Lysosomes play a major role in cellular metabolism and human health. Lysosomes are main catabolic organelles of the cell and play a central role in cellular processes including autophagy, plasma membrane repair, cellular clearance and cell signaling (1). Lysosomal dysfunction has been associated with several diseases including degenerative disorders of the brain and muscular system (1). Among other regulators of lysosome function, transcription factor EB (TFEB) stands out the most as it regulates both lysosome biogenesis and functions such as autophagy, lysosomal exocytosis, macromolecule degradation and mTOR signaling (2) (Figure 1).
Figure 1.
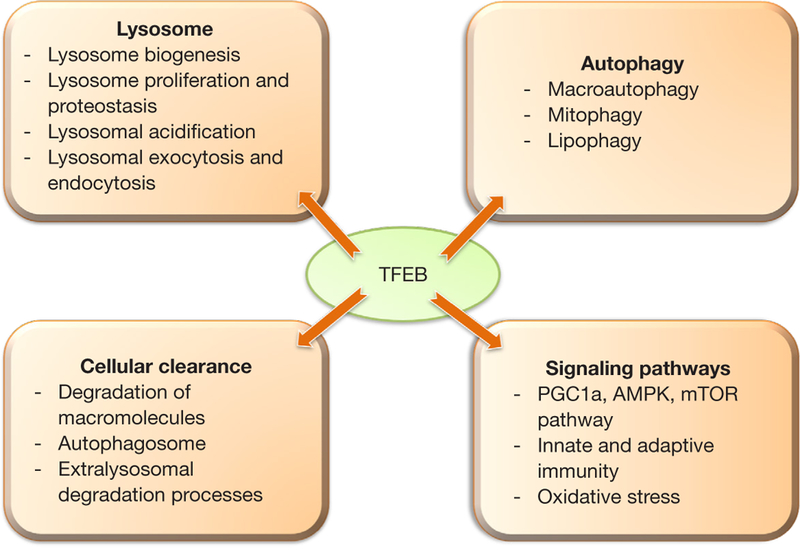
Various roles of transcription factor EB (TFEB) in cellular processes
TFEB induces the transcription of various lysosomal genes by binding to a 10-base E-box like sequence at the promoter region termed as coordinated lysosomal expression and regulation (CLEAR) (2). TFEB activity is regulated at the various levels including post-translational modifications and protein-protein interactions. TFEB is largely cytosolic and inactive in resting cells under nutrient-rich conditions (3). However, during starvation and in stress conditions such as caused by ethanol treatment or lysosomal dysfunction, TFEB translocates to the nucleus to regulate the transcription of its target genes (4).
Both TFEB sub-cellular localization and activity are regulated by the phosphorylation status of two serine residues (Ser142 and Ser211) (5). Phosphorylated TFEB remains inactive in the cytoplasm, while dephosphorylated form translocates to the nucleus to regulate gene expression of its target genes. In nutrient-rich conditions, kinases such as mechanistic target of rapamycin complex 1 (mTORC1) and extracellular signal-regulated kinase 2 are known to phosphorylate TFEB (6). Surprisingly, mTORC1 mediated phosphorylation of TFEB takes place at the lysosomal membrane (6).
Starvation studies have revealed that TFEB also controls its own transcription by binding to CLEAR motifs (3). PGC1α, a master activator of mitochondrial proliferation, regulates TFEB expression and PGC1α is also a direct target of TFEB (3). PGC1α regulates TFEB transcription via PPARα/ retinoid X receptor α (RXRα) heterodimer co-binding to the promoter region of TFEB and this phenomenon can be mimicked by the PPARα agonist, gemfibrozil (7). Other transcriptional factors that regulate TFEB transcription are the fed-state sensing nuclear receptor farnesoid X receptor (FXR) and the fasting transcriptional activator cAMP response element-binding protein (CREB) (7). Among miRNAs, miRNA-128 was shown to regulate TFEB directly. Overexpression of miR-128 caused a decrease in TFEB expression and consequently down regulation in CLEAR lysosomal genes, suggesting optimal levels of TFEB are required for lysosomal function (2).
