Summary
Multiple system atrophy (MSA) is a progressive neurodegenerative disease that affects several areas of the CNS, whose pathogenesis is still widely unclear and for which an effective treatment is lacking. We have generated induced pluripotent stem cell-derived dopaminergic neurons from four MSA patients and four healthy controls and from two monozygotic twins discordant for the disease. In this model, we have demonstrated an aberrant autophagic flow and a mitochondrial dysregulation involving respiratory chain activity, mitochondrial content, and CoQ10 biosynthesis. These defective mechanisms may contribute to the onset of the disease, representing potential therapeutic targets.
Keywords: multiple system atrophy, induced pluripotent stem cells, dopaminergic neurons, mitochondria, autophagy, MSA, neurodegeneration
Highlights
-
•
An iPSC-based neuronal model of MSA is described
-
•
Mitochondria are dysfunctional in MSA neurons
-
•
Autophagic machinery is impaired in MSA neurons
Monzio Compagnoni et al. present an iPSC-based neuronal in vitro model of multiple system atrophy. Patients' dopaminergic neurons display a dysregulation of mitochondrial functioning and autophagy, suggesting new hints for the comprehension of the pathogenesis of the disease.
Introduction
Multiple system atrophy (MSA) is a severe and progressive neurodegenerative disease. Parkinsonism, cerebellar ataxia, dysautonomia, and pyramidal features are the main clinical hallmarks. According to the predominant symptomatology at onset, either parkinsonian or cerebellar, two different subtypes of the disease can be distinguished: MSA-P and MSA-C, respectively (Fanciulli and Wenning, 2015).
Although many preclinical and clinical trials are in progress (Valera et al., 2016), an effective treatment is still lacking.
Neuropathologically, MSA is characterized by atrophy mainly in the putamen in MSA-P and in the cerebellum, middle cerebellar peduncles, and pontine basis in MSA-C. α-Synuclein accumulation is the neuropathological hallmark of the disease. Differently from other α-synucleinopathies, it occurs mainly in oligodendrocytes in the form of glial cytoplasmic inclusions. However, α-synuclein aggregates can be detected also in glial nuclei, neuronal cytoplasm, neuronal nuclei, and astroglial cytoplasm. Moreover, astrogliosis and microglial activation are common findings in MSA. Despite the peculiarity of oligodendroglial involvement, neuronal systems are strongly affected. A prominent degeneration of striatonigral pathway (both striatal medium spiny neurons and substantia nigra dopaminergic neurons) is observed in MSA-P. MSA-C displays a remarkable degeneration of Purkinje cells and cerebellopontine fibers; however, substantia nigra is also affected (Jellinger, 2014).
The role of mitochondrial dysfunction in the onset and progression of MSA has been debated. The most direct evidence supporting this scenario is the report of mutations in COQ2, a gene involved in the synthesis of CoenzymeQ10 (CoQ10), in familial and sporadic MSA cases (Multiple-System Atrophy Research Collaboration, 2013), but this finding has not been replicated in independent MSA cohorts (Sharma et al., 2014, Schottlaender et al., 2014, Ronchi et al., 2016). The assessment of the activity of respiratory chain complexes in autoptic substantia nigra and platelets of patients has not provided significant results (Gu et al., 1997). Two independent groups have recently described a reduction of CoQ10 levels selectively in cerebellum of MSA patients without mutations in COQ2, but not in striatum, frontal cortex, or occipital cortex (Schottlaender et al., 2016, Barca et al., 2016). Nevertheless, the activity level of complexes I + III and II + III, closely related to CoQ10, have not shown significant differences. Furthermore, the amount of two enzymes involved in CoQ10 synthesis (PDSS1 and COQ5) has been found to be reduced in MSA brains (Barca et al., 2016).
Upregulation of autophagy is another feature that has been observed in autoptic MSA samples (Schwarz et al., 2012, Tanji et al., 2013).
Few models of MSA have been developed, mainly represented by transgenic mice with overexpression of human α-synuclein in oligodendrocytes (Shults et al., 2005, Kahle et al., 2002, Yazawa et al., 2005). Although these models have allowed dissection of some important aspects of MSA pathogenesis, they cannot recapitulate all the aspects of human pathology. Indeed, α-synuclein is artificially overexpressed and the role of other cells, including neurons, can be underestimated.
Induced pluripotent stem cells (iPSCs), whose generation has been described only a decade ago (Takahashi and Yamanaka, 2006), have been used to model several neurodegenerative disorders, such as Alzheimer's disease, Huntington's disease, and amyotrophic lateral sclerosis (Ross and Akimov, 2014). Both idiopathic and familial (LRRK2, PINK1, PRKN, GBA, and SNCA) Parkinson's disease has been modeled as well (Torrent et al., 2015).
A single iPSC-based study has been reported so far, aiming to differentiate MSA-derived iPSCs toward oligodendrocytes (Djelloul et al., 2015). These experiments have provided evidence for α-synuclein expression in differentiating oligodendrocytes. However, this study has not explored the cellular and molecular pathogenic events in MSA neurons.
In this perspective, we have established an iPSC-based neuronal in vitro model of MSA and have demonstrated mitochondrial dysregulation and impaired autophagy in patients' neurons that can contribute to pathogenesis.
Results
Generation and Characterization of iPSCs from MSA Patients and Controls
We generated iPSCs from skin fibroblasts of four patients affected with MSA (two MSA-P and two MSA-C) and five unaffected subjects, including the healthy monozygotic twin of one of the MSA-C patients (MSA-C1/AT). Diagnosis of probable MSA was performed according to widely accepted clinical criteria (Gilman et al., 2008). The main clinical features of the subjects are described in Table 1.
Table 1.
Main Clinical Features of Subjects Involved in the Study
| Patient Code | Diagnosis | Sex | Age at Biopsy (Years) | Age at Onset (Years) | Familial History |
|---|---|---|---|---|---|
| P1 | MSA-P | F | 78 | 68 | no |
| P2 | MSA-P | M | 55 | 52 | no |
| C1 (AT) | MSA-C | F | 59 | 55 | no |
| C2 | MSA-C | F | 60 | 56 | no |
| CTR1 | – | M | 24 | – | no |
| CTR2 | – | F | 60 | – | no |
| CTR3 | – | M | 68 | – | no |
| CTR4 | – | F | 80 | – | no |
| UT | – | F | 59 | – | no |
All subjects were negative for mutations in genes commonly involved in Parkinson’s disease (LRRK2, GBA, and SNCA), multiplication of SNCA gene, and ATXN1, ATXN2, ATXN3, ATXN8, and PPP2R2B pathological repeats. Genomic rearrangements were excluded by CGH array performed on DNA from blood. As previously reported (Ronchi et al., 2016), a homozygous variant in COQ2 gene (p.A43G) was found in one patient (MSA-C2). However, the mutation did not affect respiratory chain activity in muscle and CoQ10 amount in muscle and fibroblasts.
iPSCs were generated from skin fibroblasts through a non-integrating reprogramming method based on the expression of factors OCT4, SOX2, KLF4, and C-MYC. Different clones of iPSCs were isolated and expanded.
