Abstract
The presence of high expressing epithelial cell adhesion molecule (EpCAMhigh) circulating tumor cells (CTC) enumerated by CellSearch® in blood of cancer patients is strongly associated with poor prognosis. This raises the question about the presence and relation with clinical outcome of low EpCAM expressing CTC (EpCAMlow CTC). In the EU-FP7 CTC-Trap program, we investigated the presence of EpCAMhigh and EpCAMlow CTC using CellSearch, followed by microfiltration of the EpCAMhigh CTC depleted blood. Blood samples of 108 castration-resistant prostate cancer patients and 22 metastatic breast cancer patients were processed at six participating sites, using protocols and tools developed in the CTC-Trap program. Of the prostate cancer patients, 53% had ≥5 EpCAMhigh CTC and 28% had ≥5 EpCAMlow CTC. For breast cancer patients, 32% had ≥5 EpCAMhigh CTC and 36% had ≥5 EpCAMlow CTC. 70% of prostate cancer patients and 64% of breast cancer patients had in total ≥5 EpCAMhigh and/or EpCAMlow CTC, increasing the number of patients in whom CTC are detected. Castration-resistant prostate cancer patients with ≥5 EpCAMhigh CTC had shorter overall survival versus those with <5 EpCAMhigh CTC (p = 0.000). However, presence of EpCAMlow CTC had no relation with overall survival. This emphasizes the importance to demonstrate the relation with clinical outcome when presence of CTC identified with different technologies are reported, as different CTC subpopulations can have different relations with clinical outcome.
Keywords: circulating tumor cells (CTC), EpCAM, castrate-resistant prostate cancer (CRPC), metastatic breast cancer (mBC), epithelial-to-mesenchymal transition (EMT)
INTRODUCTION
The presence of circulating tumor cells (CTC) expressing the cell surface epithelial cell adhesion molecule (EpCAM) as well as intracellular cytokeratins (CK), are associated with poor outcome in patients with metastatic as well as non-metastatic disease [1–8]. In the CellSearch® system, CTC that show no or low expression of EpCAM are discarded during magnetic isolation and their information is lost. This raises the question how many of these EpCAM-negative or EpCAM low expressing CTC (hereinafter referred to as EpCAMlow CTC) are present and whether or not their presence is also associated with poor outcome. In the FP7 EU program CTC-Trap (https://www.utwente.nl/tnw/ctctrap/) this question was investigated by collecting the blood discarded by the CellSearch system after immunomagnetic enrichment of EpCAMhigh cells. This sample is passed through a microfilter and cells remaining on the filter were fluorescently labeled for EpCAMlow CTC scoring [9]. To validate this method for detection of epithelial EpCAMlow CTC, the procedure was tested at six different sites in the CTC-Trap consortium. Healthy donor blood spiked with cells of the prostate cancer cell line PC3 (1.0 × 104 EpCAM antigens, average size 17.7 μm) and the breast cancer cell line MDA-MB-231 (1.5 × 104 EpCAM antigens, average size 15.6 μm) were tested for recovery of EpCAMhigh and EpCAMlow cells. Subsequently, the presence of EpCAMhigh and EpCAMlow CTC was investigated in 108 CRPC patients and 22 metastatic breast and their presence was related to overall survival.
RESULTS
Validation of CTC detection protocols across sites
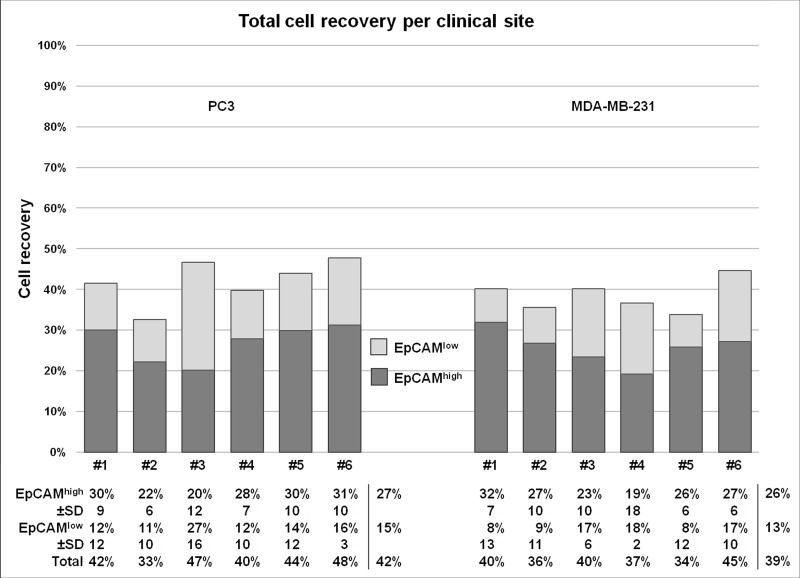
On average, 270 PC3 or MDA-MB-231 cells were spiked per tube in 36 7.5 mL blood samples of three healthy donors and sent to the six sites for processing according to the established protocols (Figure 1). The recovery of PC3 (median size 17.7 μm) and MDA-MB-231 cells (median size 15.6 μm) was determined using CellSearch (for EpCAMhigh cells) and on microsieves (for EpCAMlow cells), after filtration of the blood discarded by CellSearch. The distribution of the EpCAM density is illustrated in Supplementary Figure 1. The results are illustrated in Figure 2 and a detailed overview of cell recovery per site is provided in Supplementary Figure 2. For EpCAMhigh CTC detected by CellSearch, the recovery of PC3 cells varied between 11.9% and 40.6% (mean 26.9% ± 9 standard deviation (SD)) and for MDA-MB-231 between 2.7% and 39.0% (mean 25.7% ± 10 SD). For EpCAMlow tumor cells detected on the microsieves after filtration of the CellSearch discarded blood, the recovery of PC3 cells varied between 0.6% and 42.5% (mean 15.1% ± 11 SD) and for MDA-MB-231 between 0% and 29.3% (mean 12.8% ± 9 SD). In 18 7.5 mL blood samples obtained from three healthy donors, who donated together all 54 samples, no EpCAMhigh and no EpCAMlow cells were detected.
