Abstract
Introduction
Models characterizing intermediate disease stages of Alzheimer's disease (AD) are needed to inform clinical care and prognosis. Current models, however, use only a small subset of available biomarkers, capturing only coarse changes along the complete spectrum of disease progression. We propose the use of machine learning techniques and clinical, biochemical, and neuroimaging biomarkers to characterize progression to AD.
Methods
We used a large multimodal longitudinal data set of biomarkers and demographic and genotype information from 1624 participants from the Alzheimer's Disease Neuroimaging Initiative. Using hidden Markov models, we characterized intermediate disease stages. We validated inferred disease trajectories by comparing time to first clinical AD diagnosis. We trained an L2-regularized logistic regression model to predict disease trajectory and evaluated its discriminative performance on a test set.
Results
We identified 12 distinct disease states. Progression to AD occurred most often through one of two possible paths through these states. Paths differed in terms of rate of disease progression (by 5.44 years on average), amyloid and total-tau (t-tau) burden (by 10% and 69%, respectively), and hippocampal neurodegeneration (P < .001). On the test set, the predictive model achieved an area under the receiver operating characteristic curve of 0.85.
Discussion
Progression to AD, in terms of biomarker trajectories, can be predicted based on participant-specific factors. Such disease staging tools could help in targeting high-risk patients for therapeutic intervention trials. As longitudinal data with richer features are collected, such models will help increase our understanding of the factors that drive the different trajectories of AD.
Keywords: Machine learning, Trajectory, Longitudinal, Staging, Biomarkers
1. Introduction
Patients with Alzheimer's disease (AD) are believed to progress through intermediate disease stages, characterized by their pathophysiological and cognitive characteristics [1], [2]. Failures of multiple drug therapeutics could, in part, be attributed to heterogeneity of disease presentation, staging, and response to treatment [3], [4]. Models of disease progression could be used to assess and predict patients' intermediate disease stages in clinical trials [5] and predict potentially heterogeneous treatment response in the absence of clinical symptoms. In this study, we aimed to develop such models and characterize the complexity of AD via analyses of disease trajectories by prospectively leveraging clinical, biochemical, and neuroimaging parameters.
Several models of the temporal evolution of biomarkers [6], [7], [8], [9] in AD have been proposed. However, these models focus on only a small subset of the available biomarkers and rely on a dichotomous definition of abnormality. AD, however, is known to be heterogeneous in terms of clinical presentation [10] and pathophysiology [11], making such binary cutoffs difficult to determine [12].
Therefore, we propose a comprehensive machine learning–based approach to modeling disease progression that leverages a large multimodal subset of the available data from the underlying pathophysiological processes of neurodegeneration (magnetic resonance imaging [MRI] and fludeoxyglucose F 18 positron emission tomography [FDG-PET] scans), amyloidosis (amyloid PET, cerebrospinal fluid [CSF] amyloid levels), tauopathy (CSF tau levels), and cognitive decline (neuropsychological exams) in addition to patient demographics and risk factors. We hypothesize that leveraging multiple data modalities and machine learning techniques will lead to a more accurate characterization of disease stages. The proposed approach models disease stages as distributions over biomarkers that are learned using the data, thus obviating the need for dichotomous cutoffs to define abnormality. Furthermore, our model leverages data across participants misaligned in time and with varying follow-up durations.
We report on the application of the proposed approach to a large longitudinal cohort of participants. Such comprehensive data-driven models of disease progression can help shed light on the underlying pathophysiology of AD and are increasingly relevant as a result of the widespread availability of these biomarkers and the rapidly advancing diagnostic criteria for the disease [13], [14].
2. Methods
2.1. Study design and participants
Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). ADNI was launched in 2004 as a public-private partnership, led by a principal investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early AD.
The ADNI cohort consists of participants diagnosed as cognitively normal (with no memory complaints or significant impairment in cognitive function), MCI (some memory complaints with largely intact cognitive function and no diagnosis of dementia [15]), and AD patients [16] (based on a clinical diagnosis). Cognitively normal participants had Mini–Mental State Examination (MMSE) scores ≥24 and a Clinical Dementia Rating (CDR) of zero. Patients with MCI had MMSE scores ≥24, a rating of 0.5 or greater on the memory box score on the CDR test and objective memory loss based on delayed recall on the Wechsler Memory Scale Logical Memory II. Participants diagnosed with AD had mild AD according to the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association (now known as the Alzheimer's Association) (NINCDS-ADRDA) criteria [17] and were recruited with MMSE scores from 20–26 and a CDR of 0.5 or 1. All participants were required to have at least 6 years of education and be fluent in English or Spanish. The data were obtained from ADNI [15] on November 1, 2016. We excluded participants without at least one follow-up visit.