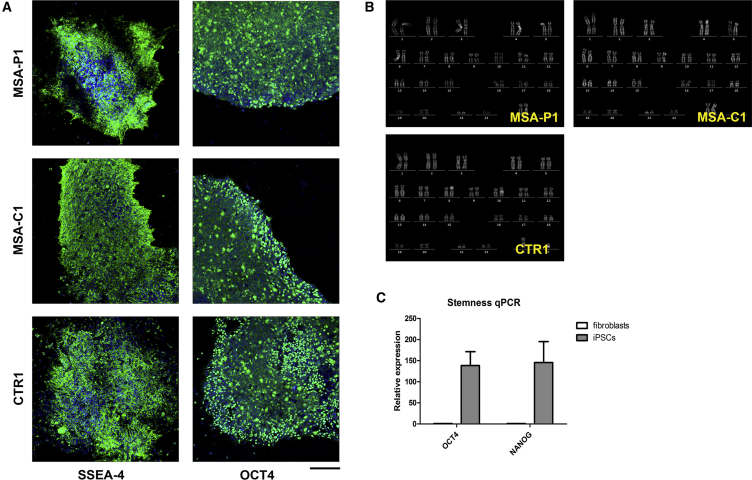
Immunocytochemistry demonstrated that most of the cells in all iPSC clones expressed the pluripotency markers SSEA-4 and OCT4 (Figure 1A). The karyotype of all the lines was assessed to exclude genetic rearrangements due to the reprogramming process (Figure 1B). RT-PCR analysis showed a high expression of the pluripotency markers OCT4 and NANOG in all iPSC lines compared with fibroblasts (Figure 1C).
Figure 1.
Characterization of iPSCs
(A) ICC showing positivity of iPSC lines for the stem cell markers SSEA4 and OCT4. Scale bar, 75 μm.
(B) Karyotype analysis of iPSC lines aimed to exclude major genetic rearrangements due to the reprogramming process.
(C) qPCR demonstrating high expression levels of stem cell-related genes OCT4 and NANOG in iPSCs compared with fibroblasts.
Data are expressed as means ± SEM.
Generation and Characterization of iPSC-Derived Dopaminergic Neurons
All iPSC lines were differentiated toward dopaminergic neurons through an already described protocol (Zhang et al., 2014), which allows the generation of tyrosine hydroxylase (TH)-positive neurons with high efficiency.
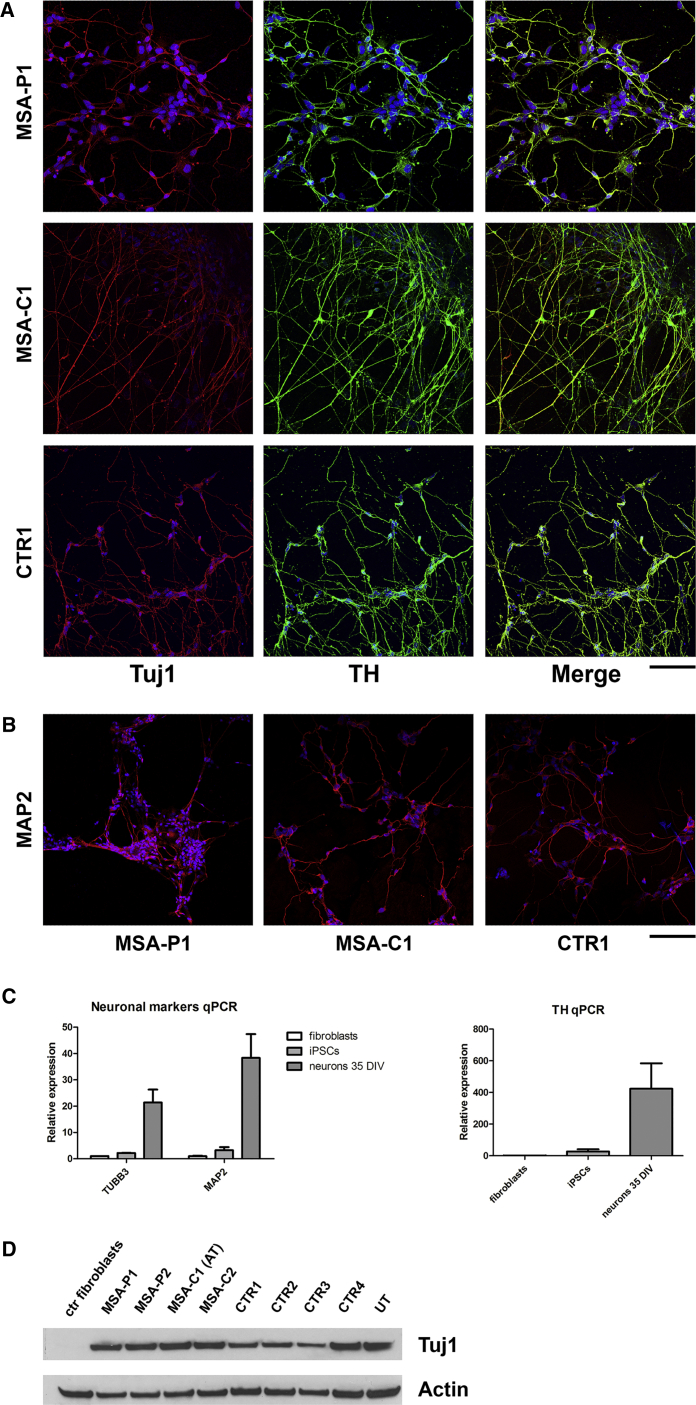
At 35 days in vitro (DIV) most of the cells were positive for the neuronal marker TUJ1 and more than 55% of TUJ1-positive cells were also positive for the catecholaminergic marker TH, as assessed by immunocytochemistry (Figure 2A and Table S1). At 50 DIV the majority of differentiated cells was also positive for the neuronal marker MAP2 (Figure 2B and Table S1). These values are in line with the ones described in the applied protocol.
Figure 2.
Characterization of iPSC-Derived Dopaminergic Neurons: First Part
(A) ICC showing positivity for the neuronal marker TUJ1 and the catecholaminergic marker TH in the majority of iPSC-derived neurons at 35 DIV. Scale bar, 50 μm.
(B) ICC showing positivity for the neuronal marker MAP2 in the majority of iPSC-derived neurons at 50 DIV. Scale bar, 75 μm.
(C) qPCR demonstrating high expression levels of the neuronal markers TUBB3 and MAP2 and of the catecholaminergic marker TH in iPSC-derived neurons at 35 DIV compared with fibroblasts and iPSCs.
(D) Western blot showing high protein amount of the neuronal marker TUJ1 in iPSC-derived dopaminergic neurons at 35 DIV compared with fibroblasts. AT, affected twin; UT, unaffected twin.
Data are expressed as means ± SEM. See also Figure S1 and Table S1.
RT-PCR was performed at 35 DIV. All the cell lines highly expressed the markers TUBB3, MAP2, and TH compared with fibroblasts and iPSCs (Figure 2C).
TUJ1 expression at 35 DIV was confirmed by a robust signal detected at western blot, which was absent in fibroblasts (Figure 2D). Western blot also demonstrated a high amount of tyrosine hydroxylase in all neuronal lines at 70 DIV (Figure S1A).
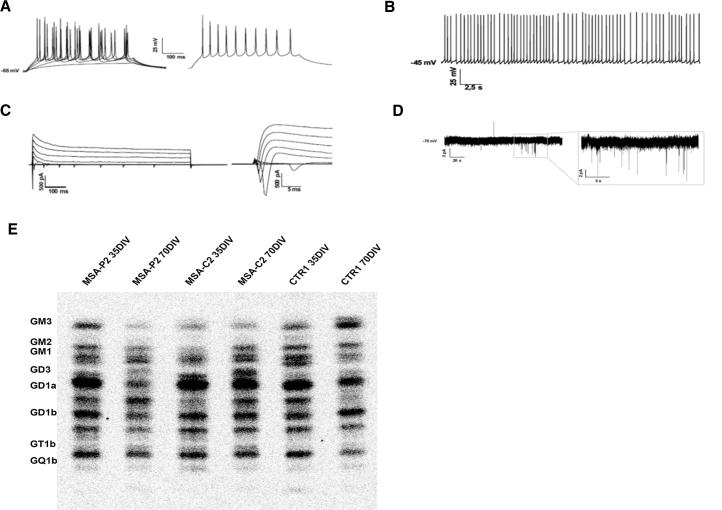
To verify a mature neuronal identity, we performed electrophysiology. At 56 DIV evoked action potentials and spontaneous firing activity in neurons were detected (Figures 3A and 3B), and inward and outward currents were recorded (Figure 3C). Post-synaptic activity was evaluated at 70 DIV, but spontaneous post-synaptic potentials were rarely observed with the exception of low amplitude spikes (Figure 3D).