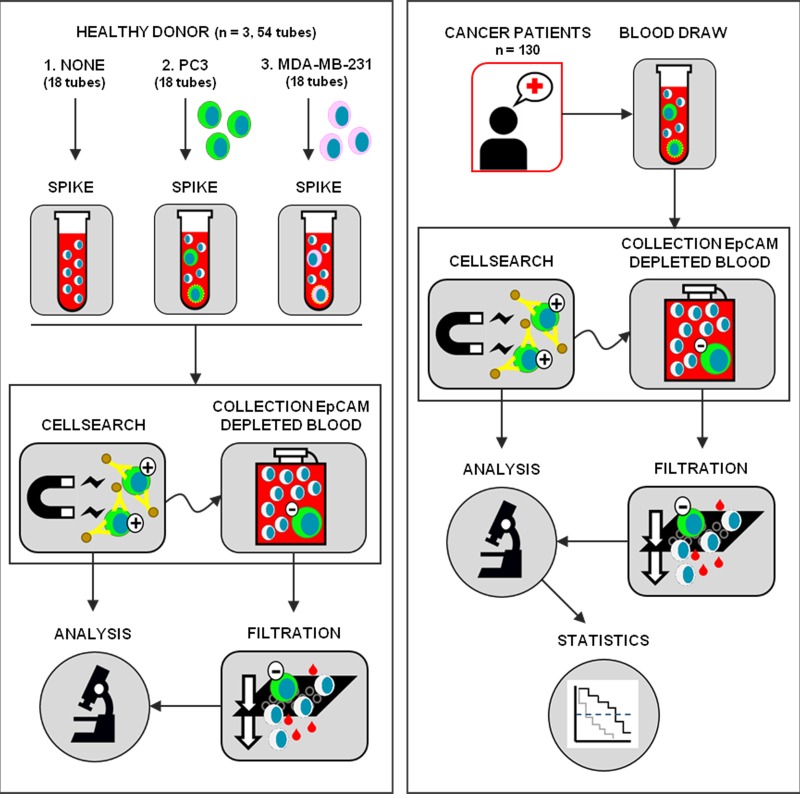
Figure 1. Schematic representation of the study.
Left panel: one site collected blood from three healthy donors and prepared three tubes for each of the six sites (Spike) to send for enumeration of EpCAMhigh cell line CTC (CellSearch) and EpCAMlow cell line CTC (Collection EpCAM Depleted Blood and Filtration): no cells were spiked in tube 1, tube 2 was spiked with on average 270 PC3 cells and tube 3 was spiked with on average 270 MDA-MB-231 cells. This process was repeated three times with three different healthy donors. Right panel: blood samples collected from 108 castrate-resistant prostate cancer patients and 22 metastatic breast cancer patients (Blood Draw) at each clinical site were processed for enumeration of EpCAMhigh CTC (CellSearch), followed by detection of EpCAMlow CTC (Collection EpCAM Depleted Blood and Filtration). CTC are detected by fluorescence microscopy and scored (Analysis) and the results are related to outcome (Statistics).
Figure 2. Recovery of PC3 and MDA-MB-231 cancer cells for each clinical site.
Recovery of on average 270 PC3 cells and MDA-MB-231 cells spiked in each blood sample, processed with the methods for EpCAMhigh and EpCAMlow CTC detection. The samples were processed with CellSearch to determine the recovery of EpCAMhigh CTC and the EpCAM depleted blood was filtered through 5 μm microsieves to determine the recovery of EpCAMlow cells. In total, per site (labeled #1–6) three samples per cell line and three negative controls were processed. The average recovery per three samples is displayed with its standard deviation, followed by an average recovery of all sites per method and cell line.
EpCAMhigh and EpCAMlow CTC in blood from metastatic breast and prostate cancer patients
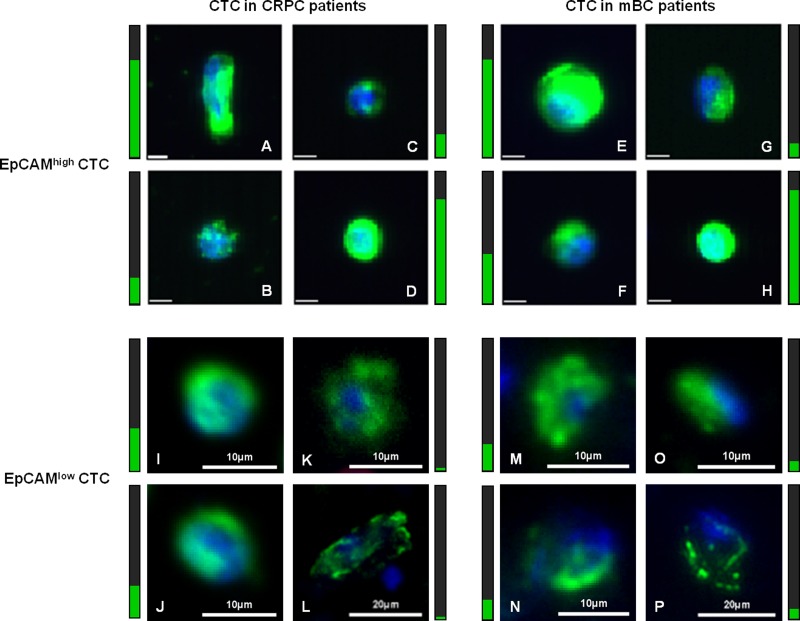
EpCAMhigh and EpCAMlow CTC were enumerated in 7.5 mL blood obtained from 108 CRPC and 22 mBC patients. A detailed overview of the number of CTC detected in the patients is provided in Table 1, whereas the actual CTC numbers for each patient is listed in Supplementary Table 1 and visualized in Supplementary Figure 3. In CRPC patients, EpCAMhigh CTC ranged from 0–3300 (median: 6, mean: 124 ± 400 SD) and EpCAMlow CTC ranged from 0–24 (median: 1, mean: 3 ± 4 SD). In mBC patients, EpCAMhigh CTC ranged from 0–208 (median: 1, mean: 13 ± 43 SD) and EpCAMlow CTC ranged from 0–35 (median: 3, mean: 3 ± 11 SD). In CRPC patients, ≥5 EpCAMhigh CTC or ≥5 EpCAMlow CTC were detected in 56 (53%) and 26 (28%) patients, respectively. In the group of mBC patients, 7 (32%) showed ≥5 EpCAMhigh CTC and 8 (36%) ≥5 EpCAMlow CTC. Summing EpCAMhigh CTC and EpCAMlow