Abstract
Summary
Errors in chromosome segregation during female meiosis I occur frequently, and aneuploid embryos account for 1/3 of all miscarriages in humans [1]. Unlike mitotic cells that require two Aurora kinase (AURK) homologs to help prevent aneuploidy (AURKA, AURKB), mammalian germ cells also require a third (AURKC)[2, 3]. AURKA is the spindle pole-associated homolog, and AURKB/C are the chromosome-localized homologs. In mitosis, AURKB has essential roles as the catalytic subunit of the chromosomal passenger complex (CPC), regulating chromosome alignment, kinetochore-microtubule attachments, cohesion, the spindle assembly checkpoint, and cytokinesis [4, 5]. In mouse oocyte meiosis, AURKC takes over as the predominant CPC kinase [6], while the requirement for AURKB remains elusive [7]. In the absence of AURKC, AURKB compensates, making defining potential non-overlapping functions difficult [6, 8]. To investigate the role(s) of AURKB and AURKC in oocytes, we analyzed oocyte-specific Aurkb and Aurkc single and double knockout (KO) mice. Surprisingly, we find that double KO female mice are fertile. We demonstrate that, in the absence of AURKC, AURKA localizes to chromosomes in a CPC- dependent manner. These data suggest that AURKC prevents AURKA from localizing to chromosomes by competing for CPC binding. This competition is important for adequate spindle length to support meiosis I. We also describe a unique requirement for AURKB to negatively regulate AURKC to prevent aneuploidy. Together, our work reveals oocyte-specific roles for the AURKs in regulating each other’s localization and activity. This inter-kinase regulation is critical to support wild type levels of fecundity in female mice.
eTOC Blurb
Nguyen et al. describe oocyte-specific functions for the three Aurora protein kinases during meiosis. The authors show, for the first time, negative inter-kinase regulation between the family members to control the localized activity of one another, and that these interactions are critical for spindle integrity and gamete euploidy.
Results and Discussion
Generation and fertility assessment of mice lacking AURKB and AURKC in oocytes
Because of the lack of specificity of AURKB inhibitors and compensation of AURKB for loss of AURKC in oocytes, distinguishing the roles of these kinases is challenging. Prior approaches used overexpression and dominant negatives to investigate AURKB/C roles in mouse oocytes [5–10]. Existing evidence supports unique AURKC roles, but the AURKB contributions during oocyte meiosis are unknown. We took a genetic approach to investigate the non-overlapping roles of AURKB/C in oocytes. We crossed an Aurkc knockout (Aurkc KO) mouse strain [7, 11] with a strain that lacked Aurkb (Aurkb cKO)[5] specifically in oocytes using Gdf9-mediated Cre excision (double KO; Aurkb cKO/Aurkc KO). Quantitative real-time PCR confirmed that Aurkb and Aurkc transcripts fell to background levels (Figures S1A-B). Western blotting using a phospho-specific AURK antibody that detects the activated kinase forms confirmed their absence (Figures S1C-D). The relative abundance of the AURKs in mouse oocytes has not previously been defined. Assuming that the phospho-specific antibody has similar affinities for each kinase, and that each kinase is phosphorylated to a similar extent, we found that pAURKA was the most abundant; pAURKC expression was ~2/3 of pAURKA; and pAURKB was the lowest in WT oocytes (Figures S1C-D). Although the levels of pAURKA increased in double KO oocytes (Figures S1C-D), total protein levels, as indicated by western blot analysis, did not change (Figures S1E-F), suggesting that in the absence of AURKB/C more AURKA can be activated.
To determine the consequence of deleting AURKB/C alone or in combination, we conducted fertility trials. Compared to WT, Aurkb cKO females were less fertile, and three of the five Aurkb cKO females failed to produce 6 litters, suggesting a premature decline in fertility (Figure 1A). These mice were significantly less fertile than Aurkc KOs [7] indicating different requirements for these kinases for fecundity and that AURKB and AURKC compensation is not reciprocal. Because a catalytic component of the CPC is essential for its function and the CPC is required to complete MI [10], it was surprising that double KO animals were not infertile (Figure 1A). Although only AURKB/C have documented CPC function in oocytes in vivo, the fertility of double KO females suggests another kinase compensates.
Figure 1.

AURKA compensates for loss of AURKB/C in oocytes.A) Litter sizes from fertility studies. Age-matched wild-type (WT), Aurkb conditional knockout (B cKO), Aurkc KO (C KO) and Aurkb/c double KO (B cKO/C KO). The number of pups/6 litters were recorded (n=4 mice/genotype). B- L) Prophase I (PI) oocytes were collected and matured to metaphase of meiosis I (Met I) or meiosis II (Met II). B) Confocal z-projections from the indicated genotypes. AURKA (red) and DNA (DAPI, blue). Asterisk denotes chromosome- localized AURKA. Each data point is the value from a single oocyte with three experimental replicates performed. C-D) Relative pixel intensity AURKA, normalized to WT, at chromosomes or spindle poles, respectively, from B. E) PI oocytes from double KO animals were microinjected with morpholino oligonucleotides of either a scrambled sequence (control) or one designed to target INCENP (INCENP KD). Shown are representative z-projections of fixed oocytes that were matured to Met I. AURKA (A, green), spindle (a-tubulin, gray), and DNA (DAPI, blue). F-G) Chromosome or spindle pole AURKA pixel intensity from E. Each data point is a single oocyte. n=6 animals/genotype/experiment; 2 experimental replicates. H) Confocal z-projections of Met I oocytes from the indicated genotypes that were matured in the presence of the indicated drug. pINCENP (green), centromeres (ACA, red), spindle (a-tubulin, grey), and DNA (DAPI, blue). n=3 animals/genotype/experiment; 3 experimental replicates I) Quantification of relative pixel intensity pINCENP from H. Values normalized to WT/DMSO. J) Percent oocytes with misaligned chromosomes from H. K) Confocal z-projections of Met II eggs from the indicated genotypes matured in the presence of the indicated drug. Spindle (a-tubulin, green), and DNA (DAPI, blue). Dotted lines outline the egg and polar body (PB). n=3 animals/genotype/experiment; 3 experimental replicates. L) Number of eggs that progressed to Met II from K. Scale bars =10 mm. Red bars indicate mean. Error bars indicate standard error of the mean. Student’s t-test was performed in C, D, F, G (*p= 0.0290, ****p = 0.00145) and One-way ANOVA was performed in A, H, I, K (****p <0.0001, **p <0.01, *p < 0.05). See also Figures S1–4.
Active CPC localizes to chromosomes in the absence of AURKB/C
To confirm that the CPC localizes to chromosomes in double KO oocytes, we assessed the localization of Survivin, the CPC chromosome docking subunit. The localization of Survivin did not change compared to WT in metaphase I (Met I) oocytes (Figures S2A-B). Next, to determine if the CPC was active, we evaluated the phosphorylation of CPC substrates INCENP, HEC1/Ndc80, and Histone H3 [12] and found that their phosphorylation in double KO oocytes was comparable to WT (Figures S2C-H). Therefore, the localization and function of the CPC is normal in double KO oocytes, despite the absence of the canonical kinase subunit.
AURKA compensates for loss of AURKB/C in an INCENP-dependent manner
The surprising normal CPC function in double KO oocytes suggests compensation. In WT oocytes, AURKA localized to spindle poles. In contrast, in double KO oocytes, AURKA localized to spindle poles and chromosomes (Figures 1B-G). This chromosome localization of AURKA is similar to AURKC localization in WT oocytes and to AURKB localization in Aurkc KO oocytes [6, 7] suggesting that it is recruited to chromosomes via the CPC. To determine if this AURKA localization depends on INCENP, the CPC scaffold, we depleted INCENP in double KO oocytes. Oocytes injected with INCENP morpholino oligonucleotides lost chromosomal-localized AURKA while retaining polar AURKA (Figures 1E-G). These data indicate that chromosomal AURKA depends on INCENP in the absence of AURKB/C.