TFEB has been shown to modulate autophagy, lipophagy and mitophagy (8). Overexpression of TFEB enhances the degradation of autophagy substrates and also shown to fasten the clearance of lipid drops and damaged mitochondria (3). TFEB also promotes lysosomal exocytosis for the cellular clearance (9). Since TFEB regulates processes like lysosomal biogenesis, autophagy and lysosomal exocytosis, therefore, it is able to modulate various cellular degradative pathways.
The liver specific loss and gain of functional studies revealed a crucial role of TFEB in lipid metabolism (3). TFEB governs lipid catabolism by regulating peroxisome proliferator-activated receptor-γ coactivator 1α (Pgc1α). Liver specific TFEB knockout mice showed impaired lipid metabolism and imbalance in the metabolic pathways in an obese mouse model (3). Mice with liver specific TFEB overexpression exhibited reduced symptoms of obesity and metabolic disorders in diet induced and genetic models of obesity (3). TFEB is also known to modulate immune responses. Depletion of TFEB and TFE3 in macrophages resulted in a decrease in the production of several pro-inflammatory cytokines such as TNF-α, IL-1β and RANTES upon LPS administration (10).
Impaired TFEB signaling plays a causal role in the pathogenesis of numerous degenerative storage diseases (2). Since TFEB is involved in various cellular clearance pathways, it is an attractive therapeutic target for various human diseases including those associated with cellular storage disorders (neurodegenerative diseases such as Parkinson’s and Alzheimer’s). Preclinical studies in mice to improve lysosomal function or biogenesis based on restoration of TFEB expression have been carried out for several diseases (1).
In a recent issue of Gastroenterology, Chao et al. demonstrated the role of TFEB in lysosome biogenesis and autophagy in a chronic ethanol plus single acute binge mouse model of alcoholic liver disease (11). Liver tissues from mice on Lieber DeCarli ethanol diet for 10 days plus a single acute binge (termed as NIAAA model) showed reduced levels of total and nuclear TFEB compared to controls. Further, hepatocytes of ethanol fed mice exhibited reduced autophagy and lysosome biogenesis and increased mTOR activation (11). Autophagy flux was studied using GFP-LC3 transgenic mice in the NIAAA binge model in the presence or absence of leupeptin, a lysosomal inhibitor. The number of GFP-LC3 puncta were found to be increased in the hepatocytes of ethanol feeding plus binge mouse group and leupeptin treated mice showed further increases in these puncta, suggesting that blocking lysosomal degradation blocks basal autophagy in the liver. Ethanol treatment showed an increase in the endogenous protein levels of LC3-II, which were further augmented in the presence of leupeptin. However, Chao et al. found that ethanol alone did not change the hepatic protein levels of p62, which were increased in leupeptin alone or leupeptin plus ethanol group. Authors postulated that enhanced p62 accumulation in leupeptin plus ethanol treated mice could be due to either impairment in the lysosomal/autolysosomal functions or reduced lysosome numbers (11). Consistent with the hypothesis, ethanol treatment caused a decrease in the lysosome numbers in hepatocytes as quantified by LAMP1 positive vesicles/puncta. An increase in the percentage of GFP-LC3 green puncta in the livers was found in the co-localization studies of GFP-LC3 puncta with Lamp1. These results suggest an insufficient number of lysosomes to fuse with autophagosomes. Because TFEB is a master regulator of lysosomal biogenesis (2), the possible involvement of TFEB in decreased lysosome numbers was examined. Chao et al. revealed that ethanol treatment not only caused a reduction in total and nuclear TFEB proteins but also decreased in its transcriptional activity as demonstrated by a decrease in the mRNA levels of its target genes (TEFB, ATP6VID and PGC1a). Nuclear TFEB staining was also decreased in the livers of alcoholic patients.