Figure 3.
Characterization of iPSC-Derived Dopaminergic Neurons: Second Part
(A) Current clamp recordings in whole-cell configuration of a representative neuron differentiated from MSA-P1 iPSCs at 56 DIV, showing evoked action potentials. On the right the maximal firing rate is shown, obtained injecting 260 pA of current.
(B) Spontaneous firing activity recorded in current clamp whole-cell configuration. Action potentials are fired at a frequency of 2–6 Hz.
(C) Representative traces of voltage-clamp recordings of inward and outward currents. Neurons were stimulated with voltage steps of 20 mV starting from 0 mV to 80 mV.
(D) Spontaneous post-synaptic potentials recorded in voltage-clamp whole-cell configuration from iPSC-derived neurons of MSAP2 at 70 DIV. Neurons were clamped at −70 mV and did not show spontaneous post-synaptic potentials, with the exception of few spikes of low amplitude.
(E) Sphingolipid composition of iPSC-derived neurons in aqueous phase. Digital autoradiography of HPTLC performed using the solvent system 0.2% chloroform/methanol/calcium chloride 50:42:11 (v/v/v).
To assess the full maturation of neurons, we evaluated three lines of iPSC-derived neurons (MSA-P2, MSA-C2, and CTR1) for sphingolipid composition (Schöndorf et al., 2014) at two differentiation steps, 35 DIV and 70 DIV (Figure 3E; Tables S2 and S3). Differentiated iPSCs contained all the polysialogangliosides found in the CNS. The radioactivity associated with gangliosides ranged from 16% to 22% of the total radioactivity associated with the sphingolipid content, and quantitatively was in the range from 1.5 to 2.7 nCi/mg of cellular protein. These data resemble those obtained from rat granule cells differentiating in culture (Prinetti et al., 2001) and suggest that the analyzed cell population is enriched with fully differentiated neurons.
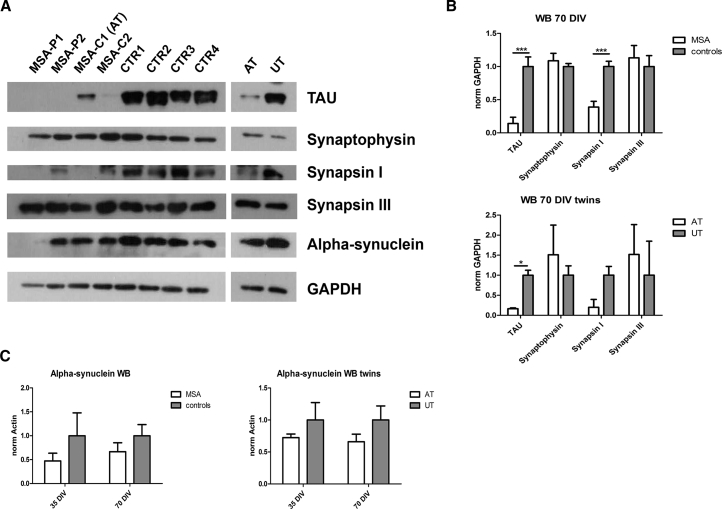
Mature MSA Neurons Display Severe Signs of Cellular Suffering
To investigate the presence of defects in neuronal maturation and survival in patients, we assessed the level of specific proteins through western blot: synapsin I, synapsin III, synaptophysin (synaptic markers), and TAU (neurite protein) (Figures 4A, 4B, and S1B). At 70 DIV MSA neurons showed a significant lower level of TAU (p < 0.001) and synapsin I (p < 0.001) compared with controls, and the twins showed a similar behavior. Data at 70 DIV were also normalized for the neuronal marker TUJ1, confirming TAU reduction in patients (p < 0.01) (Figure S1C). qPCR demonstrated that the observed Tau protein reduction in MSA was not due to low MAPT mRNA expression levels (Figure S1D).
Figure 4.
Evaluation of Neuronal Markers
(A) Western blot showing TAU, synaptophysin, synapsin I, synapsin III, α-synuclein, and GAPDH in neurons at 70 DIV.
(B) Graphs showing quantifications of TAU, synaptophysin, synapsin I, and synapsin III in neurons at 70 DIV (normalized for GAPDH).
(C) Graphs showing the amount of α-synuclein in neurons at 35 and 70 DIV measured through western blot and normalized for GAPDH. AT, affected twin; UT, unaffected twin.
Data are expressed as mean ± SEM. ∗p < 0.05; ∗∗∗p < 0.001. Three independent experiments were performed for each clone. See also Figure S1.
α-Synuclein protein amount was not different between patients and controls (Figure 4C).
Autophagy Is Impaired in MSA Neurons
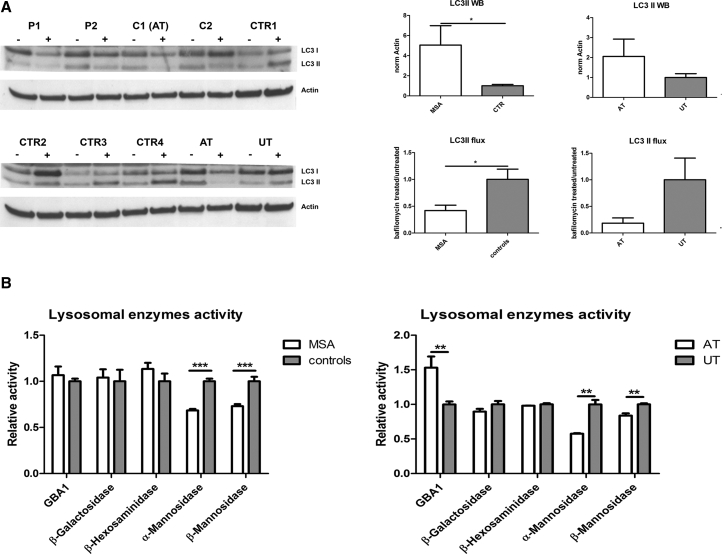
The level of autophagy-related proteins was investigated before and after treatment with bafilomycin A1, a V-ATPase inhibitor which causes block of the fusion between the autophagosome and the lysosome (Klionsky et al., 2012).
LC3-II amount was significantly higher in patients than in controls (p < 0.05), suggesting an autophagic activation. Furthermore, the autophagic flux proved to be more efficient in controls, as demonstrated by a significant difference in the ratio between LC3-II level after bafilomycin treatment and LC3-II basal level (p < 0.05). Interestingly, the two twins discordant for the disease showed a similar behavior (Figure 5A). Decreased LAMP1 and increased p62 were detected in patients (Figure S2A).
Figure 5.
Evaluation of Autophagy
(A) Western blot showing the amount of LC3I, LC3II and Actin in neurons at 35 DIV before and after treatment with bafilomycin (200 nM, 24 hr). The graphs on the right show the basal level of LC3II in neurons (normalized for actin) and the autophagic flux of LC3II (ratio treated/untreated).
(B) Graphs showing the enzymatic activities of β-glucocerebrosidase GBA1, β-galactosidase, β-hexosaminidase, α-mannosidase, and β-mannosidase measured in total cell lysates of neurons at 35 DIV. AT, affected twin; UT, unaffected twin.
Data are expressed as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Three independent experiments were performed for each clone and graphs represent pooled data from all the experiments. Two clones were used for each subject, with the exception of a few cases (single clone). Raw data showing results for each single clone are reported in Figure S5. See also Figures S2 and S3.
