Genome analyses indicate that many bacteria possess an elevated number of chemoreceptors, suggesting that these species are able to perform chemotaxis to a wide variety of compounds. The scientific community is now only beginning to explore this diversity and to elucidate the corresponding physiological relevance. The discovery of histamine chemotaxis in the human pathogen Pseudomonas aeruginosa provides insight into tactic movements that occur within the host. Since histamine is released in response to bacterial pathogens, histamine chemotaxis may permit bacterial migration and accumulation at infection sites, potentially modulating, in turn, quorum-sensing-mediated processes and the expression of virulence genes. As a consequence, the modulation of histamine chemotaxis by signal analogues may result in alterations of the bacterial virulence. As the first report of bacterial histamine chemotaxis, this study lays the foundation for the exploration of the physiological relevance of histamine chemotaxis and its role in pathogenicity.
KEYWORDS: Pseudomonas aeruginosa, chemotaxis, histamine
ABSTRACT
Histamine is a key biological signaling molecule. It acts as a neurotransmitter in the central and peripheral nervous systems and coordinates local inflammatory responses by modulating the activity of different immune cells. During inflammatory processes, including bacterial infections, neutrophils stimulate the production and release of histamine. Here, we report that the opportunistic human pathogen Pseudomonas aeruginosa exhibits chemotaxis toward histamine. This chemotactic response is mediated by the concerted action of the TlpQ, PctA, and PctC chemoreceptors, which display differing sensitivities to histamine. Low concentrations of histamine were sufficient to activate TlpQ, which binds histamine with an affinity of 639 nM. To explore this binding, we resolved the high-resolution structure of the TlpQ ligand binding domain in complex with histamine. It has an unusually large dCACHE domain and binds histamine through a highly negatively charged pocket at its membrane distal module. Chemotaxis to histamine may play a role in the virulence of P. aeruginosa by recruiting cells at the infection site and consequently modulating the expression of quorum-sensing-dependent virulence genes. TlpQ is the first bacterial histamine receptor to be described and greatly differs from human histamine receptors, indicating that eukaryotes and bacteria have pursued different strategies for histamine recognition.
INTRODUCTION
Bacteria possess different types of signal transduction systems that enable them to adapt to changes in environmental cues. In addition to one- and two-component signal transduction systems, chemosensory pathways play an important role in this process (1–3). In a canonical chemosensory pathway, signaling is initiated by the binding of signal molecules to the chemoreceptor ligand binding domain (LBD), which in turn modulates the autophosphorylation activity of the CheA histidine kinase and the transphosphorylation of the CheY response regulator, which ultimately triggers pathway output (2). While most chemoreceptors mediate chemotaxis, some also carry out alternative cellular functions, such as modulating c-di-GMP levels or type IV pilus-based motility (4–6).
Escherichia coli is the traditional model organism for the study of chemoreceptor-based signaling processes (7). It has 5 chemoreceptors, of which 4 contain a periplasmic 4-helix bundle LBD. Importantly, these chemoreceptors bind signals either directly or in complex with a periplasmic ligand binding protein. E. coli has a single chemosensory cascade that mediates chemotaxis primarily toward sugars, amino acids, or dipeptides (7, 8).
More recently, chemoreceptor-based signaling has been studied in an array of bacteria with different lifestyles (9). The existing data suggest that the typical number of chemoreceptor genes in bacteria, which can reach as high as 80, is much higher than in E. coli (10). Furthermore, sequence analyses indicate that chemoreceptors comprise more than eighty different LBD types (11). The most abundant of these are CACHE-type LBDs, which are present in either the monomodular (sCACHE) or bimodular (dCACHE) form (12). The large number of chemoreceptor genes and the diversity of LBD types suggest that bacteria can respond to a wide variety of signal molecules. The scientific community is now beginning to explore this diversity and to elucidate the corresponding physiological relevance.
Pseudomonads are important model organisms for the study of chemoreceptor function (13, 14), and the strains Pseudomonas putida KT2440 and Pseudomonas aeruginosa PAO1 have been well studied and characterized (11). The former strain is a nonpathogenic soil bacterium with a saprophytic lifestyle (15). In contrast, P. aeruginosa strains are among the most virulent opportunistic human pathogens and the leading cause of nosocomial infections, particularly in immunocompromised, cancer, burn, and cystic fibrosis patients (16).
Strains KT2440 and PAO1 have similar numbers of chemoreceptor genes: 27 and 26, respectively. The function and the corresponding ligand profiles have been established for approximately ten receptors in each strain (11, 17). Among the functionally annotated KT2440 chemotaxis receptors are several for different organic acids (18), purines (19), proteinogenic amino acids (20), and gamma-aminobutyric acid (GABA) (21). In addition, the McpU chemoreceptor of this strain was the first chemoreceptor identified that responded to the polyamines putrescine, spermidine, and cadaverine (20, 22). In contrast, PAO1 chemotaxis to proteinogenic amino acids and GABA is mediated by three paralogous receptors, namely, PctA, PctB, and PctC (23, 24). Additionally, this strain has two receptors for inorganic phosphate (25, 26) as well as receptors for malate (27, 28), α-ketoglutarate (29), and chloroethylenes (30). P. aeruginosa is also attracted to the plant hormone ethylene, and it was shown that the deletion of the gene encoding the TlpQ chemoreceptor abolished ethylene chemotaxis (31).
In this study, we provide the first report of bacterial chemotaxis toward histamine. This compound is produced by different animal tissues and is secreted by some bacteria (32). Histamine is a signal molecule with multiple functions. It is an aminergic neurotransmitter of the central and peripheral nervous systems, and it is involved in numerous biological processes (33). It is also a key modulator of local immune responses by mediating the effects on many cell types such as antigen-presenting cells, natural killer cells, and epithelial cells, as well as T and B lymphocytes (34). Bacteria have been shown to impact histamine function. For example, bacterial respiratory tract infections stimulate neutrophils to release histamine (35, 36). Also, it was shown that infection by PAO1 greatly increased neutrophil histamine content and secretion but did not alter histamine production in mast cells, which are the classical histamine reservoirs (36). Furthermore, it has been shown that histamine might play divergent roles in the immune response: it has been implicated in mediating the defense against infection (37) as well as increasing the susceptibility to infection (38). While there has been preliminary evidence that histamine is a signal molecule for bacteria, the underlying mechanisms remain largely unknown (39). The present study provides important insight into the molecular mechanisms that permit bacteria to sense and respond to histamine.
RESULTS
Identification of histamine and additional polyamines as novel ligands for the P. putida KT2440 McpU chemoreceptor.
By screening 190 compounds for binding to the purified McpU-LBD, we previously found that McpU binds to and mediates chemotaxis to putrescine, cadaverine, and spermidine (20). In the present study, we extended this screening to include 285 additional compounds. These compounds were mostly bacterial nitrogen, phosphorous, and sulfur sources (see Materials and Methods). We used a thermal shift assay to monitor changes in the midpoint of protein unfolding (Tm) caused by ligand binding (40). In the absence of ligand, McpU-LBD had a Tm of 46.5°C. Of the 95 nitrogen sources screened (Biolog plate PM3B), three additional compounds—agmatine, ethylenediamine, and histamine—caused Tm increases greater than 2°C (Fig. 1).
FIG 1.
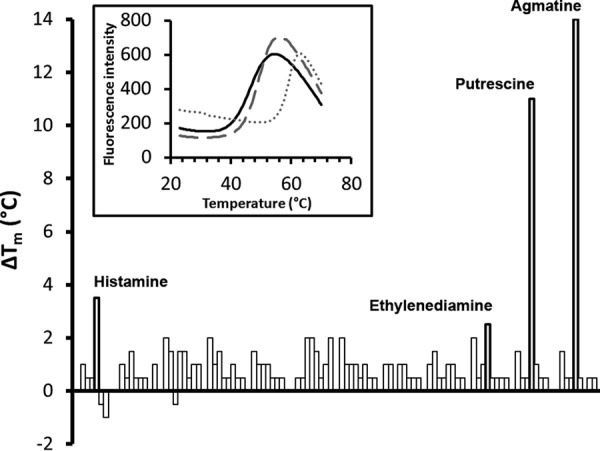
Thermal shift assays of P. putida KT2440 McpU-LBD against a library of ligands. Shown are the individual Tm changes caused by 95 compounds (Biolog array PM3B) that can serve as nitrogen sources. The inset shows the unfolding curves of McpU-LBD when free from ligand (continuous line) and in the presence of agmatine (dotted line) and histamine (dashed line).
Using isothermal titration calorimetry (ITC), we found that all three compounds bind to McpU-LBD (see Fig. S1A in the supplemental material). Very tight binding was observed for agmatine with a KD (equilibrium dissociation constant) in the nanomolar range, whereas histamine and ethylenediamine bound with much lower affinities (Table 1). It should be noted that of these three new McpU ligands and the previously identified ligands (i.e., putrescine, cadaverine, and spermidine), all except for histamine are polyamines (Fig. S1B).
TABLE 1.
