reveals recent diversification rate shifts. Proc Natl Acad Sci U S A. 2011;108:6182–6187. doi: 10.1073/pnas.1016876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Couvreur TLP, Forest F, Baker WJ. Origin and global diversification patterns of tropical rain forests: inferences from a complete genus-level phylogeny of palms. BMC Biol. 2011;9:44. doi: 10.1186/1741-7007-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahler DL, Revell LJ, Glor RE, Losos JB. Ecological opportunity and the rate of morphological evolution in the diversification of greater Antillean anoles. Evolution. 2010;64:2731–2745. doi: 10.1111/j.1558-5646.2010.01026.x. [DOI] [PubMed] [Google Scholar]
- 53.McGuire JA, Witt CC, Remsen JV, Jr, Corl A, Rabosky DL, Altshuler DL, et al. Molecular phylogenetics and the diversification of hummingbirds. Curr Biol. 2014;24:910–916. doi: 10.1016/j.cub.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 54.Sheahan MC, Chase MW. Phylogenetic relationships within Zygophyllaceae based on DNA sequences of three plastid regions, with special emphasis on Zygophylloideae. Syst Bot. 2000;25:371–384. doi: 10.2307/2666648. [DOI] [Google Scholar]
- 55.Wu SD, Lin L, Li HL, Yu SX, Zhang LJ, Wang W. Evolution of Asian interior arid-zone biota: evidence from the diversification of Asian Zygophyllum (Zygophyllaceae) PLoS One. 2015;10:e0138697. doi: 10.1371/journal.pone.0138697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Charles-Dominique T, Davies TJ, Hempson GP, Bezeng BS, Daru BH, Kabongo RM, et al. Spiny plants, mammal browsers, and the origin of African savannas. Proc Natl Acad Sci U S A. 2016;113:E5572–E5579. doi: 10.1073/pnas.1607493113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crisp MD, Cook LG. How was the Australian flora assembled over the last 65 million years? A molecular phylogenetic perspective. Annu Rev Ecol Evol Syst. 2013;44:303–324. doi: 10.1146/annurev-ecolsys-110512-135910. [DOI] [Google Scholar]
- 58.Töpel M, Antonelli A, Yesson C, Eriksen B. Past climate change and plant evolution in western North America: a case study in Rosaceae. PLoS One. 2012;7:e50358. doi: 10.1371/journal.pone.0050358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holbourn A, Kuhnt W, Clemens S, Prell W, Andersen N. Middle to late Miocene stepwise climate cooling: evidence from a high-resolution deep water isotope curve spanning 8 million years. Paleoceanography. 2013;28:688–699. doi: 10.1002/2013PA002538. [DOI] [Google Scholar]
- 60.Edwards EJ, Osborne CP, Strömberg CAE, Smith SA, C4 Grasses Consortium The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science. 2010;328:587–591. doi: 10.1126/science.1177216. [DOI] [PubMed] [Google Scholar]
- 61.Dettman DL, Kohn MJ, Quade J, Ryerson F, Ojha TP, Hamidullah S. Seasonal stable isotope evidence for a strong Asian monsoon throughout the past 10.7 my. Geology. 2001;29:31–34. doi: 10.1130/0091-7613(2001)029<0031:SSIEFA>2.0.CO;2. [DOI] [Google Scholar]
- 62.Huang Y, Clemens SC, Liu W, Wang Y, Prell WL. Large-scale hydrological change drove the late Miocene C4 plant expansion in the Himalayan foreland and Arabian peninsula. Geology. 2007;35:531–534. doi: 10.1130/G23666A.1. [DOI] [Google Scholar]
- 63.Dutton JF, Barron EJ. Miocene to present vegetation changes: a possible piece of the Cenozoic cooling puzzle. Geology. 1997;25:39–41. doi: 10.1130/0091-7613(1997)025<0039:MTPVCA>2.3.CO;2. [DOI] [Google Scholar]
- 64.Chamberlain CP, Winnick MJ, Mix HT, Chamberlain SD, Maher K. The impact of Neogene grassland expansion and aridification on the isotopic composition of continental precipitation. Global Biogeochem Cy. 2014;28:992–1004. doi: 10.1002/2014GB004822. [DOI] [Google Scholar]