In addition, we assessed the activity of five lysosomal enzymes (GBA1, β-galactosidase, β-hexosaminidase, α-mannosidase, β-mannosidase) in neurons. GBA1, β-galactosidase, and β-hexosaminidase were similar in patients and controls. By contrast, the activity of α-mannosidase and β-mannosidase proved to be reduced in patients (p < 0.001 for both the enzymes). This finding was confirmed in the two twins (Figure 5B).
Mitochondrial Functioning Is Significantly Altered in MSA Neurons
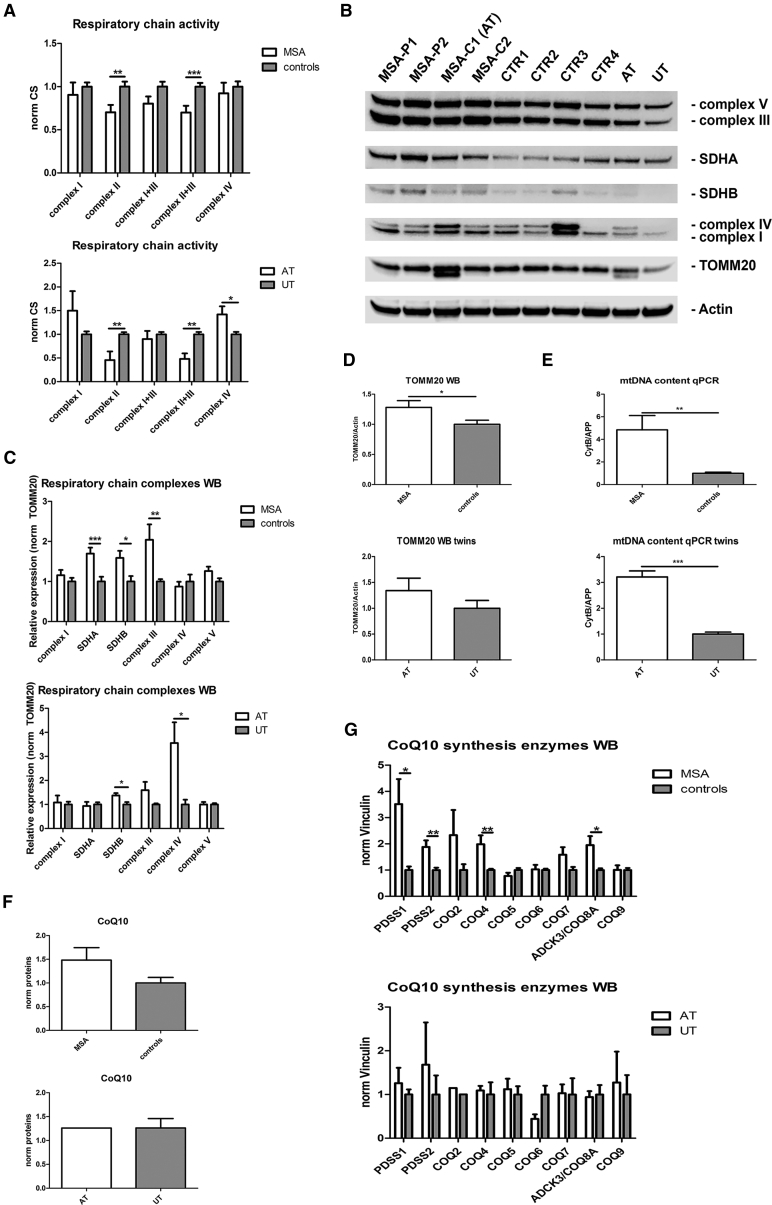
The activity of respiratory chain complexes I, II, I + III, II + III, and IV was investigated through spectrophotometric analyses. Values were normalized for the activity of citrate synthase (CS), a matrix enzyme index of mitochondrial mass. The activity level of complexes II and II + III was significantly lower in patients (p < 0.01 and p < 0.001, respectively) and a trend of reduction was observed for complexes I + III. A similar behavior was observed in the twins (Figure 6A).
Figure 6.
Evaluation of Mitochondria
(A) Graphs showing activity levels of respiratory chain complexes in neurons at 35 DIV measured through spectrophotometric analysis. Values are normalized for the activity of citrate synthase.
(B) Western blot performed on all neuronal lines at 35 DIV assessing the amount of respiratory chain complexes, TOMM20, and actin.
(C) Graphs showing the amount of the complexes of respiratory chain in neurons at 35 DIV measured through western blot and normalized for actin.
(D) Graphs showing the amount of the structural mitochondrial protein TOMM20 in neurons at 35 DIV measured through western blot and normalized for actin.
(E) Graphs showing mtDNA content in neurons at 35 DIV, measured by qPCR through the simultaneous detection of the mitochondrial gene CytB and the nuclear reference gene APP.
(F) Graphs showing CoQ10 levels in neurons at 35 DIV, measured through HPLC.
(G) Graphs showing the amount of nine enzymes involved in the synthesis of CoQ10 in neurons at 35 DIV, measured through western blot and normalized for vinculin: PDSS1, PDSS2, COQ2, COQ4, COQ5, COQ6, COQ7, ADCK3/COQ8A, and COQ9. AT, affected twin; UT, unaffected twin.
Data are expressed as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Three independent experiments were performed for each clone and graphs represent pooled data from all the experiments. Two clones were used for each subject, with the exception of a few cases (single clone). Raw data showing results for each single clone are reported in Figures S6 and S7. See also Figures S2 and S4.
To assess whether the mitochondrial respiratory enzyme activity reduction was associated with reduced protein steady-state levels, we assessed the amounts of complex I, complex II (SDHA, SDHB), complex III, complex IV, and complex V by western blot and normalized them for the mitochondrial structural protein TOMM20. The amount of complexes was not reduced, indicating that the impaired activity was not related to the level of mitochondrial protein. Interestingly, complex II (both SDHA and SDHB subunits) and complex III were significantly more expressed in patients, suggesting a possible compensatory mechanism (Figures 6B and 6C).
To explore the reason for decreased activity of complexes II + III in MSA, we assessed the amount of CoQ10 and of the enzymes involved in CoQ10 synthesis (PDSS1, PDSS2, COQ2, COQ4, COQ5, COQ6, COQ7, ADCK3/COQ8A, and COQ9). CoQ10 levels were similar between the groups, while an increased amount of CoQ10 biosynthetic enzymes (PDSS1, PDSS2, COQ4, and ADCK3/COQ8A) was detected in patients (Figures 6F, 6G, and S2B).
To assess whether the higher amount of mitochondrial respiratory enzymes and CoQ10 biosynthetic enzyme levels was also accompanied by increased mitochondrial mass, we evaluated the level of TOMM20, an outer membrane mitochondrial structural protein, which was significantly higher in patients (p < 0.05) (Figures 6B and 6D). Confirming this finding, mitochondrial DNA content, a reliable indicator of mitochondrial mass, was significantly upregulated in MSA (p < 0.01) (Figure 6E).
Interestingly, autophagy- and mitochondria-related findings were more pronounced in MSA-P, as demonstrated by reanalyzing MSA-P and MSA-C groups separately (Figures S3 and S4).
Discussion
The present study aimed to evaluate the pathogenic mechanisms of MSA, exploiting the promising model of iPSCs, so far unexplored in this disease.
The efficiency of dopaminergic differentiation was demonstrated by the high expression of markers of mature neuronal identity, the analysis of sphingolipid composition, and electrophysiological records.
At 70 DIV TAU and synapsin I were markedly decreased in MSA neurons, likely suggesting loss of neurites. Interestingly, only synapsin I was reduced, whereas synapsin III, described to be highly expressed in extrasynaptic regions (Porton et al., 2011), did not display changes. No differences were observed in the levels of TAU and synapsin I between patients and controls at 35 DIV, suggesting neuronal damage possibly correlated with aging.