2.2. Procedures
We used a large multimodal subset of the clinical, imaging, laboratory, genetic, and demographic data available for each ADNI participant in our analysis. Procedures for patients' MRI [18], PET [19], CSF [20], and neuropsychological tests [21] have been previously described. As the observed variables, we considered brain volumes extracted from MRI scans (both 1.5 T and 3.0 T), FDG-PET and amyloid standard uptake value ratio (SUVR), CSF protein levels, neuropsychological test scores, participant age, number of years of education, and the number of copies of the APOE e4 allele the participant inherited. Brain volumes from the hippocampal, entorhinal, fusiform, midtemporal, and ventricular regions were used and were normalized using the intracranial volume. Levels of amyloid β (Aβ)42, t-tau, and phosphorylated-tau (p-tau) were extracted from CSF (all CSF data were extracted from analyses performed in a single batch). The FDG-PET SUVR was averaged over the angular, temporal, and bilateral cingulate regions. The amyloid intake was averaged over the frontal, cingulate, parietal, and temporal regions and was normalized by the whole cerebellum intake to obtain the SUVR. The following clinical scores were used: Alzheimer's Disease Assessment Scale Cognitive Test, trails B, Family Activities Questionnaire, Rey Auditory Verbal Learning Test, MMSE, and the CDR-sum of boxes.
2.3. Statistical analysis
At enrollment, cognitive impairment varies among ADNI participants. Thus, we used hidden Markov models (HMMs) to align participant trajectories and estimate a model of disease progression. Given the model and a participant's trajectory of observations, one can infer the most likely disease stage at each visit.
Given the paucity of the data, we chose to discretize the continuous-valued variables (e.g., clinical scores and biological markers such as amyloid and CSF proteins) before inference. More specifically, we discretized all observed variables according to deciles and characterized each disease stage using independent categorical distributions over each biomarker. This level of abstraction can help in identifying relationships between hidden states and observations. Given the regular visit schedule, we considered discrete time steps, as opposed to continuous time steps. We assumed 6 months (with a tolerance of 2 months, consistent with the ADNI schedule) between successive visits. When a longer period elapsed between visits, we modeled intermediate visits as missing.
HMMs, which have been commonly used in speech processing [22], model data as a series of observations that depend on a series of hidden states forming a Markov chain [23]. Here, the hidden states correspond to disease stages, and the observations correspond to measurements at each visit (Supplementary Appendix A1). HMMs are parameterized by transition probabilities (i.e., the probability of transitioning from one disease stage to another) and emission probabilities (i.e., the probability of observations given the current disease stage). To estimate the HMM parameters, we used the Baum-Welch algorithm. We handled missing data, both in terms of features at particular visits or entirely missed visits, by marginalizing over these observations while training the HMM model. To select the number of hidden states (i.e., disease stages), K, we minimized the Bayesian Information Criterion. We generated confidence intervals around our model parameters using a nonparametric bootstrap approach (Supplementary Appendix A2) and show the interquartile range (IQR) with error bars. While we included a “start” stage (i.e., we modeled the probability of participant entering the study in a specific stage), since participants can drop out of the study for different reasons, we did not explicitly model an “end” stage. Additional details on initialization and model selection are included in Supplementary Appendix A3.
Given a participant's observed longitudinal biomarker data, we used the Viterbi algorithm [24] to infer the most likely trajectory through disease stages and combined stage assignments across bootstrap samples to assess positional variance.
We sorted inferred hidden states (i.e., disease stages) based on the median Alzheimer's Disease Assessment Scale Cognitive Test score of participants within each disease stage, breaking ties based on the median amyloid burden. In our analysis, we focused on transitions among the later disease stages that primarily consist of MCI or Alzheimer's participants because many of the participants in earlier stages have limited longitudinal data and these later stages exhibited less positional variance (see Supplementary Appendix A2).
We assessed the clinical predictive utility of our model for disease progression by training a Cox proportional hazards model of survival, stratified by the paths discovered in our model. We defined survival as the time between entering the study in an intermediate disease stage and the first clinical AD diagnosis or the last completed follow-up visit, whichever came earlier. Furthermore, we estimated the time taken to progress from an intermediate disease stage to the terminal stage of our model by simulating paths that begin in a particular disease stage (using our generative HMM) and measuring the time to reach the terminal stage (corresponding to advanced AD).
We estimated the ability to prospectively classify patients as following one path versus another by training an L2-regularized logistic regression model. As input to the model, we used data from participants assigned to intermediate stages, with the goal of predicting the participant's inferred trajectory. We measured discriminative performance of the trained model in terms of the area under the receiver operating characteristics curve (AUROC) on a held-out test set (20% of the data).
We used backward feature elimination to identify the most important biomarkers in predicting path of disease progression. This analysis was performed by repeatedly splitting the data into training and test sets (80/20 split), training an L2-regularized logistic regression model, and eliminating features that affected the average AUROC the least (see Supplementary Appendix A4 for more details).
We assessed statistical significance (with a significance level of 0.05) using the chi-squared test for categorical data and analysis of variance (or the Kruskal-Wallis test when appropriate) for continuous data. We used the permutation test to assess significance across multiple bootstrap samples. When assessing a significantly higher than zero value of a variable across bootstrap samples, we used the t-test (or the Wilcoxon rank-sum test when appropriate).
All preprocessing and statistical analyses were performed using MATLAB. The code is available here: https://gitlab.eecs.umich.edu/mld3/ad_profile_hmm.git. Using this code, others can apply the model to their study cohort to yield estimates of disease stage and predictions regarding which disease trajectory a participant is most likely to follow.