To determine if the CPC is active in an AURKA-dependent manner, we treated oocytes with an AURKA-specific inhibitor, MLN8237 [13]. In WT oocytes, where AURKA was polar, INCENP was phosphorylated (pINCENP) at AURK specific phosphorylation sites, and pHEC1 levels did not change (Figures 1H-I, S3)[14]. In contrast, double KO oocytes treated with MLN8237 lost pINCENP and pHEC1, indicating that this phosphorylation was dependent upon AURKA activity.
Consistent with the requirement of AURKA in regulating meiotic maturation [15], WT and double KO oocytes treated with MLN8237 failed to extrude polar bodies and reach metaphase II (Met II) (Figures 1K-L).
Further confirming the absence of AURKB/C, double KO oocytes treated with ZM447439, an AURKB/C-specific inhibitor, retained pINCENP and pHEC1 levels (Figures 1H-I, S3). These oocytes were phenotypically unaffected (Figures 1H- L). ZM447439 is an ATP-binding pocket inhibitor that does not interfere with binding of AURKB/C to the CPC [16, 17]. Therefore, phosphorylation of INCENP in double KO oocytes, but not drug-treated WT, suggests the presence of AURKB/C prevents AURKA from binding to and compensating in the CPC.
These data are consistent with a dependency on AURKA for CPC activity in double KO oocytes.
AURKA compensation is meiosis specific
AURKA compensation for loss of AURKB/C in mouse oocytes is in contrast to findings in mitotic cells in which AURKB has been depleted by RNAi [5, 18]. However, a minor population of AURKB may keep AURKA from localizing to chromosomes. Because this is the first documentation of AURKA compensation in the CPC in vivo, we tested if this mechanism is specific to meiotic cells using a knockout approach. We deleted AURKB in HeLa cells using an inducible Cas9 endonuclease and AURKB guide RNAs [19]. AURKB was absent from chromosomes upon Cas9 induction, and loss of AURKB caused abnormal metaphase figures (Figure S4)[5]. Despite the loss of AURKB, we did not detect a chromosome-associated population of AURKA at metaphase (Figure S4F). In addition, INCENP was not phosphorylated and H3S10 phosphorylation was markedly reduced, suggesting that there is not a compensatory CPC-bound kinase (Figures S4C-D, G-H). The inability of AURKA to compensate for AURKB deletion in mitosis, suggests an oocyte-specific compensation mechanism.
AURKC prevents AURKA chromosome localization via INCENP competition
AURK function is defined by localization, which is dictated by binding TPX2 or INCENP [18]. Because AURKA binds INCENP in vitro [20, 21] and AURKA chromosomal localization is INCENP-dependent in oocytes (Figures 1E-G), it is possible that competition for INCENP binding promotes AURKA’s TPX2- associated function in oocytes. We assessed AURKA localization in oocytes harboring various copy numbers of Aurkb and Aurkc. AURKA localized to spindle poles in oocytes that expressed AURKC. In contrast, oocytes that lacked AURKC displayed chromosomal AURKA (Figures 2A-B). Similar to double KO oocytes, chromosomal AURKA depended on INCENP in Aurkc KO oocytes (Figures 2C- D) while, AURKC-CPC binding and activation in Aurkb cKO oocytes was INCENP-dependent (Figures 2E-F).
Figure 2.
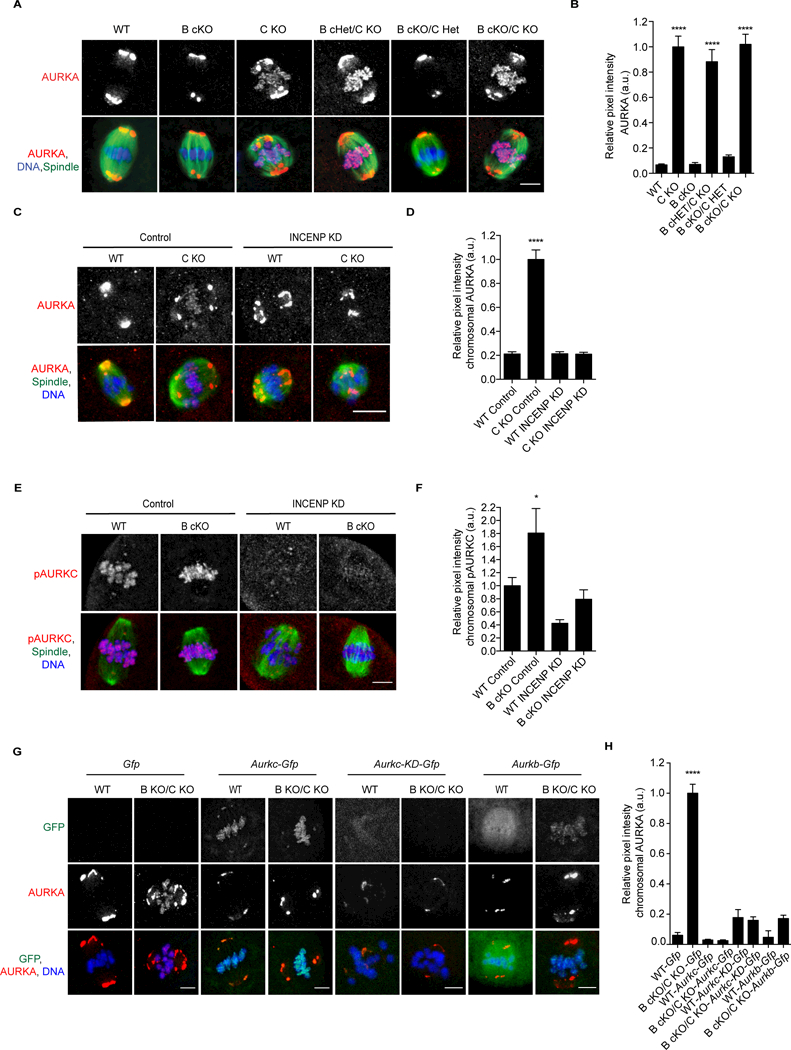
AURKC prevents AURKA from localizing to chromosomes.A) Prophase I (PI) oocytes from mice of the indicated genotypes were matured to metaphase of meiosis I (Met I) and stained to detect AURKA (red), spindle (a- tubulin, green), and DNA (DAPI, blue). Shown are representative confocal z- projections. Asterisks denote chromosome-localized AURKA. n = 1 animal/genotype/experiment; 4 experimental replicates B) Relative pixel intensity of chromosomal AURKA from A. Values normalized to B cKO/CKO. C-F) PI oocytes from the indicated genotypes were microinjected with morpholino oligonucleotides of either a scrambled sequence (control) or one designed to target INCENP (INCENP KD). Shown are representative z-projections of fixed oocytes that were matured to Met I. C) AURKA (red), spindle (a-tubulin, green), and DNA (DAPI, blue). D) Chromosome pixel intensity from C. n=4 animals/genotype/experiment; 2 experimental replicates. E) pAURKC (red), spindle (a-tubulin, green), and DNA (DAPI, blue). F) Pixel intensity of chromosomal AURKA from E. n=7 animals/genotype/experiment; 2 experimental replicates. G) WT or double KO PI oocytes were microinjected with the indicated cRNA. After ~15h, oocytes were matured to Met I and labeled to detect AURKA (red) and DNA (DAPI, blue). n = 3 animals/genotype/experiment, 3 independent experiments performed. Error bars indicate standard error of the mean. * <0.05, ****p <0.0001, One-way ANOVA. Scale bars = 10 mm. See also Figures S1, S4.
