Abstract
Aims
Sarcomeric gene mutations frequently underlie hypertrophic cardiomyopathy (HCM), a prevalent and complex condition leading to left ventricle thickening and heart dysfunction. We evaluated isogenic genome-edited human pluripotent stem cell-cardiomyocytes (hPSC-CM) for their validity to model, and add clarity to, HCM.
Methods and results
CRISPR/Cas9 editing produced 11 variants of the HCM-causing mutation c.C9123T-MYH7 [(p.R453C-β-myosin heavy chain (MHC)] in 3 independent hPSC lines. Isogenic sets were differentiated to hPSC-CMs for high-throughput, non-subjective molecular and functional assessment using 12 approaches in 2D monolayers and/or 3D engineered heart tissues. Although immature, edited hPSC-CMs exhibited the main hallmarks of HCM (hypertrophy, multi-nucleation, hypertrophic marker expression, sarcomeric disarray). Functional evaluation supported the energy depletion model due to higher metabolic respiration activity, accompanied by abnormalities in calcium handling, arrhythmias, and contraction force. Partial phenotypic rescue was achieved with ranolazine but not omecamtiv mecarbil, while RNAseq highlighted potentially novel molecular targets.
Conclusion
Our holistic and comprehensive approach showed that energy depletion affected core cardiomyocyte functionality. The engineered R453C-βMHC-mutation triggered compensatory responses in hPSC-CMs, causing increased ATP production and αMHC to energy-efficient βMHC switching. We showed that pharmacological rescue of arrhythmias was possible, while MHY7: MYH6 and mutant: wild-type MYH7 ratios may be diagnostic, and previously undescribed lncRNAs and gene modifiers are suggestive of new mechanisms.
Keywords: Hypertrophic cardiomyopathy, Disease modeling , CRISPR/Cas9 , Genome-edited human pluripotent stem cell-cardiomyocytes , R453C-βMHC
See page 3893 for the editorial comment on this article (doi: 10.1093/eurheartj/ehy388)
Translational perspectives
Affecting 1:500 people, hypertrophic cardiomyopathy (HCM) is a complex cardiovascular disease of high clinical heterogeneity, which can cause heart failure. Therapies have remained static, often involving invasive surgery. Differences in physiology and subtleties in gene expression confound use of animal models and heterologous systems, while usable human material is scarce. To create a new, high-precision model of HCM, CRISPR/Cas9 engineering produced isogenic β-MHC variants in human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs). Unrivalled molecular and function phenotyping validated HCM hPSC-CM utility, whilst adding clarity to current working hypotheses, showing potential for pharmacological rescue of arrhythmias, suggesting putative diagnostics, and pointing towards new targets for mechanistic understanding and therapeutics.
Introduction
Affecting 1:500 individuals, HCM is the most prevalent cardiac disease,1,2 often leading to sudden cardiac death at a young age (48 ± 19 years).3 Clinical spectrum varies from asymptomatic to severe cardiac dysfunction.4,5 Half of HCM patients bear mutations in one or more of >20 sarcomeric genes, leading to variable penetrance of the disease.6 This implies influence of factors beyond the single pathogenic mutation, such as genetic background7 and environmental modifiers.8 Genetic heterogeneity causes phenotypic variability, with cellular mechanisms including (i) hypertrophy, (ii) foetal gene programme initiation, (iii) energy perturbation, (iv) fibrosis, (v) contractile dysfunction, and (vi) impaired calcium cycling.9
Among the sarcomeric genes mutated in patients, MYH7 is prevalent (20–50% of genotyped cases).10MYH7 encodes beta myosin heavy chain (β-MHC), responsible for regulating actin–myosin interaction, hence cardiomyocyte contraction and ultimately cardiac function.11 Despite clinical and phenotypic heterogeneity, MYH7 mutations are associated with more severe forms of hypertrophy relative to when other sarcomeric genes are altered. This includes higher frequencies of ventricular tachycardia, greater disease penetrance, higher risk of sudden cardiac death, and earlier onset.12,13
Disease modelling of HCM using hPSC-CMs offers a pathophysiologically relevant approach to dissect the mechanics of disease and identify new targets for pharmacological intervention.14 While previous MYH7-HCM animal models provided insight into the disease,15,16 data may be misinterpreted due to species differences. Most HCM hPSC-CM modelling studies have focused on limited features17 and/or lacked isogenic controls.18,19 This confounds understanding because impact of genetic background on phenotype can exceed that caused by the pathogenic mutation.20 The only exception is an interesting, but limited, preview of the potential utility of isogenic lines in dilated cardiomyopathy via correction of a phospholamban R14del mutation patient-specific hiPSC-CMs. Impaired cardiac contractility was restored in corrected 3D engineered cardiac tissue, although this was the only phenotype assessed.21
We created a comprehensive hPSC-based model of HCM via Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR/Cas9) editing to make a c.C9123T substitution in MYH7, corresponding to a pathogenic protein change, p.R453C-βMHC. This included homozygous variants, not previously been reported for any HCM mutation. Extensive molecular and functional evaluation of isogenic hPSC-CMs phenocopied the main hallmarks of hypertrophy, showing a general association between mutation load and level of phenotypic perturbation. Key outcomes included partial phenotypic rescue of arrhythmias with ranolazine, putative diagnostics via ratiometric gene analysis, and RNAseq highlighting a potential role of several long non-coding RNAs (lncRNA) and gene modifiers. This will guide future work on mechanistic understanding, management, and treatment of HCM.
Methods
See Supplementary material online for details.
Results
CRISPR/Cas9 engineering and characterization of MYH7 variants
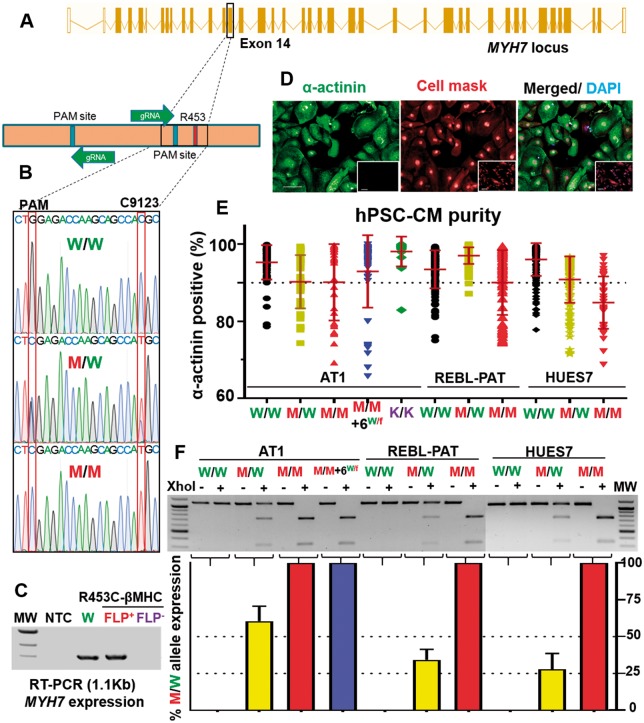
Since the c.C9123T-MYH7 mutation associates with HCM pathophysiology, we coupled a dual gRNA/Cas9-nickase/CRISPR targeting strategy with subsequent flippase-mediated cassette excision to produce 9 polymorphic variants (Figures 1A and B; Supplementary material online, Figure S1A–E and Tables S1–S3). For each of 3 hPSC lines (AT1-hiPSC, REBL-PAT-hiPSC, HUES7-hESC), isogenic sets included 9123-MYH7 parental (C/C wild-type, termed WT/WT), heterozygote (T/C, mutant - MUT/WT), and homozygote (T/T, MUT/MUT). Two additional AT1-hiPSC isogenics were included (Figure 1C; Supplementary material online, Figure S1F): Line ‘homozygote + MYH6WT/fs’ was mutant (T/T) for 9123-MYH7 with an off-target frameshift event in one allele of the homologous gene, MYH6. With the exception of this line, no other off-target events were detected; Line MYH7 ‘Knockout’ (MYH7-KO) contained the selection cassette, which disrupted RNA and protein expression; flippase-mediated cassette excision restored expression (Figure 1C). This isogenic approach minimises the genetic and epigenetic variability seen between individuals and allows impact of the R453C-βMHC mutation to be isolated.
Figure 1.
CRISPR/Cas9 engineering of MYH7. (A) Schema of MYH7 highlighting target and Protospacer Adjacent Motif (PAM) for nickase CRISPR/Cas9 editing. (B) Genotyping of MYH7 in human pluripotent stem cells, introducing c.C9123T and showing a silent mutation (TGG PAM to TCG). (C) RT-PCR showed selection cassette causes an MYH7 knockout; Flippase-mediated excision restores expression. (D) Immunostaining of cardiac α-actinin human pluripotent stem cell-cardiomyocytes and fibroblast controls (inset); cell mask and DAPI as counterstains. Bar = 100 μm. (E) Cardiomyocyte differentiation purity >90% α-actinin + cells; n = 8; Bar = 100 µm. (F) RFLP of MYH7, with ratiometric densitometry of MUT: wild-type (n = 4). FLP, Flippase; MW, molecular weight; NTC, non-template control. Data, mean ± SD.
Mono- and bi-allelic targeting frequency across the 11 lines was 16.3–25.5% and 2.6–7.1%, respectively (Supplementary material online, Figure S1D, E). Isogenics maintained pluripotency, including high efficiency cardiac differentiation; only cultures of ≥85% α-actinin positive were used (Figures 1D, E; Supplementary material online, S2A–D). β-Myosin heavy chain was expressed, except in MYH7-KO (Supplementary material online, Figure S2D). Pertinent to HCM, a ventricular cardiac subtype was predominant (Supplementary material online, Figure S3)22–24 By immunostaining, 91.3 ± 6.4% of cardiomyocytes were MLC2v+. Functional data from patch clamp of single cells or CellOPTIQ-based optical imaging of synchronous monolayers (Supplementary material online, Figure S3C, D) showed ∼60 to 90% ventricular-like morphologies using two separate analysis approaches (APD90/APD50)23 and (APD80-APD70)/APD40-APD30).24
Operator bias was reduced wherever possible by high-content and/or high-throughput approaches (Figures 1–4, and7; Supplementary material online, Figures S3, S4, S6, S9), and/or blinding the experimenter to genotype (Figures 5and6, Supplementary material online, S7 and S8). While single cell patch clamp can assess genotype-cardiac subtype correlations,25 we avoided this technique due to low technical throughput, selection bias, influence of cell density,26 and loss of electrical syncytium. Dispersal of cardiomyocytes from rabbit ventricular wedges causes high levels of single cell electrophysiological heterogeneity (personal communication, Godfrey Smith, Glasgow), an effect also seen in dispersed hPSC-CMs (Supplementary material online, Figure S3C, D).27
Figure 4.
