Abstract
An enormous number of cells in the body die by apoptosis during development and under homeostasis. Apoptotic cells are swiftly engulfed by macrophages and digested into units. This removal of apoptotic cells is called ‘efferocytosis’. For efferocytosis, macrophages recognize phosphatidylserine (PtdSer) exposed on the cell surface as an ‘eat me’ signal. In healthy cells, PtdSer is exclusively localized to the inner leaflet of the plasma membrane by the action of flippases. When cells undergo apoptosis, caspase cleaves flippases to inactivate them, while it cleaves pro-scramblases to active scramblases, which quickly translocate PtdSer to the cell surface. The PtdSer is then recognized by PtdSer-binding proteins or by PtdSer receptors on macrophages, which subsequently engulf the apoptotic cells. When efferocytosis fails, apoptotic cells can rupture, releasing cellular materials that can evoke an autoimmune response. Thus, a defect in the PtdSer-exposing or PtdSer-recognizing processes triggers autoimmunity, leading to a systemic lupus erythematosus-type autoimmune disease.
Keywords: apoptosis, macrophages, phosphatidylserine, scramblase, SLE
Efferocytosis in homeostasis and autoimmunity
Introduction: phosphatidylserine as an ‘eat me’ signal for efferocytosis
Many excess cells are generated and die during animal development (1). Most cells in the body have a finite lifespan, and billions of senescent cells die every day to maintain homeostasis (2). This cell death occurs through apoptosis, in which a cascade of cysteine proteases called caspases is activated, eventually leading to the cleavage of >500 cellular substrates (3). Although a large number of cells undergo apoptosis, it is not easy to detect the dying cells in situ, because apoptotic cells are swiftly cleared by a process called ‘efferocytosis’, in which they are engulfed by macrophages (4). Macrophages do not engulf living cells, indicating that the apoptotic cells must express an ‘eat me’ signal(s) (5). Among numerous molecules proposed to be the ‘eat me’ signal, the strongest candidate is phosphatidylserine (PtdSer) (2, 6–8). Here we describe how PtdSer is exposed on the surface of apoptotic cells, and how it is recognized by macrophages for engulfment of dead cells. We then discuss the possibility that a failure in this process activates autoimmunity.
Phospholipid flippases and scramblases
PtdSer is a glycerophospholipid. All glycerophospholipids have two fatty acids and a phosphoric acid attached to a glycerol via an ester bond. A serine, choline or ethanolamine is joined to the phosphate to produce PtdSer, phosphatidylcholine (PtdCon) or phosphatidylethanolamine (PtdEtn), respectively. Phospholipids have amphiphilic characteristics (a hydrophobic tail consisting of fatty acids, and a hydrophilic head region containing phosphoric acid) and form the bilayer structure of the plasma membrane. Interestingly, these glycerophospholipids are asymmetrically distributed between the inner and outer leaflets of the plasma membrane, probably to maintain the membrane integrity and/or for signal transduction (9). PtdSer and PtdEtn are confined to the inner leaflet, whereas PtdCon is distributed at a ratio of 6:4 between the outer and inner leaflets. This asymmetrical distribution of PtdSer and PtdEtn is maintained by flippases that translocate them from the outer to inner leaflet in an ATP-dependent manner (9, 10). We recently reported that two ubiquitously expressed members of the P4-type ATPase family (ATP11A and ATP11C) serve as flippases at the plasma membrane (11, 12).
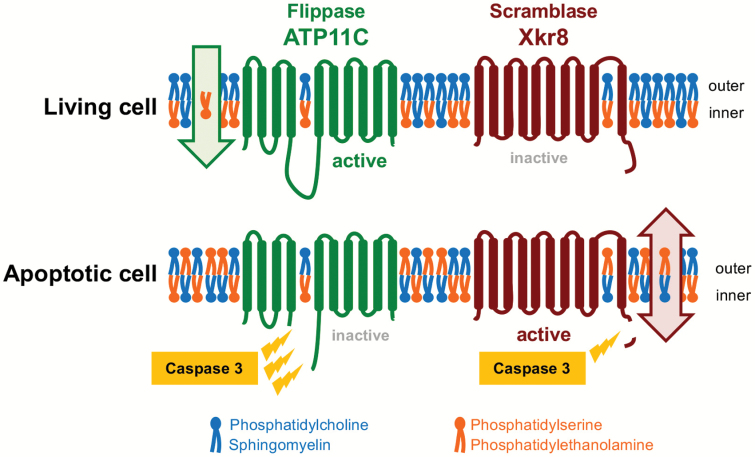
The middle of the ATP11A or ATP11C molecule contains two or three recognition sites for caspase 3, the most downstream caspase in the cascade, and these flippases are cleaved during apoptosis (11, 12) (Fig. 1). This cleavage inactivates the flippase activity, but it is not sufficient to expose PtdSer. This is because once the asymmetrical distribution of phospholipids is established, it is difficult and takes days to spontaneously break it, because the polar head group has to travel through the hydrophobic lipid layer to reach the outer leaflet (13). Therefore, to quickly expose PtdSer on the cell surface, phospholipid scramblases bidirectionally and non-specifically translocate phospholipids between the two leaflets of the plasma membrane (9).
Fig. 1.
Flippase- and scramblase-mediated regulation of apoptotic PtdSer exposure. ATP11A and ATP11C are ubiquitously expressed P4-type ATPases that contain 10 transmembrane regions. In living cells, these proteins actively translocate PtdSer from the outer to the inner leaflets, maintaining the asymmetrical distribution of PtdSer. When cells undergo apoptosis, caspase 3 cleaves ATP11A and ATP11C, and irreversibly inactivates them. Simultaneously, caspase 3 cleaves Xkr8 and activates it. Active Xkr8 seems to provide a path for phospholipids to translocate the hydrophobic layers, causing the rapid and irreversible PtdSer exposure in apoptotic cells. Blue, PtdCon and sphingomyelin; red, PtdSer and PtdEtn.