CTC to ≥5 CTC, we observed in 70% positive CRPC (n = 91) and 64% positive mBC patients, increasing the combined CTC-positivity rates by 32% and 100% respectively, in comparison to positivity rates for EpCAMhigh CTC only. In total, 37% CRPC patients and 23% mBC patients had ≥5 EpCAMhigh CTC, but <5 EpCAMlow CTC. Vice versa, <5 EpCAMhigh CTC and ≥5 EpCAMlow CTC were detected in 10% CRPC and in 18% mBC patients. Figure 3 presents a gallery of EpCAMhigh CTC (upper panels 3A–3H) and EpCAMlow CTC (lower panels 3I–3P) that were found in CRPC patients (left panels 3A–3D and 3I–3L) and mBC patients (right panels 3E–3H and 3M–3P), showing CTC of various sizes and staining of CK intensity.
Table 1. Frequency of CTC in CRPC and mBC patients.
| Prostate cancer | Breast cancer | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| % patients with EpCAMhigh CTC | % patients with EpCAMhigh CTC | ||||||||||
| # CTC | 0 | 1–4 | ≥5 | ≥10 | Total | 0 | 1–4 | ≥5 | ≥10 | Total | |
| % patients with EpCAMlow CTC | 0 | 12.1 | 9.9 | 22.0 | 18.7 | 44.0 | 18.2 | 4.5 | 9.1 | 9.1 | 31.8 |
| 1–4 | 5.5 | 6.6 | 15.4 | 14.3 | 27.5 | 9.1 | 9.1 | 13.6 | 4.5 | 31.8 | |
| ≥5 | 7.7 | 2.2 | 18.7 | 16.5 | 28.6 | 13.6 | 13.6 | 9.1 | 4.5 | 36.3 | |
| ≥10 | 1.1 | 0 | 5.5 | 4.1 | 6.6 | 9.1 | 9.1 | 9.1 | 4.5 | 27.3 | |
| Total | 25.3 | 18.7 | 56 | 49.5 | 100 | 40.9 | 27.3 | 31.8 | 18.2 | 100 | |
The frequency (%) of EpCAMhigh and EpCAMlow CTC in 7.5 mL of blood of 91 castration-resistant prostate cancer patients and 22 metastatic breast cancer patients.
Figure 3. Gallery of captured CTC in CRPC and mBC patients.
Display of EpCAMhigh CTC (panels A–H) captured with CellSearch and EpCAMlow CTC (panels I–P) captured with microsieves in castrate-resistant prostate cancer patients (left panels A–D and I–L) and metastatic breast cancer patients (right panels E–H and M–P). Cells in panel (A–B) are from the same patient, as are the cells in panels (C–D, E–F, G–H and I–J). The value for intensity of CK staining (green) is represented in the vertical bar next to the image, visualizing a very high intensity value with maximum 4095 counts as a full green bar. The nucleus is stained with DAPI (blue). The unlabelled scale bar for EpCAMhigh CTC images is 6.4 μm.
Overall survival of CRPC patients with EpCAMhigh and EpCAMlow CTC
Follow-up data from patients that were enrolled in ongoing clinical trials could not be obtained at the time this manuscript was written. Therefore, follow-up data could only be obtained from 85 out of 108 CRPC patients and 16 out of 22 mBC patients. The cohort of mBC patients that remained was too small for survival analysis and was therefore omitted.
