Summary
Fanconi anemia (FA) causes bone marrow failure early during childhood, and recent studies indicate that a hematopoietic defect could begin in utero. We performed a unique kinetics study of hematopoiesis in Fancg−/− mouse embryos, between the early embryonic day 11.5 (E11.5) to E12.5 developmental window (when the highest level of hematopoietic stem cells [HSC] amplification takes place) and E14.5. This study reveals a deep HSC defect with exhaustion of proliferative and self-renewal capacities very early during development, together with severe FA clinical and biological manifestations, which are mitigated at E14.5 due to compensatory mechanisms that help to ensure survival of Fancg−/− embryos. It also reports that a deep HSC defect is also observed during human FA development, and that human FA fetal liver (FL) HSCs present a transcriptome profile similar to that of mouse E12.5 Fancg−/− FL HSCs. Altogether, our results highlight that early mouse FL could represent a good alternative model for studying Fanconi pathology.
Keywords: HSC, Fanconi anemia, mouse embryonic development, human embryonic development, placenta, fetal liver, transcriptome
Highlights
-
•
Deep HSC defect in FL and Pl very early during Fancg−/− development
-
•
Functional and molecular compensation in Fancg−/− FL HSCs at E14.5
-
•
Deep HSC defect also observed during human FA development
-
•
E12.5 Fancg−/− FL could represent a good model for studying Fanconi pathology
A deep HSC defect is present very early during Fancg−/− mouse embryonic development, and is also reported for the first time during human Fanconi anemia development. At E14.5, functional and molecular compensation in Fancg−/− FL HSCs help ensure the survival of Fancg−/− embryos. Altogether E12.5 Fancg−/− FL could represent a good model for studying Fanconi pathology.
Introduction
In children, bone marrow failure (BMF) represents a life-threatening condition whose most common genetic cause is Fanconi anemia (FA). FA is a genetic disorder due to mutations in one of the 22 FANC complementation group genes involved in DNA repair and prevention of cellular stress (Bluteau et al., 2016, Deans and West, 2011, Dong et al., 2015, Hays, 2012, Kim et al., 2012, Knies et al., 2017, Kottemann and Smogorzewska, 2013, Kupfer, 2013, Park et al., 2016, Zhang et al., 2015). Early during childhood (median age 6.5 years) (Dokal and Vulliamy, 2008, Faivre et al., 2000, Kutler et al., 2003), 80% of the FA patients suffer from this main symptom (Kutler et al., 2003) and hematopoietic stem cell (HSC) transplantation remains the only curative treatment (Ayas et al., 2013, Peffault de Latour et al., 2013). Furthermore, FA also strongly predisposes patients to myelodysplastic syndrome and acute myeloid leukemia (AML). To date, the pathogenesis of BMF and AML remains incompletely understood (Dumitriu and Young, 2012, Garaycoechea and Patel, 2014, Muller and Williams, 2009, Nalepa and Clapp, 2014, Quentin et al., 2011, Tischkowitz and Dokal, 2004, Walter et al., 2015, Zhang et al., 2016).
Most of the time, FA patients do not show any signs of blood cytopenia at birth (Soulier, 2011). However, a deficit in the CD34+ cell numbers in FA subjects is observed very early during childhood, even before the onset of the BMF clinical symptoms (Ceccaldi et al., 2012, Kelly et al., 2007). This, added to the early developmental defects of the hematopoietic commitment described for human embryonic stem cells after knockdown of FANCA and FANCD2 genes, led us and others to hypothesize that the FA hematopoietic defect could already begin in utero (Ceccaldi et al., 2012, Kamimae-Lanning et al., 2013, Suzuki et al., 2017, Tulpule et al., 2010, Yoon et al., 2016).
Indeed, in contrast to adult HSCs, which are quiescent in the bone marrow (BM), embryonic HSCs are in active proliferation and migrate through multiple anatomical sites before they settle in the BM (Dzierzak and Speck, 2008, Mikkola and Orkin, 2006). In the mouse, between the time of their emergence in the floor of dorsal aorta in the aorta/gonad/mesonephros (AGM) region and the colonization of BM, HSCs migrate through fetal liver (FL) and placenta (Pl), where they are amplified (Dzierzak and Speck, 2008). Amplification takes place between embryonic day 11.5 (E11.5) and E15.5 in the FL (Ema and Nakauchi, 2000, Kumaravelu et al., 2002, Morrison et al., 1995), with a peak at E12.5 (30- to 50-fold increase between E11.5 and E12.5) (Gekas et al., 2005, Kumaravelu et al., 2002), and between E11.5 and E12.5 in the Pl (25-fold) (Gekas et al., 2005, Ottersbach and Dzierzak, 2005). During human embryonic development, the sequence of events leading to production of HSCs parallels that of mice, and FL represents the key organ for HSC amplification and maturation (Tavian and Peault, 2005).
In this study, we investigated whether FA pathway deficiency impairs the expansion of the HSC pool in response to high replicative stress during the first stages of HSC amplification during embryonic development, using Fancg−/− mice. Together with FANCA and FANCC, FANCG is one of the most frequently mutated gene in FA patients, and its product is part of the nuclear FA core complex that signals the cellular response to DNA damage (Dong et al., 2015). Adult Fancg−/− mice present a phenotype virtually identical to that of Fanca−/− and Fancc−/− mice (Parmar et al., 2009), with a reduction of BM HSCs and hematopoietic progenitor (HP) subsets but no BMF (Barroca et al., 2012).
We focused on early development times, when HSC amplification takes place both in FL and Pl, i.e., E11.5–E12.5. At E12.5 expansion is important in both organs, and any DNA repair defect would have a significant impact leading to an alteration of HSC pool. Our results demonstrate that an important hematopoietic defect occurs from the very first stages of hematopoietic amplification in Fancg−/− FL and Pl, and that clinical and biological compensations occur at E14.5. In addition, we show that an HSC defect is also present in an FL sample from a human FA embryo. Transcriptomic analysis of HSCs sorted from Fancg−/− E12.5 and from the human FA FLs revealed that similar biological processes are down- and upregulated in Fanconi HSCs.
Results
Fancg Deficiency Leads to Impaired Phenotype and Profound Erythrocyte Defect in Fetal Liver and Placenta during Early Mouse Embryonic Development
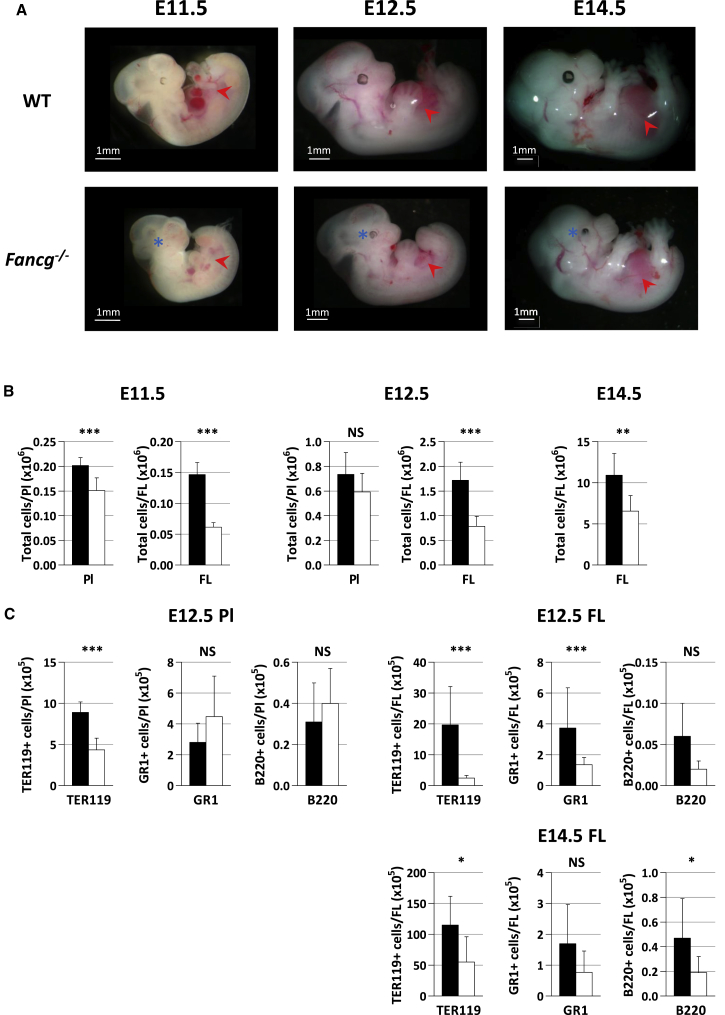
Fancg−/− homozygous mice being hypofertile (Koomen et al., 2002), embryos were obtained after timed mating of C57Bl6 Fancg+/− heterozygous breeders. Fancg−/− embryos were obtained at a lower frequency than expected at E11.5 and E12.5 (20% and 21.7%, respectively, instead of 25%, 958 embryos tested in total; p < 0.001). As observed in Fancc−/− and Fancd2−/− mice (Kamimae-Lanning et al., 2013, Suzuki et al., 2017), this frequency dramatically declined at birth (11% ± 0.8, 82 postnatal day 0 [P0] to P1 pups tested, p < 0.001), suggesting a late perinatal lethality. Interestingly, at E11.5 and E12.5 Fancg−/− embryos were systematically smaller than wild-type (WT) control littermates, while at E14.5 only half of the Fancg−/− embryos remained smaller than control (Figure 1A). This size difference between Fancg−/− and WT embryos became less pronounced around birth. Eye defects, such as anophthalmia or microphthalmia, were observed in Fancg−/− embryos at all stages of development studied (Figure 1A).