There are two types of scramblases: Ca2+-dependent and caspase-dependent (14, 15). The scramblase responsible for the apoptotic PtdSer exposure is a caspase-dependent scramblase of the XK-related (XKR) family. Three members of the Xkr protein family (Xkr4, Xkr8 and Xkr9) have been identified as caspase-dependent phospholipid scramblases (14, 16). These are membrane proteins carrying 10 transmembrane segments, and contain a caspase-recognition site at their C-terminus. Whereas Xkr8 is ubiquitously expressed, Xkr4 and Xkr9 are expressed tissue-specifically, mainly in the nervous system and intestines, respectively. Xkr8 forms a complex with basigin (BSG) or neuroplastin (NPTN), which functions as a chaperone to translocate Xkr8 from the endoplasmic reticulum to the plasma membrane (17). To gain scramblase activity, Xkr8 is cleaved by caspase 3 (Fig. 1) and the Xkr8–BSG or Xkr8–NPTN complex then forms a higher-order complex that is active.
PtdSer exposed by the activated scramblase alone does not function as an ‘eat me’ signal (18). Either the exposed PtdSer under these conditions returns quickly to the inner leaflet because of flippase activity or it has a reduced ability to bind the PtdSer receptor on macrophages. However, as mentioned above, the ATP11A and ATP11C flippases are inactivated by caspase during apoptosis. In this case, once PtdSer is exposed to the cell surface by the Xkr8 scramblase, it cannot be returned to the inner leaflet, and serves as the ‘eat me’ signal. PtdSer is exposed not only on apoptotic cells, but also on activated platelets and lymphocytes (10). In addition, some tumor cells constitutively expose PtdSer (19). These PtdSer-exposing live cells are not engulfed by macrophages, probably because the flippase is still active in them.
PtdSer-binding proteins and signal transducers for efferocytosis
Macrophages express soluble PtdSer-binding proteins or membrane-bound PtdSer receptors. In addition, there are serum PtdSer-binding proteins. Macrophages use these PtdSer-binding molecules to trap apoptotic cells and to execute the efferocytosis.
Protein S (ProS) and growth-arrest-specific 6 (Gas6) are serum proteins that share 40% identity in their amino acid sequences and have similar structural motifs (20). They specifically bind PtdSer at a Gla (gamma-carboxy glutamic acid) domain with similar affinities (21). Milk fat globule EGF factor 8 (MFG-E8) was originally discovered on the surface of milk fat globules in mammary glands and was later found to be secreted from certain macrophages and immature dendritic cells (22). MFG-E8 binds PtdSer at a Factor VIII homologous region with an affinity 5–10 times that of ProS/Gas6 (23). T-cell immunoglobulin and mucin domain-containing molecule 4 (Tim4) and its homologue Tim1 are type I transmembrane proteins expressed by resident macrophages and injured kidney, respectively (24, 25). These molecules bind PtdSer at an immunoglobulin (Ig) domain (24) with the same affinity as MFG-E8 does. Thus, there are at least three classes of proteins (Gas6/ProS; Tim1/Tim4; and MFG-E8) that use different motifs to bind PtdSer.
The soluble proteins such as ProS/Gas6 and MFG-E8 cannot support efferocytosis by themselves. Even Tim4, a type I membrane protein, cannot support efferocytosis, because its cytoplasmic region is apparently too short (40 amino acids) to mediate the signals for efferocytosis (26). ProS and Ga6 preferentially bind to one of the TAM (Tyro3, Axl and MerTK) receptors (27–29), which have tyrosine kinase activity, whereas MFG-E8 binds to the αvβ3/5-integrin complex to transduce the signal (23). Macrophages express at least one of the TAM receptors (Axl or MerTK; or both in most cases) as well as the αvβ3-integrins. Thus, ProS/Gas6 and MFG-E8 can act as a bridge between PtdSer-exposing apoptotic cells and macrophages.
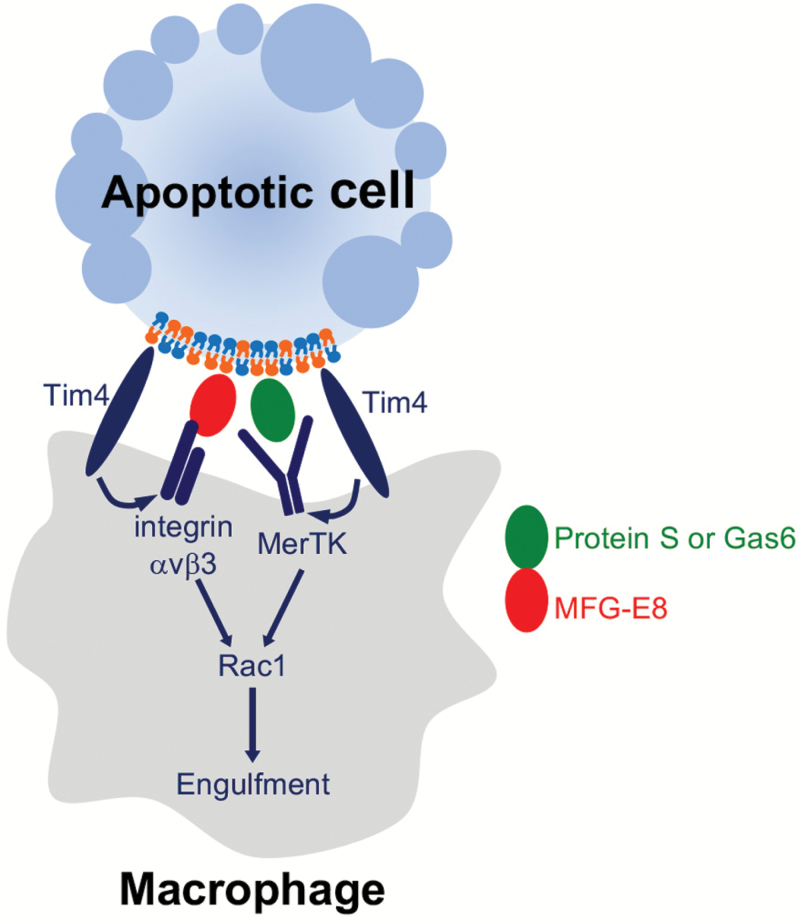
Tim4 is expressed on the resident macrophages in various tissues, including Kupffer cells, skin macrophages and marginal zone macrophages in the spleen (27, 30). Since Tim4’s affinity for PtdSer is 4–8 times that of ProS/Gas6, it seems likely that apoptotic cells are first recruited to macrophages (‘tethering’) via Tim4 and then passed to the Gas6/ProS–TAM system for engulfment (‘tickling’), at least in resident macrophages (Fig. 2). Tingible-body macrophages reside in the germinal centers of the spleen and lymph nodes, and have a strong capacity to engulf apoptotic cells, particularly activated B cells undergoing apoptosis. These macrophages express MerTK, MFG-E8, Tim4 (30–32) and probably integrin-αvβ3. However, how these molecules collaborate to elicit efferocytosis is still unclear.