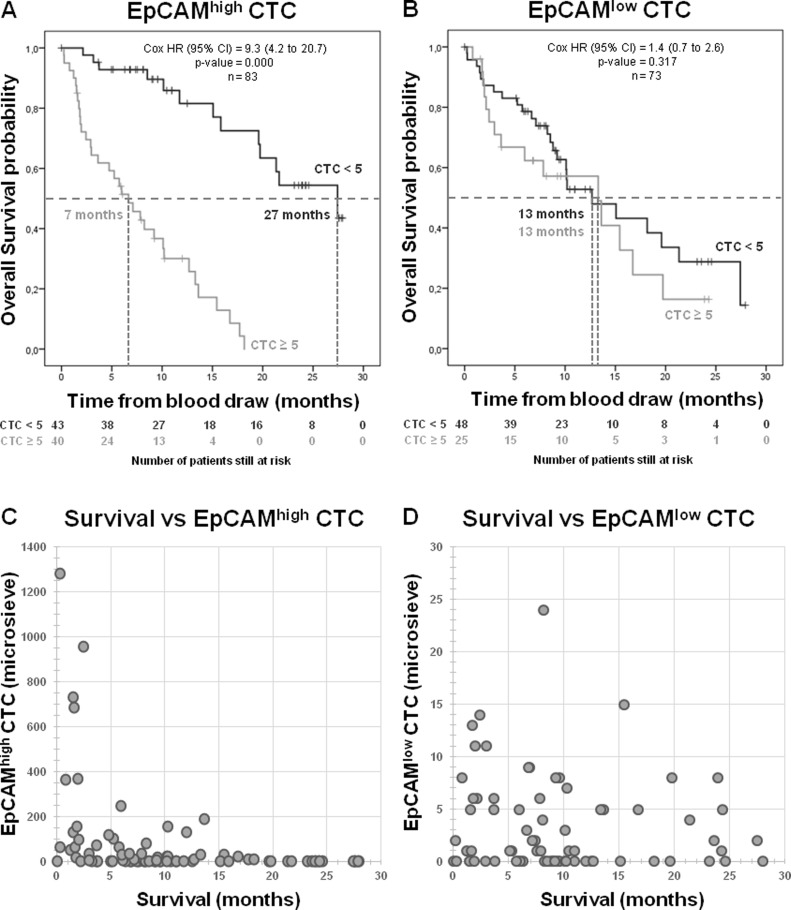
To relate the presence of CTC in CRPC patients to overall survival, the patient cohort was split into a favorable group and an unfavorable group, using 5 CTC as a threshold [2, 4]. In Figure 4 the Kaplan-Meier curves for EpCAMhigh and EpCAMlow CTC are shown. A significant difference is observed for the presence of ≥5 EpCAMhigh CTC in relation to overall survival (p = 0.000) (Figure 4A), whereas no significant difference is observed for ≥5 EpCAMlow CTC (p = 0.317) (Figure 4B). The combination of EpCAMhigh CTC and EpCAMlow CTC was related with overall survival by separating the cohort into four groups (Supplementary Figure 4A). This shows again that the strong correlation with survival can be solely contributed to EpCAMhigh CTC and not to EpCAMlow CTC (p = 0.000). Since ≥5 EpCAMlow CTC show no correlation with survival, perhaps a lower CTC cut-off value would show a correlation. However, the scatter plot of the number of EpCAMlow CTC versus survival of these patients in Figure 4D shows no trend between these two factors, whereas this trend is visible between survival and EpCAMhigh CTC (Figure 4C). A receiver operating characteristic (ROC) curve was then used to determine the highest diagnostic cut-off value for EpCAMlow CTC. Although this calculates a threshold at ≥1 EpCAMlow CTC, this value can be considered inconclusive since the separation between sensitivity and specificity is very low (Supplementary Figure 4B). Using the threshold of ≥1 EpCAMlow CTC also no relation can be observed with overall survival (p = 0.748) (Supplementary Figure 4C).
Figure 4. Overall survival for CRPC patients.
Kaplan–Meier curve of overall survival of patients with EpCAMhigh CTC from CellSearch (A), EpCAMlow CTC from microsieves (B) show a strong correlation between EpCAMhigh CTC and survival, but no correlation between EpCAMlow CTC and survival. Scatter plot of survival versus the amount of EpCAMhigh CTC (n = 83) visualizes the expected trend that most patients with high number of CTC have a short survival (C), whereas the amount EpCAMlow CTC in patients (n = 73) show no such trend (D).
DISCUSSION
In this multicenter study we determined the presence of EpCAMhigh and EpCAMlow CTC in castration resistant prostate cancer and metastatic breast cancer patients. Protocols and tools for detection were developed in the FP7-program CTC-Trap and validated at six clinical sites cooperating in the program. The current standard CellSearch method for CTC enumeration was used for the detection of CTC expressing EpCAM, followed by capturing and filtering of the sample depleted of these EpCAMhigh CTC, and stained for detection of epithelial CTC expressing no or low EpCAM (Figure 1).
To validate the procedure of CellSearch followed by filtration for detection of EpCAMlow CTC, samples spiked with cells from the PC3 or MDA-MB-231 cancer cell lines were used. In the EpCAMhigh CellSearch fraction, 71% of the PC3 total spiked cells were recovered and 74% of the spiked MDA-MB-231 cells. The actual expression levels of EpCAM on EpCAM expressing CTC is an important question. Rao et al. reported the EpCAM expression on 100 blood samples of metastatic cancer patients in which CTC were detected by flow cytometry and reported an average EpCAM density of 5.0 × 104 EpCAM antigens [10]. This density is fivefold higher than that of the PC3 (on average 1.0 × 104 EpCAM antigens) and MDA-MB-231 (on average 1.5 × 104 EpCAM antigens) cells used in this study. However, these numbers are the average density and the actual distribution of the EpCAM expression can be quite broad, as illustrated in Supplementary Figure 1 [10]. The data supports our notion that the majority of CTC with relatively high EpCAM antigen density are captured by CellSearch, while CTC expressing no EpCAM or low EpCAM levels will be present in the EpCAM depleted fraction. After passage of the EpCAM depleted blood through microsieves, only a portion of the spiked cells –and most likely also CTC – will be captured.
Relative recovery of PC3 cells and MDA-MB-231 in the EpCAMhigh fraction (CellSearch) and EpCAMlow fraction (microsieve), as shown in Figure 2, followed the expectations given the size and EpCAM distribution of both cell lines. Although in line with previous reports, the absolute number recovered cells is relatively low, thereby raising the question where and how these cells were lost [9, 11]. Only a small part of cells are destroyed during the CellSearch procedure, indicated by the 90% recovery of cells that express EpCAM at a very high density [9]. Therefore, it is likely to assume most are lost before measurement or during the filtration process. Some of the cells may be missed because they lack sufficient immunostaining for detection, or the cells have simply been degraded before or during the process, as they were not extensively fixed apart from the light fixative used during spiking the blood samples.
The observed recovery rates retrieved for each cell line includes variation introduced by EpCAM density, by different operators at each clinical site and by the three measurements for each cell line (Figure 2 and Supplementary Figure 2). Clearly, the variation in the obtained results vary a lot between the sites, stressing the need for standardization and proficiency testing to assure accurate reporting of actual CTC numbers. Variation introduced by the interpretation of the images of the CTC can be eliminated by the recently introduced ACCEPT open-source imaging program [12, 13].