Figure 1.
Dysmorphology of Fancg−/− Embryos Accompanied with Cellular Defect and Abnormal Hematopoietic Content in the FL and Pl
(A) smaller size and microphthalmia (blue asterisk) of Fancg−/− embryos between E11.5 and E14.5, accompanied with a smaller size of FL (red arrowhead). Respective numbers of Fancg−/− and WT embryos studied was n = 20 and n = 43 at E11.5, n = 186 and n = 254 at E12.5, and n = 52 and n = 53 at E14.5.
(B) Total cellularity of FL and Pl at E11.5, E12.5, and E14.5.
(C) Total number of erythroid (TER119), myeloid (GR1), and B lymphoid (B220) cells per FL and Pl at E12.5 and E14.5. Black bars, WT; white bars, Fancg−/−.
Error bars correspond to standard deviation. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; NS, not significant.
FLs of all Fancg−/− embryos were systematically smaller than their WT counterpart between E11.5 and E14.5 (Figure 1A). This was associated with a concomitant significant reduction of total FL cellularity (2.4-, 2.2-, and 1.7-fold at E11.5, E12.5, and E14.5, respectively) (Figure 1B). Some but not all E11.5 and E12.5 Fancg−/− Pl were also smaller than their WT counterparts, and this reduction in size was always accompanied with a softer structure. Furthermore, overall cellularity of Fancg−/− Pl was reduced compared with that of WT Pl (1.3-fold at E11.5 and 1.2-fold at E12.5) (Figure 1B). We next investigated hematopoietic cell content of E12.5 Fancg−/− Pl and FL, and E14.5 FL. Fluorescence-activated cell sorting (FACS) analysis markers revealed an important defect of Ter119+ erythroid cells both in Fancg−/− Pl and FL compared with WT at E12.5 (2- and 8-fold, respectively), and to a significant decrease of GR1+ myeloid and B220+ lymphoid B cells, but only in the FL (Figure 1C). This overall decrease in hematopoietic lineages in FL was less marked at later stages of development (E14.5, Figure 1C).
Early Defect in Hematopoietic Stem and Progenitor Cells in Fancg−/− FL and Pl
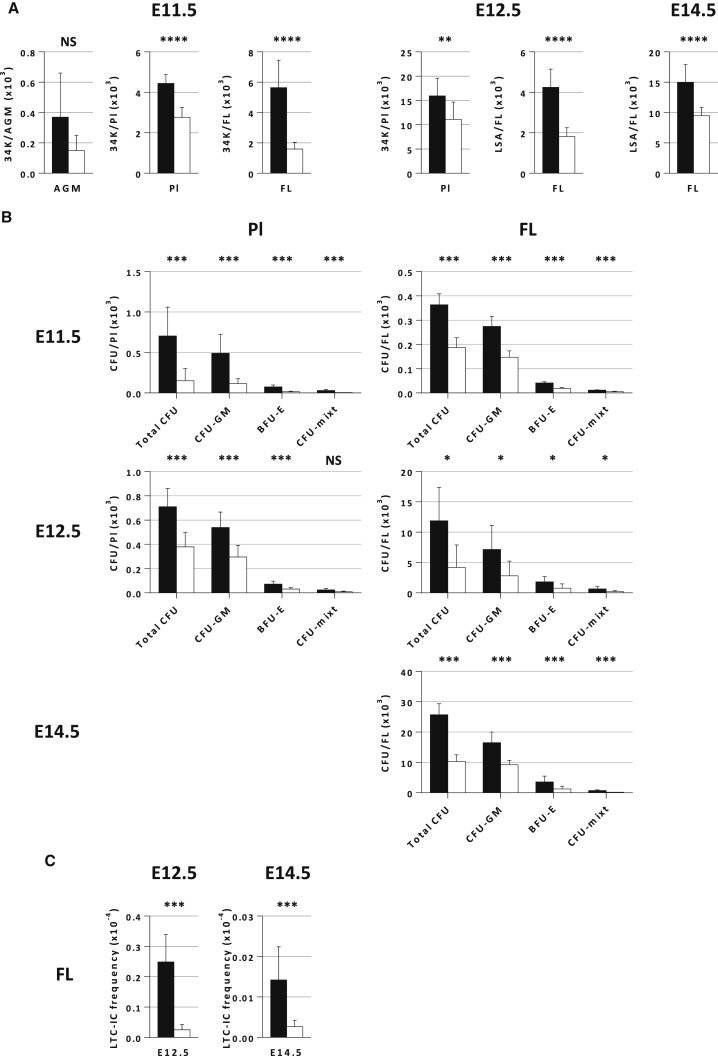
We next studied hematopoietic stem and progenitor cell (HSPC)-enriched populations in different sites of hematopoiesis during embryonic development, i.e., AGM, FL, and Pl at E11.5, FL and Pl at E12.5, and FL at E14.5. At E11.5, we chose a CD34+c-KIT+/hi (34K) population, which are enriched in HP/HSCs at this early stage of development in the AGM, FL, and Pl (M.S. and L.P., personal communication) (Gekas et al., 2005, Sanchez et al., 1996). These markers were also used for E12.5 PL (Gekas et al., 2005), while for E12.5 and E14.5 FL we used LineagenegSCA-1+AA4.1+ (LSA) as the reference HSC population (Jordan et al., 1995). At E11.5, we found an overall significant decrease of the 34K population in Fancg−/− Pl and FL compared with WT (38% and 72%, respectively), but not in the AGM (Figure 2A). This 34K defect is still present in E12.5 PL (30%). The LSA population was also found to be significantly reduced in E12.5 (57%) and E14.5 FL (37%) (Figure 2A).
Figure 2.
Defect of HSPC-Enriched Populations and CFUs in the Pl and FL of Fancg−/− Embryos Is Observed as Soon as E11.5
(A) Total number of HSPC-enriched cells in the Pl and FL at E11.5 and E12.5, and in the FL at E14.5. At E11.5 in the AGM (n = 4), Pl (n = 3), and FL (n = 3), and at E12.5 in the Pl (n = 9) the HSPC-enriched population is 34K, while in the FL at E12.5 (n = 7) and E14.5 (n = 6) it is LSA.
(B) Total CFUs and CFU-GM, BFU-E, and CFU-mixed content per FL at E11.5, E12.5, and E14.5 and per Pl at E11.5 and E12.5. Results shown represent the mean ± SE and correspond to 3–5 independent experiments, with two independent Petri dishes scored for each experiment.
(C) In vitro comparison of LTC-IC frequency in Fancg−/− and WT FL at E12.5 (n = 3) and E14.5 (n = 3). n represents distinct experiments.
Error bars correspond to standard deviation. Black bars, WT; white bars, Fancg−/−. ∗∗∗∗ p<0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; NS, not significant.
Next, we performed in vitro functional assays such as clonogenic and long-term culture-initiating cell (LTC-IC) (Petit-Cocault et al., 2007) assays to quantify progenitors and HSPC/HSCs in the FL of Fancg−/− embryos at E12.5 and E14.5. As shown on Figure 2B, clonogenic assays revealed that a significant decrease of bipotent colony-forming unit granulocyte macrophage (CFU-GM), burst-forming unit erythroid (BFU-E), and multipotent mixed colony-forming unit (CFU-mixed) was observed at all the stages studied in Fancg−/− Pl and FL compared with WT. Collectively, these results uncover a quantitative HP and HSC defect in Pl and FL early during Fancg−/− embryonic development, during stages where high HC/HSC amplification takes place.
LTC-IC assays showed that the LTC-IC frequency was 10-fold lower in FL of Fancg−/− as compared with WT embryos at E12.5 (Figure 2C). This difference in LTC-IC frequency was less marked in Fancg−/− FL at E14.5 (Figure 2C). Altogether these results indicate a severe hematopoietic defect in the immature HP/HSC compartment of Fancg−/− FL at E12.5 that gradually attenuates during embryonic development.
Qualitative and Quantitative HSC Defect of Fancg−/− FL and Pl at E12.5 Revealed by In Vivo Transplantation Assays
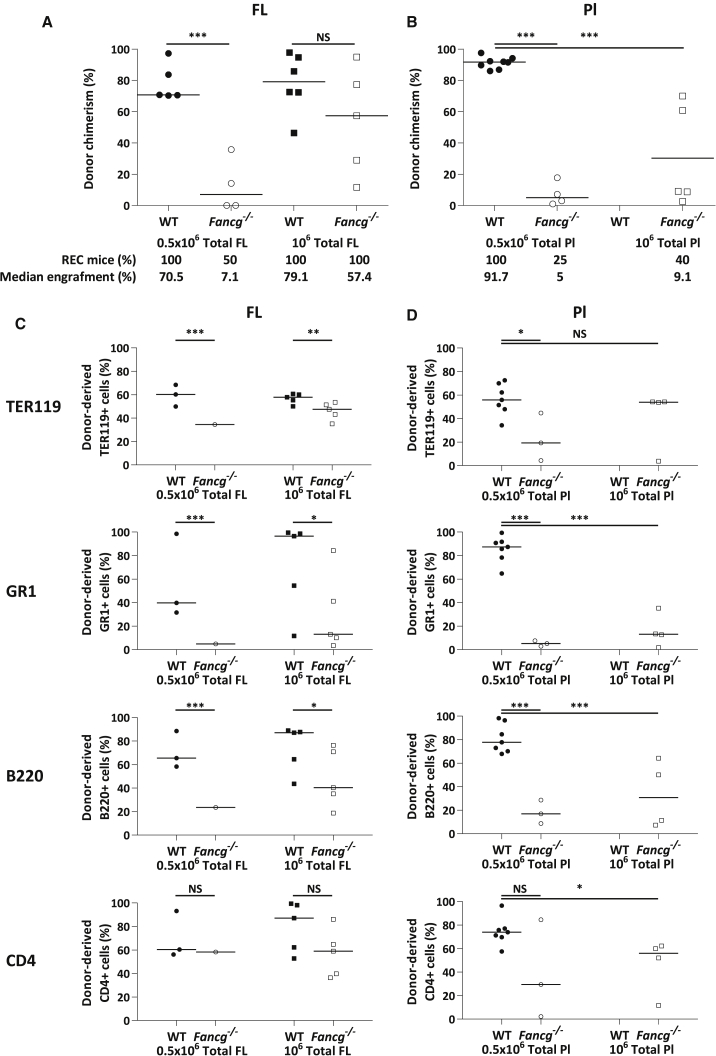
To further investigate the HSC compartment at early stages of development in Fancg−/− mice, we injected unfractionated FL and Pl cells from E12.5 Fancg−/− and WT embryos (CD45.2) into lethally irradiated congenic CD45.1 mice and tested their ability to provide long-term multilineage hematopoietic reconstitution (long-term repopulation [LTR]).
As shown on Figure 3A, when 5 × 105 unfractionated Fancg−/− and WT FL cells (which contain the same percentage of 34K and LSA HSC-enriched population [Figure S1] and equivalent to injection of 1,000 Fancg−/− or WT LSA) were implanted, only 50% of the recipients injected with Fancg−/− FL reconstituted with a low median of blood chimerism (7.1%) while all recipients injected with WT FL reconstituted with a median of engraftment of 70.5%. When 1 × 106 Fancg−/− FL cells were injected, 100% of the recipients reconstituted and the median of blood chimerism increased to 57.4%, but remained lower than that of mice injected with the same amount of WT FL cells (79.1%, Figure 3A). This reveals a qualitative defect of E12.5 FL Fancg−/− HSCs. This HSC defect is even more important in the Pl, since primary recipients barely reconstituted when 5 × 105 or 1 × 106 unfractionated E12.5 Fancg−/− Pl cells were injected (median of blood chimerism of 5% and 9.1%, respectively, versus 91.7% for mice injected with 5 × 105 WT Pl cells) (Figure 3B).
Figure 3.
High LTR Ability Defect and Impaired Myeloid Potential in E12.5 Fancg−/− Pl and FL
(A) CD45.2 chimerism in the blood of primary recipients 20–24 weeks post injection of 5 × 105 (circles) or 106 (squares) WT (black) or Fancg−/− (white) total FL cells.
(B) CD45.2 chimerism in primary recipients' blood 20–24 weeks post injection of 5 × 105 (circles) or 106 (squares) WT (black) or Fancg−/− (white) Pl cells.
(C and D) Analysis of BM hematopoietic content of primary recipients transplanted with FL (C) or Pl (D), expressed as a percentage of CD45.2+ erythroid (TER119), myeloid (GR1), B lymphoid (B220), and T-lymphoid (CD4) cells. Circles, 5 × 105 cells injected; squares, 1 × 106 cells injected; black, WT FL or Pl cells; white, Fancg−/− FL or Pl cells.
∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; NS, not significant.
Twenty to 24 weeks post injection, extensive FACS analysis of reconstituted recipient BM for co-expression of CD45.2 and specific lineage markers indicated that HSCs from FL and Pl of Fancg−/− mice were able to give rise to all hematopoietic lineages in adult recipients, although less efficiently than WT FL or Pl, especially for GR1 myeloid lineage, and to a lesser extent for B220 B lymphoid lineage (Figures 3C and 3D, respectively). This suggests a loss of myeloid potential of E12.5 Fancg−/− FL and Pl HSCs at this stage. No secondary reconstitution was observed with 5 × 105 unfractionated BM cells from primary recipients injected with Fancg−/− FL or Pl, even when BM cells injected presented a high CD45.2 chimerism (79.6%), while all mice injected with BM cells from primary recipients injected with WT FL or Pl reconstituted (Table 1). Taken together, these results suggest that early during development, when high HC/HSC amplification takes place in FL and Pl, there is an impairment of proliferative and self-renewal capacities of HSCs in Fancg−/− FL and Pl, along with an important loss of all hematopoietic lineages potential, especially myeloid.
Table 1.
Secondary Reconstitutions with BM from Primary Recipients Injected with Fancg−/− and WT Pl and FL
| Primary Transplantation | Secondary Transplantation |
|||
|---|---|---|---|---|
| No. of BM Cells Injected | Engrafted Micea/Total Mice | Median Reconstitution (%) [Lower Rec − Higher Rec] | ||
| E12.5 PL | WT | 5 × 105 | 5/5 | 11.6 [6.3 − 25.5] |
| Fancg−/− | 5 × 105 | 0/5 | 0.07 [0 − 2] | |
| E12.5 FL | WT | 5 × 105 | 3/3 | 15.9 [9.3 − 18.6] |
| Fancg−/− | 5 × 105 | 0/6 | 1.7 [0.5 − 3.6] | |
Engraftment was considered as positive when CD45.2 chimerism in the blood was >5%.
This HSC qualitative defect persists in Fancg−/− FL at E14.5, as described by others for Fancc−/− and Fancd2−/− (Kamimae-Lanning et al., 2013, Suzuki et al., 2017, Yoon et al., 2016). An HSC-enriched LSA population sorted from Fancg−/− and WT E14.5 FL was injected into lethally irradiated primary CD45.1 mice. Twenty weeks post injection, median blood chimerism was only 20.5% when 1,000 Fancg−/− LSA cells were injected, compared with 86.8% for 1,000 WT LSA cells (Figure S2A). When 5,000 LSA Fancg−/− cells were injected, high blood chimerism (≥70%) was observed in 50% of the mice (Figure S2A). However, an overall important defect in the GR1 myeloid lineage in the BM was still observed (Figure S2B), as well as a low level of blood chimerism when secondary transplantations were performed (Figure S2C), which indicates that at E14.5 Fancg−/− LSA HSCs are still defective for proliferation, self-renewal, and myeloid differentiation. Collectively, these analyses highlight a compromised function of Fancg−/− HSCs during embryonic development, which is more important at early stages when hematopoietic amplification peaks.
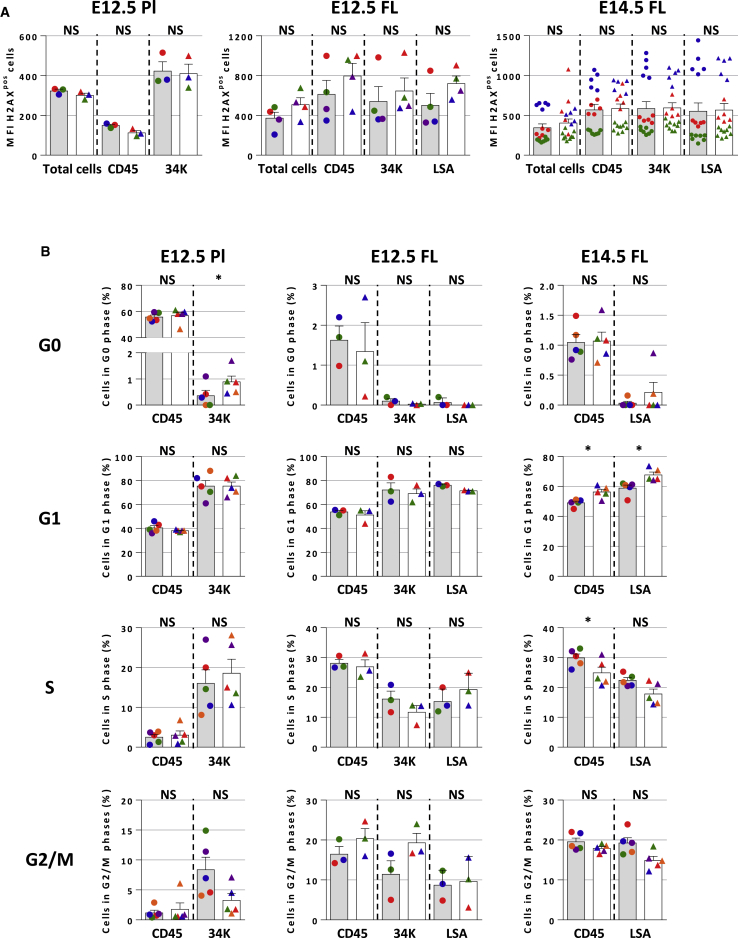
No Cell-Cycle Changes or Increased Apoptosis but Mild Increase of DNA Damage in E12.5 Fancg−/− FL HSCs
Given the high level of amplification of HSCs occurring at early stages of embryonic development, we investigated whether there was an increase in DNA damage as assessed by evaluation of γH2AX level in various E12.5 Pl and FL, and E14.5 FL populations (fractionated and unfractionated) from Fancg−/− and WT by flow cytometry. At E12.5, no difference in γH2AX level (mean fluorescence intensity [MFI]) was observed between E12.5 Fancg−/− and WT HCs (CD45) and HSCs (34K) in Pl, while in FL, although overall not statistically significant, higher levels of γH2AX were found in all Fancg−/− hematopoietic populations compared with WT for each individual experiment performed, especially in the HSC-enriched LSA population (Figure 4A). This increase in γH2AX-positive cells was no longer observed in all hematopoietic populations tested at E14.5 in Fancg−/− FL (Figure 4A). No increase in apoptosis was observed in the E12.5 FL LSA population (Figure S3A).