To determine the mechanism by which ethanol is affecting TFEB, they checked mTOR levels, a regulator of TFEB. mTOR levels were increased in the livers and in the lysosomes with a concomitant decrease in LAMP1 and TFEB after ethanol treatment. Increased mTORC1 activity was further confirmed by the phosphorylation status of its substrate proteins, S6 and 4EBP1, and increased phosphorylation of these proteins was observed in mice after ethanol treatment. The role of mTOR was further affirmed in in-vivo studies where Torin1, an mTORC1 inhibitor, improved ethanol-induced insufficient autophagy as evidenced by an increase in the numbers of hepatic GFP-LC3 puncta and endogenous LC3. Interestingly, Torin treatment only partially recovered ethanol-induced decrease in hepatic TFEB levels and its target genes (ATP6V1D, ATP6V1H and PGC1α), suggesting a potential role of other kinases in TFEB regulation. Attenuation in ethanol-induced liver injury and steatosis was observed after Torin treatment whereas leupeptin treatment augmented the disease severity.
Chao et al. further determined the casual role of TFEB in ethanol-induced liver injury in hepatocyte-specific TFEB knockout (KO) mice. Surprisingly, ALT and triglyceride levels were found to be similar between hepatocyte-specific TFEB KO mice and their WT littermates after the NIAAA alcohol diet treatment. TFEB is a member of microphthalmia-associated transcription factor (MITF) family proteins, including MITF, TFEB, TFE3 and TFEC (6) . To rule out the role of other MITF family proteins, TFE3 expression was checked in hepatocyte-specific TFEB KO mice, which were found to be normal, implying that TFE3 could compensate the function of TFEB in hepatocytes. It is also plausible that TFEB in other liver cells might compensate for the loss of TFEB in hepatocytes. Role of TEF3 was also determined using total body KO mice and similar to hepatocyte-specific TFEB KO mice, these mice showed similar ALT and triglyceride levels compared to WT mice, suggesting complex role of MITF family proteins.
To overcome this problem, authors knocked down TFEB acutely in the mouse livers using an adenovirus (Ad) shRNA approach. Mice receiving Ad-shRNA TFEB showed higher ALT and severe steatosis and increased hepatic neutrophils infiltration than mice that received control viruses, revealing a role of TFEB in ALD. Liver-specific TFEB and TFE3 knockout (DKO) mice exhibited a decrease in PGC1α protein, increased serum ALT levels and increased hepatic infiltration of neutrophils compared to their matched WT littermates after ethanol diet. These results suggest a compensatory role of TFE3 in ALD. As a gain of function, TFEB was overexpressed in the liver using an adenoviral vector approach. TFEB overexpression significantly attenuated ethanol-induced liver injury and steatosis and hepatic infiltration of neutrophils compared to control vector treated mice. Also, ethanol-induced impairment in hepatic mitochondrial bioenergetics and State 3 respiration was found to be increased in TFEB overexpressing mice. Since TFEB regulates lipid catabolism via PGC1 α, Chao et al. checked the expression of genes involved in this pathway and found that ethanol- induced decrease in the hepatic mRNA levels of fatty acid beta-oxidation genes (CPT1α, ACOX1, MACD, LCAD, and PPARα) was significantly improved in TFEB overexpressing mice. Whereas mice with TFEB knockdown of showed a further decrease in these genes after ethanol treatment. Altogether, these results suggest a fundamental role TFEB in lysosomal biogenesis and mitochondria bioenergetics in this model of ALD (Figure 2). Therapeutic strategies to maintain optimal TFEB levels to mediate normal lysosomal biogenesis might be a new avenue for ameliorating alcoholic liver disease. Further studies in other models of alcoholic liver disease, chronic alcohol administration and alcoholic hepatitis in mice as well as in-depth analysis in human alcoholic liver disease and alcoholic hepatitis should help to clarify the role of TFEB and autophagy in different forms of alcohol-induced liver damage.
Figure 2.