Impairment of two pathways, autophagy and mitochondrial functioning, was observed in MSA neurons in this study.
Autophagy
Autophagic impairment has been studied in depth in α-synucleinopathies (Xilouri et al., 2016). A possible involvement of this pathway has been described both in brains of patients with Lewy body disease and in mouse models (Klucken et al., 2012, Yu et al., 2009, Crews et al., 2010). Furthermore, an upregulation of autophagy has been described in MSA (Schwarz et al., 2012, Tanji et al., 2013).
In the present study, the assessment of LC3 II at basal level and after bafilomycin administration has demonstrated a dysregulation of autophagy in patients. Moreover, a defective activity has been found in some lysosomal enzymes. These data are corroborated by the findings observed in the two twins.
These results suggest that a generalized autophagic defect is involved in MSA. Further studies will be crucial to the understanding of whether autophagic deficiency represents a primary defect of the disease or is a consequence of other mechanisms (e.g., protein/lipid accumulation), with a particular focus on the relationship between autophagy and α-synuclein.
Mitochondria
Mitochondrial dysfunction has been widely investigated in Parkinson’s disease, and many clues strongly suggest an involvement in the pathogenesis (Schapira, 2008). Furthermore, as reported in detail in the Introduction, mitochondria have been extensively studied also in MSA.
The results of the present study support the hypothesis of mitochondrial involvement in the pathogenesis of the disease. In particular, they suggest mitochondrial dysfunction and an upregulation of several mitochondrial pathways, findings that may be closely related to each other. Indeed, the generalized mitochondrial upregulation may suggest a mitochondrial attempt to compensate the functional deficit.
Mitochondrial dysfunction is supported by spectrophotometric analyses, which show an impaired activity of respiratory chain complexes, in particular complex II and complexes II + III. On the other hand, the mitochondrial attempt to counterbalance the functional defect is suggested by the increased mitochondrial mass (assessed through two independent methods), the increased amount of respiratory chain complexes II and III, and the upregulation of several enzymes involved in CoQ10 synthesis.
CoQ10 is a component of the mitochondrial respiratory chain that shuttles electrons from complexes I and II to complex III. The involvement of CoQ10 deficiency in the pathogenesis of MSA has been previously suggested (Multiple-System Atrophy Research Collaboration, 2013, Schottlaender et al., 2016, Barca et al., 2016). Our finding of an upregulation of various CoQ10 synthesis enzymes, in particular those that catalyze the initial reactions of biosynthesis, is consistent with the hypothesis of a compensatory mechanism. In this case the mechanism is effective, as CoQ10 was not found to be decreased in MSA neurons. The result is in line with findings on autoptic samples, which show a CoQ10 reduction only in cerebellum (Schottlaender et al., 2016, Barca et al., 2016). These data may suggest that an efficient compensatory mechanism is active in most brain regions with the exception of cerebellum, which is particularly susceptible to CoQ10-related diseases (Quinzii et al., 2007).
In the attempt to provide a comprehensive hypothesis, we suggest that autophagic and mitochondrial defects may be related. Autophagic impairment may also affect mitophagy, resulting in the accumulation of senescent damaged mitochondria. These organelles do not correctly function, as the impaired complexes' activities show. Consequently, several feedback mechanisms try to overcome this functional deficit by upregulating a series of processes: mitochondrial mass, and synthesis of respiratory chain subunits and of CoQ10. However, dysfunctional mitochondria are not able to successfully carry out these tasks, the result of which is a further accumulation of malfunctioning mitochondria, which contribute to trigger the pathological compensatory mechanism in a self-propagating fashion.
This picture prompts us to further investigate the mitophagic machinery in detail in order to confirm the connection between autophagic impairment and mitochondrial defect.
Conclusions
Overall, the present study describes a comprehensive model of MSA based on iPSC-derived neurons and suggests innovative perspectives for the comprehension of the pathogenesis of this disease.
In particular, a remarkable impairment of the autophagic machinery and of specific mitochondrial pathways has emerged from the study.
These results provide an original contribution for the comprehension of a disease whose mechanisms are almost completely unknown and lay the foundations for future analyses.
Further investigation will explore α-synuclein pathological behavior, the lysosomal defect (with a particular focus on the relationship between autophagy and α-synuclein), and mitophagy, to corroborate the possible link between autophagic impairment and mitochondrial dysfunction. Furthermore, the evaluation of the same pathways also in iPSC-derived oligodendrocytes and in neuron-oligodendrocyte co-cultures may shed light on the complex pathology observed in patients and on cell-to-cell interaction.
All of these mechanisms can represent potential therapeutic targets for this still incurable disease.
Experimental Procedures
Ethical Issues
The present study, which involves human samples, was performed in compliance with Helsinki Declaration. National legislation and institutional guidelines were observed. Written informed consent was obtained from all the subjects involved in the study according to local ethical guidelines (approved by ethical committee at the IRCCS Foundation Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy).
Skin Biopsies and Fibroblast Expansion
Skin biopsies were obtained through a punch from the ventral surface of the right arm. The skin was previously disinfected with lodopovidone and anesthetized with ice spray. Fibroblasts were isolated from the biopsies and expanded. Fibroblasts were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with fetal bovine serum (15%), penicillin/streptomycin (1%), and amphotericin B (1%).
iPSC Generation and Expansion
iPSCs were generated from fibroblasts through a commercially available kit (CytoTune-iPS 2.0 Sendai Reprogramming Kit, Thermo Fisher). Fibroblasts were incubated with virus for 24 hr. After another 6 days of culture, cells were detached, plated on feeder-coated 6-well costars, and maintained in a culture medium suitable for pluripotent stem cells (Essential 8 Medium, Thermo Fisher). When colonies appeared, they were transferred to Matrigel-coated culture dishes and expanded.
Karyotype Analysis
After adding colchicine, cells were processed with a hypotonic solution (0.6% sodium citrate and 0.13% potassium chloride) and fixed with a methanol/acetic acid solution (ratio 3:1). A quinacrine solution was used to obtain Q-banding and cell metaphases were acquired under a fluorescence microscope at 100× magnification; metaphases were analyzed with a MetaSystems-Ikaros analytical system. About 30 metaphases from at least two independent cultures were analyzed according to the ISCN and to the European General Guidelines and Quality Assurance for Cytogenetics at approximately 300–400 band level.
Immunocytochemistry
Immunocytochemistry was performed following standard procedures. Slide-adherent cells were rinsed in 1× PBS (3 times), incubated in 4% paraformaldehyde for 10 min, and rinsed in 1× PBS (3 times). Cells were then incubated in blocking solution (1× PBS containing 10% BSA and 0.3% Triton) for 1 hr and in saturation solution (1× PBS containing 3% BSA) containing primary antibody at proper concentration overnight at 4°C. Cells were then incubated in saturation solution containing secondary antibody and DAPI at proper concentration for 2 hr and rinsed in 1× PBS (3 times). Finally, the slides were mounted. Images were obtained through the confocal system Leica TCS SP2 (Leica Microsystems). For details of primary and secondary antibodies, see Table S4.