Fig. 2.
Tethering and tickling processes in efferocytosis. In a set of resident macrophages, PtdSer-exposing apoptotic cells at first bind to Tim4 on the macrophages (the ‘tethering’ process). Captured apoptotic cells then bind to a TAM tyrosine kinase receptor (e.g. MerTK) with the help of ProS or Gas6. This induces phosphorylation of the TAM receptor and promotes efferocytosis by activating Rac1 (the ‘tickling’ process). In other types of macrophages and immature dendritic cells such as tingible-body macrophages and skin Langerhans cells, a secreted protein MFG-E8 functions as a bridge between PtdSer on apoptotic cells and integrins on macrophages for efferocytosis.
Signal transduction for efferocytosis
As found in the phagocytosis of bacteria, actin on macrophages polymerizes during efferocytosis, forming actin patches that establish phagocytic cups (33). Apoptotic cells then sink into the macrophage via the phagocytic cups and are transported to lysosomes (34). These processes are accompanied by the activation and inactivation of Rho family proteins such as Rac1 (Fig. 2). Classical experiments in Caenorhabditis elegans showed that Rac1 and several of its associated molecules have roles in efferocytosis and these molecules were genetically characterized (35, 36). These include CED7 (ABC transporter), CED2 (Crk), CED5 (DOCK2) and CED10 (Rac1). However, how these molecules are activated by apoptotic cells during efferocytosis is not well understood. TAM-family receptor kinases and integrin-αvβ3, which are essential signal transducers for efferocytosis, activate a variety of signaling pathways (Src, Akt, NF-κB, JAK, Fak and others) (37, 38). However, how the activation of these molecules is integrated to form the phagocytic cup for efferocytosis remains a challenging research topic.
TAM receptor-mediated efferocytosis is known to stimulate the production of anti-inflammatory cytokines such as IL-10 and TGFβ by macrophages (5, 39). In addition, TAM receptors broadly inhibit toll-like receptor (TLR)-signaling cascades and the interferon receptor (IFNR)-signaling cascades (40), damping the inflammatory response. Thus, efferocytosis has been characterized as an anti-inflammatory process.
Defective apoptotic-cell clearance in autoimmune disease
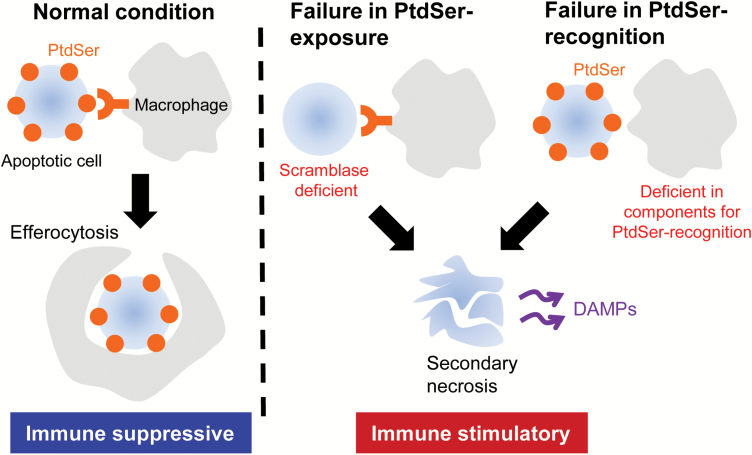
PtdSer is exposed on the surface of apoptotic cells when their plasma membrane is apparently intact. Therefore, when the PtdSer-exposing apoptotic cells are engulfed by phagocytes, intracellular materials are not released from the dead cells. On the other hand, when apoptotic cells are not promptly engulfed by macrophages, they undergo secondary necrosis, in which the plasma membrane ruptures, releasing intracellular components (41). The released cellular materials act as ‘damage-associated molecular patterns’ (DAMPs) to stimulate inflammatory and immunogenic reactions, which are likely to trigger an autoimmune response (Fig. 3).
Fig. 3.
Defects in efferocytosis and immunogenic response. Macrophages recognize PtdSer exposed on apoptotic cells, promptly engulf them (efferocytosis) and produce anti-inflammatory molecules (left) in normal conditions. Efferocytosis is thus anti-inflammatory. A deficiency of the Xkr8-scramblase causes delayed PtdSer exposure in apoptotic cells, leading to inefficient efferocytosis. A defect in the PtdSer-recognition system (Tim4, MFG-E8 and TAM receptor kinases) (right) also impairs efferocytosis. When the efferocytosis is impaired, dead cells undergo secondary necrosis and release DAMPs to stimulate inflammatory reactions.
Systemic lupus erythematosus (SLE) is an autoimmune disease that affects multiple organs, such as the skin, kidney, lungs and nervous system (42). Patients with SLE carry auto-antibodies against cellular components such as nuclei [anti-nuclear antibodies (ANA)], DNA [anti-double-stranded DNA (dsDNA) or anti-single-stranded DNA (ssDNA) antibodies] and phospholipids (anti-phospholipid antibodies). SLE can be triggered by intrinsic or environmental stimuli (43). One of these intrinsic factors is excessive apoptosis, followed by the exposure of self-antigens released from cells that have undergone secondary necrosis (44, 45). Various lines of evidence indicate that apoptotic-cell clearance is defective in SLE patients (43). For example, monocyte-derived macrophages from SLE patients have a reduced capacity to carry out efferocytosis (46). In addition, apoptotic cells are reported to accumulate in the lymph-node germinal centers (47), bone marrow (48) and UV-irradiated epidermis of SLE patients (49).