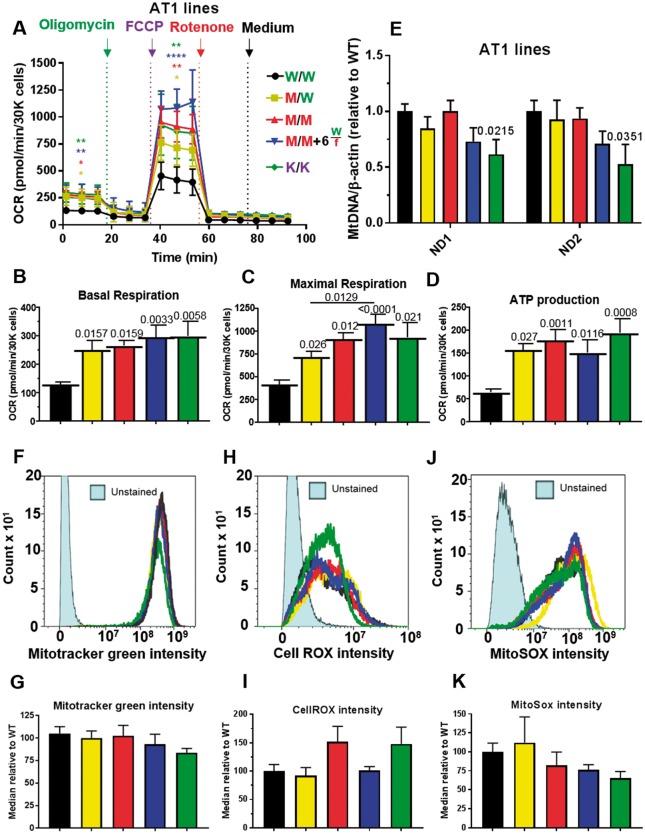
Cardiac bioenergetics analysis of AT1-hypertrophic cardiomyopathy lines. (A) Mitochondrial respiration profile of AT1 R453C-β-myosin heavy chain human pluripotent stem cell-cardiomyocytes using the Seahorse platform quantified, (B) basal respiration, (C) maximal respiration, (D) ATP production (n = 5). (E) qPCR analysis of mitochondrial (ND1-2): nuclear (β-actin) DNA ratio (n = 3). Histograms of human pluripotent stem cell-cardiomyocytes labelled with (F) mitotracker, (H) CellROX, and (J) MitoSOX obtained by flow cytometry, with corresponding quantification (G, I, K), n = 5. Data, mean ± SD. P-values are shown. One-way ANOVA test + Dunnett’s correction compared mutant lines vs. isogenic control. Student’s t-tests, treated vs. vehicle control colour-coded by inter-compared category.
Figure 7.
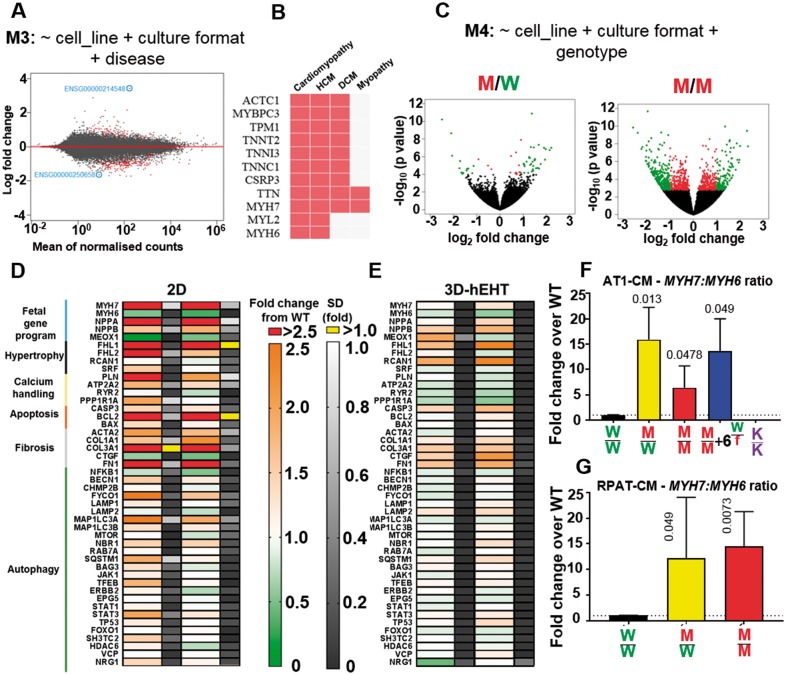
Transcriptomics analysis of hypertrophic cardiomyopathy lines. (A) MA plot of RNA-sequencing model developed showing differentially expressed genes (766, false discovery rate < 0.1) between wild-type and diseased (MYH7-mutant) conditions, using cell line and culture format as controlling factors (n = 3). (B) Genes identified were enriched for cardiomyopathy Online Mendelian Inheritance in Man (OMIM) disease. (C) Volcano plots after refinement enabled distinction between diseased states by considering the genotype showed increasing number of differentially expressed genes with mutation load (P < 0.05, red; log2 fold > 1 change, green). Fold ± SD changes in the expression of genes involved in archetypal hypertrophic cardiomyopathy pathways in the MYH7-mutant REBL-PAT-cardiomyocytes s relative to wild-type in (D) 2D cultures and (E) 3D-engineered heart tissues. q-PCR analysis of MYH7/MYH6 expression ratios in gene-edited lines in 2D cultures normalized to wild-type in (F) AT1 and (G) REBL-PAT lines (n = 4). Data, mean ± SD. P-values, one-way ANOVA test + Dunnett’s correction.
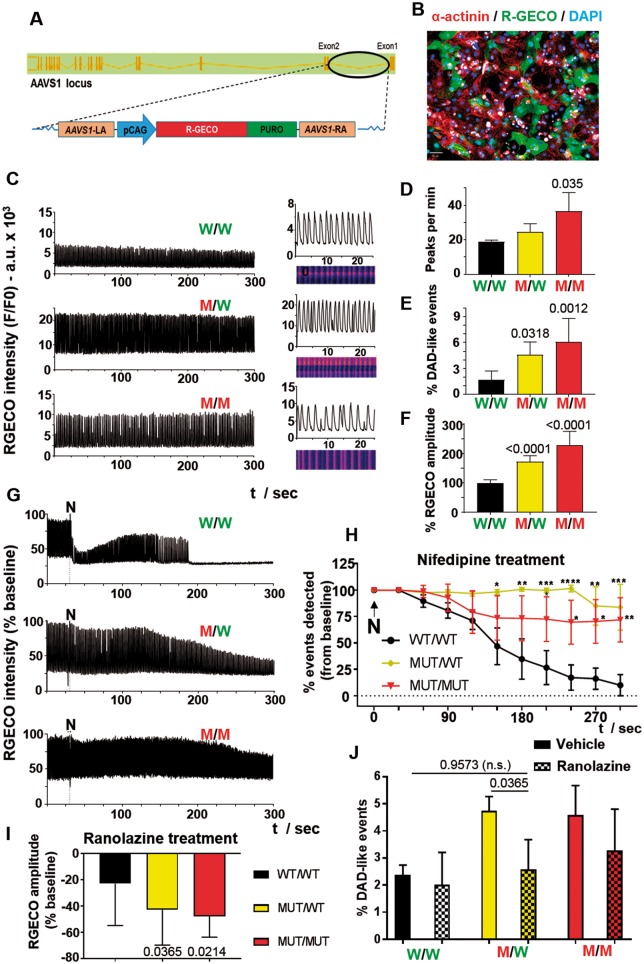
Figure 5.
Calcium handling in hypertrophic cardiomyopathy lines by optical mapping. (A) A red-genetically encoded calcium indicator expression cassette was engineered into AAVS1 of MYH7-mutant human pluripotent stem cell via nickase CRISPR/Cas9 editing. (B) α-Actinin/red-genetically encoded calcium indicator/DAPI-immunostained human pluripotent stem cell-cardiomyocytes post-gene editing demonstrates expression of the red-genetically encoded calcium indicator calcium sensor (Bar = 50 μm). (C) Optical mapping of R453C-β-myosin heavy chain REBL-PAT cardiomyocytes (n = 10) enabled quantification of (D) beat rate, (E) DAD-like abnormal events, and (F) signal amplitude (correlated with increased systolic calcium peak). Treatment of gene-edited lines with 1 μM nifedipine (n = 5) is in (G, H). Impact of 1 μM ranolazine (n = 6) on red-genetically encoded calcium indicator signal amplitude in (I) and frequency of DAD-like events in (J). Data, mean ± SD. P-values, one-way ANOVA test + Dunnett’s correction.