3. Results
Of the 1730 participants in ADNI, we excluded 106 who lacked a follow-up visit. Our final study population consisted of 1624 participants with a median follow-up period of 3 years totaling 8647 visits. The final set of observed variables consisted of 19 variables after preprocessing (i.e., discretization) the emission vector consisted of 174 dimensions (Supplementary Appendix A5). All the participants had clinical diagnoses as well as scores on neuropsychological tests. Ninety-nine percent of the participants had an MRI scan and 71% had a CSF measurement (study population characteristics and the amount of missing data for all variables are reported in Table 1).
Table 1.
Characteristics of study population at baseline enrollment
| Variable description | Study population (n = 1624) | % Patients with measurement |
|---|---|---|
| Follow-up in months, median (IQR) | 36 (24–49) | 100 |
| Age, median (IQR) | 74 (69–79) | |
| Male gender, percent | 55 | |
| Education in years, median (IQR) | 16 (14–18) | |
| MMSE, median (IQR) | 28 (26–29) | |
| ADAS-Cog, median (IQR) | 15 (9–23) | |
| APOE e4 carrier, percent | 47 | |
| Diagnosis, percent | ||
| Cognitively normal | 31 | |
| Mild cognitive impairment | 50 | |
| Alzheimer's disease | 19 | |
| Hippocampal volume: %TIV, median (IQR) | 0.44 (0.38–0.50) | 99 |
| FDG-PET SUVR, median (IQR) | 1.25 (1.14–1.34) | 75 |
| Amyloid PET SUVR, median (IQR) | 1.13 (1.02–1.38) | 52 |
| CSF t-tau in pg/mL, median (IQR) | 75.30 (52.10–113.00) | 71 |
Abbreviations: IQR, interquartile range; SUVR, standard uptake value ratio; MMSE, Mini–Mental State Examination; ADAS-Cog, Alzheimer's Disease Assessment Scale Cognitive Subscale; TIV, total intracranial volume; FDG, fludeoxyglucose F-18; PET, positron emission tomography; CSF, cerebrospinal fluid; MCI, mild cognitive impairment.
NOTE. All data pertain to patients at their baseline visit. Of the patients diagnosed as MCI at baseline, 37% were eventually diagnosed clinically with AD.
Our final model consisted of 12 disease stages. Cognitively normal and dementia participants were primarily assigned to earlier (1–4) and later (10–12) disease stages, respectively, while MCI participants fell into the intermediate disease stages (5–9) and showed some overlap with both the earlier and later disease stages (Fig. 1). Here, we focus our analysis on participants in stage 8 and onward because when participants in earlier disease stages progress to advanced disease stages, they typically do so over long periods. ADNI does not yet provide sufficient follow-up for participants in those earlier stages.
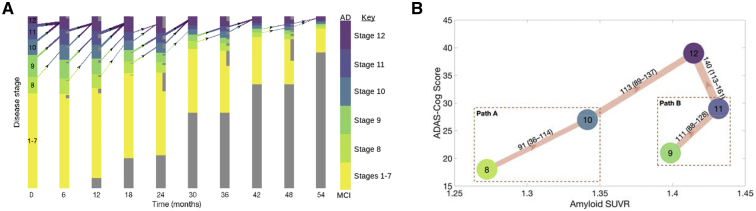
Fig. 1.
The Alzheimer's disease spectrum. Given a participant's clinical diagnosis (CN, MCI, or Alzheimer's disease), we show the empirical distribution of disease stage assignments inferred by our model. CN participants are more likely to be assigned to the earlier disease stages (79% of all CN participants in stages 1–4), and dementia participants are likely to be assigned to the later disease stages (86% of all Alzheimer's disease participants in stages 10–12). The model underscores the role of MCI as an intermediate stage, as MCI participants are assigned to stages that overlap with both normal and dementia participants (69% of all MCI participants in the intermediate stages 5–9, while 21% were in the predominantly normal stages 1–5). Error bars show the IQR. Abbreviations: MCI, mild cognitive impairment; IQR, interquartile range; CN, cognitively normal; AD, Alzheimer's disease.
We observed that participants often skip stages when progressing but that paths always consist of monotonically increasing stages. Fig. 2 illustrates transitions among disease stages in the study cohort. It reveals two parallel paths of disease progression. “Path A” progresses through stages 8→10→12, whereas “path B” progresses through stages 9→11→12. Participants had a 54% chance of taking path A versus path B. Transitions between the two paths were rare. We observed a median of 22 transitions (2.5%) between stages 9 and 10 and 8 transitions (0.8%) between stages 8 and 11.
Fig. 2.