These findings suggest that AURKC outcompetes AURKA for CPC binding, consistent with AURKB/C having a higher affinity for INCENP [20]. To test this, we microinjected Aurkc-Gfp into WT and double KO oocytes and assessed AURKA Met I localization. Similar to WT, where AURKC is CPC bound, overexpression of AURKC-GFP prevented AURKA from localizing to chromosomes in double KOs (Figures 2G-H). This competition was not dependent on the activity of AURKC (Figures 2G-H). Finally, we found that when AURKB was overexpressed, it also prevented AURKA chromosome localization (Figures 2G-H). These data indicate that in WT oocytes, where AURKC is more abundant that AURKB, that AURKC is the primary AURK preventing AURKA from localizing to chromosomes.
CPC competition is essential for spindle length and meiotic progression
Although double KO mice retained fertility, these mice were less fertile than WT (Figure 1A). To determine if this reproductive disadvantage was due to failing to keep AURKA at spindle poles, we analyzed the oocytes in vitro. The majority of WT oocytes (91.9% +/− 2.81%) completed MI, but only 43.32% +/− 10.58% of double KO oocytes did (Figures 1J-L, 3A-B). Compared to WT, double KO oocytes had significantly shorter spindles (Figures 3C-D), and they frequently contained a pole-associated bivalent (Figures 3E-F). Using live-cell imaging, we observed that these pole-associated bivalents failed to disjoin (n = 8/18) and oocytes with short spindles arrested at Met I (Figures 3E-F, Videos S1-4). Although AURKA alone can support meiotic progression in vivo, there is a reduction in oocyte quality that reduces fertility.
Figure 3.
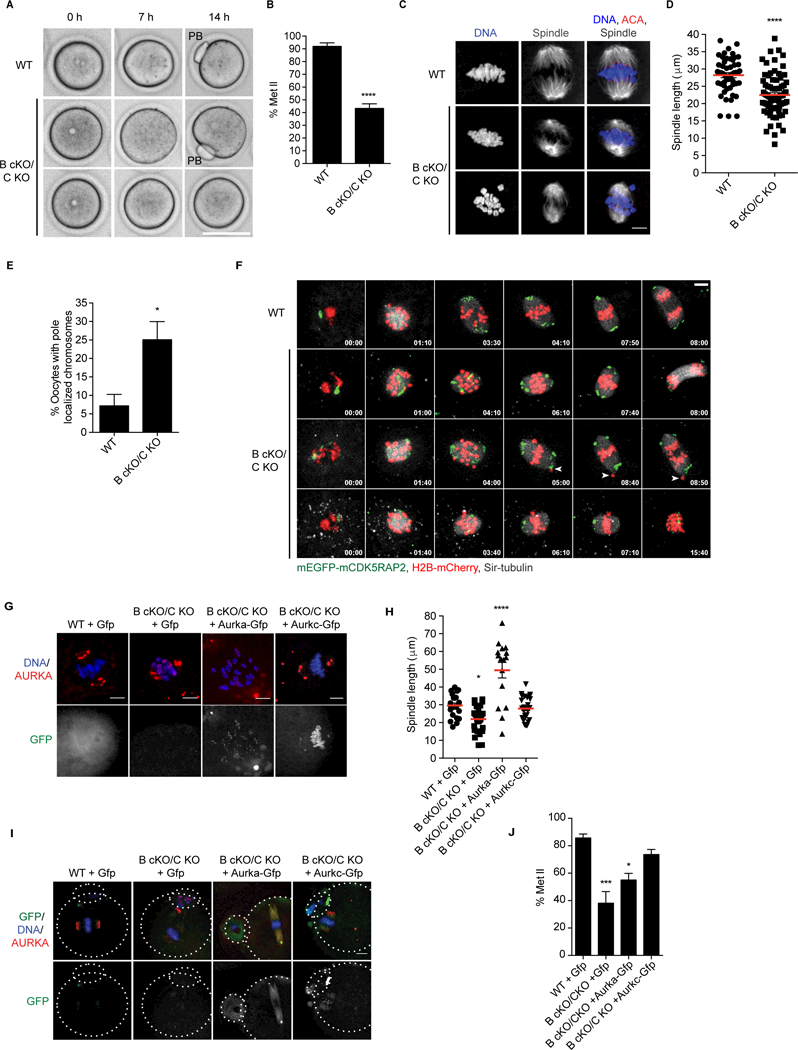
AURKC promotion of AURKA to spindle poles supports proper spindle length and meiotic progression.A) Prophase I oocytes from the indicated genotypes were matured and imaged live for 18 hours and the number of Metaphase II (Met II) eggs counted. Representative DIC images shown. Polar body (PB) indicates successful completion of metaphase of meiosis I (Met I). n = 1 animal/genotype/experiment, 4 experimental replicates. B) Percentage oocytes that reached Met II from A. C) Confocal z-projections of fixed Met I oocytes. Oocytes stained to detect centromeres (ACA, red), spindle (a-tubulin, grey), and DNA (DAPI, blue). n=3–4 animals/genotype/experiment, 4 experimental replicates. D) Quantification of spindle length from C. Each point is the spindle length from a single oocyte. E) Percentage oocytes with pole-localized chromosomes from C. (*p =0.0112). F) Montage of z-projections from WT and double KO oocytes matured live using high-resolution confocal microscopy. Oocytes microinjected with mEGFP-mCDK5RAP2 (spindle poles, green) and H2B-mCherry (DNA, red) while incubated in SiR-tubulin (spindle, grey). Time points are relative to time after nuclear envelope breakdown (hh:min). G-J) Oocytes microinjected with the indicated cRNAs were matured to either Met I or Met II and stained to detect AURKA (red) and DNA (DAPI, blue). G) Confocal z-projections of microinjected, Met I matured oocytes. Asterisks indicate spindle poles. n=3 animals/genotype/experiment, 3 experimental replicates. H) Quantification of spindle length from G. I) Microinjected, Met II matured eggs. Dotted lines outline the egg and polar body (PB). n= 1 WT and 3 double KO animals/experiment, 3 independent experiments performed. J) Quantification percent oocytes that reached Met II from I. Error bars indicate standard error of the mean. Red bars indicate mean. ****p <0.0001, *** p <0.001, *p <0.05; Student’s t-test used B, D,F and One-way ANOVA used in H, J. Scale bars are 10 mm. See also Figure S1 and Videos S1–4.
The short spindle phenotype (Figures 3C-D) is similar to depletion of Aurka in oocytes [15] and when AURKA-TPX2 interaction is perturbed in mitosis [22]. Conversely, overexpression of AURKA causes elongated spindles in oocytes [15]. We hypothesized that competition of AURKC for INCENP ensures sufficient pole-localized AURKA to achieve a robust meiotic spindle length. We overexpressed either Aurka or Aurkc in double KO oocytes and examined spindle length and meiotic progression. Although overexpression of Aurka-Gfp lengthened the spindles in double KOs (Figures 3G-H), overexpression Aurkc- Gfp produced spindles more closely resembling WT (Figures 3G-H). Completion of MI was rescued by overexpression of Aurka-Gfp or Aurkc-Gfp (Figures 3I-J). Importantly, although AURKA overexpression partially rescued these phenotypes, only when AURKC-Gfp was introduced were the phenotypes fully resolved. Therefore, AURKC likely ensures sufficient polar AURKA for full fecundity. Alternatively, AURKA/AURKC-specific targets on chromosomes or at poles may regulate spindle length.