In both CRPC and mBC patients, EpCAMhigh and EpCAMlow CTC were detected in 7.5 mL blood (Table 1). The number of patients with positive detection of CTC can be increased when both EpCAMhigh and EpCAMlow CTC are used for the ≥5 CTC threshold. This corresponds to our previous report of a small pilot study with 28 non-small cell lung cancer (NSCLC) patients [9]. Also, these findings fit in the trend of recently published filtration based studies, which are reporting large amounts of CTC detected when compared to CellSearch [6, 14–17]. A biological reason for this increase in CTC counts is by many thought to be related to epithelial-to-mesenchymal transition (EMT), which is accompanied by loss of epithelial markers, such as EpCAM and CK, and by an increase of mesenchymal markers, such as vimentin. However, co-expression of epithelial markers and mesenchymal markers on CTC also have been reported [18–25]. In this study, EpCAMlow CTC expressing cytokeratins are detected, leaving the question open on the frequency and clinical relevance of EpCAMlow CTC lacking CK and expressing mesenchymal markers [21, 22].
To determine whether the presence of EpCAMhigh and EpCAMlow CTC had similar clinical relevance in CRPC patients, we correlated their presence with the time of survival. The threshold of >5 CTC for EpCAMhigh is the threshold used in the original metastatic breast and prostate studies and was therefore also used for EpCAMlow CTC to distinguish between favorable and unfavorable patient groups [3, 23]. Whereas the presence of EpCAMhigh CTC strongly related to a poor survival (Figure 4A), the presence of EpCAMlow CTC did not (Figure 4B). A threshold of ≥1 CTC for EpCAMlow CTC also showed no significant relation to survival, as illustrated by the ROC curves and Kaplan-Meier curve presented in Supplementary Figure 4. Similar observations were made in NSCLC and this prompts the question what type of cells these EpCAMlow cells truly are [9]. Here, we define these cells as having no or low EpCAM expression, based on the effective depletion of cells expressing higher levels of EpCAM by the CellSearch system and relatively low capture of cells expressing low levels of EpCAM (Figure 2). The EpCAMlow cells have a nucleus identified by DAPI, express CK, but not CD45. The presence of CK shows that these cells are of epithelial origin, but do not proof that they are indeed tumor cells [24]. The CK expression in CTC is variable, even in the same patient (Figure 3). EpCAMlow cells that resemble the morphology of EpCAMhigh CTC (Figure 3A–3H) have a higher intensity and filamentous pattern of CK (Figure 3I, 3J), whereas low CK intensity cells show irregular CK expression (Figure 3K–3P). This may indicate that these cells are different subtypes of CTC, where each subpopulation could express its own characteristics and clinical behavior [25]. Even so, the intensity of CK staining can be very low in some patients making it difficult for operators to score CTC. Even using clearly defined criteria for operators to follow in scoring CTC, the discrepancy between operators can be high and argues for an automated approach to analyze images for cells of interest [12, 13]. Definitive proof of the cancerous nature of the EpCAMlow cells will need to come from the genetic analysis of the cells, as well as the differences between the genetic and proteomic make-up of the EpCAMhigh and EpCAMlow cells.
Concluding, in this multicenter study we validated the detection of EpCAMhigh and EpCAMlow CTC from the same blood draw tube using the CellSearch system for enumeration of the EpCAMhigh and by microscopic examination of EpCAMlow after passage of the blood depleted from EpCAMhigh cells through microsieves. We showed that the number of CTC detected in both metastatic breast and prostate cancer can be increased when considering both EpCAMhigh and EpCAMlow CTC, but also show that only the presence of EpCAMhigh CTC, and not EpCAMlow CTC, are strongly correlated with poor overall survival. This emphasizes the importance to demonstrate the relation with clinical outcome when presence of CTC identified with different technologies are reported, as different CTC subpopulations can have different relations with clinical outcome.
METHODS
Blood samples
Peripheral blood samples were drawn by venepuncture into 10 mL CellSave Preservative Tubes (Menarini Silicon Biosystems, Huntingdon Valley PA, USA) from healthy donors, metastatic breast and castration resistant prostate cancer patients. All patients and healthy donors provided written informed consent and the study protocol was approved by the medical ethical committee's at each participating site. Patient demographics of patients with known clinical outcome are provided in Table 2. The sites in the CTC-Trap consortium collaborating in this validation study are: The Institute of Cancer Research, The Royal Marsden Hospital, NHS Foundation Trust (ICR), United Kingdom; Institute Gustave Roussy (IGR), France, Instituto Oncologico Veneto (IOV), Italy; Ludwig-Maximilians-University Muenchen (LMU), Germany; Universitätsklinikum Düsseldorf (UKD), Germany; and the Universiteit Twente (UT), the Netherlands. CRPC patients (at sites ICR, IGR and IOV) and metastatic breast cancer patients (at sites LMU and UKD) were recruited and processed on site within 96 hours. A schematic overview of the study is shown in Figure 1.
Table 2. Patient demographics.
| Castration-resistant prostate cancer patients | n = 85 | Metastatic breast cancer patients | n = 16 |
|---|---|---|---|
| Age (years) | Age (years) | ||
| Median (range) | 71 (49–84) | Median (range) | 63 (37–89) |
| Unknown | 24 (28%) | Unknown | 1 (6%) |
| Status at last follow-up | Status at last follow-up | ||
| Alive | 38 (45%) | Alive | 12 (75%) |
| Dead | 47 (55%) | Dead | 4 (25%) |
| Mean follow-up time in months (min-max) | Mean follow-up time in months (min-max) | ||
| Alive | 13 (0–28) | Alive | 11 (3–17) |
| Dead | 09 (0–27) | Dead | 4 (2–8) |
| Line of therapy | Line of therapy | ||
| Before therapy | 08 (9%) | Before therapy | 5 (31%) |
| 1st line | 10 (12%) | 1st line | 2 (13%) |
| 2nd line | 19 (22%) | 2nd line | 1 (6%) |
| ≥3rd line | 24 (28%) | ≥ 3rd line | 7 (44%) |
| Unknown | 25 (29%) | Unknown | 1 (6%) |
The demographics of the patients in this study with known clinical outcome.