Figure 4.
Induced DNA Damage in E12.5 and E14.5 Fancg−/− HCs and HSCs without Apoptosis or Cell-Cycle Changes
(A) MFI of γH2AXpos cells in total, HC (CD45), and HSC (34K or SA) in E12.5 PL (n = 3), FL (n = 4), and E14.5 FL (n = 3, and each dot represents an individual mouse), measured by flow cytometry.
(B) Histograms of G0, G1, S, and G2/M phases of the cell cycle determined after Hoechst staining, for Fancg−/− and WT HC, and HSCs of E12.5 FL and Pl and E14.5 FL. Dots of same colors are related to the same experiment.
Error bars correspond to standard deviation. Gray bars, WT; white bars, Fancg−/−. ∗p < 0.05; NS, not significant.
G0, G1, S, and G2/M phases of cell cycle were analyzed by flow cytometry in Fancg−/− and WT HSCs using Hoechst 33,342/pyronine staining. At E12.5, no significant difference for any of the cell-cycle phases was observed between Fancg−/− and WT HCs or HSCs populations in FL or Pl, except an increase in G0 for 34K Pl HSCs (p < 0.05) (Figure 4B). In E14.5 Fancg−/− FL, there was an increase in G1 phase for CD45 HCs and LSA HSCs (p < 0.05) compared with WT FL, and a decrease in S phase for CD45 HCs (Figure 4B).
HSPC Defect in the FL of Human FA Embryo
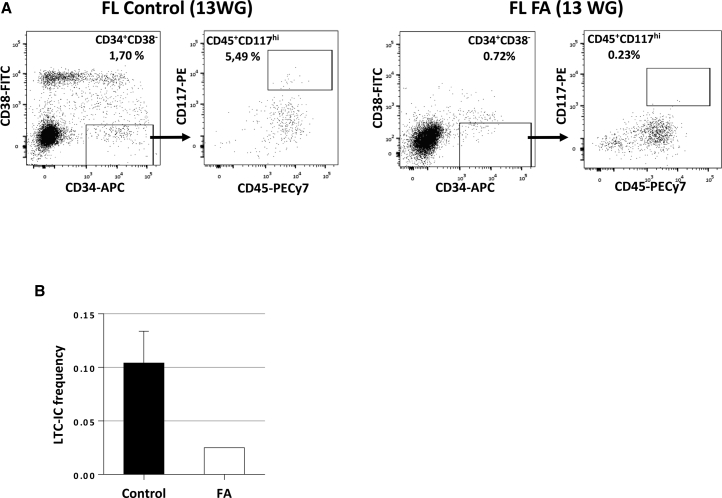
We could obtain FL samples from six human fetuses after medical abortion, strongly suspected, based on malformative signs, to have FA. Among the six, one fetus (13 weeks of gestation [WG]) was subsequently confirmed as FA and specifically diagnosed FANCB, while FA was ruled out in the others. FANCB is part of the FA core complex, as FANCG, and it has been recently shown in mouse that Fancb deficiency impairs HSC function (Du et al., 2015). We isolated a CD34+CD38−CD45+CD117(cKIT)hi cell population in normal human FL, which presents typical functional and molecular features of HSCs (L.M. et al., unpublished data). We therefore used this combination of markers to sort HSCs from the FA FL and from two normal FL of matched stages of development (13 and 17.7 WG). As shown in Figure 5A, the CD34+CD38−CD45+CD117hi population is 55-fold less frequent in FA FL than in control FL (0.0017 versus 0.093%). In vitro LTC-IC assays revealed a 3.9-fold lower frequency of LTC-IC in the CD34+CD38−CD45+CD117hi HSC-enriched population sorted from the FA FL as compared with the same population sorted from the two control FLs (Figure 5B). In addition, we were unable to amplify FA CD34+CD38−CD45+CD117hi LTC-ICs when reseeded on fresh MS5 (extended LTC-IC), while CD34+CD38−CD45+CD117hi LTC-ICs issued from a normal FL of 15 WG and cultured in the same conditions could be amplified for more than 6 months (from 102 HCs to 1014, L.M. et al., unpublished data). These data demonstrate that a hematopoietic deficit is also already present during embryonic hematopoietic development in a human FA fetus.
Figure 5.
Important Frequency and LTC-IC Defect in the CD34+CD38−CD45+CD117hi HSC-Enriched Population of a Human FA 13 WG FL
(A) Comparison of the frequency of CD34+CD38−CD45+CD117hi HSC-enriched population from an FA (FANCB) FL or from an age-matched control FL (13 WG).
(B) Comparison of LTC-IC frequency of CD34+CD38−CD45+CD117hi cells sorted from an FA (FANCB) FL (13 WG) and from two age-matched control FL (13 and 17.7 WG).
Error bars correspond to standard deviation. Black bar, control FL; white bar, FA FL.
Similar Transcriptome Profiles of E12.5 Fancg−/− and Human FA FL HSCs and Identification of Compensatory Transcriptomic Regulation in E14.5 Fancg−/− FL
To identify signaling pathways or gene regulatory networks deregulated in mouse Fancg−/− and in human FA HSCs, we performed gene expression analysis and compared Fancg−/− and WT HSCs sorted from mouse FL at E12.5 and E14.5, but also FA and normal HSCs sorted from human FL.
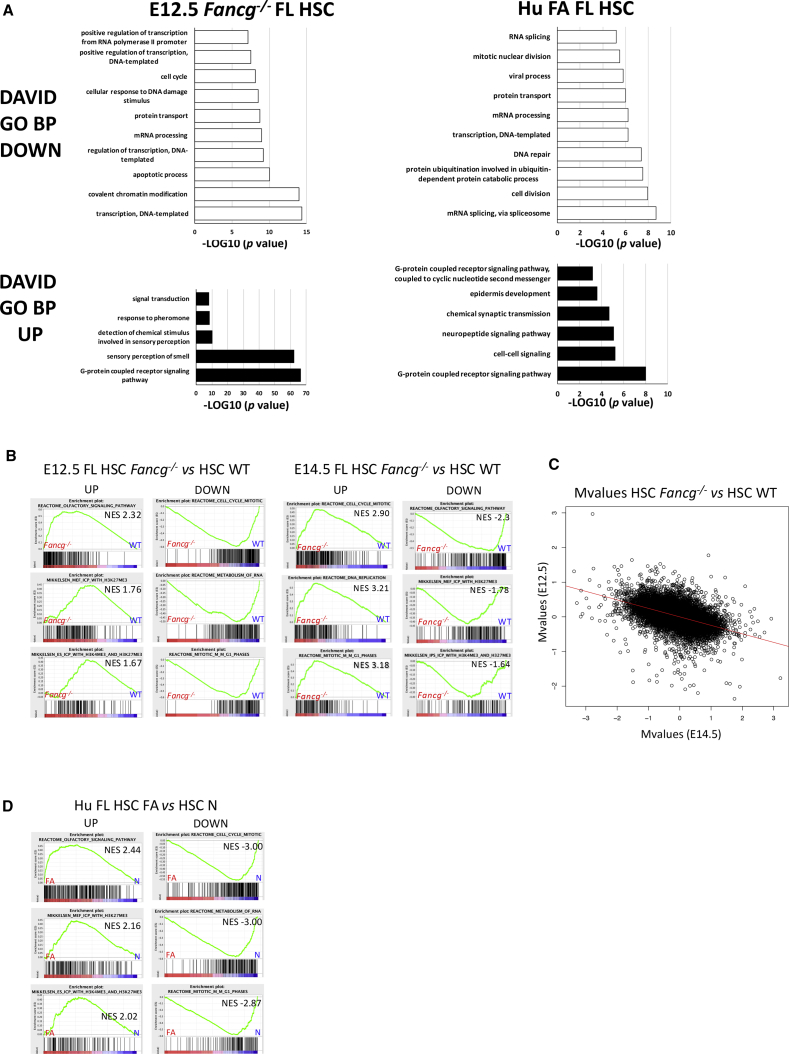
Mouse FL LSA HSC-enriched populations were sorted in three independent experiments. A total of 11,071 genes were found to be differentially expressed between Fancg−/− and WT HSCs in E12.5 FL (one-way ANOVA, p < 0.05), with 4,033 upregulated and 2,024 downregulated in Fancg−/− (fold change [FC] >1.5 or < −1.5). Gene ontology (GO) analysis indicated that the main biological processes (BP) downregulated in Fancg−/− FL HSCs at E12.5 were involved in transcription process, cell cycle, covalent chromatin modification, and DNA repair (Figure 6A and Table S1). Strikingly, for upregulated genes BP were associated with sensory perception of smell and GPCR signaling pathway (Figure 6A and Table S1). At E14.5, only 561 genes were differentially expressed (p < 0.05), with 166 upregulated and 82 downregulated in Fancg−/− (FC >1.5 or < −1.5), indicating that at this stage differences in gene expression between Fancg−/− and WT HSCs are fading. No significant BP was found for downregulated genes, and for upregulated genes the main BPs were related to angiogenesis, cell migration, and hypoxia (Table S1). Gene set enrichment analysis (GSEA) confirmed that genes upregulated in E12.5 FL Fancg−/− HSCs were most enriched in the “reactome olfactory signaling pathway,” but also in H3K4 and H3K27 trimethylation (H3K4Me3 and H3K27Me3) pathways (Figure 6B), which allows discriminating genes that are expressed, repressed, or poised for expression (Mikkelsen et al., 2007). It also supported the fact that genes downregulated are most enriched in cell cycle and metabolism of RNA reactomes (Figure 6B). Interestingly, GSEA analysis of E14.5 FL Fancg−/− HSC transcriptome revealed the exact opposite situation, i.e., that Fancg−/− LSA upregulated genes are most enriched in cell cycle and RNA metabolism, while downregulated genes are most enriched in “reactome olfactory signaling pathway” and in both H3K4 and H3K27 trimethylation (H3K4Me3 and H3K27Me3) pathways, as if at E14.5 a compensatory transcriptome regulation is accompanying the phenotypical and functional compensation that we describe. An anticorrelation study between Fancg−/− and WT HSCs at E12.5 against E14.5 shows an inversion of FC (Pearson correlation coefficient between M values [or log2(FC)] between E12.5 and E14.5 is −0.6). This result supports this compensation at E14.5 (Figure 6C).