Thermodynamic parameters for the binding of ligands to McpU-LBD and TlpQ-LBD as derived from ITC experimentsa
| Compound | McpU-LBD |
TlpQ-LBD |
KD McpU-LBD/KD TlpQ-LBD | ||
|---|---|---|---|---|---|
| KD (µM) | ΔH (kcal · mol−1) | KD (nM) | ΔH (kcal · mol−1) | ||
| Putrescine | 2 ± 0.1b | −15 ± 0.5 | 134 ± 12 | −6.8 ± 0.3 | 15 |
| Cadaverine | 22 ± 2b | −15.5 ± 0.5 | 150 ± 4 | −6.0 ± 0.1 | 147 |
| Spermidine | 4.5 ± 0.4b | −4.3 ± 0.3 | 56 ± 4 | −4.6± 0.4 | 80 |
| Agmatine | 0.48 ± 0.02 | −14.5 ± 0.2 | 150 ± 9 | −5.4 ± 0.1 | 3 |
| Ethylenediamine | 39 ± 4 | −9.7 ± 0.5 | 1,710 ± 180 | −6.3 ± 0.6 | 23 |
| Histamine | 26 ± 2 | −2.6 ± 0.3 | 639 ± 27 | −9.1 ± 0.3 | 41 |
Means and standard deviations represent data from three independent experiments.
Reported previously in reference 20.
Microcalorimetric binding studies of McpU-LBD. (A) Titrations of 17.5 µM McpU-LBD with 1 mM histamine and ethylenediamine and 30 µM McpU-LBD with 0.5 mM agmatine. (B) Chemical structure of ligands recognized by the McpU and TlpQ chemoreceptors. Download FIG S1, JPG file, 0.6 MB (615.4KB, jpg) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identification of TlpQ as a histamine receptor in Pseudomonas aeruginosa.
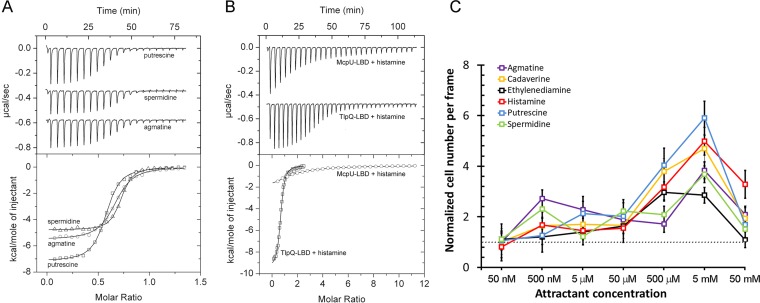
Because histamine plays an important role in the immune response, we aimed to identify McpU homologues in P. aeruginosa that may also sense and mediate chemotaxis to histamine. To this end, we carried out a sequence clustering analysis of all dCACHE-containing chemoreceptors in PAO1 and KT2440 (see Fig. S2A). This analysis revealed that the LBD of the TlpQ receptor shares 62% sequence identity with the McpU-LBD homologue (Fig. S2B). To verify TlpQ function, we purified TlpQ-LBD for ITC binding studies. The results showed that five McpU-LBD ligands bind to TlpQ-LBD with nanomolar affinities, whereas the binding of ethylenediamine was slightly weaker (Fig. 2A, Table 1). Spermidine had a KD of 56 nM, which is the highest ligand affinity ever observed for a chemoreceptor. Histamine had a KD of 639 nM, which is an affinity 41 times higher than its affinity for McpU-LBD (Table 1, Fig. 2B). Thus, the affinities of the ligands to TlpQ-LBD were 3 to 147 times higher than their affinities to the McpU-LBD (Table 1). Previous studies showed that TlpQ mediates chemotaxis to ethylene (31), but the titration of TlpQ-LBD with a saturated ethylene solution did not show any binding (data not shown).
FIG 2.
Identification and analysis of TlpQ ligands. (A) Microcalorimetric titrations of 15 µM TlpQ-LBD with 4.8 µl aliquots of 250 µM putrescine, spermidine, or cadaverine. (B) Microcalorimetric titration of 17.5 µM McpU-LBD with 9.6 µl aliquots of 1 mM histamine and titration of 15 µM TlpQ-LBD with 4.8 µl aliquots of 250 µM histamine. Upper graphs show raw titration data, while lower graphs show integrated corrected peak areas of the titration data fit using the “one binding site model.” The derived thermodynamic parameters are provided in Table 1. (C) Quantitative capillary chemotaxis assays of P. aeruginosa PAO1 toward TlpQ ligands. Shown are the ratios of cells after 2 min of exposure to the chemoattractant relative to the number of cells at the beginning of the experiment. The horizontal line marks the ratio of 1, which is indicative of no chemotaxis; n = 3.
Identification of a McpU homologue. (A) Sequence clustering of the ligand binding domains of dCACHE-containing chemoreceptors from P. putida KT2440 (blue) and P. aeruginosa PAO1 (green). The figure was produced using the Phylogeny.fr server. (B) Sequence alignment of the TlpQ and McpU chemoreceptors. Sequence identity between the two receptors is 62%. The alignment was done in the slow mode using the CLUSTALW multiple alignment tool of the NPSA suite (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_server.html). The GONNET protein weight matrix was used in the slow pairwise alignment mode using a gap opening penalty of 10 and a gap extension penalty of 0.1. Red, identical; green, highly similar; blue, weakly similar. The transmembrane regions flanking the ligand binding domain were predicted using the DAS algorithm (https://tmdas.bioinfo.se/DAS/index.html) and are highlighted in yellow. Cyan highlights amino acids that are involved in side-chain mediated interactions with bound histamine. Download FIG S2, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Characterization of histamine chemotaxis.
KT2440 and PAO1 both contain chemoreceptors that bind histamine. In initial experiments, we identified the optimal culture conditions for motility of both strains (see Fig. S3). Using these conditions, we carried out capillary chemotaxis assays of PAO1 toward the six TlpQ ligands (Fig. 2C). All ligands caused chemotaxis, with significant responses observed for some ligands at concentrations as low as 500 nM, whereas optimal responses occurred at 5 mM. In subsequent experiments, we compared the histamine dose response for KT2440 with that of PAO1 (Fig. 3A). KT2440 showed only moderate chemotaxis over the entire concentration range tested, whereas PAO1 responses were much stronger. In accordance with the different binding affinities observed by ITC, the response onsets between strains also differed. Thus, PAO1 required 500 nM histamine, while KT2440 required 5 µM.
FIG 3.

Histamine chemotaxis in different bacteria. (A) Quantitative capillary chemotaxis assays of P. aeruginosa PAO1 and P. putida KT2440 to different histamine concentrations. (B) Response of different strains to 5 mM histamine; n = 3. **, P < 0.01 (by Student’s t tests).
Assessment of motility during bacterial growth. Overnight cultures of P. aeruginosa PAO1 (continuous line) and P. putida KT2440 (dotted line) were used to inoculate LB (upper panel) and 2× YT medium (lower panel) to an OD600 of 0.01. Growth was carried out at 37°C (P. aeruginosa PAO1) or 30°C (P. putida KT2440), and bacteria were inspected microscopically. Motility scores were calculated as follows: score 1, 25% of bacteria are motile; 2, 50%; 3, 75%; and 4, 100%. Download FIG S3, TIF file, 0.1 MB (129.2KB, tif) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To assess the metabolic value of these ligands, we conducted growth experiments with PAO1 and KT2440 in minimal medium containing each of the ligands as the sole carbon or nitrogen source. We found that most of the ligands supported growth either as the carbon or nitrogen source (see Fig. S4). The exceptions were spermidine and ethylenediamine that were either not or were poor growth substrates for PAO1 and KT2440 (Fig. S4). Histamine permitted the growth of both strains as the sole C and N source.
Growth experiments with McpU/TlpQ ligands as sole carbon or nitrogen source. (A) Capacity of ligands to sustain growth of P. aeruginosa PAO1 and P. putida KT2440. Overnight cultures were used to inoculate MS minimal medium supplemented with the McpU/TlpQ ligands as either sole carbon or nitrogen source. As controls, succinate and ammonium nitrate were used. (B) Growth of Ralstonia pseudosolanacearum Ps29 in medium with histamine as the sole carbon and nitrogen source. The initial OD660 was 0.005. The composition of the RSM medium was 10 mM K2HPO3, 5.5 mM KH2PO4, 0.5 mM sodium citrate, 9.5 mM (NH4)2SO4, 1 mM MgSO4, and 28 mM glucose. In all cases, the ligands were added at a concentration of 5 mM, and shown are means and standard deviations from three biological replicates conducted in triplicates. Download FIG S4, JPG file, 0.8 MB (776.3KB, jpg) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Additional experiments were conducted to assess histamine chemotaxis in other bacteria. First, we assessed the motility of P. aeruginosa strains 227, 233, 287, 401, and 428, which were isolated from patients with urinary tract infections (41). Strains 233 and 401 exhibited motilities comparable to that of PAO1 and were therefore selected for further studies. P. aeruginosa PA14 as well as the plant pathogen Ralstonia pseudosolanacearum Ps29 were also included in these experiments. We found that all analyzed P. aeruginosa strains showed significant chemotaxis to 5 mM histamine, and their chemotactic phenotype was significantly higher than that of KT2440. On the other hand, the strain Ps29 was not attracted to histamine (Fig. 3B). Growth experiments with Ps29 in minimal medium containing histamine as the sole carbon and nitrogen source revealed no significant growth (Fig. S4B), suggesting a link between chemotaxis and the capacity to use histamine for growth.