iPSC Differentiation toward Dopaminergic Neurons
iPSCs were differentiated toward dopaminergic neurons according to the protocol described by Zhang et al. (2014) (with mild changes) which, in turn, is based on the protocol described by Kriks et al. (2011). Cells were cultured in proper media supplemented with specific factors at proper concentrations as follows. Day 0: KSR differentiation medium (81% DMEM, 15% KSR, 100× 1% non-essential amino acids, 100× 1% β-mercaptoethanol, 100× 1% penicillin/streptomycin, 100× 1% amphotericin) supplemented with 10 μM SB431542 and 100 nM LDN-193189. Days 1 and 2: KSR differentiation medium supplemented with 10 μM SB431542, 100 nM LDN-193189, 0.25 μM SAG, 2 μM purmorphamine, and 50 ng/mL fibroblast growth factor 8b (FGF8b). Days 3 and 4: KSR differentiation medium supplemented with 10 μM SB431542, 100 nM LDN-193189, 0.25 μM Smoothened agonist (SAG), 2 μM purmorphamine, 50 ng/mL FGF8b, and 3 μM CHIR99021. Days 5 and 6: 75% KSR differentiation medium and 25% N2 differentiation medium (97% DMEM, 100× 1% N2 supplement, 100× 1% penicillin/streptomycin, 100× 1% amphotericin) supplemented with 100 nM LDN-193189, 0.25 μM SAG, 2 μM purmorphamine, 50 ng/mL FGF8b, and 3 μM CHIR99021. Days 7 and 8: 50% KSR differentiation medium and 50% N2 differentiation medium supplemented with 100 nM LDN-193189 and 3 μM CHIR99021. Days 9 and 10: 25% KSR differentiation medium and 75% N2 differentiation medium supplemented with 100 nM LDN-193189 and 3 μM CHIR99021. Days 11 and 12: B27 differentiation medium (95% neurobasal medium, 50× 2% B27 supplement, 1% Glutamax, 100× 1% penicillin/streptomycin, 100× 1% amphotericin) supplemented with 3 μM CHIR99021, 10 ng/mL brain-derived neurotrophic factor (BDNF), 10 ng/mL glial cell line-derived neurotrophic factor (GDNF), 1 ng/mL transforming growth factor β3 (TGF-β3), 0.2 mM ascorbic acid, and 0.1 mM cyclic AMP. From day 13 to the end of differentiation: B27 differentiation medium supplemented with 10 ng/mL BDNF, 10 ng/mL GDNF, 1 ng/mL TGF-β3, 0.2 mM ascorbic acid, and 0.1 mM cyclic AMP. At 20 DIV cells were split using Accutase. For details of reagents, see Table S7.
Western Blot
Proteins were extracted in H2O+ protease inhibitor cocktail (P2714, Sigma), sonicated two times for 15 s at 50 W, and centrifuged for 10 min at 750 × g. Proteins were quantified through the Lowry method. Samples were prepared in Novex Bolt LDS sample buffer and Novex Bolt Reducing Agent (Thermo Fisher) and boiled for 3 min. Proteins (5–30 μg) were loaded in Precast Bolt Bis-Tris Plus Gels 4%–12% and in Novex Bolt 1× MES/SDS Running buffer at 200 V for 30 min. Trans-Blot Turbo system (Bio-Rad) was used for transfer in nitrocellulose membrane. Membranes were blocked with 5% milk (Bio-Rad) or 1% BSA + 10% horse serum. For CoQ10 synthesis enzymes, proteins were extracted from cell pellets by sonication in lysis buffer (50 nM Tris-HCl, 150 mM NaCl, 1 mM EDTA) + protease inhibitor (Complete Mini, EDTA-free, 11836170001, Roche). Proteins were quantified through the Bradford system (ThermoScience) and analyzed by electrophoresis in a Novex 10%–20% Glycine Gel (EC61355, Invitrogen) loading 10 μg of protein of each sample. After electrophoresis, proteins were transferred to a PVC transfer membrane (IPFL00010, Immobilon-FL). Membranes were blocked in PBT (PBS with Tween 20) with 5% milk before incubation.
Protein bands were visualized by chemiluminescence using ECL reagents. Intensity of the bands was quantified with ImageJ (NIH). Protein amount was normalized for actin, GAPDH, or vinculin. For details of antibodies, see Table S5.
Electrophysiology
Currents and action potentials (APs) were recorded in whole-cell configuration using an Axopatch 200B Amplifier and pClamp 10.2 software (Molecular Devices). Traces were sampled at 10 kHz using a Digidata 1322A acquisition Interface (Molecular Devices) and filtered at 1 kHz. Borosilicate glass pipettes (GB150F-8P, Science Products), with a resistance of 4–6 MΩ, were filled with an intracellular solution containing 126 mM K-gluconate, 4 mM NaCl, 10 mM HEPES, 10 mM glucose, 1 mM MgSO4, 0.5 mM CaCl2, 1 mM EGTA, 3 mM ATP (magnesium salt), and 0.1 mM GTP (sodium salt) (pH 7.2). External solution contained 140 mM NaCl, 3 mM KCl, 1.2 mM MgCl2, 2 mM CaCl2, 10 mM HEPES, and 10 mM glucose (pH 7.4). APs were recorded in current-clamp mode, by first adjusting the membrane potential (Vh) at −65 mV and then injecting five pulses of increasing intensity. Spontaneous APs were recorded at the resting membrane potential of the cell, without current injection. Voltage-dependent ionic currents were recorded in voltage-clamp mode; cell membrane potential was clamped at −70 mV and voltage steps lasting 800 ms and ranging from −80 mV to 60 mV were delivered at 20-mV increments.
Sphingolipid Analysis
[1-3H]Sphingosine (radiochemical purity over 98%; specific radioactivity 0.86 Ci/mmol) was prepared and standard lipids were extracted by bovine brain. iPSC-derived neurons were incubated with 3 × 10−8 M [1-3H]sphingosine for a 2-hr pulse followed by a 48-hr chase, dissolving the radioactive precursor of sphingolipids in culture medium. After chase, cells were harvested and cell lysates were lyophilized and subjected to lipid extraction with chloroform/methanol/water 2:1:0.1 (v/v/v). Total lipid extracts were then subjected to a two-phase partitioning, resulting in the separation of an aqueous phase containing gangliosides and an organic phase containing all other lipids. Lipids were separated by high-performance thin-layer chromatography using the solvent system 0.2% chloroform/methanol/calcium chloride 50:42:11 (v/v/v). Radioactive lipids were detected and quantified by digital autoradiography (Beta-Imager 2000 instrument, BioSpace). Identification of lipids after separation was assessed by co-migration with authentic standards. The radioactivity associated with individual lipids was determined with M3Vision software. The radioactivity associated with lipid extracts was determined by liquid scintillation counting.
DNA and RNA Extraction from Cells and Reverse Transcription
DNA and RNA were extracted through commercially available kits (FlexiGene DNA Kit [Qiagen] and RNeasy Mini Kit [Qiagen], respectively) following the manufacturer's instructions. cDNA was obtained through the commercially available kit Ready-To-Go You-Prime First-Strand Beads (GE Healthcare), using 1 μg of RNA for each sample.
qPCR
For each well, 25 μL was prepared as follows. Taqman: 12.5 μL of TaqMan Universal PCR Master Mix (Thermo Fisher), 6.5 μL of H2O, 1 μL of primer/probe assay, and 5 μL of 1:10 dilution of cDNA. SYBRgreen: 12.5 μL of Power SYBR Green PCR Master Mix (Applied Biosystems), 5 μL of 1:10 dilution of cDNA, forward and reverse primers at proper concentration, and H2O up to a total volume of 25 μL. 18S, ACTB and GAPDH were used as housekeeping genes. Data were analyzed by the ΔΔCt method. A 7500 Real-Time PCR System was used. For the evaluation of mtDNA content qPCR was performed on genomic DNA for the simultaneous detection of the mitochondrial gene CYTB and the nuclear reference gene APP (ΔΔCt method). Details of primers and probes are available in Table S6.