Knocking out genes in mice that are essential for apoptotic-cell recognition causes defective efferocytosis and induces SLE-type autoimmune diseases. Abnormalities of molecules involved in efferocytosis have also been found in human SLE patients. Thus, SLE can now be discussed in molecular terms. As described above, MFG-E8 is a bridging molecule between apoptotic cells and macrophages. A lack or excess of MFG-E8 blocks efferocytosis in vitro (50). Accordingly, mice lacking MFG-E8 accumulate apoptotic B cells in the germinal centers and develop a lupus-like autoimmune disease (31). Administering recombinant MFG-E8 induces a similar autoimmune disease in mice (51), and a high level of MFG-E8 is found in the serum of human SLE patients (50, 52).
Similarly, Mertk-deficient mice, in which efferocytosis by macrophages is significantly delayed, develop high levels of auto-antibodies in the serum (53–55). A triple mutation in all TAM receptors (Tyro3, Axl and MerTK) exaggerates the autoimmune phenotype, with respect to the titer of auto-antibodies, number of activated lymphocytes and severity of arthritis (56). In addition, the extracellular region of TAM receptors can be shed by metalloproteinases and these soluble forms can inhibit efferocytosis by trapping ProS/Gas6 (57). A high level of the shed extracellular region of TAM receptors (soluble MerTK, Axl and Tyro3) is detected in the serum of human SLE patients (58, 59).
A similar story may apply to Tim4. T and B cells are hyper-activated in Tim4-deficient mice and these mice develop auto-antibodies to dsDNA (60). Dual targeting of both the Tim4 and Mfge8 genes augments the autoimmune phenotype (61). Tim4 and Tim1 can be cleaved by metalloproteinase and the shed soluble region can bind PtdSer (62), suggesting that the soluble form of Tim4 may also trigger SLE.
Lupus-like autoimmune disease in Xkr8-deficient mice
As discussed above, defects in the PtdSer-recognition or efferocytosis system can trigger autoimmunity. It is possible that a defect in the PtdSer-exposing system has a similar effect. In fact, we recently found that mice that are defective in exposing PtdSer on apoptotic cells develop an SLE-type autoimmune disease (63).
Among the three Xkr members (Xkr4, Xkr8 and Xkr9) with apoptotic scramblase activity, mouse hematopoietic cells express only Xkr8. Notably, the thymocytes, splenocytes and neutrophils from Xkr8−/− mice show significantly delayed apoptotic PtdSer exposure and retarded PtdSer-dependent efferocytosis (63). These results confirmed that the Xkr8-mediated PtdSer exposure in apoptotic cells is essential for efficient efferocytosis by primary hematopoietic cells. Accordingly, Xkr8-deficient mice carry an increased number of TUNEL-positive (i.e. apoptotic) cells in the thymus at a young age (5 weeks old), and this phenotype is further pronounced by inducing apoptosis of the thymocytes with dexamethasone. Neutrophils have a short lifespan in the periphery (<10 h) and senescent neutrophils travel to the bone marrow, spleen or liver to be cleared by macrophages. Senescent neutrophils accumulate in the spleen of Xkr8-deficient mice (63), indicating that the caspase pathway leading to PtdSer exposure is required for the clearance of senescent neutrophils.
Xkr8-deficient mice do not develop a lupus-like phenotype in the C57BL/6 background. However, Xkr8-null female mice in the MRL background produce a large quantity of ANA and anti-dsDNA antibodies, and develop glomerulonephritis with a substantial deposition of immune complexes at the glomeruli (63).
Compensatory mechanisms that block autoimmune disease
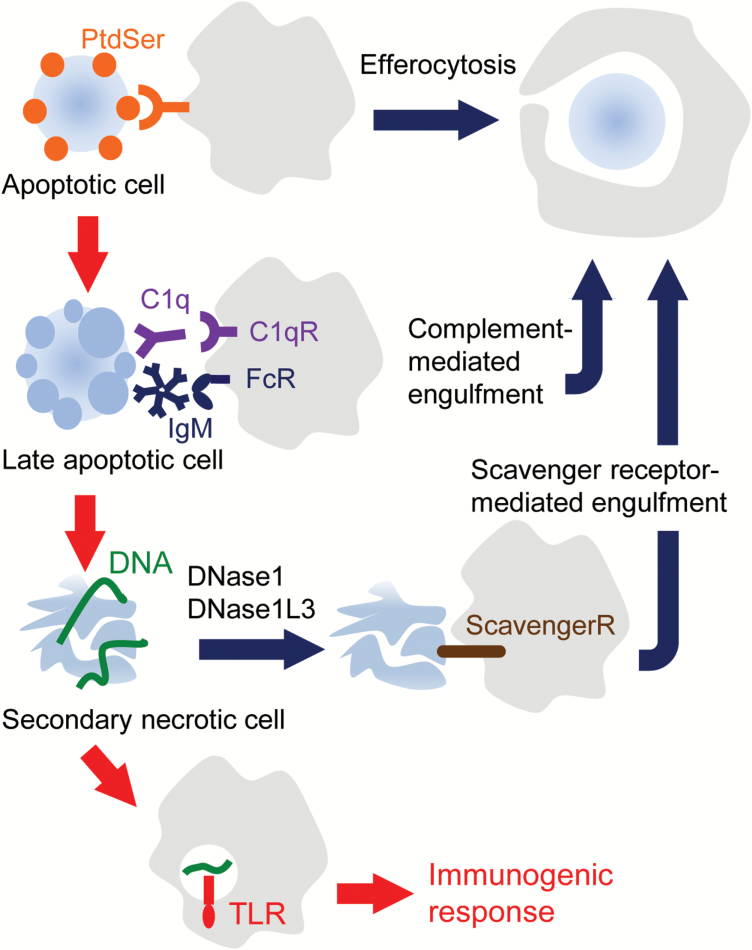
As discussed above, when apoptotic cells are not swiftly engulfed, they undergo secondary necrosis and release DAMPs to activate the immune system. However, DAMPs released from the secondary necrotic cells alone are not sufficient for SLE development (44). The SLE phenotype in efferocytosis-defective murine models strongly depends on the mouse strain. Mice deficient in MFG-E8, Xkr8 or TAM receptors develop an autoimmune disease in a 129/B6 mixed or MRL background, but not in the C57BL/6 background (31, 63). There appear to be two mechanisms—the C1q-mediated clearance of dead cell components and the degradation of DAMP-type DNA by DNase—to prevent the development of autoimmunity (Fig. 4).