Three-dimensional structure of TlpQ-LBD in complex with histamine.
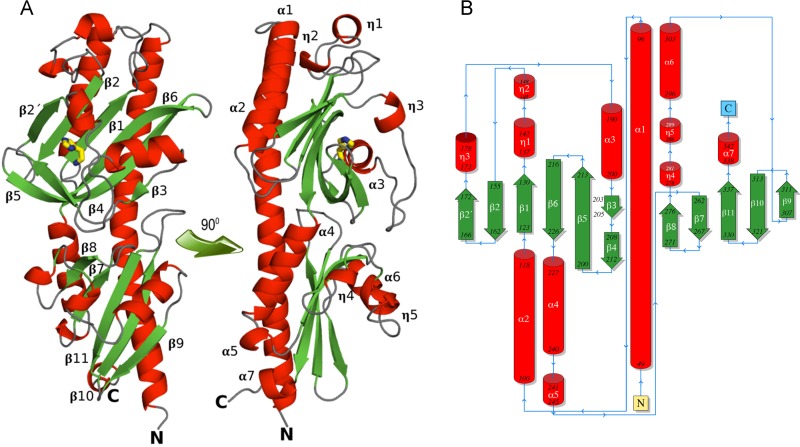
To determine the molecular determinants for histamine recognition by TlpQ, we solved the high-resolution structure of TlpQ-LBD in complex with histamine. There are four monomers in the asymmetric unit,and the superimposition of their Cα atoms resulted in root mean square deviation (RMSD) values of 0.4 to 0.8 Å, indicative of high similarity. An inspection of the structure revealed that it is a dCACHE domain (12) (Fig. 4). A long N-terminal helix is followed by two globular α/β modules, termed membrane-proximal and membrane-distal modules. The membrane-distal module contained bound histamine in all four monomers of the asymmetric unit (Fig. 4).
FIG 4.
Structure of the TlpQ chemoreceptor ligand binding domain in complex with histamine. (A) Ribbon diagram with annotated secondary structure elements. Bound histamine is shown as a stick structure. (B) Schematic representation of the secondary structure elements.
Structural alignments of TlpQ-LBD with entries in the protein data bank identified structural homologues (see Table S1). Most of the homologues are categorized as dCACHE_1 Pfam domains (12). This domain is found in histidine kinases and chemoreceptors, as well as in a novel cytosolic receptor protein (PDB identifier [ID] 5ere), and are found in different bacteria as well as in Arabidopsis. The average size of the domains, while taking into account the segment in between both transmembrane regions, is 268 ± 17 amino acids (Table S1). Of all the homologous domains that we identified, TlpQ-LBD was the largest, at 334 amino acids, namely due to particularly long inserts between β-strands 1 and 2, extended helices α1 and α2, and an extended loop between helices η3 and α3 (see Fig. S5).
Structural alignment of the Cα chain of TlpQ-LBD (in red) with a homologous structure from a histidine kinase of Shewanella oneidensis (in green). This structure is deposited in the protein data bank with ID 3lic. The arrows indicate inserts and loops in the TlpQ-LBD structure that account for its elevated size. Download FIG S5, JPG file, 0.7 MB (705.7KB, jpg) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structural alignment of TlpQ-LBD with structures deposited in the protein data bank. Shown are the structures with a Z-score above 15. Download Table S1, DOCX file, 0.03 MB (32.3KB, docx) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A well-defined electron density for histamine was observed in all four monomers, enabling the ligand placement to be determined (Fig. 5A). TlpQ ligands are present as protonated polycations at neutral pH, which explains why the ligand binding pocket is highly negatively charged (Fig. 5B). All three histamine nitrogen atoms establish hydrogen bonds (Fig. 5C). TlpQ ligands contain at least one primary amino group, and the primary amino group of histamine plays a central role in binding because it forms hydrogen bonds with the side chains of Tyr208, Asp210, and Asp239. In addition, this histamine amino group interacts with a water molecule coordinated by the main chain oxygen of Lys211 and the hydroxyl group of Tyr158. Each of the histamine imidazole nitrogen atoms forms hydrogen bonds with Asp210 and Glu170. The LBDs of TlpQ and McpU of P. putida KT2440 share approximately 50% sequence identity (Fig. S2B). When their structures containing either histamine or putrescine were superimposed (Fig. 5D), it became apparent that the primary amino groups of both ligands are coordinated in a similar manner via hydrogen bonds with Y208/D210/D239 of TlpQ-LBD or their equivalents in McpU-LBD.
FIG 5.
Ligand binding pocket of the TlpQ ligand binding domain. (A) Close-up view of the ligand binding pocket. The electron density for histamine is shown. (B) Surface charge representation of the histamine binding site; red and blue shading represent negative and positive charges, respectively. (C) Schematic representation of amino acids involved in hydrogen bonds with histamine. (D) Superimposition of the ligand binding pockets of McpU-LBD with bound putrescine (green, PDB ID 6F9G) and TlpQ-LBD with bound histamine (blue).
Histamine chemotaxis is mediated by multiple chemoreceptors in PAO1.
To assess the role of TlpQ in histamine chemotaxis, we generated a tlpQ mutant. Control experiments showed that its response to Casamino Acids was comparable to that of the wild type. However, the response of this mutant to 5 mM histamine was also similar to that of the wild type (Fig. 6A), indicating that additional chemoreceptors may be involved.
FIG 6.
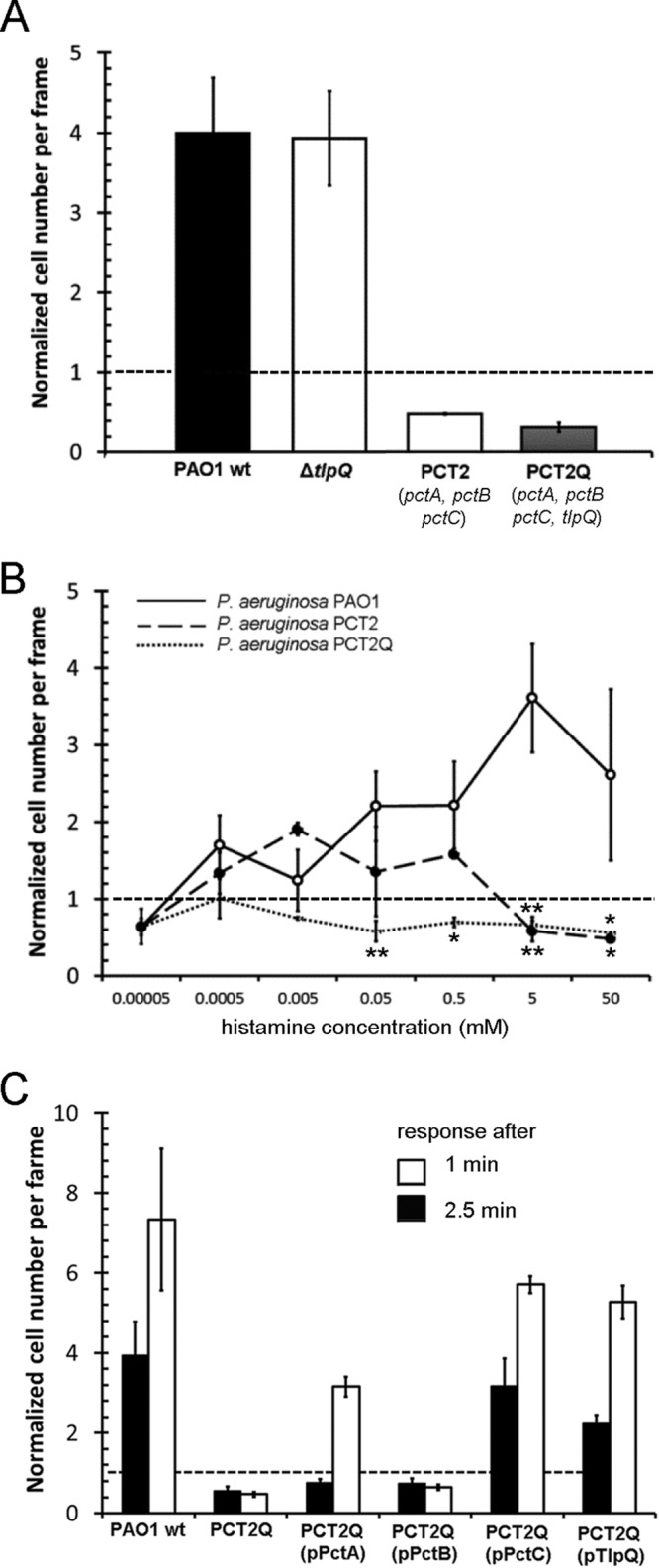
Chemotaxis to histamine is mediated by multiple chemoreceptors in P. aeruginosa PAO1. (A) Chemotactic responses to 5 mM histamine by wild-type and mutant strains. (B) Histamine dose-response chemotaxis assays for PAO1 and for PCT2 and PCT2Q mutants. (C) Chemotactic response of PAO1 and PCT2Q harboring plasmids pPctA, pPctB, pPctC, and pTlpQ to 500 µM histamine after contact times of 1 min and 2.5 min; n = 3. *, P < 0.05; **, P < 0.01 (by Student’s t tests).