Figure 6.
Highly Conserved GO Biological Processes between E12.5 Fancg−/− Pl and FL and Human FA FL HSCs
(A) Enriched GO biological process (BP) terms for downregulated (white bars) and upregulated (black bars) genes in E12.5 Fancg−/− FL HSCs and in human FA FL HSCs. GO BPs determined using DAVID annotation tool are expressed as –log10 (p value). NES, normalized enrichment score.
(B) GSEA plots for up- and downregulated genes, specifically expressed in E12.5 and E14.5 Fancg−/− FL HSCs.
(C) Anticorrelation study. Plot of log2(FC) between Fancg−/− and WT HSCs at E12.5 against E14.5.
(D) GSEA plots for up- and downregulated genes, specifically expressed in human FA FL HSCs.
We also achieved human transcriptome on sorted CD34+CD38−CD45+CD117hi HSC-enriched population from the unique FA FL (13 WG), and from two normal FLs from fetuses at the same stage of development (i.e., second trimester, 13 and 17.7 WG). Transcriptome analysis showed that 5,806 genes were differentially expressed between FA and normal FL HSCs (one-way ANOVA, p < 0.05), with 3,156 genes upregulated and 1,631 downregulated in FA FL. GO analysis indicated that as for E12.5 Fancg−/− FL CSHs, the main BP downregulated were also involved in the transcription process, cell cycle, and cell division, while the GPCR signaling pathway was the most significant upregulated BP, GSEA supporting that upregulated genes were most enriched in the “reactome olfactory signaling pathway” (162 out of 297 genes) (Figures 6A and 6D; Table S1).
Regarding a short list of genes involved in cell-cycle control, we found a dysregulation of these genes in E12.5 FL Fancg−/− HSCs and FA FL HSCs, with downregulation of both positive (CDK1, CDK6, CDC7, CDKN3) and negative (P16, P19) regulators of cell cycle (Table S2). This dysregulation is no longer observed at E14.5, and only CDKN1A (p21) is significantly upregulated (Table S2). This upregulation is also observed in FA HSCs, as described by Ceccaldi et al. (2012) for whole FA fetal liver samples. p53 gene is also significantly downregulated in E12.5 Fancg−/− FL HSCs and in human FA FL HSCs (Table S2), which was confirmed by qRT-PCR (Figure S3B). Analyses of phosphorylated p53 expression by flow cytometry did not reveal any significant difference between Fancg−/− and WT E12.5 FL HCs or HSPCs (Figure S3C).
We also investigated transcriptional regulators and markers described as being important for HSC development and maintenance. As shown on Table S3, most of these genes were significantly and highly decreased in FL Fancg−/− HSCs at E12.5, while no significant change was detected at E14.5. Decrease of Hlf, Flt3, and Mll1 in E12.5 FL Fancg−/− HSCs was confirmed by qRT-PCR (Figure S3D). Decreased expression of transcriptional regulators and markers important for HSCs was also observed in human FA FL HSCs, although to a lesser extent (Table S3).
Altogether our analysis indicates that HSCs isolated from a second trimester human FANCB FL present a transcriptomic profile very close to E12.5 Fancg−/− FL HSCs.
Discussion
In the present paper, using a Fancg−/− mouse model of FA we show that HSC defect starts very early during embryogenesis, during the earliest steps of hematopoietic amplification in FL and Pl. We also show that an HSC defect is also observed in the FL of a human FA (FANCB) fetus.
The hypothesis that FA BMF could take its origin in utero was suggested by the study of Ceccaldi et al. (2012) and was corroborated by studies of fetal hematopoiesis in E14.5 Fancc−/− and Fancd2−/− FL (Kamimae-Lanning et al., 2013, Suzuki et al., 2017, Yoon et al., 2016). In the present work, we followed hematopoiesis in Fancg−/− mice using state-of-the-art experimental hematopoiesis assays including long-term engraftment experiments on highly purified mouse and human cells. In an original way compared with previous studies, we focused on a crucial developmental window that spans from the onset of hematopoiesis in the AGM region at E11.5 until late HSC amplification in E14.5 FL, and also studied the Pl, which has been shown to be an important reservoir of HSCs during mouse development (Gekas et al., 2010, Ottersbach and Dzierzak, 2010). We paid particular attention to E12.5 when amplification peaks, both in Pl and FL (Medvinsky et al., 2011). Our study demonstrates that HSC impairment starts both in FL and Pl early during mouse development at E11.5, and is still present at E12.5 in these organs. In vivo transplantations revealed important quantitative and qualitative defect of Fancg−/− E12.5 FL and Pl HSCs, and pointed out an exhaustion in their proliferative and self-renewal capacities, with a loss of hematopoietic lineage potential, especially myeloid. This applies also to Fancg−/− FL LSA HSCs at E14.5, but to a lesser extent. Between E11.5 and E14.5 in the FL and between E11.5 and E12.5 in the Pl, HSCs are rapidly cycling, and therefore under high DNA replication stress, which could explain their compromised function in the Fancg−/− context. Actually, although not significant, an elevation of γH2AX in the hematopoietic populations (especially HSCs) studied by flow cytometry was observed in E12.5 Fancg−/− FL, indicative of a replication stress at this stage. Nevertheless, no increase in apoptosis, nor cell-cycle change, could be observed. Elevation of γH2AX was no longer detected in E14.5 FL.
All these observations indicate that during Fancg−/− embryonic development, HSC deficit is worse at E12.5 than at E14.5. Of note, external phenotypic signs of Fanconi disease such as smaller size, microphthalmia, or anophthalmia are also more pronounced early during development (E11.5–E12.5) and tend to attenuate around E14.5 (Figure 1). Indeed, as development progresses, phenotypic features of Fancg−/− embryos are more heterogeneous, some of them looking almost like WT embryos, including FL size (data not shown). Likewise, transcriptomic analysis also showed that in contrast to E12.5 FL, very few genes were significantly differentially expressed in E14.5 FL between Fancg−/− and WT HSCs. One possible explanation is that Fancg−/− embryos that are more deeply affected die around E14.5 (which could also explain the collapsed frequency of Fancg−/− mice at birth) and that a compensatory amplification takes place when proliferative stress decreases, making transcriptomic differences between Fancg−/− and WT HSCs less important, although functional defect remains. Actually, GSEA analysis of the transcriptome of E12.5 and E14.5 Fancg−/− and WT FL HSCs is in agreement with a compensatory molecular regulation at E14.5, since BP downregulated at E12.5 are upregulated at E14.5, and vice versa (Figure 6B).
Importantly, we also show that HSC function is also already impaired in utero in humans. During human development HSC amplification takes place in the FL, during the first and part of the second trimester. Our analysis of HSC-enriched CD34+CD38−CD45+CD117hi population from a 13 WG FANCB FL revealed both an important decrease in the frequency (55-fold) and functional activity (LTC-IC frequency) (4-fold) of this population compared with normal FL of matched stages of development.
Interestingly, transcriptomic analysis showed that differentially up- and downregulated genes in HSCs from human FA were found to be involved in the very same BP as in HSCs from E12.5 Fancg−/− FL, i.e., cell division, cell cycle, mRNA processing, and covalent chromatin modification for downregulated genes and GPCR pathway and olfactory signaling pathway for upregulated genes. This indicates that pathways dysregulated in fetal FA HSCs seem to be highly conserved between mouse and human during development.
Most of the genes involved in cell-cycle regulation are downregulated in Fancg−/− E12.5 FL, whether they are positive or negative regulators (Table S2). This dysregulation is also observed in human FA FL HSCs (Table S2). In line with the results described for Fancd2−/− E14.5 FL HSCs (Suzuki et al., 2017, Yoon et al., 2016), p53 gene is significantly downregulated both in Fancg−/− and FA fetal HSCs. In a genotoxic stress model, Milyavsky et al. (2010) showed that upon serial transplantations, HSCs submitted to γ-irradiation with disabled p53 accumulated DNA damage and exhibited decreased self-renewal capacity. In the same way, during the replicative stress that occurs in the FL during embryonic development, the decrease in p53 signaling pathway in HSCs could lead to the defect in self-renewal that we observed during secondary transplants of BM from primary recipients injected with Fancg−/− FL or Pl, even when BM cells injected presented a high CD45.2 chimerism.