Fig. 4.
Compensatory mechanisms for clearing dead cell components. Macrophages recognize PtdSer on apoptotic cells as an ‘eat me’ signal, and swiftly engulf them (efferocytosis). When apoptotic cells are not swiftly engulfed by efferocytosis, they can be engulfed by macrophages via the C1q receptor (C1qR) or Fc receptors (FcRs) at the late stage of apoptosis. Dead cells that escape from this engulfment then undergo necrosis and release various cellular contents through their ruptured plasma membrane. Chromosomal DNA released from the necrotic cells can be digested by DNase1 and DNase1L3, and the cellular remnants can be engulfed by phagocytes via scavenger receptors. If DNA is not efficiently cleaved by these DNases, DNA may aggregate the cellular debris to form strong antigens and/or stimulate macrophages or dendritic cells and trigger innate immunity via TLRs.
Although C1q, the first component of complement, was first reported to bind apoptotic cells and promote efferocytosis (64), recent reports indicate that it binds to cells at a late stage of apoptosis in a serum factor-dependent manner, suggesting that it is a back-up system for removing necrotic cells or DAMPs (65, 66). In addition, cellular components can be engulfed by phagocytes via scavenger receptors without inducing inflammation (67, 68). Patients with a C1q deficiency are rare, but they have the highest (>90%) prevalence of SLE (69). Furthermore, a C1q polymorphism that causes low serum C1q is associated with SLE (70). C1q-deficient mice carry excess apoptotic bodies in the glomerulus and develop a lupus-like phenotype (71). The disease severity in C1q−/− mice also largely depends on the genetic background: C1q−/− C57BL/6 mice exhibit no sign of autoimmune disease, whereas C1q deficiency in a 129 × C57BL/6 hybrid or MRL background induces an early onset and accelerated autoimmune disease (71, 72).
The recognition of nucleic acids is a mechanism for triggering an immune reaction (73). Many DNA sensors recognize not only pathological non-self DNA but also self DNA (74). Both chromosomal DNA and mitochondrial DNA can be released from cells that undergo secondary necrosis. Activated neutrophils also release DNA as ‘neutrophil extracellular traps’ (NETs) (75). Extracellular DNA is usually digested by the serum endonucleases DNase1 and DNase1-like 3 (DNase1L3, also called DNase γ). The DNase activities of these enzymes have similar biochemical characteristics except that DNase1 preferentially digests naked DNA, whereas DNase1L3 cleaves DNA that is associated with proteins (chromatin DNA or DNA in microparticles) (76, 77). Thus, when chromatin is released from dying necrotic cells, or released as microparticles from apoptotic cells, DNase1L3 is the major DNase that digests the DNA. DNase1 can cleave chromatin DNA only after the associated proteins have been degraded by proteases (77, 78).
A loss-of-function mutation of DNase1 is associated with SLE in humans and mice (79, 80). In particular, the loss of DNase1 activity in the kidney appears to contribute to glomerulonephritis in the (NZB × NZW)F1 mouse (81). Similarly, loss-of-function mutations of the DNase1L3 gene cause SLE in humans and mice (77, 82). Notably, the mouse MRL strain, in which a deficiency of Xkr8, MerTK or C1q induces the SLE phenotype, carries a missense mutation in the gene encoding DNase1L3 that reduces its ability to digest chromatin DNA (83). These results suggest that a deficiency in DNase1 or DNase1L3 increases the susceptibility of humans and mice to SLE.
Conclusions
Every day in our body, billions of cells undergo apoptosis and are swiftly removed by macrophages. This prompt clearance of apoptotic cells is essential to prevent the release of immunogenic materials from dying cells and thus the development of an autoimmune response. Current human data suggest that the accumulation of apoptotic cells owing to inefficient clearance is a causative event for SLE (43). Murine models with defective efferocytosis, such as MFG-E8-, Tim4-, TAM- or Xkr8-deficient mice, exhibit lupus-like phenotypes, supporting this hypothesis (31, 53–55, 60, 61, 63). Backup systems (complement or scavenger receptor systems) also exist to remove necrotic cells.
In addition, we now know that the accumulation of apoptotic cells, followed by the release of DAMPs from the necrotic cells alone is not sufficient to induce autoimmunity. Two DNases (DNase1 and DNase1L3) help to prevent the SLE phenotype by digesting the DNA released from dead cells. It should be noted that another DNase (DNase II) is involved in the digestion of the DNA of dying cells in lysosomes. A deficiency of DNase II causes the production of type I interferons and TNFα, and induces severe anemia and polyarthritis in mice and humans (84–86), confirming that the clearance of dead cell components, DNA in particular, is essential to maintain homeostasis in humans.
Funding
The work in our laboratory was supported in part by a Grant-in-Aid for Scientific Research (S) from the Japan Society for the Promotion of Science (15H05785); and a Grant-in-Aid from Core Research for Evolutional Science and Technology, the Japan Science and Technology Agency (JPMJCR14M4).
Acknowledgements
We thank Ms M. Fujii for secretarial assistance.
Conflicts of interest statement: The authors declared no conflicts of interest.