Cell lines and spiking
Spiking experiments were performed with cells from the prostate carcinoma cell line PC3 and the breast carcinoma cell line MDA-MB-231. These cell lines were chosen for their low expression of EpCAM, which would allow us to find a fraction in both the EpCAMhigh and EpCAMlow fraction. The median size of the MDA-MB-231 cells was 15.6 μm with an average EpCAM density of 1.5 × 104 (165 %CV) antigens. The median size of the PC3 cells was 17.7 μm with an average EpCAM density of 1.0 × 104 (89 %CV) antigens. The histograms showing the distribution of the EpCAM density of the PC3 and MDA-MB-231 cells is shown in Supplementary Figure 1. For comparison, the EpCAM density of the breast cancer cell line MCF-7, expressing EpCAM at much higher density, is shown in the figure as well. All cell lines were obtained from ATCC (Manassa VA, USA) and have not been authenticated in the past four years. They were grown at 37°C and 5% CO2. The PC3 cell line was cultured in RPMI-1640 (Life Technologies Corporation, Carlsbad CA, USA), and the MDA-MB-231 cell line was cultured in DMEM (Euroclone, Pero MI, Italy); both media were supplemented with 10% fetal bovine serum (GIBCO, Life Technologies Corporation), 2 mM UltraGlutamine (Lonza, Basel, Switzerland) and 10 mM Hepes (Lonza). The median cell size from both cell lines was determined with a Coulter counter pipette (Scepter, Millipore, Billerica, MA, USA). The EpCAM density was determined using a flow cytometer (FACS ARIA II, BD Biosciences, San Jose, CA, USA) and QuantiBrite beads (BD Biosciences).
The MDA-MB-231 cells and PC3 cells were spiked in healthy donor CellSave blood. Cell numbers for spiking were counted manually with a fluorescent microscope, using the nuclear dye Hoechst 33342 for visualization on a glass slide. Targeting a spike number of 250 cells in 7.5 mL blood volume, the exact number of cells was counted before adding the cells to the blood and subsequently used to determine the recovery in the CellSearch and on the microsieve. On average, 270 cells were spiked per tube. Unspiked blood samples from healthy volunteers were used as a negative control. Spiked samples were prepared at the IOV laboratory and distributed under temperature control to the six sites to be processed after 48 hours. Each site received a tube of unspiked blood (labeled A), one tube spiked with PC3 cells (labeled B) and one tube spiked with MDA-MB231 cells (labeled C), all from one donor. This was repeated three times with three different donors.
CTC detection by CellSearch
CTC were enumerated in aliquots of 7.5 mL of blood with the CellSearch® Circulating Tumor Cell Kit (Menarini Silicon Biosystems). Analysis was performed within 96 hours of the blood draw. Blood samples were enriched for EpCAMhigh cells and stained with DAPI, Cytokeratin-PE and CD45-APC on the CellTracks Autoprep. Image acquisition of the stained cartridges was performed on the CellTracks Analyzer II. Images of CTC candidates were identified by the CellTracks Analyzer II and presented to experienced operators for classification. Candidates were assigned as CTC when the objects were larger than 4 μm, stained with DAPI and CK, lacked CD45 staining and had morphological features consistent with that of a cell [1].
Blood sample collection after EpCAMhigh CTC enrichment
After immunomagnetic selection of EpCAMhigh cells, the CellTracks Autoprep aspirates the blood that is void of the selected cells and transports it to a waste container outside the instrument. To enable the investigation of this blood for residual tumor cells, the sample was collected manually or with the specific designed Automated Sample Collection Device (ASCD), as described previously [9]. For manual collection of the Autoprep discarded blood sample, the top of the waste container was removed. After visual inspection of presence of blood in the tubing coming out of the Autoprep, a 50 mL conical tube is placed under the outlet and the sample is collected until no blood can be observed anymore in the tubing. This is repeated for each sample. The first blood arrives at the waste container approximately 1 hour after start of the Autoprep. The protocols and tools are described in more detail at https://www.utwente.nl/tnw/mcbp/protocolsandtools/.
Filtration of the discarded CellTracks Autoprep blood sample
To filter tumor cells from the EpCAMhigh CTC depleted blood sample, microsieve membranes were used (VyCAP, Deventer, The Netherlands). Each microsieve contains 111,800 pores of 5 μm in diameter and these pores are spaced 14 μm apart in lanes with a porosity of 10% on a total surface area of 8 by 8 mm2. The specifications of the microsieves were obtained from previous experiments [9, 11, 26]. The microsieve is contained in a plastic holder that was placed in a disposable filtration unit, which can be placed on a pump unit that maintained a pressure of −100 mbar across a microsieve during filtration (VyCAP). The CellSearch discarded blood sample was transferred to the filtration unit after which the pump was switched on. The collected blood sample, varying between 25 to 40 mL, was passed through in maximum 10 minutes. After complete filtration of the sample or after 10 minutes, the pump was switched off and if there was any unfiltered sample volume remaining, this was removed with a pipette. Details of the volumes that were not filtered were used to determine how much whole blood volume was processed.