Lysosomal Enzymatic Activity Assays
The fluorogenic substrates used were purchased from Glycosynth: 4-methylumbelliferyl β-D-glucopyranoside (MUB-β-Gluc) for β-glucocerebrosidase GBA1, 4-methylumbelliferyl β-D-galactopyranoside (MUB-β-Gal) for β-galactosidase, 4-methylumbelliferyl N-acetyl-β-D-glucuronide (MUG) for β-hexosaminidase, 4-methylumbelliferyl α-D-mannopyranoside (MUB-α-Man) for α-mannosidase, and 4-methylumbelliferyl β-D-mannopyranoside (MUB-β-Man) for β-mannosidase. iPSC-derived neurons were harvested and lysed in water containing protease inhibitors. Equal amounts of cell lysates were transferred to a 96-well microplate. The activities of β-galactosidase, β-hexosaminidase, α-mannosidase, and β-mannosidase were measured in McIlvaine buffer (pH 5.2) using 0.5 mM MUB-β-Gal, 0.5 mM MUG, 0.5 mM MUB-α-Man, and 0.5 mM MUB-β-Man, respectively. For GBA1 assay, the cells were preincubated for 30 min at room temperature in McIlvaine buffer containing 5 nM AMP-DNM N-(5-adamantane-1-yl-methoxy-pentyl)-deoxynojirimycin, specific inhibitor of GBA2. After incubation with the inhibitor, MUB-β-Gluc was added at a final concentration of 6 mM. The reaction mixtures were incubated at 37°C under gentle shaking. The fluorescence was recorded after transferring 10 μL of the reaction mixtures to a microplate and adding 190 μL of 0.25 M glycine (pH 10.7).
CoQ10 Dosage
CoQ10 was isolated from pellets with the hexane/ethanol 5:2 extraction technique. The combined hexane extract was dried under slow N2 gas flow and then resuspended in 1-propanol. CoQ10 levels were determined by high-performance liquid chromatography (HPLC) on a reverse-phase Symmetry C18 3.5-mm, 4.6 × 150-mm column (Waters), using a mobile phase consisting of methanol, ethanol, 2-propanol, acetic acid (500:500:15:15), and 50 mM sodium acetate at a flow rate of 0.9 mL/min. The electrochemical detection system consisted of an ESA Coulochem III with a guard cell (upstream of the injector) at +900 mV, conditioning cell at 600 mV (downstream of the column), followed by the analytical cell at +500 mV. The CoQ10 concentration was estimated by comparison of the peak area with those of standard solutions of known concentrations.
Spectrophotometric Analysis of the Activity of Respiratory Chain Complexes
The activities of respiratory chain complexes were measured through a Lambda 2 PerkinElmer spectrophotometer. Proteins were extracted at 4°C by adding a buffer at pH 7.2 to cell pellets, sonicating three times at 50 W for 10 s, centrifuging at 750 × g for 10 min, and recovering the supernatant. Protein concentration was measured through the Lowry method. Complex I activity (NADH ubiq.1 red) was measured at 340 nm after preparing a solution containing 0.1 M K-phosphate (200 μL) (pH 7.5), 100 mM sodium azide (10 μL), 1% albumin (100 μL), 2 mM NADH (70 μL), H2O (610 μL), homogenate (10 μL), 6 mM CoQ1 (5 μL); thereafter 1 mM rotenone (5 μL) was added and the residue activity was subtracted from the total. Complex II activity (Succ. Coq. Red) was measured at 600 nm after preparing a solution containing 0.1 M K-phosphate (pH 7) (500 μL), 400 mM succinic acid (40 μL), 0.5 mM 2,6 dichloroindophenol (200 μL), 30 mM KCN (50 μL), H2O (190 μL), homogenate (20 μL), and 15 mM CoQ1 (3 μL). Complexes I + III activity (NADH cit C red) was measured at 550 nm after preparing a solution containing 0.1 M K-phosphate (pH 7.5) (250 μL), 30 mM KCN (20 μL), 2 mM NADH (120 μL), 1 mM cytochrome c (100 μL), homogenate (20 μL), and H2O (490 μL). Complexes II + III activity (Succ cit C red) was measured at 550 nm after preparing a solution containing 0.1 M K-phosphate (pH 7.5) (500 μL), 30 mM KCN (20 μL), 400 mM succinate (50 μL), 1 mM cytochrome c (100 μL), homogenate (30 μL), and H2O (300 μL). Complex IV activity (Cytochrome ox) was measured at 550 nm after preparing a solution containing 0.1 M K-phosphate (pH 7) (200 μL), 1% reduced cytochrome c (100 μL), H2O (680 μL), and homogenate (20 μL). Citrate synthase activity was measured at 412 nm after preparing a solution containing 1 mM 5,5′-dithio-bis-(2-nitrobenzoic acid) (100 μL), 10 mM oxaloacetic acid (50 μL), 10 mM acetyl-CoA (30 μL), H2O (800 μL), and homogenate (20 μL). All the experiments were performed at 30°C. For analyses we used PerkinElmer software. All the activities were normalized for the activity of citrate synthase.
Quantification and Statistical Analysis
The patients' and controls' groups were both composed of four subjects. The unaffected twin (UT) is an independent cell line and has not been included in the control group. The affected twin (AT) has been included also in the patient group, but independent experiments have been performed for comparisons with UT.
The number of replicates is indicated in the figure legends.
A two-tailed Student's t test was used for statistical analyses.
Graphs in figures represent mean ± SEM (with means of controls and UT = 1, unless otherwise indicated).
For statistical significance ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Author Contributions
Conceptualization, G.M.C. and A.D.F.; Investigation, G.M.C., G.K., M.S., M.A., G.F., A. Bellucci, D.R., A. Bordoni, M.G., S.S., F.F., E.F., E.A., C.B., R.F., S.T., M.M., G.S., M.P., M.D., R.S., M.N., N.B., G.P.C., S.C., C.M.Q., and A.D.F; Writing – Original Draft, G.M.C. and A.D.F.; Writing – Review & Editing, G.M.C., M.A., A. Bellucci, R.F., G.P.C., S.C., C.M.Q., and A.D.F.; Funding Acquisition, A.D.F.
Acknowledgments
The financial support from Intesa San Paolo to A.D.F., from Unicredit to A.D.F., from Fresco Institute to A.D.F., from Fresco Institute to G.M.C., from Joint Program Neurodegenerative Disease (JPND) Research Grant DAMNDPATHSˆ (2014) to S.C., from Fondazione Cariplo to D.R. and from Telethon Foundation to M.P. (GGP15167) are gratefully acknowledged. The project described was supported by a grant from the Fresco Parkinson Institute to New York University School of Medicine and The Marlene and Paolo Fresco Institute for Parkinson’s and Movement Disorders, which was made possible with support from Marlene and Paolo Fresco. We thank Associazione Amici del Centro Dino Ferrari for their support.
Published: October 18, 2018
Footnotes
Supplemental Information includes seven figures and seven tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.09.007.