(A) Trajectories of disease progression. Each bar is divided up according to the proportion of participants at that time step assigned to each disease stage. The darker the color in a bar, the higher the proportion of participants with AD in that disease stage (color correspondences to disease stages in our model are shown in the color bar, where the extremities in the color bar represent stages that constitute either entirely MCI or AD participants, respectively). Inset gray bars represent participants who will drop out at the next time step, while gray bars at the bottom represent the proportion of censored participants at that time step. The thickness of each arrow is proportional to the transition rate. We do not show transitions that occur <5% or self transitions. (B) The two most common distinct paths of disease progression. The width of each arrow is proportional to the median empirical count of the particular transition (with the IQR in brackets). The nodes are spatially arranged based on the median ADAS-Cog (y-axis) and amyloid burden (x-axis) of participants at each disease stage. Participants progress to AD through either path A (i.e., through stages 8→10→12) or path B (i.e., through stages 9→11→12). We do not visualize edges with fewer than 25 median transitions. Abbreviations: AD, Alzheimer's disease; MCI, mild cognitive impairment; ADAS-Cog, Alzheimer's Disease Assessment Scale Cognitive Test; IQR, interquartile range; SUVR, standard uptake value ratio.
Participants in path B had a significantly faster transition to a clinical diagnosis of AD than participants in path A (Fig. 3). The median Cox proportional hazards coefficient, relative to trajectories that began in stage 8, was 0.47 for stage 9 (IQR: [0.21–0.76], P < .001). Furthermore, participants reached the terminal disease stage, on average, 46% faster when they began in stage 9 compared to stage 8 (expected time of 11.76 vs. 17.20 years).
Fig. 3.
Dynamic characteristics of the disease paths. Participants who progress through path B (starting in stage 9) progress through disease faster than participants who progress through path A (starting in stage 8). An event is defined as a diagnosis of Alzheimer's disease. Coefficient values for a Cox proportional hazards model were found to be statistically significant for each pairwise comparison of the stages (P < .001).
In the test set, amyloid SUVR, Rey Auditory Verbal Learning Test score, age, t-tau levels, and APOE e4 allele status were most discriminative in predicting which path a participant would progress through. In the held-out test set, the classification model achieved an AUROC of 0.85 when predicting participants' progression through either path. In Table 2, we compare specific features across the first stages in either path (i.e., stage 8 vs. 9).
Table 2.
Characteristics of patients in the first stage of path A (stage 8) versus path B (stage 9)
| Measurement | Path A Median (IQR) |
Path B Median (IQR) |
P-value |
|---|---|---|---|
| Hippocampal volume (%TIV) | 0.3552 (0.3500–0.3660) | 0.4192 (0.4084–0.4275) | .117 |
| FDG-PET SUVR | 1.17 (1.16–1.19) | 1.19 (1.18–1.20) | .300 |
| CSF Aβ42 (pg/mL) | 159.0 (150.0–170.0) | 129.0 (128.0–132.0) | .035 |
| CSF t-tau (pg/mL) | 70.3 (64.8–76.5) | 121.0 (114.0–127.0) | .025 |
| CSF p-tau (pg/mL) | 29.6 (25.6–33.1) | 50.8 (47.5–53.0) | .026 |
| Amyloid PET SUVR | 1.26 (1.15–1.31) | 1.40 (1.38–1.43) | .030 |
| ADAS-Cog | 18.0 (17.7–18.7) | 21.0 (20.0–21.7) | .050 |
| TRAILS-B | 112.0 (105.0–119.0) | 104.0 (98.5–111.5) | .332 |
| FAQ | 3 (3–4) | 5 (4–6) | .224 |
| RAVLT | 33 (32–34) | 30 (29–31) | .106 |
| MMSE | 27 (27–27) | 26 (26–26) | .107 |
| CDR | 2.5 (2.0–2.5) | 2.5 (2.5–3.0) | .324 |
| Age (years) | 80.8 (79.9–81.6) | 72.4 (71.6–73.3) | .063 |
Abbreviations: SUVR, standard uptake value ratio; FDG, fludeoxyglucose F-18; PET, positron emission tomography; TIV, total intracranial volume; ADAS-Cog, Alzheimer's Disease Assessment Scale Cognitive Subscale; FAQ, Family Activities Questionnaire; RAVLT, Rey Auditory Verbal Learning Test; MMSE, Mini–Mental State Examination; CDR, clinical dementia rating; IQR, interquartile range.
NOTE. Based on these measurements, predictions about the trajectory of a patient's disease through either path A or B can be made.
A retrospective comparison of biomarker trajectories showed that, relative to participants in path A, participants in path B had on average greater hippocampal volume (by 15%, P < .001), higher levels of amyloid (by 10% P < .001), and higher levels of t-tau protein (by 69%, P < .001) (Fig. 4). Trends in amyloid burden differed significantly across paths (P < .001): amyloid burden was relatively stable in path B (median slope of −0.0037 SUVR per disease stage, P = .0486) compared to path A, which showed an increasing trend in amyloid (median slope of 0.0522 SUVR per disease stage). In addition, t-tau protein levels increased more sharply in path A (median slopes of 19.42 vs. 11.16 pg/mL per disease stage, P < .001). Finally, neurodegeneration was more rapid in path B; hippocampal volume and FDG-PET SUVR reduced at faster rates (34% faster, P < .001 and 20% faster, P < .001, respectively).
Fig. 4.