Staining of cells on microsieves
Conditions for staining on microsieves were optimized to assure uniform staining across the microsieve with a minimum of non-specific binding. After filtration, the microsieve was removed and washed once with a permeabilization buffer containing PBS, 1% bovine serum albumin (Sigma-Aldrich, St. Louis MO, USA) and 0.15% saponin (Sigma-Aldrich). Next, this buffer was placed on the sieve and removed after 15 minutes incubation at room temperature. A cocktail of fluorescently labeled antibodies was used to stain the cells on the sieve for 15 minutes at 37°C. The staining solution consisted of the following monoclonal antibodies: three CK antibody clones targeting CK 4, 5, 6, 8, 10, 13, 18 (clone C11), CK 1-8 (clone AE3) and CK 10, 14, 15, 16 and 19 (clone AE1), all conjugated to NTb575 (AcZon s.r.l., Bologna, Italy), and one antibody targeting CD45 (clone HI30) labelled with PerCP (Thermo Fisher Scientific, Waltham MA, USA). The CK-pan cocktail was diluted to a final concentration of 3.5 μg/mL and the anti-CD45 antibody was diluted to 4 μg/mL in PBS/1%BSA/0.05% saponin. After removal of the staining cocktail, the microsieve was washed once and then incubated for 5 minutes at room temperature with PBS/1%BSA. Then the sample was fixed using PBS with 1% formaldehyde (Sigma-Aldrich) for 10 minutes at room temperature. Removal of the fluid during each of the staining and washing steps was done by bringing the bottom of the microsieve in contact with an absorbing material using a staining holder (VyCAP). The microsieve was subsequently covered with ProLong® Diamond Antifade Mountant with DAPI (Thermo Fisher Scientific). A custom cut 0.85 × 0.85 cm2 glass cover slip (Menzel-Gläser, Saarbrükener, Germany) was placed on both sides of the microsieve for immediate analysis or storage in the freezer at −30°C.
Detection of cells on microsieves
Microscopes at each site were equipped with a 20× microscope objective with minimal NA0.45 and the same set of filter cubes. The following filters were used: DAPI (DAPI-50LP-A-NQF) with excitation 377/50 nm, dichroic 409 nm LP, emission 409 nm LP; PE (TRITC-B-NQF) with excitation 543/22 nm, dichroic 562 nm LP, emission 593/40 nm and PerCP (FF02-435/40, FF510-Di02 and FF01-676/29 (customized filter cube)) with excitation 435/40 nm, dichroic 510 nm LP, emission 676/29 nm. All cubes were acquired via Nikon (Semrock, Rochester, NY, USA). Images covering the entire 0.64 cm2 surface of the microsieves were acquired and stored.
Scoring of CTC
Analysis of the fluorescent images generated from the CellSearch cartridges were performed according the instructions of the manufacturer. The fluorescent images from the microsieves were analyzed using the open-source software ICY [27]. Operators were asked to annotate every DAPI+/CK+/CD45− event. In case of clogging of the microsieve, CTC counts were extrapolated to the full volume. Images of CTC were analyzed for their intensity in CK, thereby deducting the background value from the intensity value of CK staining ranging from 0 to 4095 counts.
Statistical analysis
Statistical analysis was performed in R (version 3.3.0) and SPSS (Statistics 24). For survival analysis, patients were divided in two prognostic groups: favorable for less than 5 CTC and unfavorable ≥5 CTC [2, 4]. Kaplan–Meier curves for overall survival were generated and compared using the Log-Rank test. A receiver operating characteristic curve was used to determine the EpCAMlow CTC threshold for the highest diagnostic ability. A p-value of < 0.05 was considered to indicate a significant difference.
SUPPLEMENTARY MATERIALS FIGURES AND TABLE
Abbreviations
- CK
cytokeratins
- CRPC
castration-resistant prostate cancer patients
- CTC
circulating tumor cells
- CV
coefficient of variation
- EMT
epithelial to mesenchymal transition
- EpCAM
epithelial cell adhesion molecule
- EpCAMhigh CTC
EpCAM high expressing CTC
- EpCAMlow CTC
EpCAM low expressing CTC
- mBC
metastatic breast cancer
Footnotes
Author contributions
S.W. and L.T. wrote the manuscript and prepared the figures. S.W., M.M., E.R., R.V., R.L., L.Y., B.Z., A.R., M.O., M.C., K.A., G.F. and P.F. performed the experiments. R.Z., E.T., M.T., T.F., H.N., P.R., M.F. and J.B. provided the clinical data. S.W., A.B. and M.IJ. performed the statistical analysis. R.Z., M.A., B.R., H.N., F.F., J.B., M.IJ. and L.T. are the senior co-authors. All authors reviewed and approved the manuscript.
CONFLICTS OF INTEREST
None of the authors have conflicts of interest to declare.
FUNDING
This research was made possible through FP7-HEALTH-2012-INNOVATION Circulating Tumor Cells TheRapeutic APheresis: a novel biotechnology enabling personalized therapy for all cancer patients grant #305341.