Supplemental Information
References
- Barca E., Kleiner G., Tang G., Ziosi M., Tadesse S., Masliah E., Louis E.D., Faust P., Kang U.J., Torres J. Decreased coenzyme Q10 levels in multiple system atrophy cerebellum. J. Neuropathol. Exp. Neurol. 2016;75:663–672. doi: 10.1093/jnen/nlw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews L., Spencer B., Desplats P., Patrick C., Paulino A., Rockenstein E., Hansen L., Adame A., Galasko D., Masliah E. Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of alpha-synucleinopathy. PLoS One. 2010;5:e9313. doi: 10.1371/journal.pone.0009313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Djelloul M., Holmqvist S., Boza-Serrano A., Azevedo C., Yeung M.S., Goldwurm S., Frisén J., Deierborg T., Roybon L. Alpha-synuclein expression in the oligodendrocyte lineage: an in vitro and in vivo study using rodent and human models. Stem Cell Reports. 2015;5:174–184. doi: 10.1016/j.stemcr.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanciulli A., Wenning G.K. Multiple-system atrophy. N. Engl. J. Med. 2015;372:249–263. doi: 10.1056/NEJMra1311488. [DOI] [PubMed] [Google Scholar]
- Gilman S., Wenning G.K., Low P.A., Brooks D.J., Mathias C.J., Trojanowski J.Q., Wood N.W., Colosimo C., Dürr A., Fowler C.J. Second consensus statement on the diagnosis of multiple system atrophy. Neurology. 2008;71:670–676. doi: 10.1212/01.wnl.0000324625.00404.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M., Gash M.T., Cooper J.M., Wenning G.K., Daniel S.E., Quinn N.P., Marsden C.D., Schapira A.H. Mitochondrial respiratory chain function in multiple system atrophy. Mov. Disord. 1997;12:418–422. doi: 10.1002/mds.870120323. [DOI] [PubMed] [Google Scholar]
- Jellinger K.A. Neuropathology of multiple system atrophy: new thoughts about pathogenesis. Mov. Disord. 2014;29:1720–1741. doi: 10.1002/mds.26052. [DOI] [PubMed] [Google Scholar]
- Kahle P.J., Neumann M., Ozmen L., Muller V., Jacobsen H., Spooren W., Fuss B., Mallon B., Macklin W.B., Fujiwara H. Hyperphosphorylation and insolubility of alpha-synuclein in transgenic mouse oligodendrocytes. EMBO Rep. 2002;3:583–588. doi: 10.1093/embo-reports/kvf109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucken J., Poehler A.M., Ebrahimi-Fakhari D., Schneider J., Nuber S., Rockenstein E., Schlötzer-Schrehardt U., Hyman B.T., McLean P.J., Masliah E., Winkler J. Alpha-synuclein aggregation involves a bafilomycin A 1-sensitive autophagy pathway. Autophagy. 2012;8:754–766. doi: 10.4161/auto.19371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S., Shim J.W., Piao J., Ganat Y.M., Wakeman D.R., Xie Z., Carrillo-Reid L., Auyeung G., Antonacci C., Buch A. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multiple-System Atrophy Research Collaboration Mutations in COQ2 in familial and sporadic multiple-system atrophy. N. Engl. J. Med. 2013;369:233–244. doi: 10.1056/NEJMoa1212115. [DOI] [PubMed] [Google Scholar]
- Porton B., Wetsel W.C., Kao H.T. Synapsin III: role in neuronal plasticity and disease. Semin. Cell Dev. Biol. 2011;22:416–424. doi: 10.1016/j.semcdb.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinetti A., Chigorno V., Prioni S., Loberto N., Marano N., Tettamanti G., Sonnino S. Changes in the lipid turnover, composition, and organization, as sphingolipid-enriched membrane domains, in rat cerebellar granule cells developing in vitro. J. Biol. Chem. 2001;276:21136–21145. doi: 10.1074/jbc.M010666200. [DOI] [PubMed] [Google Scholar]
- Quinzii C.M., DiMauro S., Hirano M. Human coenzyme Q10 deficiency. Neurochem. Res. 2007;32:723–727. doi: 10.1007/s11064-006-9190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchi D., Di Biase E., Franco G., Melzi V., Del Sorbo F., Elia A., Barzaghi C., Garavaglia B., Bergamini C., Fato R. Mutational analysis of COQ2 in patients with MSA in Italy. Neurobiol. Aging. 2016;45:213.e1–213.e2. doi: 10.1016/j.neurobiolaging.2016.05.022. [DOI] [PubMed] [Google Scholar]
- Ross C.A., Akimov S.S. Human-induced pluripotent stem cells: potential for neurodegenerative diseases. Hum. Mol. Genet. 2014;23(R1):R17–R26. doi: 10.1093/hmg/ddu204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira A.H. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet Neurol. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- Schöndorf D.C., Aureli M., McAllister F.E., Hindley C.J., Mayer F., Schmid B., Sardi S.P., Valsecchi M., Hoffmann S., Schwarz L.K. iPSC-derived neurons from GBA1-associated Parkinson's disease patients show autophagic defects and impaired calcium homeostasis. Nat. Commun. 2014;5:4028. doi: 10.1038/ncomms5028. [DOI] [PubMed] [Google Scholar]
- Schottlaender L.V., Houlden H., Multiple-System Atrophy (MSA) Brain Bank Collaboration Mutant COQ2 in multiple-system atrophy. N. Engl. J. Med. 2014;371:81. doi: 10.1056/NEJMc1311763. [DOI] [PubMed] [Google Scholar]
- Schottlaender L.V., Bettencourt C., Kiely A.P., Chalasani A., Neergheen V., Holton J.L., Hargreaves I., Houlden H. Coenzyme Q10 levels are decreased in the cerebellum of multiple-system atrophy patients. PLoS One. 2016;11:e0149557. doi: 10.1371/journal.pone.0149557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz L., Goldbaum O., Bergmann M., Probst-Cousin S., Richter-Landsberg C. Involvement of macroautophagy in multiple system atrophy and protein aggregate formation in oligodendrocytes. J. Mol. Neurosci. 2012;47:256–266. doi: 10.1007/s12031-012-9733-5. [DOI] [PubMed] [Google Scholar]
- Sharma M., Wenning G., Krüger R., European Multiple-System Atrophy Study Group Mutant COQ2 in multiple-system atrophy. N. Engl. J. Med. 2014;371:80–81. doi: 10.1056/NEJMc1311763. [DOI] [PubMed] [Google Scholar]
- Shults C.W., Rockenstein E., Crews L., Adame A., Mante M., Larrea G., Hashimoto M., Song D., Iwatsubo T., Tsuboi K., Masliah E. Neurological and neurodegenerative alterations in a transgenic mouse model expressing human alpha-synuclein under oligodendrocyte promoter: implications for multiple system atrophy. J. Neurosci. 2005;25:10689–10699. doi: 10.1523/JNEUROSCI.3527-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanji K., Odagiri S., Maruyama A., Mori F., Kakita A., Takahashi H., Wakabayashi K. Alteration of autophagosomal proteins in the brain of multiple system atrophy. Neurobiol. Dis. 2013;49:190–198. doi: 10.1016/j.nbd.2012.08.017. [DOI] [PubMed] [Google Scholar]
- Torrent R., De Angelis Rigotti F., Dell'Era P., Memo M., Raya A., Consiglio A. Using iPS cells toward the understanding of Parkinson's disease. J. Clin. Med. 2015;4:548–566. doi: 10.3390/jcm4040548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valera E., Monzio Compagnoni G., Masliah E. Review: novel treatment strategies targeting alpha-synuclein in multiple system atrophy as a model of synucleinopathy. Neuropathol. Appl. Neurobiol. 2016;42:95–106. doi: 10.1111/nan.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xilouri M., Brekk O.R., Stefanis L. Autophagy and alpha-synuclein: relevance to Parkinson's disease and related synucleopathies. Mov. Disord. 2016;31:178–192. doi: 10.1002/mds.26477. [DOI] [PubMed] [Google Scholar]
- Yazawa I., Giasson B.I., Sasaki R., Zhang B., Joyce S., Uryu K., Trojanowski J.Q., Lee V.M. Mouse model of multiple system atrophy alpha-synuclein expression in oligodendrocytes causes glial and neuronal degeneration. Neuron. 2005;45:847–859. doi: 10.1016/j.neuron.2005.01.032. [DOI] [PubMed] [Google Scholar]
- Yu W.H., Dorado B., Figueroa H.Y., Wang L., Planel E., Cookson M.R., Clark L.N., Duff K.E. Metabolic activity determines efficacy of macroautophagic clearance of pathological oligomeric alpha-synuclein. Am. J. Pathol. 2009;175:736–747. doi: 10.2353/ajpath.2009.080928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Xia N., Reijo Pera R.A. Directed dopaminergic neuron differentiation from human pluripotent stem cells. J. Vis. Exp. 2014:51737. doi: 10.3791/51737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.