- 65.Hernández-Hernández T, Brown JW, Schlumpberger BO, Eguiarte LE, Magallón S. Beyond aridification: multiple explanations for the elevated diversification of cacti in the New World succulent biome. New Phytol. 2014;202:1382–1397. doi: 10.1111/nph.12752. [DOI] [PubMed] [Google Scholar]
- 66.Nie ZL, Funk VA, Meng Y, Deng T, Sun H, Wen J. Recent assembly of the global herbaceous flora: evidence from the paper daisies (Asteraceae: Gnaphalieae) New Phytol. 2016;209:1795–1806. doi: 10.1111/nph.13740. [DOI] [PubMed] [Google Scholar]
- 67.Ebinger CJ, Sleep NH. Cenozoic magmatism throughout East Africa resulting from impact of a single plume. Nature. 1998;395:788–791. doi: 10.1038/27417. [DOI] [Google Scholar]
- 68.Wichura H, Jacobs LL, Lin A, Polcyn MJ, Manthi FK, Winkler DA, et al. A 17-my-old whale constrains onset of uplift and climate change in East Africa. Proc Natl Acad Sci U S A. 2015;112:3910–3915. doi: 10.1073/pnas.1421502112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang ZS, Ramstein G, Schuster M, Li C, Contoux C, Yan Q. Aridification of the Sahara desert caused by Tethys Sea shrinkage during the Late Miocene. Nature. 2014;513:401–404. doi: 10.1038/nature13705. [DOI] [PubMed] [Google Scholar]
- 70.Ruddiman WF, Kutzbach JE. Late Cenozoic plateau uplift and climate change. T Roy Soc Edin-Earth. 1990;81:301–314. doi: 10.1017/S0263593300020812. [DOI] [Google Scholar]
- 71.Antonelli A, Nylander JAA, Persson C, Sanmartín I. Tracing the impact of the Andean uplift on Neotropical plant evolution. Proc Natl Acad Sci U S A. 2009;106:9749–9754. doi: 10.1073/pnas.0811421106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoorn C, Wesselingh FP, ter Steege H, Bermudez MA, Mora A, Sevink J, et al. Amazonia through time: Andean uplift, climate change, landscape evolution. Biodivers Sci. 2010;330:927–931. doi: 10.1126/science.1194585. [DOI] [PubMed] [Google Scholar]
- 73.Liu XD, Dong BW. Influence of the Tibetan plateau uplift on the Asian monsoon-arid environment evolution. Chin Sci Bull. 2013;34:4277–4291. doi: 10.1007/s11434-013-5987-8. [DOI] [Google Scholar]
- 74.Manabe S, Broccoli AJ. Mountains and arid climates of middle latitudes. Science. 1990;247:192–195. doi: 10.1126/science.247.4939.192. [DOI] [PubMed] [Google Scholar]
- 75.Sepulchre P, Ramstein G, Fluteau F, Schuster M, Tiercelin JJ, Brunet M. Tectonic uplift and eastern Africa aridification. Science. 2006;313:1419–1423. doi: 10.1126/science.1129158. [DOI] [PubMed] [Google Scholar]
- 76.McGowran B, Holdgate GR, Li Q, Gallagher SJ. Cenozoic stratigraphic succession in southeastern Australia. Aust J Earth Sci. 2004;51:459–496. doi: 10.1111/j.1400-0952.2004.01078.x. [DOI] [Google Scholar]
- 77.Bowler JM, Kotsonis A, Lawrence CR. Environmental evolution of the mallee region, Western Murray Basin. Proc R Soc Vic. 2006;118:161–210. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Species, voucher information and GenBank accession numbers for the dataset of four markers. Table S2. The parameters of the best-fit model for each DNA region. Table S3. The crown group ages and ancestral habitat types of eleven representative plant groups in different drylands. (PDF 143 kb)
Figure S1. Phylogenetic relationships of Zygophyllaceae based on the 4-marker dataset with 164 taxa. Figure S2. Phylogenetic relationships of Zygophyllaceae based on three matrices (see Methods for details). Figure S3. Chronogram of Zygophyllaceae obtained from the BEAST analysis of the four-marker dataset. Figure S4. Ancestral habitat reconstructions for five representative plant groups inhabiting in both dryland and non-dryland floras. (PDF 2101 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding authors on reasonable request.