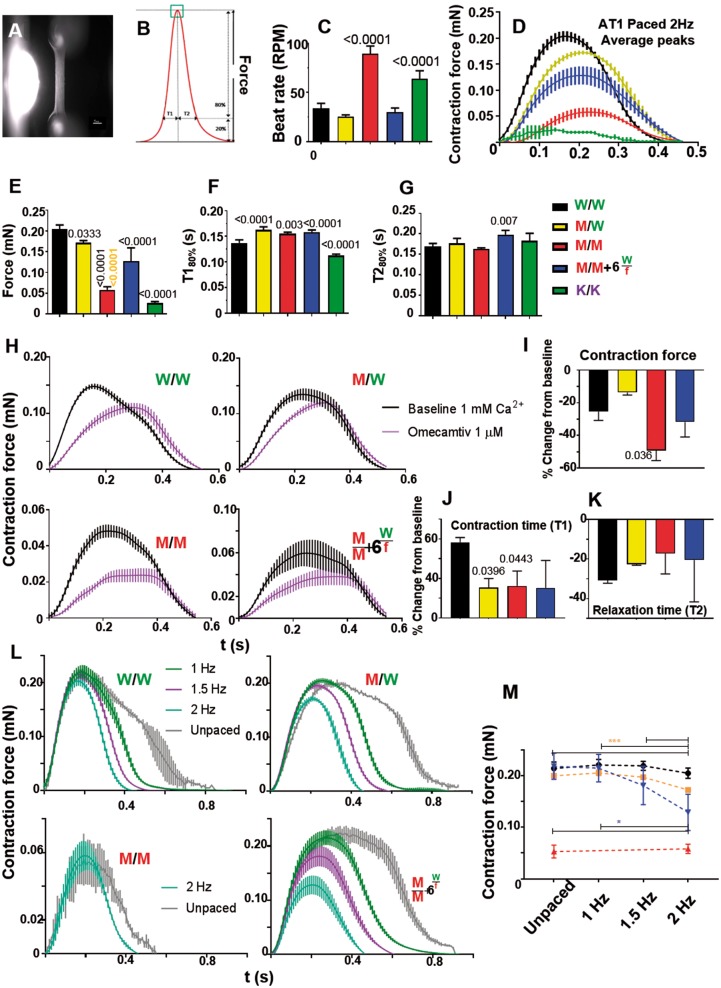
Figure 6.
Contractile force analysis in AT1-human engineered heart tissues. (A) Fibrin-based AT1-human engineered heart tissue attached to silicone posts (Bar = 1 mm). (B) Schematic contraction peak showing parameters analysed, providing data on (C) spontaneous beat rate (n = 8). Electrically paced engineered heart tissues produced average contraction peaks (D), quantified for (E) contraction force, (F) contraction time, and (G) relaxation time (n = 4). (H) 2 Hz electrically paced AT1-engineered heart tissues with or without omecamtiv mecarbil treatment produced average contraction peaks, quantified for (I) contraction force, (J) contraction time, and (K) relaxation time (n = 3). (L) Force-frequency relationship in MYH7-mutant AT1-engineered heart tissues was assessed and quantified in (M). Fast spontaneous beat rate of homozygous R453C-β-myosin heavy chain mutant meant only 2 Hz pacing was possible. (n = 4). Data, mean ± SD. P-values, one-way ANOVA test + Dunnett’s correction.
Molecular assessment of β-MHC mutant hPSC-CMs
We evaluated molecular characteristics of hPSC-CMs from MYH7 isogenic sets to determine which features of HCM were replicated and clarify outstanding or controversial questions. Unequal expression of mutant and wild-type MYH7 alleles was reported in ventricular biopsies from HCM patients.28 We used XhoI-based restriction fragment length polymorphism (RFLP) analysis on isogenic hPSC-CMs (Figure 1F). Real time- Polymerase Chain Reaction (RT-PCR) products from WT/WT lines were refractory to XhoI digestion, while MUT/MUT products were digested to 2 bands. In heterozygotes, the ratio of MUT: WT alleles ranged from 25% to 35% (HUES7, REBL-PAT) to 60% (AT1), confirming unequal expression but also variation between lines.
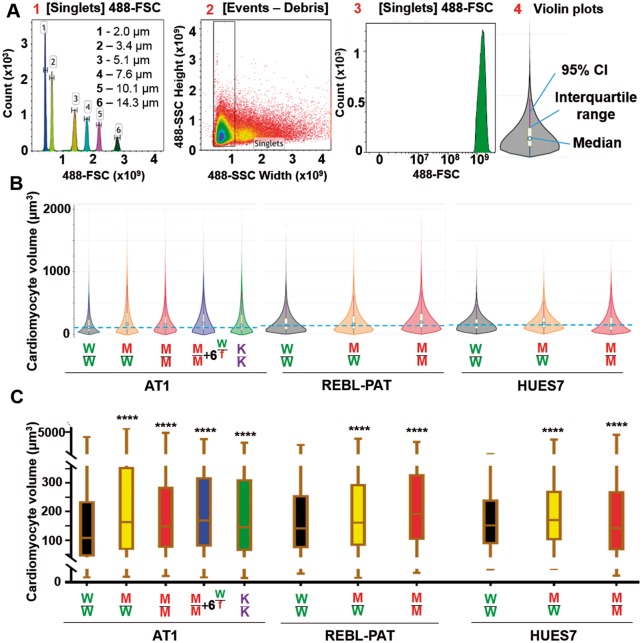
Increased cardiomyocyte size is archetypical of HCM.9 We developed a novel high-throughput (25 000 cells/sample), non-subjective and statistically powerful flow cytometry method to calculate hPSC-CM volume, whilst avoiding pitfalls of 2D analysis, including influence of cell area by substrate properties,29 time in culture,30 and serum supplementation.31 Forward scatter of calibration spheres generated a standard curve, enabling calculation of hPSC-CM size (Figure 2A). Relative to WT/WT, median volume of edited lines increased (12–51%), showing βMHC-R453C mutations cause hypertrophy in hPSC-CMs (Figure 2B, C).
Figure 2.
Flow cytometry calculation of human pluripotent stem cell-cardiomyocyte hypertrophy. (A) Calibration of forward scatter (FSC) from predefined beads sizes allowed human pluripotent stem cell-cardiomyocyte size quantification. (B) Violin plots (25 000 cells/sample) show volume of AT1 (n = 9), REBL-PAT (n = 6), and HUES7 (n = 3) lines. Dotted blue line indicates median of volume of isogenic controls. (C) Box/whiskers plots show interquartile range of volume, and highlights higher median and first quartile metrics in gene-edited lines. P-values, one-way ANOVA test + Dunnett’s correction; ****Significance all <0.0001.
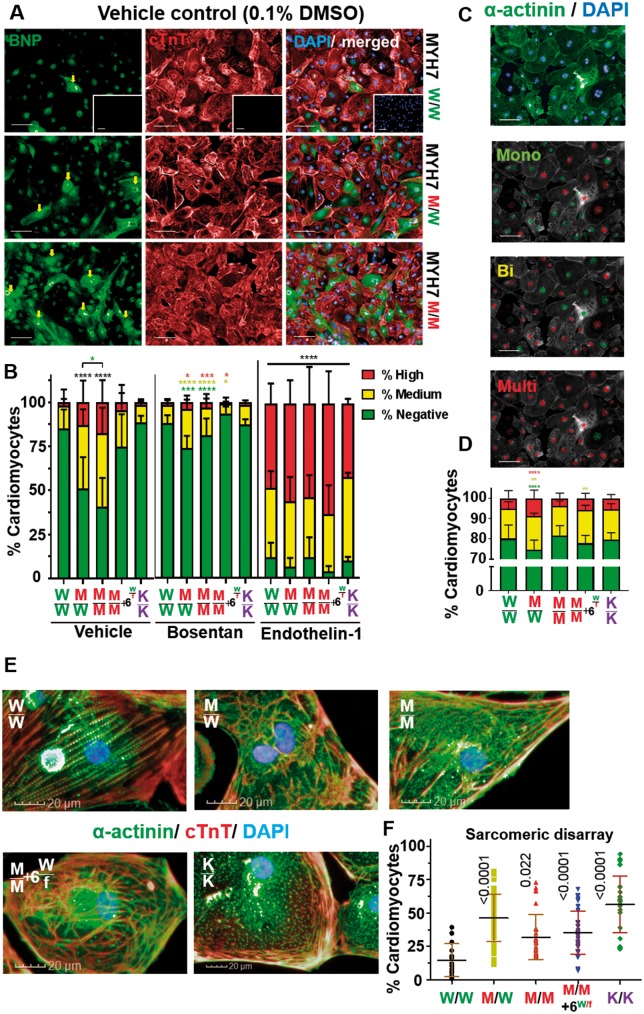
Brain natriuretic peptide (BNP) is elevated >100-fold in plasma from HCM patients.32 We adapted high-content imaging methods33 to assess ≥60 000 cells/sample for BNP expression in hPSC-CMs (Figure 3A). Data were binned into high, medium, or low/negative populations using predetermined empirical thresholds (Figure 3B; Supplementary material online, S4A–C). There was general association between percentages of hPSC-CMs expressing medium/high levels of BNP and increasing mutation load. Relative to WT/WT, WT/MUT, and MUT/MUT were ∼1.5 to 3.3 and ∼1.9- to 4-fold higher, respectively, with consistency across the three hPSC lines. Surprisingly, homozygote + MYH6WT/fs line and MYH7-KO did not show increased BNP expression, suggesting this phenotype could be specific to the βMHC-453 arginine to cysteine substitution. Expression of BNP could also be blocked by treatment with endothelin-1 (ET1) receptor antagonist, bosentan,34 or exaggerated by the known hypertrophic stimulator, ET1.33 Response of edited hPSC-CMs generally differed significantly to WT/WT (Figure 3B; Supplementary material online, S4A–E).
Figure 3.
Phenotyping hypertrophic cardiomyopathy using high-content imaging. (A) brain natriuretic peptide/cardiac Troponin T (cTnT)/DAPI-immunostained human pluripotent stem cell-cardiomyocytes (fibroblasts, negative control; inset). Arrows indicate brain natriuretic peptide positive cells. Bar = 100 μm. (B) Percentage of human pluripotent stem cell-cardiomyocytes with negative, medium, or high brain natriuretic peptide expression were binned using predetermined thresholds. Bosentan treatment (100 nM) rescued brain natriuretic peptide of mutant lines to isogenic control levels; Endothelin-1 treatment (10 nM) maximized brain natriuretic peptide expression (n = 6). (C) Images of α-actinin/DAPI-immunostained human pluripotent stem cell-cardiomyocytes analysed by the algorithm to distinguish between mono-, bi-, and multi-nucleation (Bar = 100 μm), with (D) displaying quantification (n = 8). (E) Images of human pluripotent stem cell-cardiomyocytes immunostained for sarcomeric banding (Bar = 20 μm), with (F) displaying quantification (n = 4). Data, mean ± SD. P-values shown. One-way ANOVA test + Dunnett’s correction compared mutant lines vs. their isogenic control. Student’s t-tests, treated vs. vehicle control (*P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.0001, absolute numbers in Supplementary material online, Table S4). Colour-coded by inter-compared category (black asterisks apply to all categories).