AURKB is required to negatively regulate AURKC
Aurkb cKO females were significantly less fertile than Aurkc KO and WT (Figure 1A) suggesting that non-overlapping requirements exist. We observed an increase in the active form of AURKC upon Aurkb deletion (Figures S1C-D). Because the dosage of AURKC is important for completion of oocyte meiosis [10] we evaluated fertilization competent Met II-arrested eggs. pAURKC levels increased on chromosomes in eggs isolated from Aurkb cKOs (Figures 4A-B).
Figure 4.

AURKB is required to prevent aneuploidy in female meiosis.A-D) Ovulated metaphase II-arrested (Met II) eggs of the indicated genotypes. n = 3 animals/genotype/experiment, 3 experimental replicates A, C) Confocal z- projections of eggs stained to detect A) phosphorylated AURKC (pAURKC, red) or C) phosphorylated INCENP (pINCENP, red) and DNA (DAPI, blue). B) Pixel intensity pAURKC from A. D) Pixel intensity pINCENP from C. Values normalized to WT. E) Litter sizes from fertility studies as in Figure 1A Note that the control and single KO data is from animals presented in Figureure 1A as these trials were conducted simultaneously. n=3–5 mice/genotype. F-L) Prophase-I (PI) oocytes were isolated and matured in vitro to Met I or Met II. F) In situ chromosome spreads were analyzed to determine chromosome number. Representative confocal z-projections shown. Oocytes stained to detect centromeres (ACA, red) and DNA (DAPI, grey). n= 3–6 animals/genotype/experiment; 3 experimental replicates. G) Percentage of Met II eggs that were aneuploid from F. H) Number of kinetochores per egg from F. I) Number of eggs with prematurely separated sister chromatids (PSSC) from F. J) Confocal z-projections of Met I oocytes from the indicated genotypes labeled to detect REC8 (green), and DNA (DAPI, blue). K) Relative pixel intensity REC8 from J. Values normalized to WT. L) Confocal z-projections of chromosome spreads from eggs of the indicated genotypes labeled to detect REC8 (red), centromere (ACA, green), and DNA (DAPI, blue). M) Pixel intensity REC8 from L. N) PI oocytes from the indicated genotypes were matured to Met I, fixed, and stained to detect SGO2 (green), centromeres (ACA, red), and DNA (DAPI, blue). n=1 animal/genotype/experiment, 3 experimental replicates. O) Relative pixel intensity SGO2 from N. Values normalized to WT. Boxes indicate area of optical zoom. Error bars indicate standard error of the mean. Red bars indicate mean *p <0.05, **p <0.01, ****p <0.0001; One-way ANOVA. Scale bars = 10 mm. See also Figure S1.
These levels were reduced to near WT levels when one copy of Aurkc was deleted in this genetic background (Aurkb cKO/Aurkc Het). Consistent with an increase in pAURKC, levels of pINCENP increased in Aurkb cKO eggs, and the levels were partially rescued in Aurkb cKO/Aurkc Hets (Figures 4C-D). These data suggest that AURKB is a negative regulator of AURKC.
To assess the phenotypic consequences of excess pAURKC, we evaluated the fertility of Aurkb cKO/Aurkc Het animals and observed partial rescue (Figure 4E). To determine how excess pAURKC reduces egg quality, we next analyzed chromosome numbers in Met II-arrested eggs [23]. Compared to eggs from WT and Aurkc KOs, significantly more Aurkb cKO eggs were aneuploid, exhibiting both hyper- and hypo-ploidy (Figures 4F-H). Aneuploidy was partially rescued in Aurkb cKO/Aurkc Hets (Figures 4F-H). Aurkb cKO eggs also displayed prematurely separated sister chromatids (PSSC), which was reduced in Aurkb cKO/Aurkc Het eggs (Figure 4I). These phenotypes are similar to embryos overexpressing AURKC [10, 24], but differ from those in which AURKC is overexpressed in oocytes [10]. The presence of AURKB and/or AURKC overexpression could account for these disparities.
Studies suggest that an age-related decline in the meiosis-specific cohesin subunit REC8 causes PSSC in mice and humans [25–27]. REC8 levels were unperturbed at Met I in Aurkb cKOs suggesting that loading and stability were normal [26](Figures 4J-K). Importantly, analyses of Met II chromatids revealed asignificant decrease in centromeric REC8, which was rescued in Aurkb cKO/Aurkc Hets (Figures 4L-M). Therefore, the loss of sister chromatid cohesion occurs after anaphase I onset. Centromeric cohesion of sisters is protected by SGO2 via MPS1-dependent recruitment during MI so that sister chromatids cosegregate [28]. Reduction in SGO2 by 50% causes PSSC in oocytes upon MPS1 inhibition [29]. To determine if SGO2 was perturbed in Aurkb cKOs, we evaluated its levels at Met I and found a reduction of ~60% compared to WT (Figures 4N- O). Importantly, this reduction was partially restored in oocytes from Aurkb cKO/Aurkc Hets (Figures 4N-O). This partial rescue suggests separate AURKB and AURKC functions. Because inhibition of AURKB/C with ZM447439 modestly reduces MPS1 localization and subsequently SGO2 localization, it is possible that the role for AURKB is indirect, with the loss of AURKB affecting MPS1 [29]. Alternatively, this SGO2 defect could reflect a role of AURKB in promoting the translocation of SGO2 from chromosome arms to centromeres [30, 31]. Because SGO2 is reduced, and AURKC activity is increased, a possible non-overlapping AURKC pathway could involve triggering cleavage of centromeric REC8 that is no longer protected at anaphase I onset [32]. This role is consistent with the premature loss of cohesin in oocytes that have excess active AURKC due to overexpression of Haspin [33]. These possibilities must be explored to understand the precise mechanisms of these two kinases in cohesion regulation. Taken together, these data suggest that negative regulation of AURKC by AURKB is critical for ensuring gamete euploidy.
Our data support a model in which all three Aurora kinases are required in mouse oocytes. In addition to its direct roles in spindle regulation [10, 34, 35], AURKC promotes AURKA spindle pole localization via competition to ensure a robust spindle and completion of meiosis. Additionally, oocytes require AURKB to ensure accurate chromosome segregation.
Although mammalian germ cells rely on three AURKs, organisms from other evolutionary clades rely on different homolog numbers for meiosis [2, 3]. Fungi express one AURK whereas Drosophila and nematodes express two. The AURK homologs likely evolved from a common ancestor through gene duplication [2, 3]. AURKA is the most ancestral, while the order of appearance of AURKB and AURKC is unsettled [2, 3]. The current model is that an ancestral AURKB/C hybrid from reptiles, fish, and non-placental mammals evolved from AURKA prior to AURKB and AURKC individualization in placental mammals [3]. Additional AURKs could provide an evolutionary benefit such as specialized functions and compensatory mechanisms. Our data predict that although AURKC expression may provide optimal CPC activity due to its inherent stability [7] and support proper spindle building through CPC-binding competition, its expression has required oocytes to retain AURKB expression to negatively regulate AURKC [10]. It will be of interest to determine if AURKB has other non-overlapping functions.
Classically, AURK function is spatially distinct, and determined by binding partner affinity [18]. AURK affinities can be swapped by mutation of an amino acid residue of AURKA or AURKB allowing compensation for each other’s loss, indicating that their substrate specificities are similar [20, 36]. We demonstrated that in mouse oocytes this localization is less stringent than in somatic cells because AURKA localized to chromosomes in oocytes in vivo upon deletion of Aurkc, suggesting that an oocyte-specific factor allows this localization. It is possible that we are unable to detect minor populations of AURKA-CPC that exist in HeLa cells [21, 37, 38]. Alternatively, a mechanism may prevent AURKA chromosome localization in mitotic cells. Strikingly, pups were born to double KO mothers, suggesting that AURKA compensation exists in the first mitotic divisions of preimplantation embryos before zygotic expression of Aurkb. Alternatively, the CPC may not be essential for early embryonic mitoses because INCENP and Survivin knockout embryos develop to the 8-cell stage before failing [39, 40]. These INCENP and Survivin KO findings could help explain the high level of aneuploidies that give rise to mosaic embryos in humans [41].
AURKB and AURKC mutations can cause human infertility [42, 43], highlighting their diagnostic importance in assisted reproduction. Aberrant AURKC expression also drives tumorigenesis and invasiveness in many cancer cell lines [44, 45]. Identification of the mechanism by which AURKB restricts AURKC could provide insights into how AURKC activity could be inhibited and assist in designing precision medicine approaches for patients with an AURKC-positive expression signature.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Karen Schindler (schindler@dls.rutgers.edu).
Experimental Model and Subject Details
Mice
Generation of Aurkc-’ mice were described previously [7, 11]. The floxed Aurkb mice were generated previously [5]. To generate Aurkbf/f Gdf9-Cre female mice carrying the Aurkb floxed alleles were crossed with Gdf9-Cre (Jackson Laboratories, Tg(Gdf9-icre)5092Coo/J [46]). To generate Aurkb cKO/Aurkc Het, Aurkb cHet /Aurkc KO, and double KO mice, Aurkbf/f Gdf9-Cre mice were crossed onto the Aurkch background and underwent several rounds of backcrossing. Single Aurkb cKO used for experiments were derived from the Aurkc backcrosses. Control animals used (Wildtype (WT)) are from the same genetic background as experimental transgenic animals but lack the Cre recombinase transgene. All animals were in a mixed background of C57BL/6J, 129/Sv, and CD1 and maintained following the Rutgers University Institutional Animal Use and Care Committee and National Institutes of Health guidelines. Mice were housed in 12–12 hr light-dark cycle, with constant temperature and food and water provided ad libitum. All animal experiments performed in this study were approved by the Rutgers IACUC. Rutgers Comparative Medicine Resources provided daily cage maintenance and veterinarian health checks. All oocyte experiments were conducted using healthy female mice ranging in age from 4–8 weeks. Fertility trials involved male mice starting at 8 weeks of age. Mice were not involved in other experimental procedures. Female mice were exposed to hormones to induce superovulation prior to and for purposes of experimentation.
Oocyte culture
To prevent spontaneous meiotic resumption 2.5 μM milrinone (Sigma-Aldrich) was added to bicarbonate free minimal essential medium (MEM) containing, 25 mM Hepes, pH 7.3, 3 mg/mL poyvinylpyrrolidone (PVP) for oocyte collection and microinjection. To induce meiotic resumption, oocytes were cultured in milrinone- free Chatot, Ziomek, and Bavister (CZB) medium in an atmosphere of 5% CO2 in air at 37°C[47]. Oocytes that had not undergone nuclear envelope breakdown within 2 hours of meiotic resumption were removed from the experiment. Oocytes were matured for 7.5 h for metaphase I experiments or 18 h for metaphase II analysis.
HeLa cell culture
HeLa cells (female) expressing AURKB specific gRNAs and a doxycycline- inducible Cas9 were kindly provided by Dr. Iain Cheeseman [19]. Three independent pLenti-sgRNA guides used: A4.1 A4.1 (CACCGTCTGTCGGCCGTAGGGCCA) A3.4 (CACCGCGCAGAGAG ATCGAAATCC), A4.4 (CACCGCTTTGAGATTGGGCGTCCTC).
The previously validate cells were cultured in DMEM containing 10% Tetracycline-free fetal bovine serum (GEMINi Bio-products), L-glutamine (2 mM,GIBCO), penicillin, streptomycin (100 U/ml: 100 μο/ηΙ, GIBCO), puromycin (1 mg/ml, GIBCO) and G418 (800 mg/ml, Sigma) in a humidified incubator programmed to 5% CO2 and 37Ό. To induce Cas9 expression, cells were treated with doxycycline hyclate (1 μg/ml, Sigma) for 4 days.
Method Details
Mouse genotyping
Genotyping was performed prior to weening and repeated upon use of the animals for experiments for replication and confirmation. Aurkc deletion was detected by a TaqMan copy number assay using primers/probes to detect Neo (Assay # Mr00299300_cn) and Tfrc (for normalization; Assay #4458366). The delta-delta Ct method was used to determine expression levels [48]. Genotyping for LoxP and Cre were carried out as described previously using PCR amplification [5, 6]. Primers for Aurkb LoxP (Forward: 5’ - AGGGCCTAATTGCCTCTTGT- 3’, Reverse: 5’ -GGGCATGAATTCTTGAGTCG- 3’), and Gdf9-Cre (Forward: 5’ -T CT GAT GAAGT CAGGAAGAAC C- 3’, Reverse: 5’ -GAGATGTCCTTCACTCTGATT C-3’, Internal control Forward: 5’ - CTAGGCCACAGAATTGAAAGATCT- 3’, Internal control Reverse: 5’ - GTAGGTGGA AATTCTAGCATCATC C- 3’) were used at 20 pMol using FastMix Frenche PCR beads (Bulldog Bio) following manufacturers protocol. PCR conditions are available on request.
Fertility trials
Sexually mature (6 wk) females were continuously mated to wild-type B6D2 (Jackson Laboratories) male mice (8 wk) of confirmed fertility until 6 litters were born or 11 months had passed. Cages were checked daily for pups.
Oocyte and egg collection, microinjection, and chromosome spreads
Fully grown, prophase I-arrested oocytes were collected from the ovaries of 4–8- week-old female mice 48 h after intraperitoneal injection of 5 I.U. of pregnant mare’s serum gonadotropin (PMSG) (Calbiochem). Microinjected oocytes were denuded and injected with ~10 pl of the indicated material and allowed to incubate overnight in CZB medium containing milrinone prior to maturation. For chromosome spreads, the zona pellucida was removed with acidic tyrode’s solution (Millipore) after maturation of the oocytes to metaphase II as previously described[29].
For induced ovulation and collection of metaphase II eggs, female mice (6–8 wks. age) were injected with 5 I.U. of PMSG followed by 5 I.U. of equine choronic gonadotropin (eCG) (Sigma-Aldrich) 48h later. Sixteen hours post eCG injection, cumulous-enclosed egg complexes were collected from the oviduct and cultured in hyaluronidase (3 mg/ml, Sigma) in MEM for 5 min. Denuded eggs were then washed free of hyaluronidase and allowed to recover for 5 min prior to fixation. ML8237 (Alisertib; Selleckchem), Monastrol (Sigma), and ZM447439 (Tocris) were dissolved in dimethyl sulfoxide (DMSO) (Sigma) and added to the CZB culture media at a final concentration of 40 μΜ, 100 μΜ, and 5 μΜ respectively. In vitro maturation of drug-treated oocytes was performed in organ culture dishes (Becton Dickinson) under humidified conditions.
Morpholino oligonucleotides specific for INCENP were designed and produced by Genetools as previously described [10, 34]. Morpholino sequences are INCENP 5-CGTCTTCCCGGACCACTCCTTTTCC-3’ and scrambled control, 5’- GCGCCCCCGCCGTTTCAAAGACAAA-3’. All morpholinos were injected at a concentration of 100 mM. INCENP knockdown reduces the phospho-INCENP signal by ~50% [34, 49].