GSEA analysis also uncovered epigenetic dysregulations of E12.5 Fancg−/− and human FA FL HSCs (Figures 6B and 6D), which could play an important role during the phases of high cell division and DNA replication that take place during embryogenesis. Epigenetic control is important to preserve normal homeostasis by regulating stem cell self-renewal versus differentiation in multiple tissue-specific stem cells (Beerman and Rossi, 2015). An aberrant epigenetic control of gene expression-maintaining HSC compartment has been recently hypothesized to explain defective FA hematopoiesis (Brosh et al., 2017), and our results during embryonic development are in line with this proposition.
GPCR and olfactory signaling pathways were the main BP covering upregulated genes in Fancg−/− and FA FL HSCs. Actually GPCRs contribute to hematopoietic regulation, these receptors being the direct link between HSCs and their microenvironment that hosts many hematopoietic regulators and neurotrophic factors (Ramkissoon et al., 2006). The importance of the sympathetic system has been underlined not only for maintaining homeostasis of adult BM (Mendelson and Frenette, 2014), but also in emergence of HSCs in mouse AGM (Fitch et al., 2012). A recent report also demonstrates an olfactory control of blood progenitor maintenance in Drosophila (Shim et al., 2013).
Our unique kinetics study reveals that a deep HSCs defect is already present very early during Fancg−/− mouse development, and that FA clinical and biological manifestations are more severe at early times of development (E11.5–E12.5), and are mitigated at E14.5. A deep HSC defect is also observed during human FA development, and human FA FL HSCs present a transcriptome profile similar to that of mouse E12.5 Fancg−/− FL HSCs. Our model sheds light on the interest of positioning the study of Fanc knockout hematopoiesis earlier than E14.5, during the steps of highest HSC/HC amplification in the mouse embryo. Indeed, despite the fact that FL failure seems to go through different mechanisms than adult BM failure, it might represent a good alternative model to study hematopoietic failure and certainly presents important potential clinical relevance.
Experimental Procedures
Fancg−/− Embryos
Fancg−/− mice (Koomen et al., 2002) were back-crossed on C57Bl6 background (n = 8). Fancg−/− and WT embryos were produced by natural mating of C57Bl6/J Fancg heterozygote mice. Vaginal plugs were checked in the morning, marking E0.5. The stages of the embryos used were confirmed by somite counting and/or morphological analysis. Embryos were dissected further to recover placenta and liver. Embryo tails were genotyped using the Phire PCR Kit (Thermo Scientific). All animal procedures reported in this paper were carried out in accordance with French Government regulations (Services Vétérinaires de la Santé et de la Production Animale, Ministère de l’Agriculture).
Human Fanconi Embryo
Control FL samples, as well as FL used for medical diagnosis of FA after termination of birth, were obtained from therapeutic abortions, with informed consent of the family. This was in compliance with French laws, and with a specific authorization of the French Biomedicine Agency for this project.
Cell Preparation
Single-cell suspensions of FL were prepared in DMEM medium (Fischer) with 10% fetal calf serum (FCS) (Eurobio) by repeated flushing through 23- to 26-gauge needles. Placentas were dissected and treated for 1 hr at 37°C with type I collagenase (Sigma) (0.125% in PBS with 10% FCS). After washing, cell suspension was filtered through 70-μm nylon mesh and resuspended in DMEM supplemented with 10% FCS.
Clonogenic Assays
Methylcellulose progenitor assays were performed as previously described (Petit-Cocault et al., 2007): 5 × 104 FL or 1 × 105 PL cells were plated in 1 mL of 0.8% methylcellulose in IMDM supplemented with 30% FCS, 1% BSA (Sigma), and 10−4 M β-mercaptoethanol (Sigma), in the presence of 2 U/mL erythropoietin, 10 ng/mL mouse interleukin-3, 50 ng/mL stem cell factor, and 20 ng/mL human thrombopoietin (Promocell). Samples were plated in duplicate 35-mm Petri dishes and the number of total colonies was scored after an incubation of 7 days at 37°C and 5% CO2.
Long-Term Culture-Initiating Cells Assay
LTC-IC assay was performed as previously described (Petit-Cocault et al., 2007). In brief, 3 × 103 MS5 stromal cells (Itoh et al., 1989) were seeded in LTC-IC medium in 96-well flat-bottom microplates and serial half-dilutions of test cells seeded over 24 hr later. Cultures were maintained at 37°C, 5% CO2 with weekly half-medium changes. Presence of cobblestone area forming colony (CAF-C) after 4 (mouse) or 5 (human) weeks of culture was quantified. Frequency of LTC-ICs was determined using L-Calc software (STEMCELL Technologies).
Analysis of Long-Term Repopulating Ability
Adult C57Bl6/Ly5.1 mice were lethally irradiated (9.5 Gy), and Fancg−/− or WT E12.5 total FL or Pl (C57Bl6/Ly5.2) were injected intravenously into the retro-orbital venous plexus. For E14.5 FL, lineagenegSCA-1+AA4.1+ (LSA) sorted cells were co-injected with 5 × 105 C57Bl6/Ly5.1 spleen cells for short-term radioprotection. Peripheral blood was collected at different times after injection and nucleated cells were incubated with CD45-LY5.1 (CD45.1) APC-conjugated and CD45-LY5.2 (CD45.2) PE-conjugated (Biolegend, BLE110714 and BLE109808), and analyzed by flow cytometry for the presence of donor-derived (LY5.2+) cells. The percentage of reconstitution was determined as follows: (%LY5.2/%LY5.1 + %LY5.2) × 100. A recipient mouse was considered positive when the percentage of reconstitution was above 10% for primary reconstitutions and 5% for secondary reconstitutions.
Multilineage Analysis and Secondary Transplantation
Primary recipient mice were sacrificed 20–24 weeks after primary transplant, and BM content was analyzed by flow cytometry after incubation with CD45.2 (PE), c-KIT (PE-Cy7), and APC-conjugated SCA-1, TER119, CD71, GR1, CD11B, CD41, CD61, CD19, B220, CD4, or CD8 (PE-Cy7) antibodies (Biolegend, BLE109808, BLE105814, and BLE108112; BLE116211, BLE113820, BLE108412, BLE101212, BLE133914, BLE104316, BLE115512, BLE103211, BLE100412, or BLE100722) (Petit-Cocault et al., 2007). The percentage of co-expression of CD45.2 and lineage markers was determined using the software FlowJo 10.1 (TreeStar). At the same time, 5 × 105 BM cells from primary mice transplanted with Fancg−/− and WT samples were injected into new C57Bl6/Ly5.1 irradiated mice for secondary reconstitution. Mice were bled 7–10 weeks after injection to determine the percentage of reconstitution.
Cytometry Analysis and Cell Sorting
Murine FL cells were depleted of lineage-positive cells using MACs Micro Beads (Miltenyi Biotec) with a mixture of TER119, GR1, B220, CD4. and CD8a monoclonal antibodies (Biolegend, BLE116202, BLE108402, BLE103202, BLE100402, and BLE100702). This lineageneg fraction was incubated with SCA-1 (APC) and AA4.1 (PE) antibodies (Biolegend, BLE108112 and BLE136504) and SCA-1+AA4.1+ cells (LSA) sorted on a FACSAria SORT cell sorter (BD Biosciences). Human FL cells were sorted after incubation with CD34 (APC), CD38 (FITC), CD45 (PE-Cy7) and CD117 (PE) antibodies (Biolegend, BLE343510, BLE303504, BLE304016 and BLE313204). DAPI (4′,6-diamidino-2-phenylindole) was used to exclude dead cells. For Pl, HSC CD34+c-KIThi population was sorted without lineage depletion (Biolegend, BLE343506 and BLE313212).
Cytometric analysis for FL and Pl cells used similar antibodies as for LTR studies and sorted cells, with an exclusion of dead cells by DAPI. Data were collected on LSRII (BD Biosciences) or MACSQuant analyzer 10 (Miltenyi), and analyzed using FlowJo 10.1 (TreeStar).
Cell Cycle and Apoptosis
Cell-cycle analysis was performed on E12.5 FL and Pl and E14.5 FL HC and HSC populations with Hoechst 33,342 staining protocol (Kim and Sederstrom, 2015). Cells were suspended at a concentration of 1 × 106/mL in pre-warmed medium, and Hoechst was added to a final concentration of 5 μg/mL. After incubation at 37°C for 45 min, Pyronin Y was added to a final concentration of 0.4 μg/mL and the cells were incubated at 37°C for 15 min. Cold PBS was added before incubation at +4°C with antibodies. Dead cells were excluded with 7-AAD and samples were run on a FACS-FORTESSA cytometer (BD Biosciences).
For apoptosis, immunostained cells were suspended in Annexin V buffer and stained with FITC-Annexin V (Biolegend, BLE640905) and DAPI, according to the manufacturer's guidelines. FACS analysis was performed on a FACSCanto analyzer (BD Biosciences).
DNA Damage-Induced Phosphorylation of H2AX (γH2AX) and Analysis of p53 Phosphorylation
Replicative stress induces DNA damage, which induces phosphorylation of the histone H2AX variant to γH2AX (Greve et al., 2012). E12.5 FL cells were incubated with CD45 or CD34, c-KIT, SCA-1, and AA4.1 antibodies, fixed with paraformaldehyde 4% at +4°C, washed and permeabilized in 0.25% Triton X-100, and incubated with monoclonal anti-phosphate Histone H2AX (Ser139) antibody (clone N1-431) conjugated to PerCP-Cy5.5 (BD Pharmingen, 564718). For p53 analysis, after permeabilization cells were incubated with monoclonal anti-phosphate p53 (Ser15) antibody (clone D4S1H) (Cell Signaling Technologies, 12571), then washed and incubated with Alexa Fluor 488 goat anti-rabbit immunoglobulin G (Life Technologies, A11034). Samples were analyzed on a FACSCanto analyzer (BD Bioscience).
RNA Preparation and Transcriptome Analysis
RNA of sorted cells was purified using an RNeasy Plus Micro Kit (Qiagen) according to manufacturer's protocol. In brief, cells were lysed in RLT Plus buffer supplemented with 10−4 M β-mercaptoethanol, and DNA was removed using a gDNA Eliminator spin column. RNA was purified using an RNeasy column, eluted in RNase-free water, and stored at −80°C. RNA was used for transcriptomic assay or real-time qRT-PCR assay (Supplemental Experimental Procedures). RNA concentration and integrity were evaluated with the Agilent Bioanalyzer 2100. Affymetrix Mouse Gene MTA1.0 and Hu-Gene 2.0 ST arrays were respectively used for mouse and human samples. Supervised analyses were performed to identify genes specifically up- or downregulated in Fancg−/− or FA HSCs, and lists of differentially expressed genes were analyzed for GO with DAVID (database used for annotation, visualization, and integrated discovery) (http://david.abcc.ncifcrf.gov), KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway, and reactome analyses. The global expression profile was also analyzed with GSEA with the functional (curated) dataset present in the Molecular Signature Database of the Broad Institute (www.broadinstitute.org/gsea) (Subramanian et al., 2005). An anticorrelation study was performed using R. M values (or log2(FC) between Fancg−/− and WT) of E14.5 was plotted against M values of E12.5, and Pearson correlation coefficient calculated.
Statistical Analysis
Statistical analysis was performed using the Mann-Whitney, Wilcoxon, and χ2 tests. p < 0.05 was considered significant. Data were expressed as mean or median ± SD.
Author Contributions
C.D. designed and performed experiments, and collected and analyzed data. L.M. performed experiments, collected and analyzed data. A.R., L.P., and S.S. performed experiments and collected data. D.C. performed cell sorting and set up cell-cycle analysis. S.J. performed the microarray gene expression experiment, and M.S., P.d.l.G., N.R., and F. Guidez performed analysis. F. Guimiot and J.S. provided human study material. M.P. and A.R. took care of the Fancg−/− core breeding and back cross. M.S. and J.S. conceived the project. M.S. designed the study, performed data analysis, and provided financial support. M.S., C.D., and L.M. wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
We thank Christine Chomienne and Michele Goodhardt for critical reading of the manuscript and helpful discussions. Cell sorting by Hélène Fohrer-Ting at the cytometry platform of the Centre de Recherches des Cordeliers (Paris), and support by the animal core facilities of the Institut Universitaire d’Hématologie of the Saint-Louis Hospital and of the Institut de Biologie Paris Seine are gratefully acknowledged. This work was supported by INSERM (Institut National de la Santé et de la Recherche Médicale) and CNRS (Centre National de Recherche Scientifique) and by grants from INCA (Institut du Cancer), La Ligue contre le Cancer (Comité Ile-de-France), and AFMF (Association Française de la maladie de Fanconi). C.D. was a supported by fellowships from INSERM/INCA, APPEL (Association Philantropique des Parents des Enfants Leucémiques), and 111 des Arts.
Published: October 25, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, three figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.10.001.
Accession Numbers
The GEO accession number for arrays reported in this paper is GEO: GSE120169.
Supplemental Information
Related to Figure 6 and the transcriptome analysis section in the results, reports all the GO processes and genes associated (DAVID, KEGG, and reactome analysis) in genes in Fancg−/− E12.5 and E14.5 FL and human FA FL HSCs.
References
- Ayas M., Saber W., Davies S.M., Harris R.E., Hale G.A., Socie G., LeRademacher J., Thakar M., Deeg H.J., Al-Seraihy A. Allogeneic hematopoietic cell transplantation for Fanconi anemia in patients with pretransplantation cytogenetic abnormalities, myelodysplastic syndrome, or acute leukemia. J. Clin. Oncol. 2013;31:1669–1676. doi: 10.1200/JCO.2012.45.9719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroca V., Mouthon M.A., Lewandowski D., Brunet de la Grange P., Gauthier L.R., Pflumio F., Boussin F.D., Arwert F., Riou L., Allemand I. Impaired functionality and homing of Fancg-deficient hematopoietic stem cells. Hum. Mol. Genet. 2012;21:121–135. doi: 10.1093/hmg/ddr447. [DOI] [PubMed] [Google Scholar]
- Beerman I., Rossi D.J. Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell. 2015;16:613–625. doi: 10.1016/j.stem.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluteau D., Masliah-Planchon J., Clairmont C., Rousseau A., Ceccaldi R., Dubois d'Enghien C., Bluteau O., Cuccuini W., Gachet S., Peffault de Latour R. Biallelic inactivation of REV7 is associated with Fanconi anemia. J. Clin. Invest. 2016;126:3580–3584. doi: 10.1172/JCI88010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R.M., Jr., Bellani M., Liu Y., Seidman M.M. Fanconi anemia: a DNA repair disorder characterized by accelerated decline of the hematopoietic stem cell compartment and other features of aging. Ageing Res. Rev. 2017;33:67–75. doi: 10.1016/j.arr.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R., Parmar K., Mouly E., Delord M., Kim J.M., Regairaz M., Pla M., Vasquez N., Zhang Q.S., Pondarre C. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 2012;11:36–49. doi: 10.1016/j.stem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans A.J., West S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokal I., Vulliamy T. Inherited aplastic anaemias/bone marrow failure syndromes. Blood Rev. 2008;22:141–153. doi: 10.1016/j.blre.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Dong H., Nebert D.W., Bruford E.A., Thompson D.C., Joenje H., Vasiliou V. Update of the human and mouse Fanconi anemia genes. Hum. Genomics. 2015;9:32. doi: 10.1186/s40246-015-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Amarachintha S., Erden O., Wilson A., Meetei A.R., Andreassen P.R., Namekawa S.H., Pang Q. Fancb deficiency impairs hematopoietic stem cell function. Sci. Rep. 2015;5:18127. doi: 10.1038/srep18127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitriu B., Young N.S. Damage control and its costs: BM failure in Fanconi anemia stems from overactive p53/p21. Cell Stem Cell. 2012;11:7–8. doi: 10.1016/j.stem.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E., Speck N.A. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat. Immunol. 2008;9:129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema H., Nakauchi H. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood. 2000;95:2284–2288. [PubMed] [Google Scholar]
- Faivre L., Guardiola P., Lewis C., Dokal I., Ebell W., Zatterale A., Altay C., Poole J., Stones D., Kwee M.L. Association of complementation group and mutation type with clinical outcome in fanconi anemia. European Fanconi Anemia Research Group. Blood. 2000;96:4064–4070. [PubMed] [Google Scholar]
- Fitch S.R., Kimber G.M., Wilson N.K., Parker A., Mirshekar-Syahkal B., Gottgens B., Medvinsky A., Dzierzak E., Ottersbach K. Signaling from the sympathetic nervous system regulates hematopoietic stem cell emergence during embryogenesis. Cell Stem Cell. 2012;11:554–566. doi: 10.1016/j.stem.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea J.I., Patel K.J. Why does the bone marrow fail in Fanconi anemia? Blood. 2014;123:26–34. doi: 10.1182/blood-2013-09-427740. [DOI] [PubMed] [Google Scholar]
- Gekas C., Dieterlen-Lievre F., Orkin S.H., Mikkola H.K. The placenta is a niche for hematopoietic stem cells. Dev. Cell. 2005;8:365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Gekas C., Rhodes K.E., Van Handel B., Chhabra A., Ueno M., Mikkola H.K. Hematopoietic stem cell development in the placenta. Int. J. Dev. Biol. 2010;54:1089–1098. doi: 10.1387/ijdb.103070cg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve B., Bolling T., Amler S., Rossler U., Gomolka M., Mayer C., Popanda O., Dreffke K., Rickinger A., Fritz E. Evaluation of different biomarkers to predict individual radiosensitivity in an inter-laboratory comparison—lessons for future studies. PLoS One. 2012;7:e47185. doi: 10.1371/journal.pone.0047185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays L.E. A mismatched life in the stem cell pool. Blood. 2012;120:3164–3165. doi: 10.1182/blood-2012-07-441220. [DOI] [PubMed] [Google Scholar]
- Itoh K., Tezuka H., Sakoda H., Konno M., Nagata K., Uchiyama T., Uchino H., Mori K.J. Reproducible establishment of hemopoietic supportive stromal cell lines from murine bone marrow. Exp. Hematol. 1989;17:145–153. [PubMed] [Google Scholar]
- Jordan C.T., Astle C.M., Zawadzki J., Mackarehtschian K., Lemischka I.R., Harrison D.E. Long-term repopulating abilities of enriched fetal liver stem cells measured by competitive repopulation. Exp. Hematol. 1995;23:1011–1015. [PubMed] [Google Scholar]
- Kamimae-Lanning A.N., Goloviznina N.A., Kurre P. Fetal origins of hematopoietic failure in a murine model of Fanconi anemia. Blood. 2013;121:2008–2012. doi: 10.1182/blood-2012-06-439679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly P.F., Radtke S., von Kalle C., Balcik B., Bohn K., Mueller R., Schuesler T., Haren M., Reeves L., Cancelas J.A. Stem cell collection and gene transfer in Fanconi anemia. Mol. Ther. 2007;15:211–219. doi: 10.1038/sj.mt.6300033. [DOI] [PubMed] [Google Scholar]
- Kim K.H., Sederstrom J.M. Assaying cell cycle status using flow cytometry. Curr. Protoc. Mol. Biol. 2015;111:28.6.1–28.6.11. doi: 10.1002/0471142727.mb2806s111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Yang K., Dejsuphong D., D'Andrea A.D. Regulation of Rev1 by the Fanconi anemia core complex. Nat. Struct. Mol. Biol. 2012;19:164–170. doi: 10.1038/nsmb.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knies K., Inano S., Ramirez M.J., Ishiai M., Surralles J., Takata M., Schindler D. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J. Clin. Invest. 2017;127:3013–3027. doi: 10.1172/JCI92069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koomen M., Cheng N.C., van de Vrugt H.J., Godthelp B.C., van der Valk M.A., Oostra A.B., Zdzienicka M.Z., Joenje H., Arwert F. Reduced fertility and hypersensitivity to mitomycin C characterize Fancg/Xrcc9 null mice. Hum. Mol. Genet. 2002;11:273–281. doi: 10.1093/hmg/11.3.273. [DOI] [PubMed] [Google Scholar]
- Kottemann M.C., Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaravelu P., Hook L., Morrison A.M., Ure J., Zhao S., Zuyev S., Ansell J., Medvinsky A. Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development. 2002;129:4891–4899. doi: 10.1242/dev.129.21.4891. [DOI] [PubMed] [Google Scholar]
- Kupfer G.M. Fanconi anemia: a signal transduction and DNA repair pathway. Yale J. Biol. Med. 2013;86:491–497. [PMC free article] [PubMed] [Google Scholar]
- Kutler D.I., Singh B., Satagopan J., Batish S.D., Berwick M., Giampietro P.F., Hanenberg H., Auerbach A.D. A 20-year perspective on the International Fanconi anemia registry (IFAR) Blood. 2003;101:1249–1256. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- Medvinsky A., Rybtsov S., Taoudi S. Embryonic origin of the adult hematopoietic system: advances and questions. Development. 2011;138:1017–1031. doi: 10.1242/dev.040998. [DOI] [PubMed] [Google Scholar]
- Mendelson A., Frenette P.S. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 2014;20:833–846. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola H.K., Orkin S.H. The journey of developing hematopoietic stem cells. Development. 2006;133:3733–3744. doi: 10.1242/dev.02568. [DOI] [PubMed] [Google Scholar]
- Milyavsky M., Gan O.I., Trottier M., Komosa M., Tabach O., Notta F., Lechman E., Hermans K.G., Eppert K., Konovalova Z. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell. 2010;7:186–197. doi: 10.1016/j.stem.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Hemmati H.D., Wandycz A.M., Weissman I.L. The purification and characterization of fetal liver hematopoietic stem cells. Proc. Natl. Acad. Sci. U S A. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller L.U., Williams D.A. Finding the needle in the hay stack: hematopoietic stem cells in Fanconi anemia. Mutat. Res. 2009;668:141–149. doi: 10.1016/j.mrfmmm.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalepa G., Clapp D.W. Fanconi anemia and the cell cycle: new perspectives on aneuploidy. F1000Prime Rep. 2014;6:23. doi: 10.12703/P6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottersbach K., Dzierzak E. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev. Cell. 2005;8:377–387. doi: 10.1016/j.devcel.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Ottersbach K., Dzierzak E. The placenta as a haematopoietic organ. Int. J. Dev. Biol. 2010;54:1099–1106. doi: 10.1387/ijdb.093057ko. [DOI] [PubMed] [Google Scholar]
- Park J.Y., Virts E.L., Jankowska A., Wiek C., Othman M., Chakraborty S.C., Vance G.H., Alkuraya F.S., Hanenberg H., Andreassen P.R. Complementation of hypersensitivity to DNA interstrand crosslinking agents demonstrates that XRCC2 is a Fanconi anaemia gene. J. Med. Genet. 2016;53:672–680. doi: 10.1136/jmedgenet-2016-103847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar K., D'Andrea A., Niedernhofer L.J. Mouse models of Fanconi anemia. Mutat. Res. 2009;668:133–140. doi: 10.1016/j.mrfmmm.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peffault de Latour R., Porcher R., Dalle J.H., Aljurf M., Korthof E.T., Svahn J., Willemze R., Barrenetxea C., Mialou V., Soulier J. Allogeneic hematopoietic stem cell transplantation in Fanconi anemia: the European group for blood and marrow transplantation experience. Blood. 2013;122:4279–4286. doi: 10.1182/blood-2013-01-479733. [DOI] [PubMed] [Google Scholar]
- Petit-Cocault L., Volle-Challier C., Fleury M., Peault B., Souyri M. Dual role of Mpl receptor during the establishment of definitive hematopoiesis. Development. 2007;134:3031–3040. doi: 10.1242/dev.001818. [DOI] [PubMed] [Google Scholar]
- Quentin S., Cuccuini W., Ceccaldi R., Nibourel O., Pondarre C., Pagès M.P., Vasquez N., Dubois d'Enghien C., Larghero J., Peffault de Latour R. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood. 2011;117:e161–e170. doi: 10.1182/blood-2010-09-308726. [DOI] [PubMed] [Google Scholar]
- Ramkissoon S.H., Patel H.J., Taborga M., Rameshwar P. G protein-coupled receptors in haematopoietic disruption. Expert Opin. Biol. Ther. 2006;6:109–120. doi: 10.1517/14712598.6.2.109. [DOI] [PubMed] [Google Scholar]
- Sanchez M.J., Holmes A., Miles C., Dzierzak E. Characterization of the first definitive hematopoietic stem cells in the AGM and liver of the mouse embryo. Immunity. 1996;5:513–525. doi: 10.1016/s1074-7613(00)80267-8. [DOI] [PubMed] [Google Scholar]
- Shim J., Mukherjee T., Mondal B.C., Liu T., Young G.C., Wijewarnasuriya D.P., Banerjee U. Olfactory control of blood progenitor maintenance. Cell. 2013;155:1141–1153. doi: 10.1016/j.cell.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier J. Fanconi anemia. Hematology Am. Soc. Hematol. Educ. Program. 2011;2011:492–497. doi: 10.1182/asheducation-2011.1.492. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Racine R.R., Manalo N.A., Cantor S.B., Raffel G.D. Impairment of fetal hematopoietic stem cell function in the absence of Fancd2. Exp. Hematol. 2017;48:79–86. doi: 10.1016/j.exphem.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavian M., Peault B. Embryonic development of the human hematopoietic system. Int. J. Dev. Biol. 2005;49:243–250. doi: 10.1387/ijdb.041957mt. [DOI] [PubMed] [Google Scholar]
- Tischkowitz M., Dokal I. Fanconi anaemia and leukaemia - clinical and molecular aspects. Br. J. Haematol. 2004;126:176–191. doi: 10.1111/j.1365-2141.2004.05023.x. [DOI] [PubMed] [Google Scholar]
- Tulpule A., Lensch M.W., Miller J.D., Austin K., D'Andrea A., Schlaeger T.M., Shimamura A., Daley G.Q. Knockdown of Fanconi anemia genes in human embryonic stem cells reveals early developmental defects in the hematopoietic lineage. Blood. 2010;115:3453–3462. doi: 10.1182/blood-2009-10-246694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter D., Lier A., Geiselhart A., Thalheimer F.B., Huntscha S., Sobotta M.C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520:549–552. doi: 10.1038/nature14131. [DOI] [PubMed] [Google Scholar]
- Yoon Y.M., Storm K.J., Kamimae-Lanning A.N., Goloviznina N.A., Kurre P. Endogenous DNA damage leads to p53-independent deficits in replicative fitness in fetal murine Fancd2-/- hematopoietic stem and progenitor cells. Stem Cell Reports. 2016;7:840–853. doi: 10.1016/j.stemcr.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.S., Benedetti E., Deater M., Schubert K., Major A., Pelz C., Impey S., Marquez-Loza L., Rathbun R.K., Kato S. Oxymetholone therapy of fanconi anemia suppresses osteopontin transcription and induces hematopoietic stem cell cycling. Stem Cell Reports. 2015;4:90–102. doi: 10.1016/j.stemcr.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kozono D.E., O'Connor K.W., Vidal-Cardenas S., Rousseau A., Hamilton A., Moreau L., Gaudiano E.F., Greenberger J., Bagby G. TGF-β inhibition rescues hematopoietic stem cell defects and bone marrow failure in Fanconi anemia. Cell Stem Cell. 2016;18:668–681. doi: 10.1016/j.stem.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 6 and the transcriptome analysis section in the results, reports all the GO processes and genes associated (DAVID, KEGG, and reactome analysis) in genes in Fancg−/− E12.5 and E14.5 FL and human FA FL HSCs.