Multi-nucleation is a controversial feature of HCM in human hearts. While some reports found bi-nucleated cardiomyocytes increased in hypertrophied hearts (28.7% vs. 13.5% for healthy),35 others observed no differences.36 We performed dual α-actinin/4′,6-diamidino-2-phenylindole (DAPI) staining of hPSC-CMs from isogenic sets and developed a high-content imaging algorithm (40 000 cells/line) to distinguish mono-, bi-, and multi-nucleation (Figure 3C, D; Supplementary material online, Figure S4F–G). Strikingly, across all isogenic sets, WT/MUT showed a significant increase in bi- and multi-nucleation (1.9–3.6% and 1.1–1.9%) relative to WT/WT, suggesting an association with ratio of healthy: mutant βMHC proteins.
We explored sarcomeric structure, since myofibrillar disarray was reported in HCM hiPSC-CM lines17 and associates with cardiomyocyte dysfunction.37 To overcome commonly reported subjective methods,38,39 we developed a novel high-content, machine learning approach to identify morphology and texture of ≥40 000 cells/line (Figure 3E; Supplementary material online, Figure S4H, I). While disarrayed sarcomeres were detected in 14.0 ± 2.6% of WT/WT hPSC-CMs, significant increases were observed in all mutant lines, with a four-fold increase (to 56.4 ± 4.3%) in MYH7-KO (Figure 3F). Thus, disruption trended as a function of mutation load in R453C-βMHC hPSC-CMs.
Since βMHC is known to interact with multiple proteins in, or associated with, the sarcomere, we explored the impact of the R453C mutation. Previously, elegant experiments using yeast-two-hybrids showed that mutations in cMyBP-C abolished interaction with βMHC S2 (tail) domains.40 However, in yeast, expression of full length proteins of >1500 amino acids, such as βMHC is challenging.41 As an alternative, we used in silico modelling. Current structural homology models42–45 indicate the head domain (S1) of βMHC interacts with: itself (via S1–S2); cMyBP-C; actin; ATP.
R453 is located between the HCM loop and Switch-2 of βMHC S1 and interacts with the proximal S2 region when folded back. The change to cysteine is predicted to interfere with this S1–S2 βMHC interaction, by disrupting the hydrogen bond established between R453 and Q882 (Supplementary material online, Figure S5A–C). R453 is located close to the interface of cMyBP-C (Supplementary material online, Figure S5D) and, in molecular dynamics simulations, can form contacts with its C1 domain (Supplementary material online, Figure S5E). Contrastingly, R453 is located away from the predicted interface of S1 with actin (Supplementary material online, Figure S5F) and ATP-binding region (Supplementary material online, Figure S5G), so the mutation is not predicted to interfere directly with these interactions.45 However, targeted molecular dynamics simulations have shown R453C to cause changes in the flexibility of the loop between the motor domain and the actin binding site.46
Mitochondrial respiration rates are perturbed in βMHC mutant hPSC-CMs
Although controversial, one working hypothesis states that mutant proteins within the sarcomere cause inefficient sarcomeric ATP utilization, energy depletion, increased oxygen consumption, and cardiac dysfunction.47,48 To determine whether or not our model supported this hypothesis, we analysed the isogenic hPSC-CMs with the Seahorse platform (Figure 4A; Supplementary material online, Figure S6A). This profiled oxygen consumption rates (OCR) during sequential addition of electron transport chain inhibitors, enabling calculation of basal and maximal respiration rates. The Seahorse also measures ATP production in a manner that correlates to outputs from other direct approaches, such as Luciferase ATP assay.49 There was a positive association between these parameters and mutation load, such that MYH7-KO and homozygote + MYH6WT/fs had the highest values followed in order by MUT/MUT, WT/MUT, and WT/WT (Figure 4B–D; Supplementary material online, Figure S6B–D). This was most striking in AT1-hiPSC-CMs, where basal respiration, maximal respiration and ATP production increased by ∼3-, 2.75-, and ∼3-fold, respectively.
Surprisingly, these changes were not due to greater mitochondrial content, since mitochondrial: nuclear DNA ratio50 showed little difference (Figure 4E; Supplementary material online, Figure S6E). This was supported by flow cytometry using Mitotracker® (Figure 4F, G; Supplementary material online, Figure S6F, G), which reports on mitochondrial content and function.51 We speculated that similar mitochondria content in the mutant lines necessitated harder work to meet the energy demands, hence lead to increased reactive oxygen species (ROS) and cell stress. However, flow cytometry calculation of total cell ROS or mitochondrial-specific ROS showed little interline difference (Figure 4H–K; Supplementary material online, S6H–K). Thus, the isogenic lines supported the energy depletion model but did not suggest any dramatic increases in cell stress via ROS production, at least under these test conditions.
βMHC mutant hPSC-CMs show altered calcium handling
Calcium handling is central in excitation–contraction regulation, hence development of HCM. We used nickase CRISPR/Cas9 to knock-in a red genetically encoded calcium indicator (R-GECO1) expression cassette into the safe AAVS1 locus of isogenic REBL-PAT-hiPSC-CM trio (Figure 5A, B; Supplementary material online, Figure S7A–D). Calcium imaging was analysed by confocal line scans (Figure 5C–F). Relative to WT/WT, there was an upward trend in beat rate (50% in MUT/MUT), frequency of delayed after depolarization (DAD)-like events interspersing the main peaks (>9-fold increase in MUT/MUT) and signal amplitude/higher systolic calcium peak (WT/MUT, ∼1.72-fold; MUT/MUT, ∼2.27-fold). Availability of cytosolic calcium to trigger beating in diseased lines for longer was corroborated by treatment of cardiomyocytes with 1 μM nifedipine, an L-type calcium channel blocker (Figure 5G, H).
These findings suggested that higher cytosol calcium concentrations caused DAD-like arrhythmias in mutant hPSC-CMs. To explore whether pharmacological rescue was possible, hPSC-CMs were treated with 1 μM ranolazine, which acts as an enhancer of the outward mode of sodium-calcium exchanger (NCX) by blocking late sodium currents, hence indirectly promotes Ca2+ efflux.52 This led to reduced R-GECO1 signal amplitude and frequency of DAD-like events in diseased lines, particularly WT/MUT (Figure 5I, J, Supplementary material online, Figure S7E). Thus, altered calcium handling and arrhythmogenesis were identified in R453C-βMHC lines, and partial rescue could be achieved with ranolazine.
3D engineering unveils hypo-contractility, negative clinotropy, and an exacerbated negative force–frequency relationship
Contraction is the fundamental purpose of the heart. Human engineered heart tissues (hEHTs) directly measure contraction force by partially recapitulating the 3D architecture of cardiac tissue by imposing the auxotonic tension present in vivo.53,54 We produced hEHTs from AT1 and REBL-PAT isogenics (Figure 6A, B), wherein cardiomyocytes exhibited excellent alignment (Supplementary material online, Figure S8A). Beat rate was fast in MUT/MUT and MYH7-KO (Figure 6C). Analysis of force under 2 Hz pacing (Figure 6D–G; Supplementary material online, Figure S8C–F), contraction time (T180%), and relaxation time (T280%) produced trends similar to the 2D assays; mutation load associated with poorer functional output. There was a predominance of hypocontractility and negative clinotropy (increased T180%), although little change in T280%. Contraction in MYH7-KO was so compromised (∼ten-fold lower force than WT/WT), measurements were almost impossible (Figure 6D, E).
We attempted pharmacological rescue of reduced contraction force and increased contraction time in hEHTs formed from mutant hPSC-CMs. Omecamtiv mecarbil is a cardiac myosin activator that acts by prolonging the actin–myosin interaction state, thereby extending systolic ejection time and increasing cardiac contractility.55 AT1- and REBL-PAT-hEHTs treated with 1 µM omecamtiv mecarbil enhanced negative clinotropy but, unexpectedly, decreased contractile force, apparently exacerbating impact of the 453-βMHC mutation (Figure 6H–K; Supplementary material online, Figure S8G–J).
The inability of HCM-afflicted hearts to produce more force during exercise-induced increases in beat rate contributes to sudden cardiac death.56 We simulated this scenario in hEHTs. In AT1-hiPSC-CM-EHTs, force declined as a function of pacing frequency in WT/MUT and homozygous + MYH6WT/fs lines (Figure 6L, M). Measurement was not possible in AT1-MUT/MUT due to the high spontaneous beat rate. Unlike AT1-EHTs, decline in force was not seen during stepped pacing of hEHTs formed from REBL-PAT-hiPSC-CMs (Supplementary material online, Figure S8K). This may be due to their high baseline beat rates (∼1.5 to 2 Hz) and/or low intrinsic force production (0.04–0.06 mN) (Supplementary material online, Figure S8B), or may relate to the higher expression of the mutant allele in AT1 vs. REBL-PAT heterozygotes (60% vs. 35%, respectively) (Figure 1F).
RNA-seq highlights lncRNAs as potential therapeutic candidates for HCM
To provide new insight into HCM, we used global transcriptome analysis of isogenic hPSC-CM cultured as 2D monolayers (AT1, REBL-PAT, HUES7) and hEHTs in 3D (AT1, REBL-PAT). Principal component analysis (PCA) showed that hPSC culture format and cell line origin were the main variance factors (Supplementary material online, Figure S9A), reinforcing the importance of isogenic controls to model disease.20 Analysis from all variants in 2D and 3D identified 290 differentially expressed genes [<10% false discovery rate (FDR) Supplementary material online, Figure S9B, C]. Two layers of refinement were applied: first, correction for cell line of origin and culture format returned 766 genes (FDR < 0.1; Figure 7A). These included loci associated with several cardiomyopathies (Figure 7B, Supplementary material online, Figure S9D), but also previously unidentified lncRNAs as top hits (2- to 8-fold change); Second, sub-classifying genotypes within the diseased category into volcano plots showed number of differentially expressed genes associated with increasing mutation load (Figure 7C), following the trend of the phenotypic assays.