Empirical distribution of biomarker values at various disease stages for participants in path A (green) versus path B (red). Includes the data pertaining to all time points during which a participant was classified as belonging to a particular stage. The left splits of the violins (shown in green) represent disease stages in path A, whereas the right splits (shown in red) represent disease stages in path B. The violin at stage 12 is split for participants who arrive from path A versus path B. The area of each split violin is proportional to the sample size of the data that it represents. Hippocampal volume is in mm3, amyloid (whole cerebellum normalized) load is the SUVR, t-tau level is in pg/mL, and age is in years. The horizontal axis represents the disease stage, while the vertical axis represents the value of each biomarker. The solid line in each split represents the median of the data, and the dashed lines represent the median ± the median absolute deviation. Abbreviation: SUVR, standard uptake value ratio.
Compared to participants in path B, participants in path A were older (median age of 81 [IQR: 76–85] at stage 8 vs. 73 [IQR: 69–78] at stage 9, P < .001), had higher education levels (odds ratio 1.27 of having at least 15 years of education, P < .001), and were less likely to have two copies of the APOE ε4 (odds ratio 0.33, P < .001).
4. Discussion
Patients with AD progress through the disease at different rates (i.e., time to clinical diagnosis of AD) and with different pathophysiological responses (e.g., rate and extent of neurodegeneration) [4]. Clinically, such heterogeneity among participants could inform studies pertaining to participant-specific risk/protective factors that affect disease progression and estimates of participant response to drugs or therapies. In light of the failure of many promising treatments for AD when tested in heterogeneous cohorts in phase 3 clinical trials [3], delineating tracks of functional progression may allow clinical trials of interventions targeted to specific AD subtypes. Furthermore, such data could be used in the development of potential treatments targeting specific processes associated with AD such as amyloidosis and cognitive reserve, among others [25]. Along with participant-specific therapies, comprehensive models of disease progression could lead to more specific prognoses.
Leveraging a longitudinal cohort of 1624 ADNI participants, we characterized the intermediate stages of AD based on 19 variables that included biomarkers, demographics, and genotype. Using a comprehensive machine learning approach, we identified two distinct paths of progression to AD. Participants in path B progressed to our model's terminal stage faster and were more likely to be clinically diagnosed with AD sooner than those in path A. In a test set, we showed that by using the observed variables, we could accurately predict participant progression through paths A versus B (AUROC of 0.85).
Our work builds upon existing longitudinal studies of biomarkers in AD. Past work has relied on cutoff scores, such as Alzheimer's Disease Assessment Scale Cognitive Test or CDR-SB, to define abnormality [26], [27], [28] and has used a small subset of biomarkers [27], [29], [30]. Previous attempts to stage of AD participants have focused on dichotomizing amyloidosis and neurodegeneration [2], [31], [32] and, more recently, tauopathy [8]. Our study, by contrast, considers a variety of biomarkers such as glucose metabolism (as measured by FDG-PET), CSF protein levels, a number of neuropsychological tests, as well as demographic characteristics and uses a distribution over each biomarker instead of a binary cutoff. In addition, previous work has required strong assumptions about biomarker trajectories [29], [33], [34], while ours does not, where examples of strong assumptions include representing biomarker trajectories as linear or sigmoidal curves. Our approach provides a more comprehensive pathophysiological picture of disease progression. Such a comprehensive approach will become particularly important as amyloid and tau burden measures become more commonly available at diagnostic centers.
The proposed approach to disease staging naturally exploits variations in demographics and genotype, grouping participants into distinct paths of progression. The concept of heterogeneity in AD, that is, variations in its clinical presentation and biomarker trajectories, has been investigated previously, and several potential subtypes have been identified [35], [36], [37], [38]. However, these studies are typically based on cross-sectional data from a small subset of biomarkers. In comparison, we characterized heterogeneity in participant trajectories as they progress through the disease. Finally, previous work has sought to model disease progression between clinical disease states, such as cognitively normal, MCI, and AD [39], or focused on the MCI to AD progression only [40]. While we focused our analyses on the later stages, the proposed approach models disease progression between and within all three clinical disease states. We included cognitively normal participants for completeness. The model/approach can be applied to any individual at a clinic or in a study to get an idea of where that individual lies along the disease progression spectrum. In addition, the first four stages of our model that describe cognitively normal participants are consistent with previous studies, which suggest having at least three groups for such participants [41].
A data-driven analysis such as ours can be used to generate hypotheses regarding the role of genetic or environmental risk factors toward the clinical presentation of AD. The comparatively higher education level of participants in path A suggests that higher education acts as a slowing mechanism for both the age of onset and the rate of progression. This protective effect of higher education has previously been referred to as higher cognitive reserve [42], [43]. Despite the smaller hippocampal volumes, participants in path A progress slower relative to those in path B, indicating potential resistance to the damage of the disease. More longitudinal analysis is required to better understand the complex relationship among cognitive reserve, education, and hippocampal volume [44]. The increased risk of amyloidosis in APOE ε4 carriers has been investigated in the literature [45]. Carriers of APOE ε4 were found to be at a higher risk of amyloidosis [8] and faster cognitive decline in the presence of amyloidosis [32]. However, we investigate factors such as cognitive reserve and APOE ε4 genotype longitudinally while using a large subset of biomarker data and accounting for heterogeneity in biomarker trajectories. This allows us to characterize the slight decline in amyloid burden that occurs in participants in path B as they reach the terminal disease stage, compared to participants in path A, who show an increasing trend in amyloid burden throughout disease progression. Furthermore, our results show an increased risk from having two copies of the APOE ε4 allele [46] compared to analyses that define patients broadly as APOE ε4 carriers.