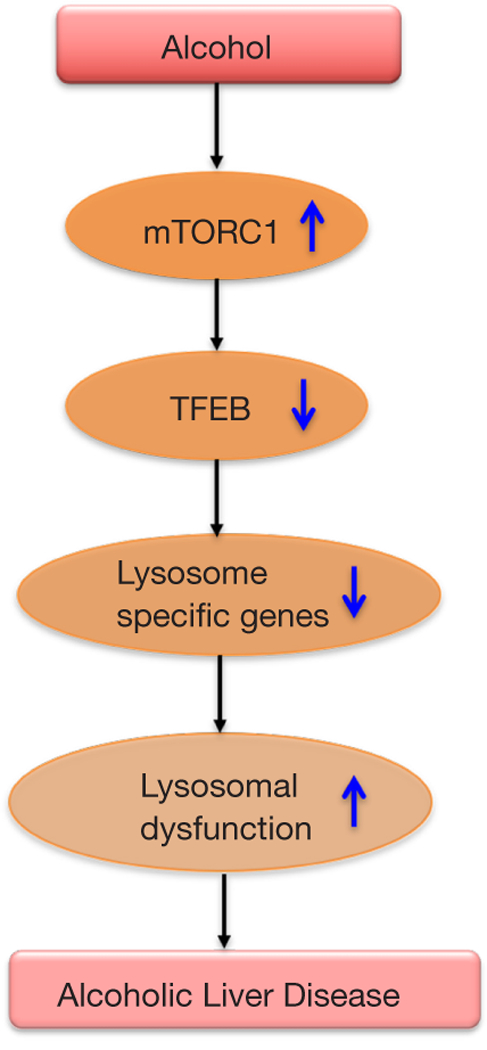
Impaired transcription factor EB (TFEB) signaling leads to alcoholic liver disease.
Acknowledgements
The authors thank Candice Dufour for expert assistance in preparation of this manuscript.
Funding: This work was supported by National Institutes of Health Grant R01 AA020744 (to G.S.).
Footnotes
Provenance: This is an invited Editorial commissioned by Guest Section Editor Kaiping Zhang (Academic Director, AME Publishing Company).
Comment on: Chao X, Wang S, Zhao K, et al. Impaired TFEB-Mediated Lysosome Biogenesis and Autophagy Promote Chronic Ethanol-Induced, Liver Injury and Steatosis in Mice. Gastroenterology 2018;155:865–79.e12.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Sardiello M Transcription factor EB: from master coordinator of lysosomal pathways to candidate therapeutic target in degenerative storage diseases. Ann N Y Acad Sci 2016;1371:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sardiello M, Palmieri M, di Ronza A, et al. A gene network regulating lysosomal biogenesis and function. Science 2009;325:473–7. [DOI] [PubMed] [Google Scholar]
- 3.Settembre C, De Cegli R, Mansueto G, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 2013;15:647–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomes PG, Trambly CS, Fox HS, et al. Acute and Chronic Ethanol Administration Differentially Modulate Hepatic Autophagy and Transcription Factor EB. Alcohol Clin Exp Res 2015;39:2354–63. [DOI] [PubMed] [Google Scholar]
- 5.Martina JA, Chen Y, Gucek M, et al. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012;8:903–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Napolitano G, Ballabio A. TFEB at a glance. J Cell Sci 2016;129:2475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh A, Jana M, Modi K, et al. Activation of peroxisome proliferator-activated receptor alpha induces lysosomal biogenesis in brain cells: implications for lysosomal storage disorders. J Biol Chem 2015;290:10309–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science 2011;332:1429–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medina DL, Fraldi A, Bouche V, et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev Cell 2011;21:421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastore N, Brady OA, Diab HI, et al. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy 2016;12:1240–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao X, Wang S, Zhao K, et al. Impaired TFEB-Mediated Lysosome Biogenesis and Autophagy Promote Chronic Ethanol-Induced Liver Injury and Steatosis in Mice. Gastroenterology 2018;155:865–79.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]