References
- 1. Fuchs Y. and Steller H. 2011. Programmed cell death in animal development and disease. Cell 147:742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nagata S. 2018. Apoptosis and clearance of apoptotic cells. Annu. Rev. Immunol. 36:489. [DOI] [PubMed] [Google Scholar]
- 3. Nagata S. and Tanaka M. 2017. Programmed cell death and the immune system. Nat. Rev. Immunol. 17:333. [DOI] [PubMed] [Google Scholar]
- 4. deCathelineau A. M. and Henson P. M. 2003. The final step in programmed cell death: phagocytes carry apoptotic cells to the grave. Essays Biochem. 39:105. [DOI] [PubMed] [Google Scholar]
- 5. Ren Y. and Savill J. 1998. Apoptosis: the importance of being eaten. Cell Death Differ. 5:563. [DOI] [PubMed] [Google Scholar]
- 6. Fadok V. A., Voelker D. R., Campbell P. A., Cohen J. J., Bratton D. L. and Henson P. M. 1992. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 148:2207. [PubMed] [Google Scholar]
- 7. Medina C. B. and Ravichandran K. S. 2016. Do not let death do us part: ‘find-me’ signals in communication between dying cells and the phagocytes. Cell Death Differ. 23:979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henson P. M. 2017. Cell removal: efferocytosis. Annu. Rev. Cell Dev. Biol. 33:127. [DOI] [PubMed] [Google Scholar]
- 9. Leventis P. A. and Grinstein S. 2010. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39:407. [DOI] [PubMed] [Google Scholar]
- 10. Bevers E. M. and Williamson P. L. 2016. Getting to the outer leaflet: physiology of phosphatidylserine exposure at the plasma membrane. Physiol. Rev. 96:605. [DOI] [PubMed] [Google Scholar]
- 11. Segawa K., Kurata S. and Nagata S. 2016. Human type IV P-type ATPases that work as plasma membrane phospholipid flippases and their regulation by caspase and calcium. J. Biol. Chem. 291:762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Segawa K., Suzuki J. and Nagata S. 2014. Flippases and scramblases in the plasma membrane. Cell Cycle 13:2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kornberg R. D. and McConnell H. M. 1971. Lateral diffusion of phospholipids in a vesicle membrane. Proc. Natl Acad. Sci. USA 68:2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suzuki J., Denning D. P., Imanishi E., Horvitz H. R. and Nagata S. 2013. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341:403. [DOI] [PubMed] [Google Scholar]
- 15. Suzuki J., Umeda M., Sims P. J. and Nagata S. 2010. Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468:834. [DOI] [PubMed] [Google Scholar]
- 16. Suzuki J., Imanishi E. and Nagata S. 2014. Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J. Biol. Chem. 289:30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suzuki J., Imanishi E. and Nagata S. 2016. Xkr8 phospholipid scrambling complex in apoptotic phosphatidylserine exposure. Proc. Natl Acad. Sci. USA 113:9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Segawa K., Suzuki J. and Nagata S. 2011. Constitutive exposure of phosphatidylserine on viable cells. Proc. Natl Acad. Sci. USA 108:19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Birge R. B., Boeltz S., Kumar S., et al. 2016. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ. 23:962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hafizi S. and Dahlbäck B. 2006. Gas6 and protein S. Vitamin K-dependent ligands for the Axl receptor tyrosine kinase subfamily. FEBS J. 273:5231. [DOI] [PubMed] [Google Scholar]
- 21. Anderson H. A., Maylock C. A., Williams J. A., Paweletz C. P., Shu H. and Shacter E. 2003. Serum-derived protein S binds to phosphatidylserine and stimulates the phagocytosis of apoptotic cells. Nat. Immunol. 4:87. [DOI] [PubMed] [Google Scholar]
- 22. Hanayama R., Miyasaka K., Nakaya M. and Nagata S. 2006. MFG-E8-dependent clearance of apoptotic cells, and autoimmunity caused by its failure. Curr. Dir. Autoimmun. 9:162. [DOI] [PubMed] [Google Scholar]
- 23. Hanayama R., Tanaka M., Miwa K., Shinohara A., Iwamatsu A. and Nagata S. 2002. Identification of a factor that links apoptotic cells to phagocytes. Nature 417:182. [DOI] [PubMed] [Google Scholar]
- 24. Miyanishi M., Tada K., Koike M., Uchiyama Y., Kitamura T. and Nagata S. 2007. Identification of Tim4 as a phosphatidylserine receptor. Nature 450:435. [DOI] [PubMed] [Google Scholar]
- 25. Ichimura T., Asseldonk E. J., Humphreys B. D., Gunaratnam L., Duffield J. S. and Bonventre J. V. 2008. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J. Clin. Invest. 118:1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park D., Hochreiter-Hufford A. and Ravichandran K. S. 2009. The phosphatidylserine receptor TIM-4 does not mediate direct signaling. Curr. Biol. 19:346. [DOI] [PubMed] [Google Scholar]
- 27. Yanagihashi Y., Segawa K., Maeda R., Nabeshima Y. I. and Nagata S. 2017. Mouse macrophages show different requirements for phosphatidylserine receptor Tim4 in efferocytosis. Proc. Natl Acad. Sci. USA 114:8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsou W. I., Nguyen K. Q., Calarese D. A., et al. 2014. Receptor tyrosine kinases, TYRO3, AXL, and MER, demonstrate distinct patterns and complex regulation of ligand-induced activation. J. Biol. Chem. 289:25750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lew E. D., Oh J., Burrola P. G., et al. 2014. Differential TAM receptor-ligand-phospholipid interactions delimit differential TAM bioactivities. eLife 3:e03385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wong K., Valdez P. A., Tan C., Yeh S., Hongo J. A. and Ouyang W. 2010. Phosphatidylserine receptor Tim-4 is essential for the maintenance of the homeostatic state of resident peritoneal macrophages. Proc. Natl Acad. Sci. USA 107:8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hanayama R., Tanaka M., Miyasaka K., , et al. 2004. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science 304:1147. [DOI] [PubMed] [Google Scholar]
- 32. Rahman Z. S., Shao W. H., Khan T. N., Zhen Y. and Cohen P. L. 2010. Impaired apoptotic cell clearance in the germinal center by Mer-deficient tingible body macrophages leads to enhanced antibody-forming cell and germinal center responses. J. Immunol. 185:5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakaya M., Kitano M., Matsuda M. and Nagata S. 2008. Spatiotemporal activation of Rac1 for engulfment of apoptotic cells. Proc. Natl Acad. Sci. USA 105:9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kitano M., Nakaya M., Nakamura T., Nagata S. and Matsuda M. 2008. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature 453:241. [DOI] [PubMed] [Google Scholar]
- 35. Pinto S. M. and Hengartner M. O. 2012. Cleaning up the mess: cell corpse clearance in Caenorhabditis elegans. Curr. Opin. Cell Biol. 24:881. [DOI] [PubMed] [Google Scholar]
- 36. Reddien P. W. and Horvitz H. R. 2004. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 20:193. [DOI] [PubMed] [Google Scholar]
- 37. Graham D. K., DeRyckere D., Davies K. D. and Earp H. S. 2014. The TAM family: phosphatidylserine sensing receptor tyrosine kinases gone awry in cancer. Nat. Rev. Cancer 14:769. [DOI] [PubMed] [Google Scholar]
- 38. Clark E. A. and Brugge J. S. 1995. Integrins and signal transduction pathways: the road taken. Science 268:233. [DOI] [PubMed] [Google Scholar]
- 39. Savill J., Dransfield I., Gregory C. and Haslett C. 2002. A blast from the past: clearance of apoptotic cells regulates immune responses. Nat. Rev. Immunol. 2:965. [DOI] [PubMed] [Google Scholar]
- 40. Rothlin C. V., Ghosh S., Zuniga E. I., Oldstone M. B. and Lemke G. 2007. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 131:1124. [DOI] [PubMed] [Google Scholar]
- 41. Silva M. T., do Vale A. and dos Santos N. M. 2008. Secondary necrosis in multicellular animals: an outcome of apoptosis with pathogenic implications. Apoptosis 13:463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rahman A. and Isenberg D. A. 2008. Systemic lupus erythematosus. N. Engl. J. Med. 358:929. [DOI] [PubMed] [Google Scholar]
- 43. Muñoz L. E., Lauber K., Schiller M., Manfredi A. A. and Herrmann M. 2010. The role of defective clearance of apoptotic cells in systemic autoimmunity. Nat. Rev. Rheumatol. 6:280. [DOI] [PubMed] [Google Scholar]
- 44. Bondanza A., Zimmermann V. S., Dell’Antonio G., et al. 2004. Requirement of dying cells and environmental adjuvants for the induction of autoimmunity. Arthritis Rheum. 50:1549. [DOI] [PubMed] [Google Scholar]
- 45. Bondanza A., Zimmermann V. S., Rovere-Querini P., et al. 2004. Inhibition of phosphatidylserine recognition heightens the immunogenicity of irradiated lymphoma cells in vivo. J. Exp. Med. 200:1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Herrmann M., Voll R. E., Zoller O. M., Hagenhofer M., Ponner B. B. and Kalden J. R. 1998. Impaired phagocytosis of apoptotic cell material by monocyte-derived macrophages from patients with systemic lupus erythematosus. Arthritis Rheum. 41:1241. [DOI] [PubMed] [Google Scholar]
- 47. Baumann I., Kolowos W., Voll R. E., et al. 2002. Impaired uptake of apoptotic cells into tingible body macrophages in germinal centers of patients with systemic lupus erythematosus. Arthritis Rheum. 46:191. [DOI] [PubMed] [Google Scholar]
- 48. Hepburn A. L., Lampert I. A., Boyle J. J., et al. 2007. In vivo evidence for apoptosis in the bone marrow in systemic lupus erythematosus. Ann. Rheum. Dis. 66:1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kuhn A., Herrmann M., Kleber S., et al. 2006. Accumulation of apoptotic cells in the epidermis of patients with cutaneous lupus erythematosus after ultraviolet irradiation. Arthritis Rheum. 54:939. [DOI] [PubMed] [Google Scholar]
- 50. Yamaguchi H., Takagi J., Miyamae T., et al. 2008. Milk fat globule EGF factor 8 in the serum of human patients of systemic lupus erythematosus. J. Leukoc. Biol. 83:1300. [DOI] [PubMed] [Google Scholar]
- 51. Asano K., Miwa M., Miwa K., , et al. 2004. Masking of phosphatidylserine inhibits apoptotic cell engulfment and induces autoantibody production in mice. J. Exp. Med. 200:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kishi C., Motegi S. I. and Ishikawa O. 2017. Elevated serum MFG-E8 level is possibly associated with the presence of high-intensity cerebral lesions on magnetic resonance imaging in patients with systemic lupus erythematosus. J. Dermatol. 44:783. [DOI] [PubMed] [Google Scholar]
- 53. Scott R. S., McMahon E. J., Pop S. M., et al. 2001. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature 411:207. [DOI] [PubMed] [Google Scholar]
- 54. Cohen P. L., Caricchio R., Abraham V., et al. 2002. Delayed apoptotic cell clearance and lupus-like autoimmunity in mice lacking the c-mer membrane tyrosine kinase. J. Exp. Med. 196:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Khan T. N., Wong E. B., Soni C. and Rahman Z. S. 2013. Prolonged apoptotic cell accumulation in germinal centers of Mer-deficient mice causes elevated B cell and CD4+ Th cell responses leading to autoantibody production. J. Immunol. 190:1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu Q. and Lemke G. 2001. Homeostatic regulation of the immune system by receptor tyrosine kinases of the Tyro 3 family. Science 293:306. [DOI] [PubMed] [Google Scholar]
- 57. Sather S., Kenyon K. D., Lefkowitz J. B., , et al. 2007. A soluble form of the Mer receptor tyrosine kinase inhibits macrophage clearance of apoptotic cells and platelet aggregation. Blood 109:1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ballantine L., Midgley A., Harris D., Richards E., Burgess S. and Beresford M. W. 2015. Increased soluble phagocytic receptors sMer, sTyro3 and sAxl and reduced phagocytosis in juvenile-onset systemic lupus erythematosus. Pediatr. Rheumatol. Online J. 13:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu J., Ekman C., Jönsen A., et al. 2011. Increased plasma levels of the soluble Mer tyrosine kinase receptor in systemic lupus erythematosus relate to disease activity and nephritis. Arthritis Res. Ther. 13:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rodriguez-Manzanet R., Sanjuan M. A., Wu H. Y., et al. 2010. T and B cell hyperactivity and autoimmunity associated with niche-specific defects in apoptotic body clearance in TIM-4-deficient mice. Proc. Natl Acad. Sci. USA 107:8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Miyanishi M., Segawa K. and Nagata S. 2012. Synergistic effect of Tim4 and MFG-E8 null mutations on the development of autoimmunity. Int. Immunol. 24:551. [DOI] [PubMed] [Google Scholar]
- 62. Schweigert O., Dewitz C., Möller-Hackbarth K., , et al. 2014. Soluble T cell immunoglobulin and mucin domain (TIM)-1 and -4 generated by a disintegrin and metalloprotease (ADAM)-10 and -17 bind to phosphatidylserine. BBA Mol. Cell Res. 1843:275. [DOI] [PubMed] [Google Scholar]
- 63. Kawano M. and Nagata S. 2018. Lupus-like autoimmune disease caused by a lack of Xkr8, a caspase-dependent phospholipid scramblase. Proc. Natl Acad. Sci. USA 280:2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Galvan M. D., Greenlee-Wacker M. C. and Bohlson S. S. 2012. C1q and phagocytosis: the perfect complement to a good meal. J. Leukoc. Biol. 92:489. [DOI] [PubMed] [Google Scholar]
- 65. Liang Y. Y., Arnold T., Michlmayr A., , et al. 2014. Serum-dependent processing of late apoptotic cells for enhanced efferocytosis. Cell Death Dis. 5:e1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Colonna L., Parry G. C., Panicker S. and Elkon K. B. 2016. Uncoupling complement C1s activation from C1q binding in apoptotic cell phagocytosis and immunosuppressive capacity. Clin. Immunol. 163:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shichita T., Ito M., Morita R., et al. 2017. MAFB prevents excess inflammation after ischemic stroke by accelerating clearance of damage signals through MSR1. Nat. Med. 23:723. [DOI] [PubMed] [Google Scholar]
- 68. Ramirez-Ortiz Z. G., Pendergraft W. F., Prasad A., et al. 2013. The scavenger receptor SCARF1 mediates the clearance of apoptotic cells and prevents autoimmunity. Nat. Immunol. 14:917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Botto M. and Walport M. J. 2002. C1q, autoimmunity and apoptosis. Immunobiology 205:395. [DOI] [PubMed] [Google Scholar]
- 70. Martens H. A., Zuurman M. W., de Lange A. H., et al. 2009. Analysis of C1q polymorphisms suggests association with systemic lupus erythematosus, serum C1q and CH50 levels and disease severity. Ann. Rheum. Dis. 68:715. [DOI] [PubMed] [Google Scholar]
- 71. Botto M., Dell’Agnola C., Bygrave A. E., et al. 1998. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat. Genet. 19:56. [DOI] [PubMed] [Google Scholar]
- 72. Mitchell D. A., Pickering M. C., Warren J., et al. 2002. C1q deficiency and autoimmunity: the effects of genetic background on disease expression. J. Immunol. 168:2538. [DOI] [PubMed] [Google Scholar]
- 73. Roers A., Hiller B. and Hornung V. 2016. Recognition of endogenous nucleic acids by the innate immune system. Immunity 44:739. [DOI] [PubMed] [Google Scholar]
- 74. Marshak-Rothstein A. 2006. Toll-like receptors in systemic autoimmune disease. Nat. Rev. Immunol. 6:823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Apel F., Zychlinsky A. and Kenny E. F. 2018. The role of neutrophil extracellular traps in rheumatic diseases. Nat. Rev. Rheumatol. 14:467. [DOI] [PubMed] [Google Scholar]
- 76. Napirei M., Wulf S., Eulitz D., Mannherz H. G. and Kloeckl T. 2005. Comparative characterization of rat deoxyribonuclease 1 (Dnase1) and murine deoxyribonuclease 1-like 3 (Dnase1l3). Biochem. J. 389:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sisirak V., Sally B., D’Agati V., et al. 2016. Digestion of chromatin in apoptotic cell microparticles prevents autoimmunity. Cell 166:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Napirei M., Ludwig S., Mezrhab J., Klöckl T. and Mannherz H. G. 2009. Murine serum nucleases–contrasting effects of plasmin and heparin on the activities of DNase1 and DNase1-like 3 (DNase1l3). FEBS J. 276:1059. [DOI] [PubMed] [Google Scholar]
- 79. Martínez Valle F., Balada E., Ordi-Ros J. and Vilardell-Tarres M. 2008. DNase 1 and systemic lupus erythematosus. Autoimmun. Rev. 7:359. [DOI] [PubMed] [Google Scholar]
- 80. Napirei M., Karsunky H., Zevnik B., Stephan H., Mannherz H. G. and Möröy T. 2000. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat. Genet. 25:177. [DOI] [PubMed] [Google Scholar]
- 81. Seredkina N. and Rekvig O. P. 2011. Acquired loss of renal nuclease activity is restricted to DNaseI and is an organ-selective feature in murine lupus nephritis. Am. J. Pathol. 179:1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Al-Mayouf S. M., Sunker A., Abdwani R., et al. 2011. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat. Genet. 43:1186. [DOI] [PubMed] [Google Scholar]
- 83. Wilber A., O’Connor T. P., Lu M. L., Karimi A. and Schneider M. C. 2003. Dnase1l3 deficiency in lupus-prone MRL and NZB/W F1 mice. Clin. Exp. Immunol. 134:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kawane K., Fukuyama H., Kondoh G., , et al. 2001. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science 292:1546. [DOI] [PubMed] [Google Scholar]
- 85. Kawane K., Ohtani M., Miwa K., et al. 2006. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 443:998. [DOI] [PubMed] [Google Scholar]
- 86. Rodero M. P., Tesser A., Bartok E., et al. 2017. Type I interferon-mediated autoinflammation due to DNase II deficiency. Nat. Commun. 8:2176. [DOI] [PMC free article] [PubMed] [Google Scholar]