Focused transcriptomics reveals core pathways triggered by HCM
Focused analysis of 2D and 3D samples for AT1 and REBL-PAT isogenic sets via a ∼50 genes nanoString RNA chip (Figure 7D, E; Supplementary material online, S9F–G) enabled querying of genes involved in (i) foetal gene programme, (ii) hypertrophy, (iii) calcium handling, (iv) apoptosis, (v) fibrosis, and (vi) autophagy. Data from 2D hPSC-CMs identified increased expression of genes involved in the Foetal programme (NPPA/B, validating BNP data), hypertrophic responses (FHL1/2), apoptosis (CASP3), and fibrosis (FN1). While changes were sustained in 3D (mainly fibrosis and hypertrophy), certain opposing trends were seen, corroborating conclusions from RNAseq data on the importance of culture format. In 3D, decreases in the genes involved in calcium handling machinery were found, while changes in apoptosis and autophagy were less pronounced.
From the transcriptional data, we noted changes in expression of MYH7 and MYH6. These were confirmed by qRT-PCR analysis, showing ∼5- to 15-fold increases in the MYH7:MYH6 ratio in the diseased lines across all three isogenic groups (Figure 7F, G; Supplementary material online, Figure S9E). This is consistent with the 3.5-fold change in MYH7:MYH6 ratio caused by hypertrophy of human hearts.57 These observations suggest a compensatory feedback loop, whereby sarcomere inefficiency downregulates the ‘energy hungry’ fast αMHC isoform in favour of the normally ‘energy-efficient’ βMHC isoform. Altogether, transcriptomic analyses highlighted foetal gene programme initiation, hypertrophic responses, and αMHC to βMHC isoform switching as the main pathways triggered in HCM.
Discussion
New investigative tools are needed for HCM. Few pharmacological treatments exist, and the condition can necessitate surgery and/or heart transplantation.58 Advancements are confounded by heterogeneity, wherein reproducibility of genotype–phenotype correlations are challenging because human material is limited and frequency of the same ‘natural’ mutations within families is low, compromising statistical power.59 We overcame these issues by creating 11 isogenic variants in three different hPSC lines centred on a c.C9123T-MYH7 (p.R453C-βMHC) substitution; until now, engineered homozygotes have not been reported for human-based HCM. Our data demonstrated unequivocally that a single R453C-βMHC mutation causes a severe and penetrant pathophysiology independent of genetic background.60
Our use of 12 different phenotyping approaches far exceeds previous studies,21 showing salient features of HCM were recreated in the hPSC-CMs expressing the mutant βMHC. This validation is essential since hPSC-CMs are often cited as being immature and hence not representative of the adult cardiomyocyte or intact myocardium. Our comprehensive approach, coupled with other evidence that hPSC-CMs replicate morphology,53 contractility,53 electrophysiology,61 signalling,62 and metabolism,63 gives confidence that the outcomes we observed in vitro are also relevant for HCM in vivo. hPSC-CM immaturity may even be advantageous by modelling early disease stages, which is particularly relevant for R453-βMHC patients who typically show an early onset of heart failure.60 This is when treatment is most likely to be effective, hence will be useful for further mechanistic dissection, development of diagnostics, and drug testing. A next logical step will be further refinement by generating complex tissues or organ-on-a-chip. This will require production of other cells types found in the heart, such as cardiac fibroblasts, endothelial cells, and smooth muscle cells, although robust protocols for hPSC-based differentiation of these lineages are currently at various stages of development.64,65
For many of the molecular assays and functional phenotyping, the level of dysfunction associated well with mutation load but sometimes differed between the three hPSC lines, mirroring HCM complexity.59 Closer inspection showed association with the ratio of MUT:WT MYH7 allele expression. Heterozygote AT1 showed the most severe phenotypes, and had a MUT:WT ratio of 60%, followed by REBL-PAT (35%) and HUES7 (25%). This raises the intriguing possibility of whether this ratio could be a diagnostic predictor of severity of pathophysiology in patients.28,66
Isogenic sets will add clarity to the field. The impact of HCM on mitochondrial respiration is controversial. Explants of human hypertrophied hearts showed ∼two-fold higher OCR,48 whereas skinned muscle bundles obtained from myocardium of explanted human hearts showed no difference relative to healthy controls.67 These previous studies have been constrained by variability and scarceness of the material, limitations overcome by isogenic hPSC-CMs Our data on mitochondrial function/content, calcium handling and the transcriptome support the energy depletion model of HCM, which states disorganised sarcomere causes inefficient ATP usage and imposes increased energetic demands on the cardiomyocyte.47 This compromises the energy available to reduce cytosolic calcium levels back to baseline, precipitating arrhythmogenesis. Interestingly, stress cues, such as energy deficits, are known to trigger ploidy-activated genes, promoting the α-MHC to β-MHC isoform switch, which leads to enhanced production of ATP and allows the cardiomyocytes to be more energy-efficient.57,68 Our observation of cell stress via energy depletion, increased multi-nucleation (potentially leading to polyploidy) and isoform switch supports this as an underlying mechanism of R453C-βMHC mediated HCM.
The isogenic sets supported the notion that mitochondrial content in failing hearts is not increased.69 We expected that increased demand from the same number of mitochondria in cardiomyocytes with inefficient sarcomeres would lead to increased ROS and cell stress, as reported in mitochondrial cardiomyopathies.70 At least under baseline spontaneous beating, this proved incorrect. High frequency pacing (2–4 Hz) may unveil further phenotypes. While this is not possible on the Seahorse platform, the advent of optogenetics-based methods for pacing hPSC-CMs71 may provide a future route of enquiry.
Our data on contractility are compatible with literature investigating HCM in human cells and tissues, but contradict rodent studies. In R453C-βMHC hPSC-CMs, we observed sarcomeric disarray and hypo-contractility, despite increased metabolic demands. Force generation in R723G-βMHC human heart biopsies or E848-βMHC hPSC-CM myofibrils was 30–66% lower.37,72 Tension was ∼35% lower in several βMHC-mutant tissues,73,74 whereas tension-cost was higher in R403Q-βMHC75 and R403W-βMHC76 variants. Contrastingly, murine C2C12 myoblasts expressing recombinant R453C-βMHC showed increased force generation and reduced maximum ATPase activity.77 Rodent cardiomyocyte-derived EHTs bearing missense mutations in sarcomeric genes (FHL2,78ANKRD1,79 and MYBPC380,81) showed hypercontractile phenotypes.
These differences may underscore the delicate balance between α-MHC and β-MHC expression, which is impacted on by species differences and/or transgenic overexpression. Isoform switch from β-MHC to α-MHC occurs in ventricles of mice during development,82 but β-MHC is always predominant in human ventricles.83 Hypercontractile phenotypes associated with α-MHC mutations in mice84,85 may cause opposing effects in the human predominant ventricular counterpart (β-MHC). This may explain why the homozygous + MYH6fs/wt we describe showed a higher contractile force than the homozygous line that lacks the off-target MYH6 event. This is supported the observation that multi-nucleation, BNP expression and foetal gene programme initiation were not exhibited by hPSC-CMs bearing the additional MYH6 mutation. Subtlety in levels of α-MHC and β-MHC isoforms may also explain why only heterozygous R453C-βMHC hPSC-CMs showed increased proportion of multi-nucleated cells.
The isogenic sets of hPSC-CMs described here will be useful to evaluate new therapies, building upon current strategies.86 Ranolazine reduced the number of DAD-like arrhythmias in heterozygous R453C-βMHC cardiomyocytes, which is the genotype that reflects most closely the clinical situation for patients. This suggests that further evaluation of drugs that modulate calcium within this model system will be warranted. Omecamtiv mecarbil caused negative clinotropy, consistent with the known mode of action of this drug,55 but did not rescue the hypocontractility. We speculate this may be due to disruption of sarcomeric interactions by the R453-βMHC mutation, as predicted by in silico modelling (Supplementary material online, Figure S5), high beat rates or to lack of t-tubules, which could prevent drug-induced activation of myosin.
In summary, we generated a scalable human model of HCM by using CRISPR/Cas9 to produce isogenic sets of C9123T-MYH7 (R453C-βMHC) mutants in hPSC-CMs. The utility of the model was validated and now points towards routes for pharmacological rescue and diagnostics. Identification of novel lncRNAs and putative gene modifiers provide an avenue for new mechanistic and functional understanding via knockout, overexpression and pathways analysis, whereas suggesting new putative diagnostic biomarkers and targets for therapy. This model (Figure 8) will pave the way in evaluating single or combined drug- and/or gene-based therapeutics for HCM.
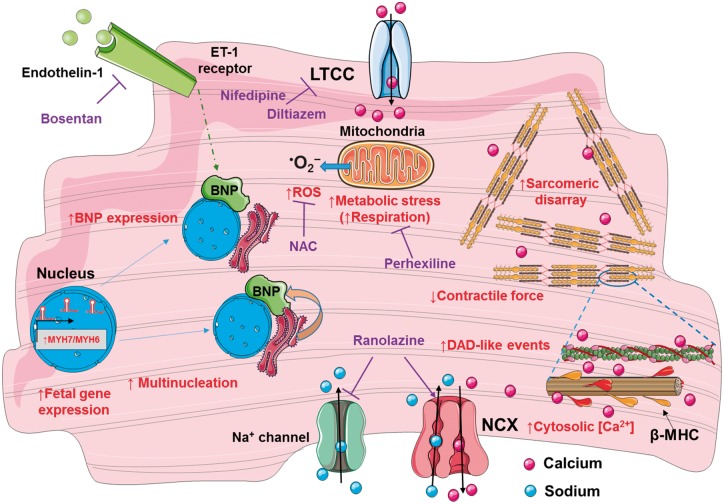
Figure 8.