REFERENCES
- 1.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 2.Bidard FC, Peeters DJ, Fehm T, Nolé F, Gisbert-Criado R, Mavroudis D, Grisanti S, Generali D, Garcia-Saenz JA, Stebbing J, Caldas C, Gazzaniga P, Manso L, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 2014;15:406–14. doi: 10.1016/S1470-2045(14)70069-5. [DOI] [PubMed] [Google Scholar]
- 3.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–21. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 4.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–09. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 5.Hiltermann TJ, Pore MM, van den Berg A, Timens W, Boezen HM, Liesker JJ, Schouwink JH, Wijnands WJ, Kerner GS, Kruyt FA, Tissing H, Tibbe AG, Terstappen LW, Groen HJ. Circulating tumor cells in small-cell lung cancer: a predictive and prognostic factor. Ann Oncol. 2012;23:2937–42. doi: 10.1093/annonc/mds138. [DOI] [PubMed] [Google Scholar]
- 6.Krebs MG, Sloane R, Priest L, Lancashire L, Hou JM, Greystoke A, Ward TH, Ferraldeschi R, Hughes A, Clack G, Ranson M, Dive C, Blackhall FH. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol. 2011;29:1556–63. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 7.Janni WJ, Rack B, Terstappen LW, Pierga JY, Taran FA, Fehm T, Hall C, de Groot MR, Bidard FC, Friedl TW, Fasching PA, Brucker SY, Pantel K, Lucci A. Pooled Analysis of the Prognostic Relevance of Circulating Tumor Cells in Primary Breast Cancer. Clin Cancer Res. 2016;22:2583–93. doi: 10.1158/1078-0432.CCR-15-1603. [DOI] [PubMed] [Google Scholar]
- 8.van Dalum G, Stam GJ, Scholten LF, Mastboom WJ, Vermes I, Tibbe AG, De Groot MR, Terstappen LW. Importance of circulating tumor cells in newly diagnosed colorectal cancer. Int J Oncol. 2015;46:1361–68. doi: 10.3892/ijo.2015.2824. [DOI] [PubMed] [Google Scholar]
- 9.de Wit S, van Dalum G, Lenferink AT, Tibbe AG, Hiltermann TJ, Groen HJ, van Rijn CJ, Terstappen LW. The detection of EpCAM(+) and EpCAM(-) circulating tumor cells. Sci Rep. 2015;5:12270. doi: 10.1038/srep12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao CG, Chianese D, Doyle GV, Miller MC, Russell T, Sanders RA, Jr, Terstappen LW. Expression of epithelial cell adhesion molecule in carcinoma cells present in blood and primary and metastatic tumors. Int J Oncol. 2005;27:49–57. doi: 10.3892/ijo.27.1.49. [DOI] [PubMed] [Google Scholar]
- 11.Coumans FA, van Dalum G, Beck M, Terstappen LW. Filter characteristics influencing circulating tumor cell enrichment from whole blood. PLoS One. 2013;8:e61770. doi: 10.1371/journal.pone.0061770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeune LL, de Wit S, Berghuis AMS, IJzerman MJ, Terstappen LWMM, Brune C. How to Agree on a CTC: Evaluating the Consensus in Circulating Tumor Cell Scoring. Cytometry A. 2018 Sep 24; doi: 10.1002/cyto.a.23576. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeune L, van Dalum G, Decraene C, Proudhon C, Fehm T, Neubauer H, Rack B, Alunni-Fabbroni M, Terstappen LW, van Gils SA, Brune C. Quantifying HER-2 expression on circulating tumor cells by ACCEPT. PLoS One. 2017;12:e0186562. doi: 10.1371/journal.pone.0186562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F, Laplanche A, Chauchereau A, Lacroix L, Planchard D, Le Moulec S, André F, Fizazi K, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer. 2011;105:847–53. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pecot CV, Bischoff FZ, Mayer JA, Wong KL, Pham T, Bottsford-Miller J, Stone RL, Lin YG, Jaladurgam P, Roh JW, Goodman BW, Merritt WM, Pircher TJ, et al. A novel platform for detection of CK+ and CK− CTCs. Cancer Discov. 2011;1:580–86. doi: 10.1158/2159-8290.CD-11-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneck H, Gierke B, Uppenkamp F, Behrens B, Niederacher D, Stoecklein NH, Templin MF, Pawlak M, Fehm T, Neubauer H, Disseminated Cancer Cell Network (DCC Net) Duesseldorf EpCAM-Independent Enrichment of Circulating Tumor Cells in Metastatic Breast Cancer. PLoS One. 2015;10:e0144535. doi: 10.1371/journal.pone.0144535. Erratum in: Correction: EpCAM-Independent Enrichment of Circulating Tumor Cells in Metastatic Breast Cancer. [PLoS One. 2016] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lampignano R, Yang L, Neumann MH, Franken A, Fehm T, Niederacher D, Neubauer H. A Novel Workflow to Enrich and Isolate Patient-Matched EpCAMhigh and EpCAMlow/negative CTCs Enables the Comparative Characterization of the PIK3CA Status in Metastatic Breast Cancer. Int J Mol Sci. 2017;18:1885. doi: 10.3390/ijms18091885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krebs MG, Hou JM, Sloane R, Lancashire L, Priest L, Nonaka D, Ward TH, Backen A, Clack G, Hughes A, Ranson M, Blackhall FH, Dive C. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol. 2012;7:306–15. doi: 10.1097/JTO.0b013e31823c5c16. [DOI] [PubMed] [Google Scholar]
- 19.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–84. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kallergi G, Papadaki MA, Politaki E, Mavroudis D, Georgoulias V, Agelaki S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011;13:R59. doi: 10.1186/bcr2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alix-Panabières C, Mader S, Pantel K. Epithelial-mesenchymal plasticity in circulating tumor cells. J Mol Med (Berl) 2017;95:133–42. doi: 10.1007/s00109-016-1500-6. [DOI] [PubMed] [Google Scholar]
- 22.Willipinski-Stapelfeldt B, Riethdorf S, Assmann V, Woelfle U, Rau T, Sauter G, Heukeshoven J, Pantel K. Changes in cytoskeletal protein composition indicative of an epithelial-mesenchymal transition in human micrometastatic and primary breast carcinoma cells. Clin Cancer Res. 2005;11:8006–14. doi: 10.1158/1078-0432.CCR-05-0632. [DOI] [PubMed] [Google Scholar]
- 23.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 24.Moll R, Franke WW, Schiller DL, Geiger B, Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- 25.Adams DL, Stefansson S, Haudenschild C, Martin SS, Charpentier M, Chumsri S, Cristofanilli M, Tang CM, Alpaugh RK. Cytometric characterization of circulating tumor cells captured by microfiltration and their correlation to the CellSearch(®) CTC test. Cytometry A. 2015;87:137–44. doi: 10.1002/cyto.a.22613. [DOI] [PubMed] [Google Scholar]
- 26.Coumans FA, van Dalum G, Beck M, Terstappen LW. Filtration parameters influencing circulating tumor cell enrichment from whole blood. PLoS One. 2013;8:e61774. doi: 10.1371/journal.pone.0061774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ICY Available from: http://icy.bioimageanalysis.org/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.