Constructs and cRNA generation
Generation of Aurka, Aurkc, Aurkb, and Aurkc-KD constructs were previously described [6, 9, 50]. Aurka and Aurkc were PCR amplified from a C57BL/6 12.5d embryo cDNA library and cloned into pIVT-GFP. Mutagenesis was performed using a site-directed Mutagenesis kit per manufacturer’s instructions (Agilent, QuikChange II site-directed mutagenesis kit). mEGFP-Cdk5rap2 cRNA was produced from a pGEMHE vector containing mouse full-length Cdk5rap2 (NM_145990.4) N-terminally fused to EGFP (a gift from Dr. Tomoya Kitajima, RIKEN). To generate cRNA, plasmids were linearized with Nde I, purified (Qiagen, QIAquick PCR Purification), and in vitro transcribed using an mMessage mMachine T7 kit (Ambion), according to manufacturer’s protocol. The synthesized cRNAs were then purified using an RNAeasy kit (Qiagen) and stored at −80°C. All cRNAs were injected at 500 ng/ μΐ.
Real-Time PCR
Twenty prophase-I arrested oocytes were collected and frozen before processing. Gfp mRNA (2 ng) was added to all samples and used as the reference gene. Total RNA for each mixture was purified using the Picopure RNA isolation kit (Thermo Fisher) per the manufacturer’s protocol. cDNA was generated by reverse transcription utilizing random hexamers and Superscript II (Thermo Fisher). Expression levels of the Aurks were measured by quantitative real-time PCR analysis acquired using an ABI Prism 7000 (Life Technologies) according to the manufacturer’s instructions. Taqman probes specific for Aurkb, and Aurkc (MM01718140_m1, and MM03039426_g1, respectively, Life Technologies, Grand Island, NY) were used for gene expression detection and the comparative Ct method was used to determine the difference in expression between samples [48].
Western blotting
A total of 100 prophase-I arrested oocytes,100 Met II eggs, or 20 mg HeLa DNA were mixed with SDS sample buffer (1% SDS, 1% β-mercaptoethanol, 20% glycerol, 50 mM Tris-HCl (pH 6.8)), and denatured at 95°C for 10 min. Western blotting was performed as previously described [49] followed by incubation of the membrane overnight with primary antibodies. The membranes were incubated with horseradish peroxidase-conjugated secondary antibodies GE Healthcare Biosciences, #NA931) for 1 h. Protein bands were detected using the ECL Select western blotting detection reagents (Amersham) following the manufacturer’s protocol.
Immunocytochemistry
Following meiotic maturation, oocytes were fixed in PBS-containing paraformaldehyde (PFA: Sigma) at room temperature (ACA and pINCENP: 2% PFA for 20 mins; Survivin, pAURKA; pHECI, pAURKC, REC8, and SGO2: 2% PFA + 0.1% Triton-X for 20 mins; TPX2: 3.7% PFA 1 h) or 100% methanol at −20 “C (AURKA: 10 min) followed by 2 consecutive washes through blocking buffer (PBS + 0.3% (wt/vol) BSA + 0.1% (vol/vol) Tween-20). Prior to immunostaining, oocytes were permeabilized for 20 min in PBS containing 0.1% (vol/vol) Triton X- 100 and 0.3% (wt/vol) BSA followed by 10 min in blocking buffer. Immunostaining was performed by incubating cells in primary antibody for 1 h at room temperature followed by 3 consecutive 10 min incubations in blocking buffer.
After washing, secondary antibodies were diluted 1:200 in blocking solution and the sample was incubated for 1 h at room temperature followed by 3 consecutive 10 min incubations in blocking buffer. The cells were next mounted in VectaShield (Vector Laboratories) with 4’, 6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI; Life Technologies; 1:170).
For immunofluorescence of meiotic spreads, a clean microscope slide was dipped in Milli-Q water (pH 9.2) containing 1% PFA, 0.15%, Triton X-100 (Sigma), and 3 mM dithiothreitol (Sigma). Zona pellucida-free metaphase II eggs were pipetted along the length of the slide and dried. Prior to immunostaining, slides were washed with PBS followed by incubation in blocking solution (PBS + 3% BSA) for 10 min. Immunostaining was performed at room temperature by incubating slides in primary antibody diluted in blocking buffer for 3 h followed by 3 subsequent 10 min incubations in blocking buffer. After washes, slides were incubated in secondary antibodies diluted in blocking buffer for 1 h followed by 3 subsequent 10 min incubations in blocking buffer. Slides were mounted in Vectashield with DAPI.
For immunofluorescence of HeLa cell lines, on day 3 of doxycycline treatment, 150 X105 cells were plated in 6 well plates containing coated poly-L-Lysine coated coverslips and were fixed 24 h after plating with PBS containing 3.7% paraformaldehyde for 10 min at room temperature. After washing, cells were permeabilized using PBS-0.5% Triton-X 100 for 10 min at room temperature and were subsequently blocked in PBS-1% Bovine Serum Albumin (Sigma) for 1 h at room temperature. Samples were then incubated primary antibody diluted in PBS-0.1% Triton-X 100 for 1 hour in a humidified chamber at room temperature. Slides then were incubated in subsequent PBS washes, 5 min each. Next, cells were incubated with secondary antibodies diluted in PBS-0.1% Triton-X 100 for 30 min in a humidified dark chamber at room temperature. Slides were then incubated in subsequent PBS washes, 5 min each. Coverslips were mounted on microscope slides with Vectashield DAPI (1:177).
Antibodies
The following primary antibodies were used for immunoblot (IB) and immunofluorescence (IF) (mitotic cells (MIF), chromosomes spreads (CSIF), intact oocytes (OIF)) experiments: anti-a-tubulin (WB: 1:10,000; Sigma-Aldrich). Mouse anti a-tubulin Alexa-fluor 488 conjugated (OIF: 1:100, MIF: 1:200, Life Technologies), rabbit anti a-tubulin Alexa-fluor 488 conjugated (OIF: 1:100, Cell Signaling Technologies) AURKA (OIF: 1:500, IB: 1:500; Bethyl), IAK1(AURKA) (MIF: 1:500, BD Biosciences), Phosphorylated AURKA (pAURKA Thr288) (OIF: 1:100; Cell Signaling Technology), AURKB (MIF: 1:1000, Abcam), phosphorylated AURKC (pAURKC Thr171) (OIF:1:500; gift from Dr. T.K. Tang, Institute of Biomedical Sciences), pAURKA/B/C (phosphorylated AURKA Thr288, AURKB Thr223, and AURKC Thr198, respectively) (IB: 500; Cell Signaling Technology); phosphorylated INCENP (pINCENP Ser893/Ser894, AURK specific phosphorylation sites on INCENP) (OIF/MIF: 1:1000; gift from Dr. M. Lampson, University of Pennsylvania)[14], phosphorylated HEC1 (pHEC1 Ser55) (OIF: 1:100; Genetex), Phosphorylated Histone H3 (pH3 Ser10) (OIF/MIF: 1:500, Millipore), Crest/ACA (OIF: 1:30, CSIF/MIF: 1:100, Antibodies Incorporated), SGO2 (OIF: 1:50, a gift from J.L. Barbero, Biological Research Center), REC8 (OIF: 1:1000, CSIF 1:100, a gift from R. Schultz, University of Pennsylvania)[33], anti-MSY2 (WB:1:10,000, gift from R. Schultz). The following secondary antibodies were used at 1:200 for OIF experiments and 1:800 for MIF: Anti- human-Alexa-633 (Life Technologies), anti-human-Alexa-488 (Life Technologies), anti-mouse-Alexa-488 (Life Technologies) anti-mouse-Alexa568 (Life Technologies), anti-rabbit-Alexa568 (Life Technologies).
Microscopy
Images were captured using a Zeiss 510 Meta laser-scanning confocal microscope with 40x objective. For each image, optical z-slices were obtained using a 0.5–1.5 mm step with a zoom setting of 2. For comparison of pixel intensities, the laser power was kept constant for each oocyte in an experiment. Images used for in situ chromosome spreads were obtained using a Zeiss Axiovert 200M epifluorescence microscope with a 63x objective. Z-slices were obtained using a 0.25 mm step.
Live cell imaging
Oocytes expressing mEGFP-mCDK5RAP2, H2B-mCHERRY from microinjected cRNAs and stained with SiR-tubulin were imaged by confocal microscope Leica TCS SP5 as described previously [34]. Oocytes were microinjected with 125 ng/DL mEGFP-Cdk5rap2 and 50 ng/ml of H2B-mCherry cRNAs and incubated at prophase of meiosis I for 2 h. Oocytes were then incubated in 100 nM SiR- tubulin in MEM medium for live imaging. For analysis of Metaphase II maturation competency, prophase-I-arrested oocytes were matured on an EVOS FL Auto Imaging System (Life Technologies) with a 10X objective. The microscope stage was heated to 37°C and 5% CO 2 was maintained using the EVOS Onstage Incubator. Brightfield images were acquired every 20 min for 24 h.
Quantification and Statistical Analysis
Image Analysis
Image J software was used to process all images (NIH). For analysis, z-slices for each image were merged into a projection. Chromosome alignment analysis was performed as previously described [33, 51]. The spindle length was first measured in a single z-plane using the Image J line tool. This value was next divided in half to determine the spindle midzone. Chromatids 4 mm from the spindle midzone were considered unaligned[52]. Spindle length was measured by marking the x,y, and z coordinates for each of the two spindle poles per image and using the Pythagorean theorem to determine distance between the two points. For in situ chromosome spreads, the numbers of kinetochores were counted in Met II eggs using ACA immunoreactivity as a marker[23]. Eggs were considered euploid if they contained 40 chromatids. Deviations from this number were noted as aneuploid. The distances between sister kinetochores were measured using the line tool in Image J. Only sister kinetochores visible in the same plane were measured. For pixel intensity analyses the average pixel intensity was recorded using the measurement tool. To define the region of the chromosomes for intensity measurements, the DNA channel (DAPI) was used as a mask. MTOC markers, including AURKA and a-tubulin were used to define spindle poles for pixel intensity measurements.
Statistical Analysis
One-way analysis of variance (Anova) and Student’s T-test were used to evaluate the difference between and among groups using Prism software (GraphPad Software) as indicated in the Figure legends. Statistical details for each experiment can be found in Figure legends and the Results section. “Experimental n” refers to the number of animals used per experiment with an average of 25 oocytes per animal. Data is shown as the mean ± the standard error of the mean (SEM). p < 0.05 was considered significant.
Supplementary Material
Highlights.
· AURKA supports meiosis in the absence of AURKB/C
· AURKC promotes AURKA spindle function by preventing AURKA chromosome localization
· AURKB is required for meiosis I
· AURKB negatively regulates AURKC activity to ensure gamete euploidy
Acknowledgments
The authors thank Drs. Cecilia Blengini, Iain Cheeseman, Francesca Duncan, Greg FitzHarris, Philip Jordan, Michael Lampson, Patricia Melloy, Kim McKim, Richard Schultz, and Ruth Steward for helpful discussions. PS and DD were supported by Inter-Excellence program by project LTAUSA17097 and LO1609 from National Sustainability Programme of the Czech Ministry of Education, Youth and Sports. Work in the M.M. laboratory was supported by MINECO (SAF2015–69920-R co-funded by ERDF-EU) and Worldwide Cancer Research (WCR15–0278) grants. This work was supported by NIH grants (F31 HD089591 (A.L.N.) and R01 GM112801 (K.S.)).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Hassold T, and Hunt P (2001). To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2, 280–291. [DOI] [PubMed] [Google Scholar]
- 2.Brown JR, Koretke KK, Birkeland ML, Sanseau P, and Patrick DR (2004). Evolutionary relationships of Aurora kinases: Implications for model organism studies and the development of anti-cancer drugs. BMC Evolutionary Biology 4, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seeling JM, Farmer AA, Mansfield A, Cho H, and Choudhary M (2017). Differential Selective Pressures Experienced by the Aurora Kinase Gene Family. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams RR, Carmena M, and Earnshaw WC (2001). Chromosomal passengers and the (aurora) ABCs of mitosis. Trends in Cell Biology 11. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Miranda G, Trakala M, Martin J, Escobar B, Gonzalez A, Ghyselinck NB, Ortega S, Canamero M, Perez de Castro I, and Malumbres M (2011). Genetic disruption of aurora B uncovers an essential role for aurora C during early mammalian development. Development 138, 2661–2672. [DOI] [PubMed] [Google Scholar]
- 6.Balboula AZ, and Schindler K (2014). Selective disruption of aurora C kinase reveals distinct functions from aurora B kinase during meiosis in mouse oocytes. PLoS Genet 10, e1004194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schindler K, Davydenko O, Fram B, Lampson MA, and Schultz RM (2012). Maternally recruited Aurora C kinase is more stable than Aurora B to support mouse oocyte maturation and early development. Proc Natl Acad Sci U S A 109, E2215–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasai K, Katayama H, Hawke DH, and Sen S (2016). Aurora-C Interactions with Survivin and INCENP Reveal Shared and Distinct Features Compared with Aurora-B Chromosome Passenger Protein Complex. PLoS One 11, e0157305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen HL, Tang CJ, Chen CY, and Tang TK (2005).Overexpression of an Aurora-C kinase-deficient mutant disrupts the Aurora-B/INCENP complex and induces polyploidy. J Biomed Sci 12, 297–310. [DOI] [PubMed] [Google Scholar]
- 10.Sharif B, Na J, Lykke-Hartmann K, McLaughlin SH, Laue E, Glover DM, and Zernicka-Goetz M (2010). The chromosome passenger complex is required for fidelity of chromosome transmission and cytokinesis in meiosis of mouse oocytes. J Cell Sci 123, 4292–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimmins S, Crosio C, Kotaja N, Hirayama J, Monaco L, Hoog C, van Duin M, Gossen JA, and Sassone-Corsi P (2007). Differential functions of the Aurora-B and Aurora-C kinases in mammalian spermatogenesis. Mol Endocrinol 21, 726–739. [DOI] [PubMed] [Google Scholar]
- 12.Lampson MA, and Cheeseman IM (2011). Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol 21, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manfredi MG, Ecsedy JA, Chakravarty A, Silverman L, Zhang M, Hoar KM, Stroud SG, Chen W, Shinde V, Huck JJ, et al. (2011). Characterization of Alisertib (MLN8237), an investigational small-molecule inhibitor of aurora A kinase using novel in vivo pharmacodynamic assays. Clin Cancer Res 17, 7614–7624. [DOI] [PubMed] [Google Scholar]
- 14.Salimian KJ, Ballister ER, Smoak EM, Wood S, Panchenko T, Lampson MA, and Black BE (2011). Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr Biol 21, 1158–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saskova A, Solc P, Baran V, Kubelka M, Schultz RM, and Motlik J (2008). Aurora kinase A controls meiosis I progression in mouse oocytes. Cell Cycle 7, 2368–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, and Taylor SS (2003). Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane SI, Chang HY, Jennings PC, and Jones KT (2010). The Aurora kinase inhibitor ZM447439 accelerates first meiosis in mouse oocytes by overriding the spindle assembly checkpoint. Reproduction 140, 521–530. [DOI] [PubMed] [Google Scholar]
- 18.Carmena M, Ruchaud S, and Earnshaw WC (2009). Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol 21, 796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKinley KL, and Cheeseman IM (2017). Large-Scale Analysis of CRISPR/Cas9 Cell-Cycle Knockouts Reveals the Diversity of p53- Dependent Responses to Cell-Cycle Defects. Dev Cell 40, 405–420 e402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu J, Bian M, Liu J, Jiang Q, and Zhang C (2009). A single amino acid change converts Aurora-A into Aurora-B-like kinase in terms of partner specificity and cellular function. Proc Natl Acad Sci U S A 106, 6939–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLuca KF, Meppelink A, Broad AJ, Mick JE, Peersen OB, Pektas S, Lens SMA, and DeLuca JG (2018). Aurora A kinase phosphorylates Hec1 to regulate metaphase kinetochore-microtubule dynamics. J Cell Biol 217, 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bird AW, and Hyman AA (2008). Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J Cell Biol 182, 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein P, and Schindler K (2011). Mouse oocyte microinjection, maturation and ploidy assessment. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Wang P, Zhang B, Zhang J, Ming J, Xie W, and Na J(2017). Differential regulation of H3S10 phosphorylation, mitosis progression and cell fate by Aurora Kinase B and C in mouse preimplantation embryos. Protein Cell 8, 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang T, Duncan FE, Schindler K, Schultz RM, and Lampson MA (2010). Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr Biol 20, 1522–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burkhardt S, Borsos M, Szydlowska A, Godwin J, Williams SA, Cohen PE, Hirota T, Saitou M, and Tachibana-Konwalski K (2016). Chromosome Cohesion Established by Rec8-Cohesin in Fetal Oocytes Is Maintained without Detectable Turnover in Oocytes Arrested for Months in Mice. Curr Biol 26, 678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsutsumi M, Fujiwara R, Nishizawa H, Ito M, Kogo H, Inagaki H, Ohye T, Kato T, Fujii T, and Kurahashi H (2014). Age-related decrease of meiotic cohesins in human oocytes. PLoS One 9, e96710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitajima TS, Kawashima SA, and Watanabe Y (2004). The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427, 510–517. [DOI] [PubMed] [Google Scholar]
- 29.El Yakoubi W, Buffin E, Cladiere D, Gryaznova Y, Berenguer I, Touati SA, Gomez R, Suja JA, van Deursen JM, and Wassmann K (2017). Mps1 kinase-dependent Sgo2 centromere localisation mediates cohesin protection in mouse oocyte meiosis I. Nat Commun 8, 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, and Watanabe Y (2008). Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol 10, 42–52. [DOI] [PubMed] [Google Scholar]
- 31.Rattani A, Ballesteros Mejia R, Roberts K, Roig MB, Godwin J, Hopkins M, Eguren M, Sanchez-Pulido L, Okaz E, Ogushi S, et al. (2017). APC/C(Cdh1) Enables Removal of Shugoshin-2 from the Arms of Bivalent Chromosomes by Moderating Cyclin-Dependent Kinase Activity. Curr Biol 27, 1462–1476 e1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrandiz N, Barroso C, Telecan O, Shao N, Kim HM, Testori S, Faull P, Cutillas P, Snijders AP, Colaiacovo MP, et al. (2018). Spatiotemporal regulation of Aurora B recruitment ensures release of cohesion during C. elegans oocyte meiosis. Nat Commun 9, 834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen AL, Gentilello AS, Balboula AZ, Shrivastava V, Ohring J, and Schindler K (2014). Phosphorylation of threonine 3 on histone H3 by haspin kinase is required for meiosis I in mouse oocytes. J Cell Sci 127, 5066–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balboula AZ, Nguyen AL, Gentilello AS, Quartuccio SM, Drutovic D, Solc P, and Schindler K (2016). Haspin kinase regulates microtubule-organizing center clustering and stability through Aurora kinase C in mouse oocytes. J Cell Sci 129, 3648–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lane SI, and Jones KT (2014). Non-canonical function of spindle assembly checkpoint proteins after APC activation reduces aneuploidy in mouse oocytes. Nat Commun 5, 3444. [DOI] [PubMed] [Google Scholar]
- 36.Hans F, Skoufias DA, Dimitrov S, and Margolis RL (2009). Molecular distinctions between Aurora A and B: a single residue change transforms Aurora A into correctly localized and functional Aurora B. Mol Biol Cell 20, 3491–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eot-Houllier G, Magnaghi-Jaulin L, Fulcrand G, Moyroud FX, Monier S, and Jaulin C (2018). Aurora A-dependent CENP-A phosphorylation at inner centromeres protects bioriented chromosomes against cohesion fatigue. Nat Commun 9, 1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLuca JG (2018). Aurora A Kinase Function at Kinetochores. Cold Spring Harb Symp Quant Biol. [DOI] [PubMed] [Google Scholar]
- 39.Cutts SM, Fowler KJ, Kile BT, Hii LL, O’Dowd RA, Hudson DF, Saffery R, Kalitsis P, Earle E, and Choo KH (1999). Defective chromosome segregation, microtubule bundling and nuclear bridging in inner centromere protein gene (Incenp)-disrupted mice. Hum Mol Genet 8, 1145–1155. [DOI] [PubMed] [Google Scholar]
- 40.Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, and Choo KH (2000). Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol 10, 1319–1328. [DOI] [PubMed] [Google Scholar]
- 41.van Echten-Arends J, Mastenbroek S, Sikkema-Raddatz B, Korevaar JC, Heineman MJ, van der Veen F, and Repping S (2011). Chromosomal mosaicism in human preimplantation embryos: a systematic review. Hum Reprod Update 17, 620–627. [DOI] [PubMed] [Google Scholar]
- 42.Lopez-Carrasco A, Oltra S, Monfort S, Mayo S, Rosello M, Martinez F, and Orellana C (2013). Mutation screening of AURKB and SYCP3 in patients with reproductive problems. Mol Hum Reprod 19, 102–108. [DOI] [PubMed] [Google Scholar]
- 43.Dieterich K, Soto Rifo R, Faure AK, Hennebicq S, Ben Amar B, Zahi M, Perrin J, Martinez D, Sele B, Jouk PS, et al. (2007). Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat Genet 39, 661–665. [DOI] [PubMed] [Google Scholar]
- 44.Zekri A, Lesan V, Ghaffari SH, Tabrizi MH, and Modarressi MH (2012). Gene amplification and overexpression of Aurora-C in breast and prostate cancer cell lines. Oncol Res 20, 241–250. [DOI] [PubMed] [Google Scholar]
- 45.Khan J, Ezan F, Cremet JY, Fautrel A, Gilot D, Lambert M, Benaud C, Troadec MB, and Prigent C (2011). Overexpression of active Aurora-C kinase results in cell transformation and tumour formation. PLoS One 6, e26512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lan ZJ, Xu X, and Cooney AJ (2004). Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol Reprod 71, 1469–1474. [DOI] [PubMed] [Google Scholar]
- 47.Chatot CL, Ziomek CA, Bavister BD, Lewis JL, and Torres I (1989). An improved culture medium supports development of random- bred 1-cell mouse embryos in vitro. J Reprod Fertil 86, 679–688. [DOI] [PubMed] [Google Scholar]
- 48.Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 49.Fellmeth JE, Gordon D, Robins CE, Scott RT Jr., Treff NR, and Schindler K (2015). Expression and characterization of three Aurora kinase C splice variants found in human oocytes. Mol Hum Reprod 21, 633–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shuda K, Schindler K, Ma J, Schultz RM, and Donovan PJ (2009). Aurora kinase B modulates chromosome alignment in mouse oocytes. Mol Reprod Dev 76, 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lane SI, Yun Y, and Jones KT (2012). Timing of anaphase-promoting complex activation in mouse oocytes is predicted by microtubule- kinetochore attachment but not by bivalent alignment or tension. Development 139, 1947–1955. [DOI] [PubMed] [Google Scholar]
- 52.Marin D, Nguyen AL, Scott RT Jr., and Schindler K (2018). Using Mouse Oocytes to Assess Human Gene Function During Meiosis I. J Vis Exp.(134), e57442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