To identify these additional chemoreceptors, we screened a number of mutants, in which 3 to 7 chemoreceptor genes had been deleted. Our results showed that the deletion of the pctA, pctB, and pctC chemoreceptor genes (strain PCT2) abolished chemotaxis to 5 mM histamine (Fig. 6A). PctA, PctB, and PctC are chemoreceptors for l-amino acids (23, 24), while PctC also mediates chemotaxis toward GABA (21).
To clarify the roles of PctA, PctB, PctC, and TlpQ in histamine chemotaxis, we conducted dose-response experiments using wild-type PCT2 as well as a mutant in which the pctABC as well as the tlpQ gene had been deleted, named PCT2Q (Fig. 6B). The latter mutant was devoid of histamine chemotaxis over the entire concentration range (50 nM to 50 mM), whereas significant chemotaxis was observed for the PCT2 mutant at a concentration range between 500 nM and 500 µM. This indicates that TlpQ mediates chemotaxis to low histamine concentrations, which is in agreement with the very high affinity observed in vitro. In contrast, one or more of the PctA, PctB, and PctC receptors mediate chemotaxis to elevated histamine concentrations.
To assess the role of the individual chemoreceptors, the PCT2Q mutant devoid of histamine chemotaxis was complemented with plasmids containing one of the four chemoreceptors—an approach that has previously proven effective to study complex chemotactic processes (42). To confirm the phenotypes of these strains, chemotaxis was measured toward previously identified ligands, namely l-Ile (PctA), l-Arg (PctB), and GABA (PctC) (23, 24), and the three complemented strains responded to these ligands. Histamine chemotaxis measurements revealed that the pctC and tlpQ genes in trans recovered histamine chemotaxis using an exposure time of 1 min. At 2.5 min, complementation with pctA, pctC, and tlpQ resulted in significant chemotaxis (Fig. 6C). Thus, these data reveal that histamine chemotaxis is mediated by the concerted action of PctA, PctC, and TlpQ. To assess the role of these receptors in another strain, we generated a triple mutant in the homologous receptors of P. aeruginosa PA14. As shown in Fig. S6, the deletion of these receptors also abolished histamine chemotaxis.
Quantitative capillary chemotaxis assays of P. aeruginosa PA14 and a mutant defective in the pctA, pctC, and tlpQ genes towards histamine. Data have been corrected with the number of bacteria (6,900 ± 142) that swam into buffer-containing capillaries. Shown are means and standard deviations from three individual experiments conducted in duplicates. Download FIG S6, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TlpQ, PctA, and PctC employ different mechanisms to mediate histamine chemotaxis.
To determine the mechanism by which PctA and PctC respond to histamine, microcalorimetric binding studies with purified PctA-LBD and PctC-LBD were conducted. Whereas the proteins bound l-Ala and l-Gln (24), respectively, histamine did not bind. Direct microcalorimetric titrations can only provide information on high-affinity binding events because of the limitations presented by ligand dilution heats. To assess the possibility of low-affinity histamine binding, we conducted a competition assay. PctA-LBD was titrated with l-Ala in the presence and absence of 20 mM histamine. However, the resulting titration curves were almost identical (see Fig. S7), confirming that histamine does not bind directly to PctA-LBD.
Assessment of potential low-affinity binding of histamine to PctA-LBD by microcalorimetric competition experiments. Shown are microcalorimetric titrations of 39 µM PctA-LBD with 1 mM l-alanine either in the absence or presence of 20 mM histamine. The resulting integrated peak areas are almost identical indicating that histamine does not compete with l-alanine for binding at PctA-LBD. Download FIG S7, JPG file, 0.2 MB (220.4KB, jpg) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To assess whether PctA and PctC may be activated by histamine-containing periplasmic binding proteins, pulldown experiments with immobilized PctA-LBD and PctC-LBD as well as PAO1 protein extracts were conducted using previously verified protocols (25). However, our results provided no evidence for binding partners to either domain.
DISCUSSION
The elevated numbers of chemoreceptors in many bacteria suggest that this abundance confers chemotactic capabilities to many different stimuli, and the scientific community is only beginning to explore the diversity of these responses. In general, chemoeffectors can be classified into three groups according to their physiological role. First, the majority of chemoattractants are important nutritional sources, as evidenced by numerous receptors that respond to different organic or amino acids (11). Second, chemoattraction has been observed for signal molecules such as plant hormones (31, 43), neurotransmitters (44), and quorum sensing signals (45), which inform bacteria about their environment. Lastly, chemoreceptors can signal the presence of compounds, such as histamine, that are involved in multiple functions.
Thus, what is the physiological relevance of chemotaxis toward histamine? One possibility is certainly that, like most of the other McpU/TlpQ ligands, histamine supports growth as the sole C and N source. However, chemotaxis to host signals has been shown for many different pathogens to be essential for efficient infection and virulence (46). Importantly, P. aeruginosa PAO1 was shown to greatly increase neutrophil histamine content and secretion in mouse models (36), and chemotaxis to this host-derived signal will result in an accumulation of bacterial cells at the infection site. This increase in bacterial cell density likely alters the expression of quorum-sensing-controlled genes, including those responsible for the production of virulence determinants and biofilm formation in P. aeruginosa (47). Nonetheless, the precise assessment of the role of histamine chemotaxis in the virulence of P. aeruginosa is technically a difficult undertaking, since it is unfeasible to generate a mutant that is deficient in histamine chemotaxis without impairing taxis to the remaining identified ligands for PctA (17 amino acids), PctC (GABA and 2 amino acids), and TlpQ (5 polyamines) (24).
The interference with motility and chemotaxis is an alternative strategy to block bacterial pathogens (48). Previous work has shown that some chemoreceptors recognize chemoattractants and antagonists (27, 49, 50), and the identification of antagonists that specifically interfere with histamine chemotaxis may thus be an alternative approach to modulate the virulence properties of P. aeruginosa. Remarkably, the identification of these antagonists may be facilitated by the resolution of the three-dimensional structure of TlpQ-LBD in complex with histamine (Fig. 4 and 5).
High sensitivity histamine responses are mediated by the TlpQ chemoreceptor, which binds histamine directly. TlpQ is in many aspects an atypical chemoreceptor. First, its LBD, which is 334 amino acids, is larger than any other known chemoreceptor LBD (11). Second, it has the highest affinity ever observed for the binding of a chemoattractant to the recombinant LBD of a chemoreceptor. Histamine binding occurred with an affinity of 639 nM, which is among the highest affinities observed for chemoattractants. This unusually high affinity permits responses to very low histamine concentrations, and the onset of chemotactic response occurred at the unusually low concentration of 500 nM (Fig. 2C).
Frequently, the deletion of a chemoreceptor abolishes taxis to a given compound, indicating that there is a single receptor for a given chemoattractant (18, 29). However, histamine chemotaxis is mediated by the concerted action of three receptors. Thus, what is the advantage of having multiple receptors for the same chemoattractant? In this context, close similarities exist between histamine chemotaxis and the mechanisms by which P. aeruginosa is attracted to inorganic phosphate (Pi). Pi is a key signaling molecule that controls the expression of many virulence genes (51, 52). Chemotaxis to a low Pi concentration is mediated by the CtpL receptor, whereas CtpH is responsible for responses to high concentrations (25, 26). Whereas CtpH recognizes Pi directly at its LBD, CtpL is stimulated by the Pi-loaded periplasmic binding protein PstS (25). A chemoreceptor, either stimulated by direct or indirect signal recognition, is characterized by a response range (53). The combined action of multiple chemoreceptors with different sensing abilities permits the microorganism to expand its response range to a given chemoattractant. The presence of multiple receptors for a given chemoeffector may suggest that a compound is particularly physiologically relevant.
Four human histamine receptors have been described, termed H1, H2, H3, and H4 (54). Histamine was shown to mediate the chemotaxis of mast cells via the H4 receptor, and this mechanism might be responsible for mast cell accumulation in allergic tissues (55). However, the topology of eukaryotic histamine receptors differs entirely from that of their bacterial counterparts. All four receptor types form a barrel composed of seven transmembrane helices (54). The three-dimensional structure of the human H1 receptor has been solved (56), which revealed that ligands bind within this transmembrane barrel. Therefore, the evolutionary strategies to sense histamine greatly differ between bacteria and humans.