Relative to path B, path A is associated with lower levels of Alzheimer's pathology (i.e., tauopathy and amyloidosis) but greater neurodegeneration. We hypothesize that, due to their lower age, participants in path B are resistant to neurodegeneration, at least for a while. Alternatively, participants in path A might follow a heterogeneous biomarker trajectory, where neurodegeneration precedes amyloidosis. Such hypotheses could by tested by observing participants as they progress through the presymptomatic disease stages. For example, future studies could make use of data sets that follow participants through the presymptomatic stages of the disease (e.g., the Dominantly Inherited Alzheimer Network database [47]). While such studies collect longitudinal evaluations of presymptomatic participants, it may be some time before enough data have been collected to apply the proposed approach.
Our study has a number of limitations. First, a smaller proportion of the population has CSF collections and amyloid scans compared to the proportion with MRI and FDG-PET scans. Second, the effect of education on disease progression is merely an association rather than a causal effect; further causal inference and potentially additional data are needed to characterize any potential causal relationships. Third, since the ADNI data set focuses on those with a classical AD clinical profile, we were unable to investigate the heterogeneity that occurs with comorbid diseases during the AD state. Finally, our study population consists of volunteers who were examined in a clinical research setting. Still, because our study did not rely on ADNI-specific components, the proposed approach can be applied to other data sets. Future work should validate our findings on a cohort with greater diversity in terms of race, comorbidities, and so on. In addition, there are many directions in which others may build on the work presented here. In particular, researchers may focus on subsets of the ADNI population (e.g., only MCI participants), characterizing group-specific heterogeneity.
In summary, our model characterizes intermediate stages of AD and the heterogeneous biomarker trajectories associated with its progression. As longitudinal data with richer features such as clinical histories, omics data, and ecologic parameters are collected, such models will help increase our understanding of the factors that drive the different trajectories of AD. Clinically, such models will aid in matching patients with potential therapies designed to target pathophysiological processes of the disease, facilitating the development of effective patient-specific drugs/therapies.
Research in Context.
-
1.
Systematic review: We searched the literature for reports on disease staging and biomarker trajectories in Alzheimer's disease. Previous research has analyzed transitions between disease stages defined by a simple two or three biomarker construct, where each biomarker is dichotomized into a normal or abnormal state.
-
2.
Interpretation: We model trajectories of disease progression using a large multimodal longitudinal data set. Our use of longitudinal data allows us to infer patients' trajectories as they progress through intermediate disease stages. Our model identifies two common but distinct paths of disease progression and characterizes them in terms of biomarker trajectories and clinical outcomes.
-
3.
Future directions: The data suggest that most patients who progress from mild cognitive impairment to Alzheimer's disease do so through one of two paths that have distinct biomarker characteristics. Progression through either of these paths is predictable and could affect treatment response.
Acknowledgments
This research program is supported by the NIH/NIA funded Michigan Alzheimer’s Disease Center (5P30AG053760) and the National Science Foundation (IIS-1553146). The views and conclusions in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the NSF or the NIH.
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; Euroimmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
The authors have no conflicts of interest to report.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.dadm.2018.06.007.
Supplementary data
Appendices
References
- 1.Young A.L., Oxtoby N.P., Daga P., Cash D.M., Fox N.C., Ourselin S. A data-driven model of biomarker changes in sporadic Alzheimer's disease. Brain. 2014;137:2564–2577. doi: 10.1093/brain/awu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jack C.R., Therneau T.M., Wiste H.J., Weigand S.D., Knopman D.S., Lowe V.J. Transition rates between amyloid and neurodegeneration biomarker states and to dementia: a population-based, longitudinal cohort study. Lancet Neurol. 2016;15:56–64. doi: 10.1016/S1474-4422(15)00323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehta D., Jackson R., Paul G., Shi J., Sabbagh M. Why do trials for Alzheimer's disease keep failing? A discontinued drug perspective for 2010-2015. Expert Opin Investig Drugs. 2017;26:735–739. doi: 10.1080/13543784.2017.1323868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam B., Masellis M., Freedman M., Stuss D.T., Black S.E. Clinical, imaging, and pathological heterogeneity of the Alzheimer's disease syndrome. Alzheimers Res Ther. 2013;5 doi: 10.1186/alzrt155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sperling R.A., Karlawish J., Johnson K.A. Preclinical Alzheimer disease—the challenges ahead. Nat Rev Neurol. 