Proposed hypertrophic cardiomyopathy mechanisms. Phenotypes investigated (red) in the developed model of hypertrophic cardiomyopathy of R453C-β-myosin heavy chain human pluripotent stem cell-cardiomyocytes; putative rescue drugs (purple). LTCC, L-type calcium channels; NAC, N-acetyl-cysteine; NCX, sodium-calcium exchange pump.
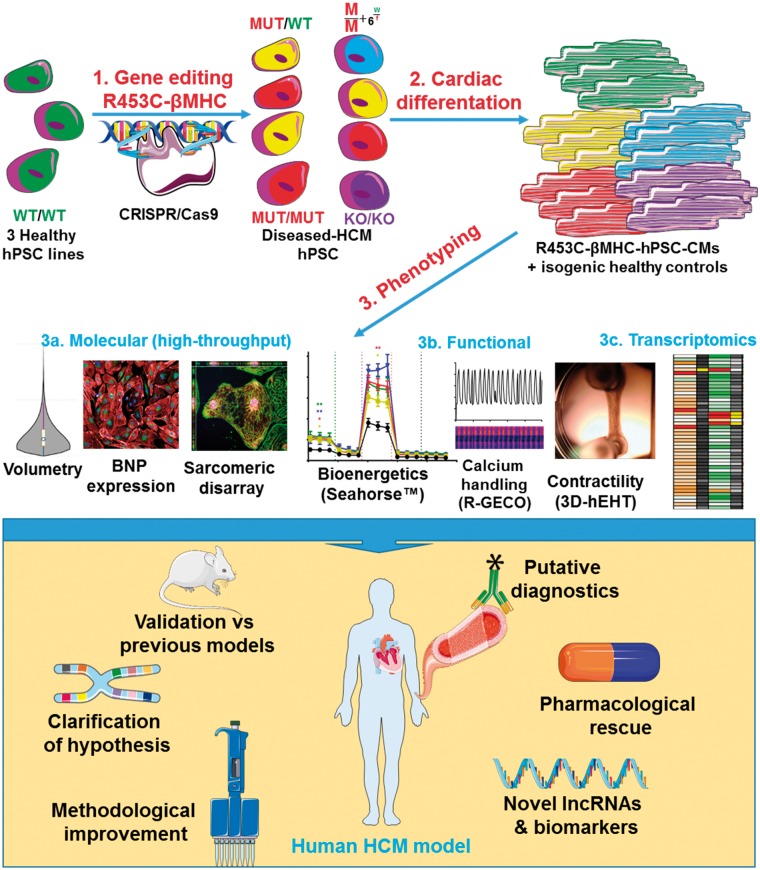
Take home figure.
1) The R453C-betaMHC pathological change was introduced in three independent healthy hPSC lines using CRISPR/Cas9. 2) The gene edited hPSC lines were differentiated to generate isogenic sets of hPSC-cardiomyocytes. 3) Phenotyping of hPSC-CMs in terms of a) molecular, b) functional and c) transcriptomics analyses has validated the human HCM model generated, leading to new mechanistic and pharmacological understanding of the disease.
Supplementary Material
Acknowledgements
Authors thank BHF, BIRAX, MRC, NC3Rs, Heart Research UK, German Research Foundation, ERC, EC, DZHK, German Ministry of Education and Research, and the Freie und Hansestadt Hamburg.
Funding
This work was supported by the British Heart Foundation [grant numbers SP/15/9/31605, RG/15/6/31436, PG/14/59/31000, RG/14/1/30588, RM/13/30157, P47352/CRM]; Britain Israel Research and Academic Exchange Partnership [04BX14CDLG]; the Medical Research Council [grant number MR/M017354/1: MICA: Development of Metrics and Quality Standards for Scale up of Human Pluripotent Stem Cells]; the National Centre for the Replacement, Refinement & Reduction of Animals in Research [grant numbers CRACK-IT:35911-259146, NC/K000225/1]; the German Research Foundation [DFG-Es-88/12-1, HA3423/5-1]; European Research Council [ERC-AG-IndivuHeart]; European Commission [FP7-Biodesign]; German Centre for Cardiovascular Research (DZHK) and the German Ministry of Education and Research, the Freie und Hansestadt Hamburg.
Conflict of interest: none declared.
References
- 1. Elliott PM, Anastasakis A, Borger MA, Borggrefe M, Cecchi F, Charron P, Hagege AA, Lafont A, Limongelli G, Mahrholdt H, McKenna WJ, Mogensen J, Nihoyannopoulos P, Nistri S, Pieper PG, Pieske B, Rapezzi C, Rutten FH, Tillmanns C, Watkins H.. ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy. The Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J 2014;35:2733–2779. [DOI] [PubMed] [Google Scholar]
- 2. Semsarian C, Ingles J, Maron MS, Maron BJ.. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol 2015;65:1249–1254. [DOI] [PubMed] [Google Scholar]
- 3. Harris KM, Spirito P, Maron MS, Zenovich AG, Formisano F, Lesser JR, Mackey-Bojack S, Manning WJ, Udelson JE, Maron BJ.. Prevalence, Clinical profile, and significance of left ventricular remodeling in the end-stage phase of hypertrophic cardiomyopathy. Circulation 2006;114:216–225. [DOI] [PubMed] [Google Scholar]
- 4. Sedaghat-Hamedani F, Kayvanpour E, Tugrul OF, Lai A, Amr A, Haas J, Proctor T, Ehlermann P, Jensen K, Katus HA, Meder B.. Clinical outcomes associated with sarcomere mutations in hypertrophic cardiomyopathy: a meta-analysis on 7675 individuals. Clin Res Cardiol 2018;107:30–41. [DOI] [PubMed] [Google Scholar]
- 5. Melacini P, Basso C, Angelini A, Calore C, Bobbo F, Tokajuk B, Bellini N, Smaniotto G, Zucchetto M, Iliceto S, Thiene G, Maron BJ.. Clinicopathological profiles of progressive heart failure in hypertrophic cardiomyopathy. Eur Heart J 2010;31:2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cahill TJ, Ashrafian H, Watkins H.. Genetic cardiomyopathies causing heart failure. Circ Res 2013;113:660–675. [DOI] [PubMed] [Google Scholar]
- 7. Marian AJ. Modifier genes for hypertrophic cardiomyopathy. Curr Opin Cardiol 2002;17:242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ingles J, Burns C, Bagnall RD, Lam L, Yeates L, Sarina T, Puranik R, Briffa T, Atherton JJ, Driscoll T, Semsarian C.. Nonfamilial hypertrophic cardiomyopathy—prevalence, natural history, and clinical implications. Circ Cardiovasc Genet 2017;10:e001620.. [DOI] [PubMed] [Google Scholar]
- 9. Harvey PA, Leinwand LA.. The cell biology of disease: cellular mechanisms of cardiomyopathy. J Cell Biol 2011;194:355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keren A, Syrris P, McKenna WJ.. Hypertrophic cardiomyopathy: the genetic determinants of clinical disease expression. Nat Clin Pract Cardiovasc Med 2008;5:158–168. [DOI] [PubMed] [Google Scholar]
- 11. Stehle R, Iorga B.. Kinetics of cardiac sarcomeric processes and rate-limiting steps in contraction and relaxation. J Mol Cell Cardiol 2010;48:843–850. [DOI] [PubMed] [Google Scholar]
- 12. Van Driest SL, Jaeger MA, Ommen SR, Will ML, Gersh BJ, Tajik AJ, Ackerman MJ.. Comprehensive analysis of the beta-myosin heavy chain gene in 389 unrelated patients with hypertrophic cardiomyopathy. J Am Coll Cardiol 2004;44:602–610. [DOI] [PubMed] [Google Scholar]
- 13. Marian AJ, Roberts R.. The molecular genetic basis for hypertrophic cardiomyopathy. J Mol Cell Cardiol 2001;33:655–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Denning C, Borgdorff V, Crutchley J, Firth KSA, George V, Kalra S, Kondrashov A, Hoang MD, Mosqueira D, Patel A, Prodanov L, Rajamohan D, Skarnes WC, Smith JGW, Young LE.. Cardiomyocytes from human pluripotent stem cells: from laboratory curiosity to industrial biomedical platform. Biochim Biophys Acta 2016;1863:1728–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nagueh SF, Chen S, Patel R, Tsybouleva N, Lutucuta S, Kopelen HA, Zoghbi WA, Quiñones MA, Roberts R, Marian AJ.. Evolution of expression of cardiac phenotypes over a 4-year period in the β-myosin heavy chain-Q403 transgenic rabbit model of human hypertrophic cardiomyopathy. J Mol Cell Cardiol 2004;36:663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lowey S, Lesko LM, Rovner AS, Hodges AR, White SL, Low RB, Rincon M, Gulick J, Robbins J.. Functional effects of the hypertrophic cardiomyopathy R403Q mutation are different in an α- or β-myosin heavy chain backbone. J Biol Chem 2008;283:20579–20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tanaka A, Yuasa S, Mearini G, Egashira T, Seki T, Kodaira M, Kusumoto D, Kuroda Y, Okata S, Suzuki T, Inohara T, Arimura T, Makino S, Kimura K, Kimura A, Furukawa T, Carrier L, Node K, Fukuda K.. Endothelin‐1 induces myofibrillar disarray and contractile vector variability in hypertrophic cardiomyopathy-induced pluripotent stem cell–derived cardiomyocytes. J Am Heart Assoc 2014;3:e001263.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lan F, Lee Andrew S, Liang P, Sanchez-Freire V, Nguyen Patricia K, Wang L, Han L, Yen M, Wang Y, Sun N, Abilez Oscar J, Hu S, Ebert Antje D, Navarrete Enrique G, Simmons Chelsey S, Wheeler M, Pruitt B, Lewis R, Yamaguchi Y, Ashley Euan A, Bers Donald M, Robbins Robert C, Longaker Michael T, Wu Joseph C.. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 2013;12:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han L, Li Y, Tchao J, Kaplan AD, Lin B, Li Y, Mich-Basso J, Lis A, Hassan N, London B, Bett GCL, Tobita K, Rasmusson RL, Yang L.. Study familial hypertrophic cardiomyopathy using patient-specific induced pluripotent stem cells. Cardiovasc Res 2014;104:258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sala L, Bellin M, Mummery CL.. Integrating cardiomyocytes from human pluripotent stem cells in safety pharmacology: has the time come? Br J Pharmacol 2017;174:3749–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stillitano F, Turnbull IC, Karakikes I, Nonnenmacher M, Backeris P, Hulot J-S, Kranias EG, Hajjar RJ, Costa KD.. Genomic correction of familial cardiomyopathy in human engineered cardiac tissues. Eur Heart J 2016;37:3282–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chuva de Sousa Lopes SM, Hassink RJ, Feijen A, van Rooijen MA, Doevendans PA, Tertoolen L, Brutel de la Rivière A, Mummery CL.. Patterning the heart, a template for human cardiomyocyte development. Dev Dyn 2006;235:1994–2002. [DOI] [PubMed] [Google Scholar]
- 23. Anderson D, Self T, Mellor IR, Goh G, Hill SJ, Denning C.. Transgenic enrichment of cardiomyocytes from human embryonic stem cells. Mol Ther 2007;15:2027–2036. [DOI] [PubMed] [Google Scholar]
- 24. Ma J, Guo L, Fiene SJ, Anson BD, Thomson JA, Kamp TJ, Kolaja KL, Swanson BJ, January CT.. High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. Am J Physiol 2011;301:H2006–H2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, Denning C.. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J 2011;32:952–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Du David TM, Hellen N, Kane C, Terracciano Cesare MN.. Action potential morphology of human induced pluripotent stem cell-derived cardiomyocytes does not predict cardiac chamber specificity and is dependent on cell density. Biophys J 2015;108:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rajamohan D, Kalra S, Duc Hoang M, George V, Staniforth A, Russell H, Yang X, Denning C.. Automated electrophysiological and pharmacological evaluation of human pluripotent stem cell-derived cardiomyocytes. Stem Cells Dev 2016;25:439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tripathi S, Schultz I, Becker E, Montag J, Borchert B, Francino A, Navarro-Lopez F, Perrot A, Özcelik C, Osterziel K-J, McKenna WJ, Brenner B, Kraft T.. Unequal allelic expression of wild-type and mutated β-myosin in familial hypertrophic cardiomyopathy. Basic Res Cardiol 2011;106:1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Engler AJ, Carag-Krieger C, Johnson CP, Raab M, Tang H-Y, Speicher DW, Sanger JW, Sanger JM, Discher DE.. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J Cell Sci 2008;121:3794–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lundy SD, Zhu W-Z, Regnier M, Laflamme MA.. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev 2013;22:1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dambrot C, Braam SR, Tertoolen LGJ, Birket M, Atsma DE, Mummery CL.. Serum supplemented culture medium masks hypertrophic phenotypes in human pluripotent stem cell derived cardiomyocytes. J Cell Mol Med 2014;18:1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gardner DG. Natriuretic peptides: markers or modulators of cardiac hypertrophy? Trends Endocrinol Metab 2003;14:411–416. [DOI] [PubMed] [Google Scholar]
- 33. Carlson C, Koonce C, Aoyama N, Einhorn S, Fiene S, Thompson A, Swanson B, Anson B, Kattman S.. Phenotypic screening with human iPS cell-derived cardiomyocytes. J Biomol Screen 2013;18:1203–1211. [DOI] [PubMed] [Google Scholar]
- 34. Harada M, Itoh H, Nakagawa O, Ogawa Y, Miyamoto Y, Kuwahara K, Ogawa E, Igaki T, Yamashita J, Masuda I, Yoshimasa T, Tanaka I, Saito Y, Nakao K.. Significance of ventricular myocytes and nonmyocytes interaction during cardiocyte hypertrophy: evidence for endothelin-1 as a paracrine hypertrophic factor from cardiac nonmyocytes. Circulation 1997;96:3737–3744. [DOI] [PubMed] [Google Scholar]
- 35. Schneider R, Pfitzer P.. Number of nuclei in isolated human myocardial cells. Virchows Arch B Cell Pathol 1972;12:238–258. [PubMed] [Google Scholar]
- 36. Vliegen HW, van der Laarse A, Cornelisse CJ, Eulderink F.. Myocardial changes in pressure overload-induced left ventricular hypertrophy. A study on tissue composition, polyploidization and multinucleation. Eur Heart J 1991;12:488–494. [DOI] [PubMed] [Google Scholar]
- 37. Kraft T, Witjas-Paalberends ER, Boontje NM, Tripathi S, Brandis A, Montag J, Hodgkinson JL, Francino A, Navarro-Lopez F, Brenner B, Stienen GJM, van der Velden J.. Familial hypertrophic cardiomyopathy: functional effects of myosin mutation R723G in cardiomyocytes. J Mol Cell Cardiol 2013;57:13–22. [DOI] [PubMed] [Google Scholar]
- 38. Wyles SP, Hrstka SC, Reyes S, Terzic A, Olson TM, Nelson TJ.. Pharmacological modulation of calcium homeostasis in familial dilated cardiomyopathy: an in vitro analysis from an RBM20 patient-derived iPSC model. Clin Transl Sci 2016;9:158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC, Wu JC.. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med 2012;4:130ra47–130ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Lange WJ, Grimes AC, Hegge LF, Spring AM, Brost TM, Ralphe JC.. E258K HCM-causing mutation in cardiac MyBP-C reduces contractile force and accelerates twitch kinetics by disrupting the cMyBP-C and myosin S2 interaction. J Gen Physiol 2013;142:241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakayama M, Kikuno R, Ohara O.. Protein–protein interactions between large proteins: two-hybrid screening using a functionally classified library composed of long cDNAs. Genome Res 2002;12:1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nag S, Trivedi DV, Sarkar SS, Adhikari AS, Sunitha MS, Sutton S, Ruppel KM, Spudich JA.. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat Struct Mol Biol 2017;24:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Spudich James A. The myosin mesa and a possible unifying hypothesis for the molecular basis of human hypertrophic cardiomyopathy. Biochem Soc Trans 2015;43:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Colegrave M, Peckham M.. Structural implications of β-cardiac myosin heavy chain mutations in human disease. Anat Rec 2014;297:1670–1680. [DOI] [PubMed] [Google Scholar]
- 45. Lorenz M, Holmes KC.. The actin-myosin interface. Proc Natl Acad Sci USA 2010;107:12529–12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bloemink M, Deacon J, Langer S, Vera C, Combs A, Leinwand L, Geeves MA.. The hypertrophic cardiomyopathy myosin mutation R453C alters ATP binding and hydrolysis of human cardiac β-myosin. J Biol Chem 2014;289:5158–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ashrafian H, Redwood C, Blair E, Watkins H.. Hypertrophic cardiomyopathy: a paradigm for myocardial energy depletion. Trends Genet 2003;19:263–268. [DOI] [PubMed] [Google Scholar]
- 48. Lindenmayer GE, Sordahl LA, Harigaya S, Allen JC, Besch HR, Schwartz A.. Some biochemical studies on subcellular systems isolated from fresh recipient human cardiac tissue obtained during transplantation. Am J Cardiol 1971;27:277–283. [DOI] [PubMed] [Google Scholar]
- 49. de Moura MB, Van Houten B, Bioenergetic analysis of intact mammalian cells using the seahorse XF24 extracellular flux analyzer and a luciferase ATP assay In: Keohavong P, Grant SG, eds. Molecular Toxicology Protocols. Totowa, NJ: Humana Press; 2014. p589–602. [DOI] [PubMed] [Google Scholar]
- 50. Burkart AM, Tan K, Warren L, Iovino S, Hughes KJ, Kahn CR, Patti M-E.. Insulin resistance in human iPS cells reduces mitochondrial size and function. Sci Rep 2016;6:22788.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Warnes G. Flow cytometric assays for the study of autophagy. Methods 2015;82:21–28. [DOI] [PubMed] [Google Scholar]
- 52. Coppini R, Ferrantini C, Yao L, Fan P, Del Lungo M, Stillitano F, Sartiani L, Tosi B, Suffredini S, Tesi C, Yacoub M, Olivotto I, Belardinelli L, Poggesi C, Cerbai E, Mugelli A.. Late sodium current inhibition reverses electromechanical dysfunction in human hypertrophic cardiomyopathy. Circulation 2013;127:575–584. [DOI] [PubMed] [Google Scholar]
- 53. Mannhardt I, Breckwoldt K, Letuffe-Brenière D, Schaaf S, Schulz H, Neuber C, Benzin A, Werner T, Eder A, Schulze T, Klampe B, Christ T, Hirt Marc N, Huebner N, Moretti A, Eschenhagen T, Hansen A.. Human engineered heart tissue: analysis of contractile force. Stem Cell Rep 2016;7:29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eder A, Vollert I, Hansen A, Eschenhagen T.. Human engineered heart tissue as a model system for drug testing. Adv Drug Deliv Rev 2016;96:214–224. [DOI] [PubMed] [Google Scholar]
- 55. Liu LCY, Dorhout B, van der Meer P, Teerlink JR, Voors AA.. Omecamtiv mecarbil: a new cardiac myosin activator for the treatment of heart failure. Expert Opin Investig Drugs 2016;25:117–127. [DOI] [PubMed] [Google Scholar]
- 56. Maron BJ, Spirito P, Ackerman MJ, Casey SA, Semsarian C, Estes NAM, Shannon KM, Ashley EA, Day SM, Pacileo G, Formisano F, Devoto E, Anastasakis A, Bos JM, Woo A, Autore C, Pass RH, Boriani G, Garberich RF, Almquist AK, Russell MW, Boni L, Berger S, Maron MS, Link MS.. Prevention of sudden cardiac death with implantable cardioverter-defibrillators in children and adolescents with hypertrophic cardiomyopathy. J Am Coll Cardiol 2013;61:1527–1535. [DOI] [PubMed] [Google Scholar]
- 57. Lowes BD, Minobe W, Abraham WT, Rizeq MN, Bohlmeyer TJ, Quaife RA, Roden RL, Dutcher DL, Robertson AD, Voelkel NF, Badesch DB, Groves BM, Gilbert EM, Bristow MR.. Changes in gene expression in the intact human heart. Downregulation of alpha-myosin heavy chain in hypertrophied, failing ventricular myocardium. J Clin Invest 1997;100:2315–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Maron BJ, Ommen SR, Semsarian C, Spirito P, Olivotto I, Maron MS.. Hypertrophic cardiomyopathy: present and future, with translation into contemporary cardiovascular medicine. J Am Coll Cardiol 2014;64:83–99. [DOI] [PubMed] [Google Scholar]
- 59. Lopes LR, Rahman MS, Elliott PM.. A systematic review and meta-analysis of genotype–phenotype associations in patients with hypertrophic cardiomyopathy caused by sarcomeric protein mutations. Heart 2013;99:1800–1811. [DOI] [PubMed] [Google Scholar]
- 60. Watkins H, Rosenzweig A, Hwang D-S, Levi T, McKenna W, Seidman CE, Seidman JG.. Characteristics and prognostic implications of myosin missense mutations in familial hypertrophic cardiomyopathy. N Engl J Med 1992;326:1108–1114. [DOI] [PubMed] [Google Scholar]
- 61. Lemoine MD, Mannhardt I, Breckwoldt K, Prondzynski M, Flenner F, Ulmer B, Hirt MN, Neuber C, Horváth A, Kloth B, Reichenspurner H, Willems S, Hansen A, Eschenhagen T, Christ T.. Human iPSC-derived cardiomyocytes cultured in 3D engineered heart tissue show physiological upstroke velocity and sodium current density. Sci Rep 2017;7:5464.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Uzun AU, Mannhardt I, Breckwoldt K, Horváth A, Johannsen SS, Hansen A, Eschenhagen T, Christ T.. Ca2+-currents in human induced pluripotent stem cell-derived cardiomyocytes effects of two different culture conditions. Front Pharmacol 2016;7:300.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Horváth A, Lemoine MD, Löser A, Mannhardt I, Flenner F, Uzun AU, Neuber C, Breckwoldt K, Hansen A, Girdauskas E, Reichenspurner H, Willems S, Jost N, Wettwer E, Eschenhagen T, Christ T.. Low resting membrane potential and low inward rectifier potassium currents are not inherent features of hiPSC-derived cardiomyocytes. Stem Cell Rep 2018;10:822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Granata A, Serrano F, Bernard WG, McNamara M, Low L, Sastry P, Sinha S.. An iPSC-derived vascular model of Marfan syndrome identifies key mediators of smooth muscle cell death. Nat Genet 2017;49:97–109. [DOI] [PubMed] [Google Scholar]
- 65. Iyer D, Gambardella L, Bernard WG, Serrano F, Mascetti VL, Pedersen RA, Sinha S, Talasila A.. Robust derivation of epicardium and its differentiated smooth muscle cell progeny from human pluripotent stem cells. Development 2016;143:904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Montag J, Syring M, Rose J, Weber A-L, Ernstberger P, Mayer A-K, Becker E, Keyser B, dos Remedios C, Perrot A, van der Velden J, Francino A, Navarro-Lopez F, Ho CY, Brenner B, Kraft T.. Intrinsic MYH7 expression regulation contributes to tissue level allelic imbalance in hypertrophic cardiomyopathy. J Muscle Res Cell Motil 2017;38:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sharov VG, Todor AV, Silverman N, Goldstein S, Sabbah HN.. Abnormal mitochondrial respiration in failed human myocardium. J Mol Cell Cardiol 2000;32:2361–2367. [DOI] [PubMed] [Google Scholar]
- 68. Anatskaya OV, Vinogradov AE.. Somatic polyploidy promotes cell function under stress and energy depletion: evidence from tissue-specific mammal transcriptome. Funct Integr Genomics 2010;10:433–446. [DOI] [PubMed] [Google Scholar]
- 69. Scheubel RJ, Tostlebe M, Simm A, Rohrbach S, Prondzinsky R, Gellerich FN, Silber R-E, Holtz J.. Dysfunction of mitochondrial respiratory chain complex I in human failing myocardium is not due to disturbed mitochondrial gene expression. J Am Coll Cardiol 2002;40:2174–2181. [DOI] [PubMed] [Google Scholar]
- 70. Sebastiani M, Giordano C, Nediani C, Travaglini C, Borchi E, Zani M, Feccia M, Mancini M, Petrozza V, Cossarizza A, Gallo P, Taylor RW, D’Amati G.. Induction of mitochondrial biogenesis is a maladaptive mechanism in mitochondrial cardiomyopathies. J Am Coll Cardiol 2007;50:1362–1369. [DOI] [PubMed] [Google Scholar]
- 71. Björk S, Ojala EA, Nordström T, Ahola A, Liljeström M, Hyttinen J, Kankuri E, Mervaala E.. Evaluation of optogenetic electrophysiology tools in human stem cell-derived cardiomyocytes. Front Physiol 2017;8:884.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pioner JM, Racca AW, Klaiman JM, Yang K-C, Guan X, Pabon L, Muskheli V, Zaunbrecher R, Macadangdang J, Jeong MY, Mack DL, Childers MK, Kim D-H, Tesi C, Poggesi C, Murry CE, Regnier M.. Isolation and mechanical measurements of myofibrils from human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Rep 2016;6:885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Witjas-Paalberends ER, Piroddi N, Stam K, van Dijk SJ, Oliviera VS, Ferrara C, Scellini B, Hazebroek M, ten Cate FJ, van Slegtenhorst M, dos Remedios C, Niessen HWM, Tesi C, Stienen GJM, Heymans S, Michels M, Poggesi C, van der Velden J.. Mutations in MYH7 reduce the force generating capacity of sarcomeres in human familial hypertrophic cardiomyopathy. Cardiovasc Res 2013;99:432–441. [DOI] [PubMed] [Google Scholar]
- 74. Lankford EB, Epstein ND, Fananapazir L, Sweeney HL.. Abnormal contractile properties of muscle fibers expressing beta-myosin heavy chain gene mutations in patients with hypertrophic cardiomyopathy. J Clin Invest 1995;95:1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Witjas-Paalberends ER, Ferrara C, Scellini B, Piroddi N, Montag J, Tesi C, Stienen GJM, Michels M, Ho CY, Kraft T, Poggesi C, van der Velden J.. Faster cross-bridge detachment and increased tension cost in human hypertrophic cardiomyopathy with the R403Q MYH7 mutation. J Physiol 2014;592:3257–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Keller DI, Coirault C, Rau T, Cheav T, Weyand M, Amann K, Lecarpentier Y, Richard P, Eschenhagen T, Carrier L.. Human homozygous R403W mutant cardiac myosin presents disproportionate enhancement of mechanical and enzymatic properties. J Mol Cell Cardiol 2004;36:355–362. [DOI] [PubMed] [Google Scholar]
- 77. Sommese RF, Sung J, Nag S, Sutton S, Deacon JC, Choe E, Leinwand LA, Ruppel K, Spudich JA.. Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human β-cardiac myosin motor function. Proc Natl Acad Sci USA 2013;110:12607–12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Friedrich FW, Reischmann S, Schwalm A, Unger A, Ramanujam D, Münch J, Müller OJ, Hengstenberg C, Galve E, Charron P, Linke WA, Engelhardt S, Patten M, Richard P, van der Velden J, Eschenhagen T, Isnard R, Carrier L.. FHL2 expression and variants in hypertrophic cardiomyopathy. Basic Res Cardiol 2014;109:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Crocini C, Arimura T, Reischmann S, Eder A, Braren I, Hansen A, Eschenhagen T, Kimura A, Carrier L.. Impact of ANKRD1 mutations associated with hypertrophic cardiomyopathy on contraction parameters of engineered heart tissue. Basic Res Cardiol 2013;108:349. [DOI] [PubMed] [Google Scholar]
- 80. Stöhr A, Friedrich FW, Flenner F, Geertz B, Eder A, Schaaf S, Hirt MN, Uebeler J, Schlossarek S, Carrier L, Hansen A, Eschenhagen T.. Contractile abnormalities and altered drug response in engineered heart tissue from Mybpc3-targeted knock-in mice. J Mol Cell Cardiol 2013;63:189–198. [DOI] [PubMed] [Google Scholar]
- 81. Wijnker PJM, Friedrich FW, Dutsch A, Reischmann S, Eder A, Mannhardt I, Mearini G, Eschenhagen T, van der Velden J, Carrier L.. Comparison of the effects of a truncating and a missense MYBPC3 mutation on contractile parameters of engineered heart tissue. J Mol Cell Cardiol 2016;97:82–92. [DOI] [PubMed] [Google Scholar]
- 82. Lyons GE, Schiaffino S, Sassoon D, Barton P, Buckingham M.. Developmental regulation of myosin gene expression in mouse cardiac muscle. J Cell Biol 1990;111:2427–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Reiser PJ, Portman MA, Ning X-H, Moravec CS.. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Am J Physiol Heart Circ Physiol 2001;280:H1814–H1820. [DOI] [PubMed] [Google Scholar]
- 84. Tyska MJ, Hayes E, Giewat M, Seidman CE, Seidman JG, Warshaw DM.. Single-molecule mechanics of R403Q cardiac myosin isolated from the mouse model of familial hypertrophic cardiomyopathy. Circ Res 2000;86:737–744. [DOI] [PubMed] [Google Scholar]
- 85. Debold EP, Schmitt JP, Patlak JB, Beck SE, Moore JR, Seidman JG, Seidman C, Warshaw DM.. Hypertrophic and dilated cardiomyopathy mutations differentially affect the molecular force generation of mouse α-cardiac myosin in the laser trap assay. Am J Physiol Heart Circ Physiol 2007;293:H284–H291. [DOI] [PubMed] [Google Scholar]
- 86. Tardiff JC, Carrier L, Bers DM, Poggesi C, Ferrantini C, Coppini R, Maier LS, Ashrafian H, Huke S, van der Velden J.. Targets for therapy in sarcomeric cardiomyopathies. Cardiovasc Res 2015;105:457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.