Here, we provide the first report of bacterial chemotaxis toward histamine. Histamine is vital to cellular processes in mammals, and the initial evidence suggests that histamine also functions as a bacterial signal molecule. This study expands the range of known bacterial chemoeffectors and lays the foundation for deciphering the molecular mechanisms underlying histamine chemotaxis and its role in bacterial virulence.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used are listed in Table 2, and oligonucleotides are listed in Table S2 in the supplemental material.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics1 | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli BL21(DE3) | F− ompI hsdSB(rB− mB−) gal dam met | 69 |
| DH5α | supE44 lacU169(φ80lacZΔM15) hsdR17 (rK− mK−) recA1 endA1 gyrA96 thi-1 relA1 | 70 |
| HB101 | F− Δ(gpt-proA)62 leuB6 supE44 ara-14 galK2 lacY1 Δ(mcrC-mrr) rpsL20 (Smr) xyl-5 mtl-1 recA13 thi-1 | 71 |
| JM109 | F′ traD36 proA+B+ lacIq Δ(lacZ)M15 Δ(lac-proAB) glnV44 e14− gyrA96 recA1 relA1 endA1 thi hsdR17 | 72 |
| S17-1 λpir | Tpr Smr (recA thi pro hsdR)− M+RP4: 2-Tc:Mu: Km Tn7 λpir | 73 |
| Ralstonia pseudosolanacearum Ps29 | Wild-type strain race 1, biovar 3, phylotype I | 74 |
| Pseudomonas putida KT2440 | Wild type | 15 |
| KT2440R | Rifampicin-resistant derivative of KT2440 | 75 |
| KT2440R-McpU | KT2440R transposon mutant pp1228::mini-Tn5-Km; Rifr, Kmr | 76 |
| Pseudomonas aeruginosa PAO1 | Wild-type strain | 77 |
| PAO1 ΔpctA | PAO1 derivative, pctA gene deletion mutant | This study |
| PCTB1 | PAO1 derivative, pctB::Km; Km- | 23 |
| PCTC1 | PAO1 derivative, pctC::Km; Km- | 23 |
| PAO1 ΔtlpQ | PAO1 derivative, pa2654 gene deletion mutant | This study |
| PCT2 | PAO1 derivative; ΔpctB-pctA-pa4308-pctC::km; Kmrb | 23 |
| PCTAQ | PAO1 derivative; ΔpctA ΔtlpQ | This study |
| PCT2Q | PAO1 derivative; ΔpctB-pctA-pa4308-pctC::Km, ΔtlpQ; Kmr | This study |
| PCT2QP | PAO1 derivative; ΔpctB-pctA-pa4308-pctC::Km, ΔtlpQ ΔtlpP; Kmr | J. Kato lab |
| PCT2QART | PAO1 derivative; ΔpctB-pctA-pa4308-pctC, ΔtlpQ ΔtlpA (pa1646), ΔtlpR(pa2652) ΔtlpT(pa1930); Kmr | J. Kato lab |
| P. aeruginosa PA14 | Wild-type strain; human clinical isolate that elicits disease in plants, nematodes, insects, and mice | 78 |
| PA14-ACQ | PA14 derivative; ΔpctA ΔpctC ΔtlpQ Δpa4308 | This study |
| P. aeruginosa isolate 227 | Isolated clinical strain from patients with urinary tract infections | 41 |
| P. aeruginosa isolate 233 | Isolated clinical strain from patients with urinary tract infections | 41 |
| P. aeruginosa isolate 287 | Isolated clinical strain from patients with urinary tract infections | 41 |
| P. aeruginosa isolate 401 | Isolated clinical strain from patients with urinary tract infections | 41 |
| P. aeruginosa isolate 428 | Isolated clinical strain from patients with urinary tract infections | 41 |
| Plasmids | ||
| pUCP18 | Escherichia-Pseudomonas shuttle vector; Apr | 79 |
| pPctA | pUCP18 with a PCR fragment containing pctA; Apr | 80 |
| pPctB | pUCP18 with a PCR fragment containing pctB; Apr | 80 |
| pPctC | pUCP18 with a PCR fragment containing pctC; Apr | This study |
| pTlpQ | pUCP18 with a PCR fragment containing tlpQ; Apr | 27 |
| pK18mobsacB | Plasmid for allelic exchange; pK18 oriVE.c.lacZα mob sacB; Kmr | 81 |
| pK18mobsacB-pctA | pK18mobsacB containing a deletion of the pctA gene; Kmr | This study |
| pK18mobsacB-tlpQ | pK18mobsacB containing a deletion of the tlpQ gene; Kmr | This study |
| pK18mobsacB-pctABC | pK18mobsacB containing a deletion of pctB, pctA, pa4308, pctC; Kmr | This study |
| pK18mobsacB-pctC | pK18mobsacB containing a deletion of pctC and pa4308; Kmr | This study |
| pET28b(+) | Protein expression plasmid; Kmr | Novagen |
| pET28b-McpU | pET28b derivative used to produce His-tagged McpU-LBD; Kmr | 20 |
| pET28b-TlpQ | pET28b derivative used to produce His-tagged TlpQ-LBD; Kmr | This study |
Ap, ampicillin; Km, kanamycin; Rif, rifampin.
The pa4308 gene (orf-1), which forms part of the pctABC operon, encodes a hypothetical protein that is not involved in chemotaxis (23).
Oligonucleotides used in this study. Download Table S2, DOCX file, 0.02 MB (16KB, docx) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Construction of bacterial mutant strains and plasmids.
The pctA and tlpQ genes were deleted in different mutant strains by unmarked gene deletion. Plasmids pK18mobsacB-pctA and pK18mobsacB-tlpQ were generated by amplifying 0.6- to 1.2-kb regions up- and downstream of the target gene. PCR products were digested with the restriction enzymes listed in Table S2 and cloned into pK18mobsacB. The resulting plasmids were introduced into E. coli S17-1 λpir by electroporation. Plasmids were transferred to PAO1 by conjugation, and cells were selected in Simmons citrate (BBL; Becton, Dickinson) agar plates supplemented with kanamycin. For plasmid excision, LB medium was inoculated with a kanamycin-resistant colony and grown for 12 h, and then spread on LB plates containing 20% (wt/vol) sucrose. To construct the triple deletion mutant in pctA, pctC, and tlpQ in P. aeruginosa PA14, pK18mobsacB-pctA and pK18mobsacB-tlpQ were consecutively conjugated into PA14 to generate the double mutant. Subsequently, the plasmid pK18mobsacB-pctC, generated by amplifying regions up- and downstream of pctC, was conjugated into PA14 ΔpctA ΔtlpQ. Kanamycin-resistant colonies were grown on LB agar plates supplemented with 20% (wt/vol) sucrose for selection. For complementation purposes, the pctC gene was amplified by PCR and cloned into plasmid pUCP18 using the restriction enzymes listed in Table S2. The ligation mixture was electroporated into E. coli JM109, and transformants were selected on carbenicillin-containing LB plates. The resulting plasmid pPctC was transferred to PCTC1 and PCT2Q by electroporation.
Construction of the TlpQ-LBD expression plasmid.
The DNA fragment encoding the LBD of TlpQ was amplified, digested with NdeI and BamHI, and cloned into pET28b(+) linearized with the same enzymes.
Overexpression and purification of proteins.
PctA-LBD and PctC-LBD were overexpressed and purified as described in reference 24. McpU-LBD and TlpQ-LBD were generated as reported in reference 20.
Thermal shift assay.
Thermal shift experiments were conducted as reported in reference in 57. McpU-LBD in polybuffer {5 mM Tris, 5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 5 mM MES [morpholineethanesulfonic acid], 10% glycerol [vol/vol], 150 mM NaCl, pH 7.0} was used at a final concentration of 10 µM. Biolog (Hayward, CA, USA) compound arrays PM3B (nitrogen sources), PM4A (phosphorous and sulfur sources), and PM5 (nutrient supplements) were used for screening. The compositions of these arrays are provided at http://208.106.130.253/pdf/pm_lit/PM1-PM10.pdf.
Isothermal titration calorimetry.
Titrations were carried out in a VP microcalorimeter (MicroCal, Northampton, MA, USA) at 25°C. Proteins dialyzed into polybuffer were titrated with ligands in dialysis buffer. Typically, 15 to 30 µM protein was titrated with 0.25 to 1 mM ligand solutions. For ethylene binding studies, TlpQ-LBD was titrated with 12-µl aliquots of a saturated ethylene solution in polybuffer, prepared as reported in reference 31. The mean enthalpies from the injection of ligands into the buffer were subtracted from titration data prior to data fitting using the “one binding site model” of ORIGIN.
Chemotaxis assays. (i) Soft agar plate assays.
Strains were grown overnight in M9 minimal medium containing 0.1% glucose (wt/vol), diluted to an optical density at 600 nm (OD660) of 1 with fresh medium, and washed twice with M9 medium. The pellet was resuspended in 1 ml M9 medium. Ten-microliter aliquots of 5 mM chemoattractant solutions were placed onto plates containing M9 medium, 2.5 mM glucose, and 0.25% (wt/vol) agar. Two-microliter aliquots of bacterial suspensions were placed horizontally to each of the chemoattractant spots. The plates were incubated at 30°C for 16 to 20 h.
(ii) Quantitative capillary chemotaxis assays.
Two protocols were used that differed in the ways the cells were counted. The first protocol was used to generate the data shown Fig. S6, whereas the second protocol was used for the remaining chemotaxis experiments. In the first protocol, overnight cultures of strains were diluted to an OD660 of 0.05 in MS medium (58) supplemented with 6 mg·liter−1 Fe citrate, trace elements, and 15 mM glucose and grown at 37°C. At an OD660 of 0.4, the cultures were centrifuged at 1,700 × g for 5 min and the pellet was washed twice with chemotaxis buffer (50 mM potassium phosphate, 20 mM EDTA, 0.05% [vol/vol] glycerol, pH 7.0). The cells were resuspended in this buffer and adjusted to an OD660 of 0.1, and 230-µl aliquots were placed into 96-well plates. Capillary tubes (P1424, Microcaps; Drummond Scientific) were heat sealed at one end and filled with chemotaxis buffer or chemotaxis chemoattractant solution. The capillaries were then immersed in bacterial suspensions at their open ends. After 30 min at room temperature, the capillaries were removed and rinsed with sterile water, and the content was expelled into 1 ml of M9 medium. Serial dilutions were plated on LB medium, and the CFU were determined. In all cases, the data were corrected to the number of cells that swam into the buffer-containing capillaries. In the second protocol, we used computer-assisted image analysis as reported previously (59). Briefly, capillaries were filled with chemoattractant solutions in 10 mM HEPES buffer (pH 7.0) containing 1% (wt/vol) agarose and heat sealed on one side. The cells were grown in 2× yeast extract-tryptone (YT), medium and 10-μl aliquots of the cell suspension were placed onto a microscope slide within the U-shaped spacer, which was then covered by a coverslip. The chemoattractant-filled capillaries were introduced into the chemotaxis chamber, and cell movement was videotaped, with images taken at the beginning and at different time intervals. If not otherwise stated, the contact time between the cell and the chemoattractant was 2 min. The Bioinformatics Assistant Icy Sport detector software (60) was used to determine the number of cells per image. The magnitude of chemotaxis was expressed as the number of cells after a given time over the number of cells at the beginning of the experiment. The data shown are the means and standard deviations from three experiments conducted in triplicates.
Growth experiments.
PAO1 was grown overnight in MS minimal medium (29) containing 20 mM d-glucose. Cultures were diluted to an OD600 of 0.02 in MS medium supplemented with 5 mM carbon or nitrogen source. The assays were performed in 100-well polystyrene plates and incubated at 30°C (KT2440) or 37°C (PAO1) in a Bioscreen microbiological growth analyzer. The data represent the means and standard deviations from three biological replicates conducted in triplicates.
Assessment of motility.
To assess bacterial motility, PAO1 and KT2440 were used to inoculate LB and 2× YT medium to an OD600 of 0.01. Growth was carried out at 37°C (PAO1) or 30°C (KT2440), and the bacteria were inspected microscopically. According to their motility, they were given different scores: score 1, 25% of bacteria are motile; 2, 50%; 3, 75%; and 4, 100%.
Crystallization and structure resolution.
Crystallization trials were carried out with TlpQ-LBD in the absence and presence of histamine. TlpQ-LBD in polybuffer was incubated with a 2-fold molar excess of histamine on ice for 30 min. Unbound ligand was removed by buffer exchange using 10-kDa-cutoff filters (Amicon) and polybuffer. The apo- (6 mg/ml) and ligand-bound (26 mg/ml) proteins were loaded into 0.3-mm-diameter capillaries for counter diffusion crystallization using screen kits from Triana S & T (Granada, Spain). Only the LBD-TlpQ–histamine complex produced crystals in 1.5 M ammonium phosphate and 0.1 M sodium citrate pH 5.6. The same protocol was used to crystallize the Se-Met TlpQ-LBD. The capillaries were emptied into mother solution containing 10% to 25% (vol/vol) glycerol as the cryoprotectant. Crystals were diffracted at the European Synchrotron Radiation Facility and the Spanish Synchrotron ALBA. The data were indexed and integrated with XDS (61) and scaled with SCALA (62). All attempts to obtain a molecular replacement solution failed. Phases were obtained from the Se-methionine derivative by combining SAD data, at the selenium peak, with an initial model generated by Phyre2 (63), as input files for Auto-Rickshaw (64). All 26 expected heavy atom positions were identified by SHELXD (65) using the data to 3.5 Å. The model generated was refined with phenix.refine (66). Further refinement was performed against the best data set (2.45 Å) with phenix.refine (66) using Coot (67). Model quality was checked using MolProbity (68). The refinement statistics and quality indicators of the final model are summarized in Table S3. The structure was deposited at the protein data bank with identifier (ID) 6fu4.
Data collection and refinement statistics of the three-dimensional structure of TlpQ-LBD (values in parentheses are for highest resolution shell). Download Table S3, DOCX file, 0.01 MB (15.3KB, docx) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We acknowledge the European Synchrotron Radiation Facility (beamlines ID23-1 & 2, ID30B, and ID30A-3) and the Spanish Synchrotron ALBA (beamline XALOC) and thank the staff for their invaluable support.
This work was supported by FEDER funds and Fondo Social Europeo through grants held by T. Krell from the Spanish Ministry for Economy and Competitiveness (grants BIO2013-42297 and BIO2016-76779-P) and by grant BIO2016-74875-P to J. A. Gavira.
Footnotes
Citation Corral-Lugo A, Matilla MA, Martín-Mora D, Silva Jiménez H, Mesa Torres N, Kato J, Hida A, Oku S, Conejero-Muriel M, Gavira JA, Krell T. 2018. High-affinity chemotaxis to histamine mediated by the TlpQ chemoreceptor of the human pathogen Pseudomonas aeruginosa. mBio 9:e01894-18. https://doi.org/10.1128/mBio.01894-18.
Contributor Information
Gerald L. Hazelbauer, University of Missouri-Columbia.
Tarek Msadek, Institut Pasteur.
REFERENCES
- 1.Galperin MY. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol 5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hazelbauer GL, Falke JJ, Parkinson JS. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci 33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu Rev Genet 41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 4.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal 3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitchurch CB, Leech AJ, Young MD, Kennedy D, Sargent JL, Bertrand JJ, Semmler AB, Mellick AS, Martin PR, Alm RA, Hobbs M, Beatson SA, Huang B, Nguyen L, Commolli JC, Engel JN, Darzins A, Mattick JS. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol 52:873–893. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 7.Parkinson JS, Hazelbauer GL, Falke JJ. 2015. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matilla MA, Krell T. 2017. Chemoreceptor-based signal sensing. Curr Opin Biotechnol 45:8–14. doi: 10.1016/j.copbio.2016.11.021. [DOI] [PubMed] [Google Scholar]
- 9.Bardy SL, Briegel A, Rainville S, Krell T. 2017. Recent advances and future prospects in bacterial and archaeal locomotion and signal transduction. J Bacteriol 199:e00203-17. doi: 10.1128/JB.00203-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexandre G, Greer-Phillips S, Zhulin IB. 2004. Ecological role of energy taxis in microorganisms. FEMS Microbiol Rev 28:113–126. doi: 10.1016/j.femsre.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Ortega A, Zhulin IB, Krell T. 2017. Sensory repertoire of bacterial chemoreceptors. Microbiol Mol Biol Rev 81:e00033-17. doi: 10.1128/MMBR.00033-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB. 2016. Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol 12:e1004862. doi: 10.1371/journal.pcbi.1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sampedro I, Parales RE, Krell T, Hill JE. 2015. Pseudomonas chemotaxis. FEMS Microbiol Rev 39:17–46. doi: 10.1111/1574-6976.12081. [DOI] [PubMed] [Google Scholar]
- 14.Kato J, Kim HE, Takiguchi N, Kuroda A, Ohtake H. 2008. Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J Biosci Bioeng 106:1–7. doi: 10.1263/jbb.106.1. [DOI] [PubMed] [Google Scholar]
- 15.Belda E, van Heck RGA, José Lopez-Sanchez M, Cruveiller S, Barbe V, Fraser C, Klenk H-P, Petersen J, Morgat A, Nikel PI, Vallenet D, Rouy Z, Sekowska A, Martins dos Santos VAP, de Lorenzo V, Danchin A, Médigue C. 2016. The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolic chassis. Environ Microbiol 18:3403–3424. doi: 10.1111/1462-2920.13230. [DOI] [PubMed] [Google Scholar]
- 16.Juhas M. 2015. Pseudomonas aeruginosa essentials: an update on investigation of essential genes. Microbiology 161:2053–2060. doi: 10.1099/mic.0.000161. [DOI] [PubMed] [Google Scholar]
- 17.Ortega DR, Fleetwood AD, Krell T, Harwood CS, Jensen GJ, Zhulin IB. 2017. Assigning chemoreceptors to chemosensory pathways in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 114:12809–12814. doi: 10.1073/pnas.1708842114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia V, Reyes-Darias JA, Martin-Mora D, Morel B, Matilla MA, Krell T. 2015. Identification of a chemoreceptor for C2 and C3 carboxylic acids. Appl Environ Microbiol 81:5449–5457. doi: 10.1128/AEM.01529-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez M, Morel B, Corral-Lugo A, Krell T. 2016. Identification of a chemoreceptor that specifically mediates chemotaxis toward metabolizable purine derivatives. Mol Microbiol 99:34–42. doi: 10.1111/mmi.13215. [DOI] [PubMed] [Google Scholar]
- 20.Corral-Lugo A, De la Torre J, Matilla MA, Fernández M, Morel B, Espinosa-Urgel M, Krell T. 2016. Assessment of the contribution of chemoreceptor-based signaling to biofilm formation. Environ Microbiol 18:3355–3372. doi: 10.1111/1462-2920.13170. [DOI] [PubMed] [Google Scholar]
- 21.Reyes-Darias JA, García V, Rico-Jiménez M, Corral-Lugo A, Lesouhaitier O, Juárez-Hernández D, Yang Y, Bi S, Feuilloley M, Muñoz-Rojas J, Sourjik V, Krell T. 2015. Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol Microbiol 97:488–501. doi: 10.1111/mmi.13045. [DOI] [PubMed] [Google Scholar]
- 22.Gavira JA, Ortega A, Martín-Mora D, Conejero-Muriel MT, Corral-Lugo A, Morel B, Matilla MA, Krell T. 2018. Structural basis for polyamine binding at the dCACHE domain of the McpU chemoreceptor from Pseudomonas putida. J Mol Biol 430:1950–1963. doi: 10.1016/j.jmb.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Taguchi K, Fukutomi H, Kuroda A, Kato J, Ohtake H. 1997. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 143:3223–3229. doi: 10.1099/00221287-143-10-3223. [DOI] [PubMed] [Google Scholar]
- 24.Rico-Jimenez M, Munoz-Martinez F, Garcia-Fontana C, Fernandez M, Morel B, Ortega A, Ramos JL, Krell T. 2013. Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA). Mol Microbiol 88:1230–1243. doi: 10.1111/mmi.12255. [DOI] [PubMed] [Google Scholar]
- 25.Rico-Jiménez M, Reyes-Darias JA, Ortega Á, Díez Peña AI, Morel B, Krell T. 2016. Two different mechanisms mediate chemotaxis to inorganic phosphate in Pseudomonas aeruginosa. Sci Rep 6:28967. doi: 10.1038/srep28967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H, Kato J, Kuroda A, Ikeda T, Takiguchi N, Ohtake H. 2000. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J Bacteriol 182:3400–3404. doi: 10.1128/JB.182.12.3400-3404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin-Mora D, Ortega A, Perez-Maldonado FJ, Krell T, Matilla MA. 2018. The activity of the C4-dicarboxylic acid chemoreceptor of Pseudomonas aeruginosa is controlled by chemoattractants and antagonists. Sci Rep 8:2102. doi: 10.1038/s41598-018-20283-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alvarez-Ortega C, Harwood CS. 2007. Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl Environ Microbiol 73:7793–7795. doi: 10.1128/AEM.01898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Mora D, Ortega A, Reyes-Darias JA, García V, López-Farfán D, Matilla MA, Krell T. 2016. Identification of a chemoreceptor in Pseudomonas aeruginosa that specifically mediates chemotaxis towards alpha-ketoglutarate. Front Microbiol 7:1937. doi: 10.3389/fmicb.2016.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim HE, Shitashiro M, Kuroda A, Takiguchi N, Ohtake H, Kato J. 2006. Identification and characterization of the chemotactic transducer in Pseudomonas aeruginosa PAO1 for positive chemotaxis to trichloroethylene. J Bacteriol 188:6700–6702. doi: 10.1128/JB.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H-E, Shitashiro M, Kuroda A, Takiguchi N, Kato J. 2007. Ethylene chemotaxis in Pseudomonas aeruginosa and other Pseudomonas species. Microb Environ 22:186–189. doi: 10.1264/jsme2.22.186. [DOI] [Google Scholar]
- 32.Barcik W, Wawrzyniak M, Akdis CA, O’Mahony L. 2017. Immune regulation by histamine and histamine-secreting bacteria. Curr Opin Immunol 48:108–113. doi: 10.1016/j.coi.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 33.De Benedetto A, Yoshida T, Fridy S, Park JE, Kuo IH, Beck LA. 2015. Histamine and skin barrier: are histamine antagonists useful for the prevention or treatment of atopic dermatitis? J Clin Med 4:741–755. doi: 10.3390/jcm4040741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Mahony L, Akdis M, Akdis CA. 2011. Regulation of the immune response and inflammation by histamine and histamine receptors. J Allergy Clin Immunol 128:1153–1162. doi: 10.1016/j.jaci.2011.06.051. [DOI] [PubMed] [Google Scholar]
- 35.Xu X, Zhang D, Zhang H, Wolters PJ, Killeen NP, Sullivan BM, Locksley RM, Lowell CA, Caughey GH. 2006. Neutrophil histamine contributes to inflammation in mycoplasma pneumonia. J Exp Med 203:2907–2917. doi: 10.1084/jem.20061232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X, Zhang H, Song Y, Lynch SV, Lowell CA, Wiener-Kronish JP, Caughey GH. 2012. Strain-dependent induction of neutrophil histamine production and cell death by Pseudomonas aeruginosa. J Leukoc Biol 91:275–284. doi: 10.1189/jlb.0711356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metz M, Doyle E, Bindslev-Jensen C, Watanabe T, Zuberbier T, Maurer M. 2011. Effects of antihistamines on innate immune responses to severe bacterial infection in mice. Int Arch Allergy Immunol 155:355–360. doi: 10.1159/000321614. [DOI] [PubMed] [Google Scholar]
- 38.Beghdadi W, Porcherie A, Schneider BS, Dubayle D, Peronet R, Huerre M, Watanabe T, Ohtsu H, Louis J, Mecheri S. 2008. Inhibition of histamine-mediated signaling confers significant protection against severe malaria in mouse models of disease. J Exp Med 205:395–408. doi: 10.1084/jem.20071548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kyriakidis DA, Theodorou MC, Tiligada E. 2012. Histamine in two component system-mediated bacterial signaling. Front Biosci (Landmark ed) 17:1108–1119. [DOI] [PubMed] [Google Scholar]
- 40.Krell T. 2015. Tackling the bottleneck in bacterial signal transduction research: high-throughput identification of signal molecules. Mol Microbiol 96:685–688. doi: 10.1111/mmi.12975. [DOI] [PubMed] [Google Scholar]
- 41.Oura H, Tashiro Y, Toyofuku M, Ueda K, Kiyokawa T, Ito S, Takahashi Y, Lee S, Nojiri H, Nakajima-Kambe T, Uchiyama H, Futamata H, Nomura N. 2015. Inhibition of Pseudomonas aeruginosa swarming motility by 1-naphthol and other bicyclic compounds bearing hydroxyl groups. Appl Environ Microbiol 81:2808–2818. doi: 10.1128/AEM.04220-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ni B, Huang Z, Fan Z, Jiang CY, Liu SJ. 2013. Comamonas testosteroni uses a chemoreceptor for tricarboxylic acid cycle intermediates to trigger chemotactic responses towards aromatic compounds. Mol Microbiol 90:813–823. doi: 10.1111/mmi.12400. [DOI] [PubMed] [Google Scholar]
- 43.Antunez-Lamas M, Cabrera E, Lopez-Solanilla E, Solano R, Gonzalez-Melendi P, Chico JM, Toth I, Birch P, Pritchard L, Liu H, Rodriguez-Palenzuela P. 2009. Bacterial chemoattraction towards jasmonate plays a role in the entry of Dickeya dadantii through wounded tissues. Mol Microbiol 74:662–671. doi: 10.1111/j.1365-2958.2009.06888.x. [DOI] [PubMed] [Google Scholar]
- 44.Pasupuleti S, Sule N, Cohn WB, MacKenzie DS, Jayaraman A, Manson MD. 2014. Chemotaxis of Escherichia coli to norepinephrine (NE) requires conversion of NE to 3,4-dihydroxymandelic acid. J Bacteriol 196:3992–4000. doi: 10.1128/JB.02065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laganenka L, Colin R, Sourjik V. 2016. Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat Commun 7:12984. doi: 10.1038/ncomms12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matilla MA, Krell T. 2018. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol Rev 42:fux052. doi: 10.1093/femsre/fux052. [DOI] [PubMed] [Google Scholar]
- 47.Azam MW, Khan AU. 20 July 2018. Updates on the pathogenicity status of Pseudomonas aeruginosa. Drug Discov Today doi: 10.1016/j.drudis.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 48.Erhardt M. 2016. Strategies to block bacterial pathogenesis by interference with motility and chemotaxis. Curr Top Microbiol Immunol 398:185–205. doi: 10.1007/82_2016_493. [DOI] [PubMed] [Google Scholar]
- 49.Bi S, Yu D, Si G, Luo C, Li T, Ouyang Q, Jakovljevic V, Sourjik V, Tu Y, Lai L. 2013. Discovery of novel chemoeffectors and rational design of Escherichia coli chemoreceptor specificity. Proc Natl Acad Sci U S A 110:16814–16819. doi: 10.1073/pnas.1306811110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu D, Ma X, Tu Y, Lai L. 2015. Both piston-like and rotational motions are present in bacterial chemoreceptor signaling. Sci Rep 5:8640. doi: 10.1038/srep08640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bains M, Fernandez L, Hancock RE. 2012. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl Environ Microbiol 78:6762–6768. doi: 10.1128/AEM.01015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaborin A, Romanowski K, Gerdes S, Holbrook C, Lepine F, Long J, Poroyko V, Diggle SP, Wilke A, Righetti K, Morozova I, Babrowski T, Liu DC, Zaborina O, Alverdy JC. 2009. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci U S A 106:6327–6332. doi: 10.1073/pnas.0813199106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neumann S, Hansen CH, Wingreen NS, Sourjik V. 2010. Differences in signalling by directly and indirectly binding ligands in bacterial chemotaxis. EMBO J 29:3484–3495. doi: 10.1038/emboj.2010.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panula P, Chazot PL, Cowart M, Gutzmer R, Leurs R, Liu WL, Stark H, Thurmond RL, Haas HL. 2015. International Union of Basic and Clinical Pharmacology. XCVIII. Histamine receptors. Pharmacol Rev 67:601–655. doi: 10.1124/pr.114.010249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hofstra CL, Desai PJ, Thurmond RL, Fung-Leung WP. 2003. Histamine H4 receptor mediates chemotaxis and calcium mobilization of mast cells. J Pharmacol Exp Ther 305:1212–1221. doi: 10.1124/jpet.102.046581. [DOI] [PubMed] [Google Scholar]