2013;9:54–58. doi: 10.1038/nrneurol.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jack C.R., Knopman D.S., Jagust W.J., Petersen R.C., Weiner M.W., Aisen P.S. Tracking pathophysiological processes in Alzheimer's disease: An updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jack C.R., Vemuri P., Wiste H.J., Weigand S.D., Aisen P.S., Trojanowski J.Q. Evidence for ordering of Alzheimer disease biomarkers. Arch Neurol. 2011;68:1526–1535. doi: 10.1001/archneurol.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jack C.R., Wiste H.J., Weigand S.D., Therneau T.M., Knopman D.S., Lowe V. Age-specific and sex-specific prevalence of cerebral β-amyloidosis, tauopathy, and neurodegeneration in cognitively unimpaired individuals aged 50–95 years: A cross-sectional study. Lancet Neurol. 2017;16:435–444. doi: 10.1016/S1474-4422(17)30077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jack C.R., Holtzman D.M. Biomarker modeling of Alzheimer's disease. Neuron. 2013;80:1347–1358. doi: 10.1016/j.neuron.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheltens N.M.E., Tijms B.M., Koene T., Barkhof F., Teunissen C.E., Wolfsgruber S. Cognitive subtypes of probable Alzheimer's disease robustly identified in four cohorts. Alzheimers Dement. 2017;13:1226–1236. doi: 10.1016/j.jalz.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cummings J.L., Vinters H.V., Cole G.M., Khachaturian Z.S. Alzheimer's disease Etiologies, pathophysiology, cognitive reserve, and treatment opportunities. Neurology. 1998;51:S2–S17. doi: 10.1212/wnl.51.1_suppl_1.s2. [DOI] [PubMed] [Google Scholar]
- 12.Bartlett J.W., Frost C., Mattsson N., Skillbäck T., Blennow K., Zetterberg H. Determining cut-points for Alzheimer's disease biomarkers: Statistical issues, methods and challenges. Biomark Med. 2012;6:391–400. doi: 10.2217/bmm.12.49. [DOI] [PubMed] [Google Scholar]
- 13.Dubois B., Feldman H.H., Jacova C., Dekosky S.T., Barberger-Gateau P., Cummings J. Research criteria for the diagnosis of Alzheimer's disease: Revising the NINCDS–ADRDA criteria. Lancet Neurol. 2007;6:734–746. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- 14.Frisoni G.B., Boccardi M., Barkhof F., Blennow K., Cappa S., Chiotis K. Strategic roadmap for an early diagnosis of Alzheimer's disease based on biomarkers. Lancet Neurol. 2017;16:661–676. doi: 10.1016/S1474-4422(17)30159-X. [DOI] [PubMed] [Google Scholar]
- 15.ADNI Study Information. 2004. Avaialble at: http://www.adni-info.org/. Accessed November 1, 2016.
- 16.Mueller S.G., Weiner M.W., Thal L.J., Petersen R.C., Jack C.R., Jagust W. Ways toward an early diagnosis in Alzheimer's disease: The Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimer's Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease Report of the NINCDS-ADRDA Work Group* under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 18.Jack C.R., Bernstein M.A., Fox N.C., Thompson P., Alexander G., Harvey D. The Alzheimer's disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jagust W.J., Bandy D., Chen K., Foster N.L., Landau S.M., Mathis C.A. The Alzheimer's Disease Neuroimaging Initiative positron emission tomography core. Alzheimer's Dement. 2010;6:221–229. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trojanowski J.Q., Vandeerstichele H., Korecka M., Clark C.M., Aisen P.S., Petersen R.C. Update on the biomarker core of the Alzheimer's Disease Neuroimaging Initiative subjects. Alzheimer's Dement. 2010;6:230–238. doi: 10.1016/j.jalz.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aisen P.S., Petersen R.C., Donohue M.C., Gamst A., Raman R., Thomas R.G. Clinical Core of the Alzheimer's Disease Neuroimaging Initiative: Progress and plans. Alzheimer's Dement. 2010;6:239–246. doi: 10.1016/j.jalz.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gales M., Young S. The application of hidden Markov models in speech recognition. Found Trends Signal Process. 2008;1:195–304. [Google Scholar]
- 23.Rabiner L.R. A tutorial on hidden Markov models and selected applications in speech recognition. Proc of the IEEE. 1989;77:257–286. [Google Scholar]
- 24.Viterbi A. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans Information Theory. 1967;13:260–269. [Google Scholar]
- 25.Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer's disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 26.Caroli A., Frisoni G.B. The dynamics of Alzheimer's disease biomarkers in the Alzheimer's Disease Neuroimaging Initiative cohort. Neurobiol Aging. 2010;31:1263–1274. doi: 10.1016/j.neurobiolaging.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang E., Farnum M., Lobanov V., Schfultz T., Raghavan N., Samtani M.N. Quantifying the pathophysiological timeline of Alzheimer's disease. J Alzheimers Dis. 2011;26:745–753. doi: 10.3233/JAD-2011-110551. [DOI] [PubMed] [Google Scholar]
- 28.Donohue M.C., Jacqmin-Gadda H., Le Goff M., Thomas R.G., Raman R., Gamst A.C. Estimating long-term multivariate progression from short-term data. Alzheimers Dement. 2014;10:S400–S410. doi: 10.1016/j.jalz.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schiratti J.B., Allassonniere S., Routier A., Colliot O., Durrleman S. A mixed- effects model with time reparametrization for longitudinal univariate manifold-valued data. Inf Process Med Imaging. 2015;9123:564–575. doi: 10.1007/978-3-319-19992-4_44. [DOI] [PubMed] [Google Scholar]