- 56.Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Han GW, Kobayashi T, Stevens RC, Iwata S. 2011. Structure of the human histamine H1 receptor complex with doxepin. Nature 475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernandez M, Ortega A, Rico-Jimenez M, Martin-Mora D, Daddaoua A, Matilla MA, Krell T. 2018. High-throughput screening to identify chemoreceptor ligands. Methods Mol Biol 1729:291–301. doi: 10.1007/978-1-4939-7577-8_23. [DOI] [PubMed] [Google Scholar]
- 58.Abril MA, Michan C, Timmis KN, Ramos JL. 1989. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol 171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nikata T, Sumida K, Kato J, Ohtake H. 1992. Rapid method for analyzing bacterial behavioral responses to chemical stimuli. Appl Environ Microbiol 58:2250–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Chaumont F, Dallongeville S, Chenouard N, Herve N, Pop S, Provoost T, Meas-Yedid V, Pankajakshan P, Lecomte T, Le Montagner Y, Lagache T, Dufour A, Olivo-Marin JC. 2012. Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods 9:690–696. doi: 10.1038/nmeth.2075. [DOI] [PubMed] [Google Scholar]
- 61.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 63.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 64.Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. 2005. Auto-rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr D Biol Crystallogr 61:449–457. doi: 10.1107/S0907444905001307. [DOI] [PubMed] [Google Scholar]
- 65.Schneider TR, Sheldrick GM. 2002. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr 58:1772–1779. doi: 10.1107/S0907444902011678. [DOI] [PubMed] [Google Scholar]
- 66.Afonine PV, Mustyakimov M, Grosse-Kunstleve RW, Moriarty NW, Langan P, Adams PD. 2010. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 66:1153–1163. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi SH, Couloux A, Lee SW, Yoon SH, Cattolico L, Hur CG, Park HS, Segurens B, Kim SC, Oh TK, Lenski RE, Studier FW, Daegelen P, Kim JF. 2009. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J Mol Biol 394:644–652. doi: 10.1016/j.jmb.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 70.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res 17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boyer HW, Roulland-Dussoix D. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol 41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 72.Zylstra GJ, Wackett LP, Gibson DT. 1989. Trichloroethylene degradation by Escherichia coli containing the cloned Pseudomonas putida F1 toluene dioxygenase genes. Appl Environ Microbiol 55:3162–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Phornphisutthimas S, Thamchaipenet A, Panijpan B. 2007. Conjugation in Escherichia coli: a laboratory exercise. Biochem Mol Biol Educ 35:440–445. doi: 10.1002/bmb.113. [DOI] [PubMed] [Google Scholar]
- 74.Hida A, Oku S, Kawasaki T, Nakashimada Y, Tajima T, Kato J. 2015. Identification of the mcpA and mcpM genes, encoding methyl-accepting proteins involved in amino acid and l-malate chemotaxis, and involvement of McpM-mediated chemotaxis in plant infection by Ralstonia pseudosolanacearum (formerly Ralstonia solanacearum phylotypes I and III). Appl Environ Microbiol 81:7420–7430. doi: 10.1128/AEM.01870-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Espinosa-Urgel M, Ramos JL. 2004. Cell density-dependent gene contributes to efficient seed colonization by Pseudomonas putida KT2440. Appl Environ Microbiol 70:5190–5198. doi: 10.1128/AEM.70.9.5190-5198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duque E, Molina-Henares AJ, de la Torre J, Molina-Henares MA, del Castillo T, Lam J, Ramos JL. 2007. Towards a genome-wide mutant library of Pseudomonas putida strain KT2440, p 227–251. In Ramos JL, Filloux A (ed), Pseudomonas: a model system in biology, volV Springer, Dorchester, the Netherlands. [Google Scholar]
- 77.Holloway BW, Krishnapillai V, Morgan AF. 1979. Chromosomal genetics of Pseudomonas. Microbiol Rev 43:73–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 79.Schweizer HP. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 80.Shitashiro M, Tanaka H, Hong CS, Kuroda A, Takiguchi N, Ohtake H, Kato J. 2005. Identification of chemosensory proteins for trichloroethylene in Pseudomonas aeruginosa. J Biosci Bioeng 99:396–402. doi: 10.1263/jbb.99.396. [DOI] [PubMed] [Google Scholar]
- 81.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microcalorimetric binding studies of McpU-LBD. (A) Titrations of 17.5 µM McpU-LBD with 1 mM histamine and ethylenediamine and 30 µM McpU-LBD with 0.5 mM agmatine. (B) Chemical structure of ligands recognized by the McpU and TlpQ chemoreceptors. Download FIG S1, JPG file, 0.6 MB (615.4KB, jpg) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identification of a McpU homologue. (A) Sequence clustering of the ligand binding domains of dCACHE-containing chemoreceptors from P. putida KT2440 (blue) and P. aeruginosa PAO1 (green). The figure was produced using the Phylogeny.fr server. (B) Sequence alignment of the TlpQ and McpU chemoreceptors. Sequence identity between the two receptors is 62%. The alignment was done in the slow mode using the CLUSTALW multiple alignment tool of the NPSA suite (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_server.html). The GONNET protein weight matrix was used in the slow pairwise alignment mode using a gap opening penalty of 10 and a gap extension penalty of 0.1. Red, identical; green, highly similar; blue, weakly similar. The transmembrane regions flanking the ligand binding domain were predicted using the DAS algorithm (https://tmdas.bioinfo.se/DAS/index.html) and are highlighted in yellow. Cyan highlights amino acids that are involved in side-chain mediated interactions with bound histamine. Download FIG S2, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Assessment of motility during bacterial growth. Overnight cultures of P. aeruginosa PAO1 (continuous line) and P. putida KT2440 (dotted line) were used to inoculate LB (upper panel) and 2× YT medium (lower panel) to an OD600 of 0.01. Growth was carried out at 37°C (P. aeruginosa PAO1) or 30°C (P. putida KT2440), and bacteria were inspected microscopically. Motility scores were calculated as follows: score 1, 25% of bacteria are motile; 2, 50%; 3, 75%; and 4, 100%. Download FIG S3, TIF file, 0.1 MB (129.2KB, tif) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth experiments with McpU/TlpQ ligands as sole carbon or nitrogen source. (A) Capacity of ligands to sustain growth of P. aeruginosa PAO1 and P. putida KT2440. Overnight cultures were used to inoculate MS minimal medium supplemented with the McpU/TlpQ ligands as either sole carbon or nitrogen source. As controls, succinate and ammonium nitrate were used. (B) Growth of Ralstonia pseudosolanacearum Ps29 in medium with histamine as the sole carbon and nitrogen source. The initial OD660 was 0.005. The composition of the RSM medium was 10 mM K2HPO3, 5.5 mM KH2PO4, 0.5 mM sodium citrate, 9.5 mM (NH4)2SO4, 1 mM MgSO4, and 28 mM glucose. In all cases, the ligands were added at a concentration of 5 mM, and shown are means and standard deviations from three biological replicates conducted in triplicates. Download FIG S4, JPG file, 0.8 MB (776.3KB, jpg) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structural alignment of the Cα chain of TlpQ-LBD (in red) with a homologous structure from a histidine kinase of Shewanella oneidensis (in green). This structure is deposited in the protein data bank with ID 3lic. The arrows indicate inserts and loops in the TlpQ-LBD structure that account for its elevated size. Download FIG S5, JPG file, 0.7 MB (705.7KB, jpg) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structural alignment of TlpQ-LBD with structures deposited in the protein data bank. Shown are the structures with a Z-score above 15. Download Table S1, DOCX file, 0.03 MB (32.3KB, docx) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quantitative capillary chemotaxis assays of P. aeruginosa PA14 and a mutant defective in the pctA, pctC, and tlpQ genes towards histamine. Data have been corrected with the number of bacteria (6,900 ± 142) that swam into buffer-containing capillaries. Shown are means and standard deviations from three individual experiments conducted in duplicates. Download FIG S6, TIF file, 1.5 MB (1.5MB, tif) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Assessment of potential low-affinity binding of histamine to PctA-LBD by microcalorimetric competition experiments. Shown are microcalorimetric titrations of 39 µM PctA-LBD with 1 mM l-alanine either in the absence or presence of 20 mM histamine. The resulting integrated peak areas are almost identical indicating that histamine does not compete with l-alanine for binding at PctA-LBD. Download FIG S7, JPG file, 0.2 MB (220.4KB, jpg) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides used in this study. Download Table S2, DOCX file, 0.02 MB (16KB, docx) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data collection and refinement statistics of the three-dimensional structure of TlpQ-LBD (values in parentheses are for highest resolution shell). Download Table S3, DOCX file, 0.01 MB (15.3KB, docx) .
Copyright © 2018 Corral-Lugo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.