- 30.Gordon B.A., Blazey T.M., Su Y., Hari-Raj A., Dincer A., Flores S. Spatial patterns of neuroimaging biomarker change in individuals from families with autosomal dominant Alzheimer's disease: a longitudinal study. Neurology. 2018;17:241–250. doi: 10.1016/S1474-4422(18)30028-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jack C.R., Wiste H.J., Weigand S.D., Rocca W.A., Knopman D.S., Mielke M.M. Age-specific population frequencies of cerebral β-amyloidosis and neurodegeneration among people with normal cognitive function aged 50–89 years: A cross-sectional study. Lancet Neurol. 2014;13:997–1005. doi: 10.1016/S1474-4422(14)70194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cburnham S., Bourgeat P., Doré V., Savage G., Brown B., Laws S. Clinical and cognitive trajectories in cognitively healthy elderly individuals with suspected non-Alzheimer's disease pathophysiology (SNAP) or Alzheimer's disease pathology: A longitudinal study. Lancet Neurol. 2016;15:1044–1053. doi: 10.1016/S1474-4422(16)30125-9. [DOI] [PubMed] [Google Scholar]
- 33.Jedynak B.M., Lang A., Liu B., Katz E., Zhang Y., Wyman B.T. A computational neurodegenerative disease progression score: Method and results with the Alzheimer's disease Neuroimaging Initiative cohort. NeuroImage. 2012;63:1478–1486. doi: 10.1016/j.neuroimage.2012.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bilgel M., Prince J.L., Wong D.F., Resnick S.M., Jedynak B.M. A multivariate nonlinear mixed effects model for longitudinal image analysis: Application to amyloid imaging. Neuroimage. 2016;134:658–670. doi: 10.1016/j.neuroimage.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J.Y., Na H.K., Kim S., Kim H., Kim H.J., Seo S.W. Robust Identification of Alzheimer's Disease subtypes based on cortical atrophy patterns. Sci Rep. 2017;7 doi: 10.1038/srep43270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferreira D., Verhagen C., Hernández-Cabrera J.A., Cavallin L., Guo C.J., Ekman U. Distinct subtypes of Alzheimer's disease based on patterns of brain atrophy: Longitudinal trajectories and clinical applications. Sci Rep. 2017;7 doi: 10.1038/srep46263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byun M.S., Kim S.E., Park J., Yi D., Choe Y.M., Sohn B.K. Heterogeneity of regional brain atrophy patterns associated with distinct progression rates in Alzheimer's disease. PLoS One. 2015;10:e0142756. doi: 10.1371/journal.pone.0142756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray M.E., Graff-Radford N.R., Ross O.A., Petersen R.C., Duara R., Dickson D.W. Neuropathologically defined subtypes of Alzheimer's disease with distinct clinical characteristics: A retrospective study. Lancet Neurol. 2011;10:785–796. doi: 10.1016/S1474-4422(11)70156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt-Richberg A., Guerrero R., Ledig C., Molina-Abril H., Frangi A.F., Rueckert D. Multi- stage biomarker models for progression estimation in Alzheimer's disease. Inf Process Med Imaging. 2015;9123:387–398. doi: 10.1007/978-3-319-19992-4_30. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt-Richberg A., Ledig C., Guerrero R., Molina-Abril H., Frangi A.F., Rueckert D. Learning biomarker models for progression estimation of Alzheimer's Disease. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nettiksimmons J., Harvey D., Brewer J., Carmichael O., DeCarli C., Jack C.R., Jr. Subtypes based on cerebrospinal fluid and magnetic resonance imaging markers in normal elderly predict cognitive decline. Neurobiol Aging. 2010;31:1419–1428. doi: 10.1016/j.neurobiolaging.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo R.Y., Jagust W.J. Effect of cognitive reserve markers on Alzheimer pathological progression. Alzheimer Dis Assoc Disord. 2013;27 doi: 10.1097/WAD.0b013e3182900b2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern Y. Cognitive reserve in ageing and Alzheimer's disease. Lancet Neurol. 2012;11:1006–1012. doi: 10.1016/S1474-4422(12)70191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang D.W., Lim H.K., Joo S.H., Lee N.R., Lee C.U. The association between hippocampal subfield volumes and education in cognitively normal older adults and amnestic mild cognitive impairment patients. Neuropsychiatr Dis Treat. 2018;14:143–152. doi: 10.2147/NDT.S151659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vemuri P., Knopman D.S., Lesnick T.G., Przybelski S.A., Mielke M.M., Graff-Radford J. Evaluation of amyloid protective factors and Alzheimer disease neurodegeneration protective factors in elderly individuals. JAMA Neurol. 2017;74:718–726. doi: 10.1001/jamaneurol.2017.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maestre G., Ottman R., Stern Y., Gurland B., Chun M., Tang M.X. Apolipoprotein E and Alzheimer's disease: Ethnic variation in genotypic risks. Ann Neurol. 1995;37:254–259. doi: 10.1002/ana.410370217. [DOI] [PubMed] [Google Scholar]
- 47.DIAN Study Information. 2008. Available at: https://dian.wustl.edu/. Accessed May 1, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendices