Abstract
A key question in biology is to understand how interspecies morphological diversities originate. Plant roots present a huge interspecific phenotypical variability, mostly because roots largely contribute to adaptation to different kinds of soils. One example is the interspecific cortex layer number variability, spanning from one to several. Here, we review the latest advances in the understanding of the mechanisms expanding and/or restricting cortical layer number in Arabidopsis thaliana and their involvement in cortex pattern variability among multi-cortical layered species such as Cardamine hirsuta or Oryza sativa.
Keywords: ground tissue, plants, root development, asymmetric cell division, Arabidopsis thaliana, cortex
1. Introduction
How different morphologies originate in nature is a fundamental question in biology. Researchers in evolutionary developmental biology are trying to shed light on the molecular mechanisms underlying the differences in shape and anatomy that allow organisms to cope with the large diversity of environments on land. In this view, one challenge is the identification of feasible model systems to study differences in development. A breakthrough approach to isolate the genetic mechanisms at the basis of phenotypical differences is the use of comparative studies [1–3]. Most of the success of this strategy consists in the identification of genetic differences underlying phenotypical diversity over a short evolutionary scale, such as differences in gene activity and/or expression.
Plant roots represent an ideal model system for comparative anatomy studies: (i) plant root anatomy largely varies among species; and (ii) roots are transparent and have simple anatomy, allowing fine and precise microscopy analysis of differences in anatomical traits between plant species. The roots of most plants have a radial symmetry and can be represented by a series of concentric cylinders. Briefly, the outer cylinder represents the epidermis, and the inner cylinders represent the cortex layer(s), the endodermis, the pericycle and the vascular bundle [4,5]. Cortex(es) and the endodermis form all together the ground tissue (GT) (figure 1) [4,5]. All root tissues originate from a set of initials/stem cells located in specialized region at the tip of the root, called root apical meristem (RAM) [6–8].
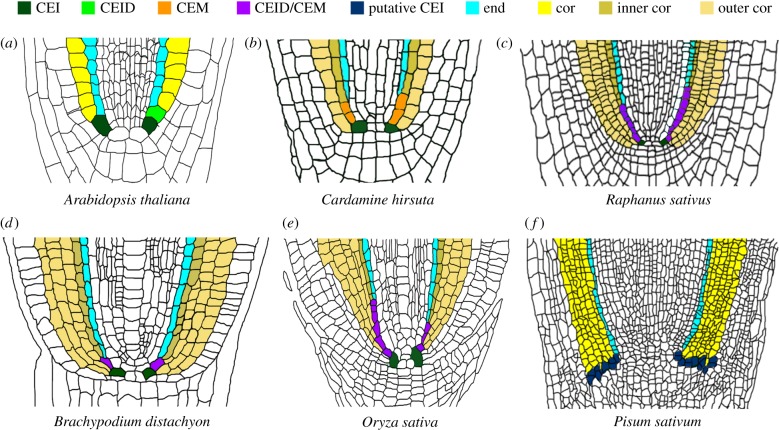
Figure 1.
GT cell fate in different species. Cartoon representing longitudinal sections of (a) Arabidopsis thaliana, (b) Cardamine hirsuta, (c) Raphanus sativum, (d) Brachypodium distachyon, (e) Oryza sativa and (f) Pisum sativum root meristem. Abbreviations in colour key at top of figure: end, endodermis; cor, cortex; CEM, mixed cortex and endodermis identity cell. Note that the number of cortex layers varies among species.
The extraordinary diffusion of plants in multiple different environments is partly due to the fundamental role of roots in anchoring plants to soil and allowing the uptake of water and nutrients. Root cortical tissue plays a fundamental role in permitting plants to cope with a variety of environments. In plants living in wet soils, such as rice, the control of water/air ratio is determined by the formation of the aerenchyma, a specialized tissue derived from root cortex secondary growth [9]. In plants such as turnip or horseradish living in adverse weather conditions, root cortex originates storage parenchyma, a tissue where simple carbohydrates are converted in starch [9]. In legumes such as Medicago sativa and truncatula, the dedifferentiation processes of cortex cells give rise to the symbiotic nodule, where the symbiosis with nitrogen-fixing rhyzobia takes place [10,11]. Moreover, it was recently shown that plants such as Arabidopsis thaliana and Hakea actites uptake proteins from soil and transfer them into cortex as nitrogen source [12]. Hence, modulating the number of cortical layers, plants can model their resistance to stresses and changes in the environment; therefore, the understanding of the molecular mechanism controlling cortex layer number is of primary interest in plant biology.
Cortex origin and architecture largely vary depending on the phyletic origin (monocots versus dicots) and on the type of RAM: open RAM is characterized by the absence of boundaries between specific tissues in the growing tip and closed RAM has distinct boundaries between apical regions that can be identified [13,14]. Some species of plants presenting a closed RAM (e.g. the model system A. thaliana) have one cortex layer at seedling stage, whereas some other species show multiple cortical layers (e.g. Cardamine hirsuta, Raphanus sativus, Hordeum vulgare, Brachypodium distachyon or Oryza sativa; figure 1) [15,16]. Furthermore, cortical layer number can also vary in the same organism. Arabidopsis acquires one cortex layer more, called middle cortex (MC), during development [17,18], whereas O. sativa shows a complicated cortex layer number pattern: rice embryo primary roots (radicule) show five cortex layers, whereas the lateral and crown roots show a variable number between 1 and 10 [15,19,20]. In species presenting an open meristem such as Pisum sativum (pea), there are no specific tiers of cortical initials [21]. Morphological analysis suggests that cortical initials in pea are distributed in continuous layers located over the columella [13] (figure 1).
Such interspecific variability of cortex layer number epitomizes an extraordinary resource for comparative anatomy studies. The tremendous technological advance of recent years made more approachable research on a huge variety of model systems, enhancing the knowledge on root patterning. In this review, we aim to shed light on the differences in molecular pathways subtending GT variability among species based on (i) the most recent advances in understanding the GT patterning in Arabidopsis and (ii) the latest findings in understanding the mechanisms controlling cortical layer number in Arabidopsis close and distant relatives.
2. Root cortex patterning in Arabidopsis
The GT (cortex and the endodermis) originates from the RAM, a region located at the tip of the root where a set of self-renewing stem cells divide producing all the root tissues. In Arabidopsis, a stem cell, called cortex endodermis initial (CEI), gives rise to the GT (figure 2). An asymmetric anticlinal division of CEI gives rise to a self-renewed stem cell (CEI) and to a daughter cell (CEID) (figure 2). Subsequently, a periclinal division occurs in the CEID generating endodermis and cortex [4] (figure 2). The CEID periclinal division occurs already in late embryo development, determining the formation of an embryonic cortex and endodermis and, therefore, defining the tissue organization of the primary root [4]. During post-embryonic development, an additional periclinal division occurs in the meristematic endodermis giving rise to a novel cortex layer, the MC [17] (figure 2).
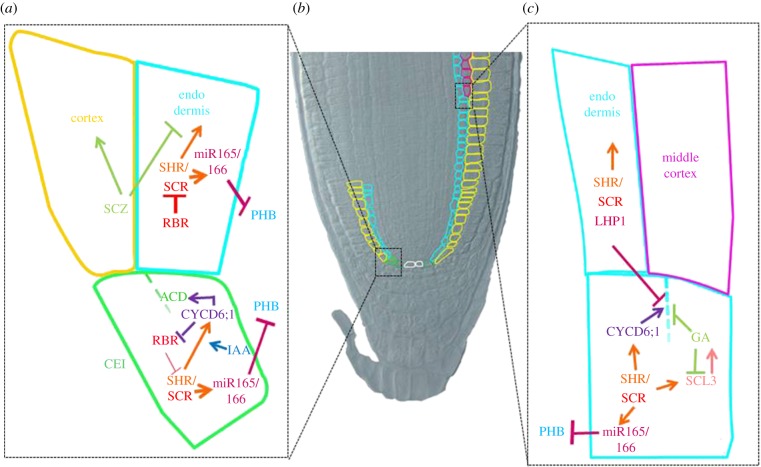
Figure 2.
Model depicting the main pathways contributing to asymmetric cell division in Arabidopsis root. (a) Cartoon representing Arabidopsis CEI and endodermis and cortex. In Arabidopsis CEI (in green), a SHR/SCR complex promotes ACD via activation of CYCD6;1 (CYCD6;1). High levels of Auxin (IAA) in the CEI contribute to CYCD6 activation, whereas CYCD6 inactivates RBR, promoting SCR activity. In the endodermis cells (light blue), RBR binds to SCR inhibiting cell division, whereas SHR instructs endodermis fate. In the cortex (yellow), SCZ promotes cortex fate and inhibits endodermis identity. SHR/SCR complex promotes miR165/6 expression restricting PHB to the stele. (b) Nomarski differential contrast interference image depicting 10 days after germination of Arabidopsis root meristem. GT cells are coloured: green (CEI), light blue (endodermis), yellow (cortex), white (QC), purple (MC). (c) Cartoon representing Arabidopsis endodermis and MC. In Arabidopsis endodermis (light blue), SCR/LHP1 complex inhibits division. When MC (purple) is forming, SCR in complex with SHR induces CYCD6 and, hence, ACD. SCL3, a target of SCR and GA, inhibits ACD and sustains GA activity. GA inhibits ACD (dashed line).
(a). A genetic network controls endodermis and cortex patterning
In recent years, several genetic pathways controlling the CEID periclinal division have been discovered in Arabidopsis. GRAS family transcription factors SHORT-ROOT (SHR) and SCARECROW (SCR) are the main fate determinant of the GT, as suggested by monolayered GT in shr and scr mutants [22,23]. Consistent with their fundamental role in GT development, SHR is expressed in the vascular tissue, but moves to the CEI, CEID and endodermis via plasmodesmata (PD) [24–27], whereas SCR is expressed in the CEI, CEID and endodermis, and sequesters SHR to the nucleus in those cells [22,24].
In the CEID, SHR directly activates SCR, increasing its expression and regulating the periclinal division of the CEID [28–31] (figure 2). Once that SCR sequesters SHR to the nucleus, they form an active transcriptional complex and together they regulate CEID division promoting the expression of CYCLIND6;1 (CYCD6;1) [29,30]. The SHR/SCR complex induces the expression of the BIRD Zinc finger proteins NUTCRACKER (NUT), JACKDAW (JKD), MAGPIE (MGP) and BALDIBIS (BIB) that act in concert with SCR to reduce SHR movements, thus establishing and maintaining the boundaries between stele and GT [26,30,32].
SHR/SCR module controls also the CEI to CEID transition acting via a bi-stable circuit that integrates radial and longitudinal information and regulates cell cycle progression [28,32–34].
Indeed, SCR directly interacts with the orthologue of the animal cell cycle regulator RETINOBLASTOMA-RELATED (RBR) protein. The RBR/SCR/SHR interaction reduces SHR/SCR complex formation and hence inhibits CEI division [32]. The molecular mechanism guiding the periclinal division of the CEID relies on the negative activity of the CYCD6;1 on RBR: CYCD6;1 mediates RBR phosphorylation, and hence its inactivation. CEID asymmetric cell division (ACD) and GT fate specification depend also on hormone activity. The longitudinal gradient of auxin, a plant hormone with morphogen-like characteristics [35], positively influences CYCD6;1 transcription supporting SHR/SCR complex formation and RBR phosphorylation. Subsequently, the decreased auxin levels reduce CYCD6;1 expression, allowing RBR to negatively regulate cell division and sustaining the acquirement of cortical and endodermal cell fate.
Interestingly, whereas SHR is required for determining endodermis specification, SCR is not involved in this process; indeed, scr monolayered GT shows both cortical and endodermal identity, whereas the shr one shows only cortex identity. Recently, it has been proposed that SHR promotes the expression of genes involved in Casparian strip formation independently of SCR, confirming the central role of SHR in determining endodermis identity [36].
Recent findings have also involved auxin in GT initiation during embryogenesis, as the auxin-dependent transcription factor MONOPTEROS (MP/AUXIN RESPONSIVE FACTOR 5/ARF5) promotes the establishment of the GT acting directly on the progenitor cells of the GT [37]. Intriguingly, MP does not require SHR/SCR module for initiating the GT, suggesting that these genes are involved mostly in the regulation of the formative divisions of a pre-formed GT layer rather than in the formation of this tissue. Further studies will be required to elucidate the molecular factors through which MP regulates GT initiation. Also, Moller et al. [37] suggest that MP activity is necessary for promoting SHR and SCR expression, as those genes are strongly downregulated in mp mutants, thus involving auxin also in the maintenance of GT patterning. Nonetheless, lack of the GT itself in mp mutants makes it difficult to understand whether MP genuinely maintains GT patterning or only regulates GT establishment.
(b). Restriction mechanism of ground tissue proliferation
The genetic mechanisms described above are necessary and sufficient to initiate and maintain formative divisions. Nevertheless, other independent mechanisms determine the confinement of the GT and fate division. One of the best-known transcription factors involved in GT fate separation is SCHIZORIZA (SCZ). SCZ is a transcription factor member of the Heat Shock family genes necessary for root cell fate separation and cortex identity (figure 2), as scz loss-of-function mutants show extra layers in the root expressing both epidermal, endodermal and cortical markers, whereas SCZ overexpressors show ectopic cortex identity specification [38,39]. Expression of SCZ in the cortex is sufficient to rescue the cortex extra layer formation in scz mutant background, suggesting that SCZ is required for cortex cell fate specification [39]. Future studies will help to clarify how SCZ controls fate separation and define cortex identity.
microRNAs (miRs), small RNA fragments acting as a repressor, have largely been linked to the GT patterning [40–42]. Mutants in genes involved in miRs biogenesis and function, such as HYPONASTIC LEAVES (HYL1) and ARGONAUTE1 (AGO1) involved in miRNA cleavage and target recognition, respectively, exhibit additional layers in the GT [41,43], suggesting an active role for miRs in GT boundary definition. In Arabidopsis microRNA165/6 family, members regulate the spatial distribution of HD-ZIPIII (HOMEO-DOMAIN LEUCINE ZIPPER III) family transcription factors such as PHABULOSA (PHB) and PHAVOLUTA (PHV). miR165/6/PHB/PHV module is involved in GT development, as transgenics with reduced miR165/6 activity and miR165/6 insensitive PHB and PHV mutants show additional cortical layers. It was shown that SHR regulates the expression of the four miR165/6 loci expressed in the GT (figure 2) [40]. Once produced in this tissue, miR165/6 migrates towards the stele via PD generating a radial gradient with a maximum in the GT and a minimum in the stele [40]. This movement results in a miR165/6 dose-dependent restriction of PHB expression that specify both xylem differentiation and GT patterning (figure 2) [40,41]. The SHR-dependent miR165/6 expression does not rule out the possibility of a parallel activity of miR165/6 to SHR and SCR in GT specification as residual expression of miR165/6 is detectable in shr mutant root [40–42]. Identification of additional components regulating miR165/6 in the GT will permit to better clarify the involvement of these genes in GT development. Moreover, it has been shown that PHB modulates CYCD6;1 expression independently on SHR [43]. Nevertheless, how the two pathways interact in the GT patterning is still an object of study.
Interestingly, small signalling molecules, such as peptides, also contribute to boundaries formation via robust transcription factor activity confinement [44]. In root, CLV3/EMBRYO SURROUNDING REGION-RELATED (CLE) peptides control root meristem size as their ectopic applications cause root meristem size reductions and additional cortical layer production [45]. PUB4, a plant U-box E3 ubiquitin ligase, was isolated as a downstream factor of CLV peptides in controlling ACDs timing [46]. Hence, cortical layer number restriction is based on the establishment and maintenance of positional information whose variation leads to a GT patterning alteration. How all those mechanisms interact is still a matter of discussion.
(c). Mechanisms controlling middle cortex development
In Arabidopsis, an additional cortex layer, called MC for its anatomical position, is formed between 7 and 10 days after germination [17,18,47]. Instead of originating from a periclinal division occurring in the CEID, the MC originates from a periclinal asynchronous division of an endodermis cell far from the QC. This division gives rise to a new layer with cortex identity [17] (figure 2).
SHR and SCR play a major role in MC formation, despite acting antagonistically in this context [15,18,48]. SHR-dependent reactivation of CYCD6;1 in the endodermis is necessary and sufficient to drive a formative endodermal division that will give rise to the MC (figure 2) [29]. On the contrary, SCR represses MC development (figure 2), as shown by in scr hypomorphic mutants [18].
Interestingly, SCR presents a dichotomous behaviour depending on its different interactors in the CEI and MC. In MC development, the interaction between SCR and the chromodomain-containing protein LIKE HETEROCHROMATIN PROTEIN1 (LHP1) determines the repression of the ACD in the GT responsible for MC formation. In accordance, lhp1 mutants show premature formation of the second longitudinal ACD similarly to scr mutants and several SHR/SCR targets are repressed by this gene. Hence, SCR might induce or repress formative divisions depending on the amount of SCR interacting with SHR or with LHP1, respectively [49]. Interestingly, LHP1 might act both as a positive and negative regulator, as in the shoot it promotes the expression of the auxin synthesis gene YUCCA4 [50]. Recent findings have shown that other epigenetic factors are also involved in GT development, for example the histone deacetylase (HDAC) family HDA19 interacts with SCR in the CEI and thus affects cortical cell fate [51]. LHP1 and HDA19 involvement in GT patterning highlights the major contribution of epigenetic regulation in post-embryonic development. In future, it will be interesting to understand how epigenetic control interferes with patterning, and whether it is related to plant ageing and/or growth environment.
Among the specific mechanisms for MC development, the plant hormone gibberellin (GA) was found to have a significant role in MC formation timing. Indeed, GA treatments are sufficient to delay the formative division of the endodermis from which the MC originates, whereas plants treated with the GA inhibitor Paclobutrazol show premature formation of MC [18,49]. SCR/SHR and GA pathways convey on the regulation of the transcriptional regulator SCARECROW LIKE 3 (SCL3) in MC development. Indeed, SCL3 is activated by SHR/SCR and repressed by GA [28,52] (figure 2). Interestingly, SCL3 regulates positively GA activity, controlling the timing of MC formation (figure 2).
Recently, a vacuolar sorting protein involved in protein recycling and interacting with SHR, SHRUBBY (SHBY), was shown to play a role in integrating SCR/SHR and GA pathways [53]. SHBY inhibits SHR activity in the MC and positively regulates GA signalling via an unidentified mechanism, preventing the formative division generating the MC [53]. This interesting finding suggests that MC formation is not only subject to a tight transcriptional regulation, but it is also finely regulated by protein turnover.
An interesting case in MC patterning is represented by the role of SPINDLY (SPY). SPY is an O-linked glucosamine acetyltransferase with GA response–repressive functions [54]. Although spy mutants present high GA levels, premature MC formation can be observed in this background [52]. Because SPY homologues in animals interact with histone deacetylase [55], recent theories posit that SPY might control MC formation epigenetically. Nevertheless, the lack of direct evidence of SPY involvement in epigenetic MC control does not exclude the possibility of additional molecular pathways controlled by SPY. For instance, it was recently shown that SPY has a role in maintaining cellular redox homeostasis and that oxidative stress induces MC formation [56]. In particular, premature MC formation in the spy mutant is suppressed by a reducing agent, while it is induced by H2O2 treatment [54]. This suggests that the increase in the number of cortex layers is a developmental response to oxidative stress. In this way, this regulation of cortex proliferation would result in a protective response carried out by plants to limit the entry of harmful elements and maintain a healthy redox state of the cell. In accordance, spy mutant is more tolerant to high salt concentration in the soil [57], supporting the idea that the cortex contributes to counteracting soil-based abiotic stress [58].
(d). Mechanisms of cortex proliferation in species with multiple cortex layer
In nature cortex, proliferation represents a chance for plants to adapt to their ecological niches. In rice roots, overexpression of the NAC domain protein NAC10 results in enhanced root diameter due to increased cortical, epidermal and stele size. Intriguingly, those plants are better adapted to stress than the wild-type ones, most probably thanks to their cortical system [55]. Most of the species, from ferns to angiosperms, present several cortical layers. The additional formative divisions, at the basis of those multiple cortex layer formation, happen in precise positions early in development [13]. As the model system Arabidopsis develops only one cortical layer, the isolation of a feasible model system for multiple cortex layer development is mandatory. In O. sativa, the most diffused monocot model system, root architecture, is compounded by a series of adventitious roots, called crown roots, surrounding a primary root (radicule), carrying several lateral roots [56]. All the roots of rice present similar anatomy with the exception of different cortex layer number (one in the lateral roots and over 10 in the crown roots) [15,20,59]. In the rice stem cell niche, the CEI gives rise to the epidermis and the CEID. An additional formative division of the CEID originates endodermis and cortex [15]. Subsequently, other periclinal asymmetric divisions occur, giving rise to multiple cortex layers [15]. Usage of immunohistochemical markers suggests a different identity of inner and outer rice cortical layers [20]. Future studies will clarify the physiological and developmental differences among the two tissues.
In recent years, several findings have suggested an active role for SHR in cortical layer number determination in rice. Two orthologues of SHR and SCR are present in the rice genome. In situ hybridization and two hybrid system experiments on these genes support the idea that the OsSHR/SCR module controls endodermis development similarly to Arabidopsis, where SHR movements are limited to the endodermis by the interaction with SCR determining the identity of this tissue [28,60]. Nevertheless, it is still debated whether SHR plays a central role in cortical layer number. Corroborating this hypothesis, OsSHR2 or a B. distachyon orthologue of SHR (BdSHR) in Arabidopsis is sufficient to generate extra layers with cortex identity in Arabidopsis [61]. In Arabidopsis, OsSHR1/OsSHR2/BdSHR moves from the stele to the cortex, triggering the SCR/CYCD6;1 circuit and, hence, causing extra divisions in the GT [61]. Moreover, transgenic rice plants overexpressing SHR show an increase in outer cortical layer number [62]. Immunolocalizations of SHR2 in rice have shown that SHR2 protein is detectable in both endodermis and outer cortical layers. These data suggest that SHR might play a key role in multi-layered cortical patterning; nevertheless, the lack of OsSHR1/2 and OsSCR1/2 mutants and tissue-specific complementation makes it difficult to understand the specific role of SHR/SCR in rice GT development.
(e). Cardamine hirsuta, a model system for comparative development studies: a future perspective
The usage of closely related species has emerged as a successful strategy to understand the molecular differences that underline interspecific variability [1,63–65]. Among the close relative of Arabidopsis exhibiting multiple cortical layers, C. hirsuta represents a breakthrough in our understanding of the genetic basis of root anatomical diversity. In recent years, Cardamine emerged as a powerful system to identify molecular mechanisms at the base of biological diversity in leaf morphology and petal and fruit development [66–73]. Cardamine suits most of the characteristic of a model system. It is diploid, it possesses a completely sequenced and annotated small genome (196 mega bases) [74,75], it is self-compatible and it has a short life cycle (about four months) [74]. Cardamine can be transformed via Agrobacterium tumefaciens-based floral dip methodology with a high efficiency [74]. The readiness of the genetic tractability of Cardamine permits the exploitation of genetic screens and gene expression analysis and manipulation. Cardamine and Arabidopsis present several morphological divergent traits in the root [74]. Macroscopically, Cardamine primary root emerges from seed and produces several adventitious roots about 6 days after germination. Microscopically, Cardamine primary root presents anatomical traits divergent from Arabidopsis such as root meristem size, statoliths distribution and number of cortical layers [74]. Accurate analysis of Cardamine root GT patterning demonstrated that Cardamine show two cortical layers (an outer and an inner one) originating from a developmental domain of mix cortex and endodermis identity (CEM) absent in Arabidopsis. In this species, the CEI firstly divides asymmetrically giving rise to a cortex layer and a CEM that subsequently divides periclinally originating the endodermis and an inner cortex (figure 2b) [43]. The Cardamine inner and outer cortical layers are patterned by stereotypical division happening during embryogenesis. We recently found out that HD-ZIPIII members pattern Cardamine GT. In Cardamine, as in Arabidopsis, five loci encoding five HD-ZIPIII transcription factors (ChPHABULOSA; ChPHAVALUTA; ChCORONA; ChREVOLUTA; ChHB8) are present. As in Arabidopsis, their expression is modulated by the activity of miR165/6. In Cardamine, knockdown of those transcription factors results in the absence of the inner cortical layer, suggesting that the activity of those genes is necessary for Cardamine additional cortex formation [43]. Intriguingly, in Cardamine, miR165/6 activity is low in the CEM, generating a broader expression domain of PHB that is therefore expressed in this tissue [43]. As in Arabidopsis PHB directs CYCD6;1 expression, it is tempting to speculate that PHB directs formative divisions enriching Cardamine GT anatomy via cell cycle regulation. However, further studies will elucidate how PHB and the other HD-ZIPIII are involved in Cardamine GT patterning. Also, it will be interesting to understand whether HD-ZIPIII are necessary for CEM formation or whether their function is required only for formative division regulation in the CEM. Future research will shed light on the conservation of both patterning and molecular processes underlying GT variability. Whether the development of multiple cortical layers in plants is dependent on the presence of additional stem cells, CEM or both is still a matter for discussion. One possibility is that in species showing several cortical layers, the outer cortex depends on the activity of extra stem cells, whereas the inner cortical layer originates from CEM (figure 1). From this perspective, different molecular mechanisms might act in controlling cortex proliferation. On the one hand, SHR/SCR circuit might regulate additional CEI activity, hence, generating the outer layers; on the other hand, HD-ZIPIII might regulate additional divisions of the inner cortical layers. This hypothesis is also supported by recent findings showing that overexpression of SHR in rice leads to an increment in outer cortical layer formation [62]. Hence, it is fundamental to understand how SHR/SCR and HD-ZIPIIIs coordinate their activity to determine plant cortical layer variability. More studies on novel root monocot and dicot model system showing multiple cortical layers will permit to better elucidate the mechanisms at the basis of the variability of cortex patterning. Also, whether the knowledge acquired by studying close meristem species such as rice and Cardamine is applicable also in open meristem species such as pea is still completely unexplored.
Acknowledgements
We acknowledge A. Rocchetti, S. Sabatini, E. Salvi, E. Pacifici, G. Bertolotti and E. Pierdonati for constructive discussions on the manuscript. We also acknowledge the referees for the valuable comments and suggestions which have led to significant improvement on the presentation and quality of this paper.
Data accessibility
No original data were reported.
Authors' contributions
All of the authors were involved in the development and writing of this article.
Competing interests
We claim no competing interests.
Funding
This work was supported by an FIRB (Futuro in Ricerca 2013) project grant from the Ministero dell'Istruzione, dell'Università e della Ricerca (FIRB2013-RBFR13DCDS to G.D.R. and R.D.I.).
References
- 1.Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433, 481–487. ( 10.1038/nature03235) [DOI] [PubMed] [Google Scholar]
- 2.Wittkopp PJ, Stewart EE, Arnold LL, Neidert AH, Haerum BK, Thompson EM, Akhras S, Smith-Winberry G, Shefner L. 2009. Intraspecific polymorphism to interspecific divergence: genetics of pigmentation in Drosophila. Science 326, 540–544. ( 10.1126/science.1176980) [DOI] [PubMed] [Google Scholar]
- 3.Nikolov LA, Tsiantis M. 2017. Using mustard genomes to explore the genetic basis of evolutionary change. Curr. Opin. Plant Biol. 36, 119–128. ( 10.1016/j.pbi.2017.02.005) [DOI] [PubMed] [Google Scholar]
- 4.Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84. [DOI] [PubMed] [Google Scholar]
- 5.Scheres B. 2000. Non-linear signaling for pattern formation? Curr. Opin Plant Biol. 3, 412–417. ( 10.1016/S1369-5266(00)00105-9) [DOI] [PubMed] [Google Scholar]
- 6.Weigel D, Jurgens G. 2002. Stem cells that make stems. Nature 415, 751–754. ( 10.1038/415751a) [DOI] [PubMed] [Google Scholar]
- 7.Steeves TA, Sussex IM. 1989. Patterns in plant development, 2nd edn Cambridge, UK: Cambridge University Press. [Google Scholar]
- 8.Scheres B. 2007. Stem-cell niches: nursery rhymes across kingdoms. Nat. Rev. Mol. Cell Biol. 8, 345–354. ( 10.1038/nrm2164) [DOI] [PubMed] [Google Scholar]
- 9.Fahn A. 1990. Plant anatomy, 4th edn Oxford, UK: Pergamon. [Google Scholar]
- 10.Timmers AC, Auriac MC, Truchet G. 1999. Refined analysis of early symbiotic steps of the Rhizobium–Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126, 3617–3628. [DOI] [PubMed] [Google Scholar]
- 11.Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T. 2014. Fate map of Medicago truncatula root nodules. Development 141, 3517–3528. ( 10.1242/dev.110775) [DOI] [PubMed] [Google Scholar]
- 12.Paungfoo-Lonhienne C, et al. 2008. Plants can use protein as a nitrogen source without assistance from other organisms. Proc. Natl Acad. Sci. USA 105, 4524–4529. ( 10.1073/pnas.0712078105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heimsch C, Seago JL Jr. 2008. Organization of the root apical meristem in angiosperms. Am. J. Bot. 95, 1–21. ( 10.3732/ajb.95.1.1) [DOI] [PubMed] [Google Scholar]
- 14.Clowes FAL. 1994. Origin of the epidermis in root meristems. New Phytol. 127, 335–347. ( 10.1111/j.1469-8137.1994.tb04284.x) [DOI] [PubMed] [Google Scholar]
- 15.Pauluzzi G, Divol F, Puig J, Guiderdoni E, Dievart A, Perin C. 2012. Surfing along the root ground tissue gene network. Dev. Biol. 365, 14–22. ( 10.1016/j.ydbio.2012.02.007) [DOI] [PubMed] [Google Scholar]
- 16.Kirschner GK, Stahl Y, Von Korff M, Simon R. 2017. Unique and conserved features of the barley root meristem. Front. Plant Sci. 8, 1240 ( 10.3389/fpls.2017.01240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baum SF, Dubrovsky JG, Rost TL. 2002. Apical organization and maturation of the cortex and vascular cylinder in Arabidopsis thaliana (Brassicaceae) roots. Am. J. Bot. 89, 908–920. ( 10.3732/ajb.89.6.908) [DOI] [PubMed] [Google Scholar]
- 18.Paquette AJ, Benfey PN. 2005. Maturation of the ground tissue of the root is regulated by gibberellin and SCARECROW and requires SHORT-ROOT. Plant Physiol. 138, 636–640. ( 10.1104/pp.104.058362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark LH, Harris WH. 1981. Observations on the root anatomy of rice (Oryza sativa L.). Am. J. Botany 68, 154–161. ( 10.2307/2442846) [DOI] [Google Scholar]
- 20.Henry S, Divol F, Bettembourg M, Bureau C, Guiderdoni E, Perin C, Dievart A. 2015. Immunoprofiling of rice root cortex reveals two cortical subdomains. Front. Plant Sci. 6, 1139 ( 10.3389/fpls.2015.01139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Popham RA. 1966. Laboratory manual for plant anatomy. Ann Arbor, MI: CV Mosby, University of Michigan. [Google Scholar]
- 22.Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. 1996. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433. ( 10.1016/S0092-8674(00)80115-4) [DOI] [PubMed] [Google Scholar]
- 23.van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. 1995. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378, 62–65. ( 10.1038/378062a0) [DOI] [PubMed] [Google Scholar]
- 24.Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. 2000. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567. ( 10.1016/S0092-8674(00)80865-X) [DOI] [PubMed] [Google Scholar]
- 25.Nakajima K, Sena G, Nawy T, Benfey PN. 2001. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413, 307–311. ( 10.1038/35095061) [DOI] [PubMed] [Google Scholar]
- 26.Sena G, Jung JW, Benfey PN. 2004. A broad competence to respond to SHORT ROOT revealed by tissue-specific ectopic expression. Development 131, 2817–2826. ( 10.1242/dev.01144) [DOI] [PubMed] [Google Scholar]
- 27.Vaten A, et al. 2011. Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell 21, 1144–1155. ( 10.1016/j.devcel.2011.10.006) [DOI] [PubMed] [Google Scholar]
- 28.Cui H, et al. 2007. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316, 421–425. ( 10.1126/science.1139531) [DOI] [PubMed] [Google Scholar]
- 29.Sozzani R, et al. 2010. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466, 128–132. ( 10.1038/nature09143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long Y, et al. 2017. In vivo FRET-FLIM reveals cell-type-specific protein interactions in Arabidopsis roots. Nature 548, 97–102. ( 10.1038/nature23317) [DOI] [PubMed] [Google Scholar]
- 31.Clark NM, Hinde E, Winter CM, Fisher AP, Crosti G, Blilou I, Gratton E, Benfey PN, Sozzani R. 2016. Tracking transcription factor mobility and interaction in Arabidopsis roots with fluorescence correlation spectroscopy. eLife 5, e14770 ( 10.7554/eLife.14770) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B. 2007. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 21, 2196–2204. ( 10.1101/gad.440307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heidstra R, Welch D, Scheres B. 2004. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 18, 1964–1969. ( 10.1101/gad.305504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrell JE., Jr 2002. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin Cell Biol. 14, 140–148. ( 10.1016/S0955-0674(02)00314-9) [DOI] [PubMed] [Google Scholar]
- 35.Di Mambro R, et al. 2017. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc. Natl Acad. Sci. USA 114, E7641–E7649. ( 10.1073/pnas.1705833114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li P, et al. 2018. Construction of a functional casparian strip in non-endodermal lineages is orchestrated by two parallel signaling systems in Arabidopsis thaliana. Curr. Biol. 28, 2777–2786. ( 10.1016/j.cub.2018.07.028) [DOI] [PubMed] [Google Scholar]
- 37.Moller BK, Ten Hove CA, Xiang D, Williams N, Lopez LG, Yoshida S, Smit M, Datla R, Weijers D. 2017. Auxin response cell-autonomously controls ground tissue initiation in the early Arabidopsis embryo. Proc. Natl Acad. Sci. USA 114, E2533–E2539. ( 10.1073/pnas.1616493114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mylona P, Linstead P, Martienssen R, Dolan L. 2002. SCHIZORIZA controls an asymmetric cell division and restricts epidermal identity in the Arabidopsis root. Development 129, 4327–4334. [DOI] [PubMed] [Google Scholar]
- 39.ten Hove CA, Willemsen V, de Vries WJ, van Dijken A, Scheres B, Heidstra R. 2010. SCHIZORIZA encodes a nuclear factor regulating asymmetry of stem cell divisions in the Arabidopsis root. Curr. Biol. 20, 452–457. ( 10.1016/j.cub.2010.01.018) [DOI] [PubMed] [Google Scholar]
- 40.Carlsbecker A, et al. 2010. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321. ( 10.1038/nature08977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyashima S, Koi S, Hashimoto T, Nakajima K. 2011. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138, 2303–2313. ( 10.1242/dev.060491) [DOI] [PubMed] [Google Scholar]
- 42.Miyashima S, Honda M, Hashimoto K, Tatematsu K, Hashimoto T, Sato-Nara K, Okada K, Nakajima K. 2013. A comprehensive expression analysis of the Arabidopsis MICRORNA165/6 gene family during embryogenesis reveals a conserved role in meristem specification and a non-cell-autonomous function. Plant Cell Physiol. 54, 375–384. ( 10.1093/pcp/pcs188) [DOI] [PubMed] [Google Scholar]
- 43.Di Ruocco G, Bertolotti G, Pacifici E, Polverari L, Tsiantis M, Sabatini S, Costantino P, Dello Ioio R. 2018. Differential spatial distribution of miR165/6 determines variability in plant root anatomy. Development 145, 153858 ( 10.1242/dev.153858) [DOI] [PubMed] [Google Scholar]
- 44.Wang G, Fiers M. 2010. CLE peptide signaling during plant development. Protoplasma 240, 33–43. ( 10.1007/s00709-009-0095-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fiers M, Golemiec E, Xu J, van der Geest L, Heidstra R, Stiekema W, Liu CM. 2005. The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17, 2542–2553. ( 10.1105/tpc.105.034009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kinoshita A, et al. 2015. A plant U-box protein, PUB4, regulates asymmetric cell division and cell proliferation in the root meristem. Development 142, 444–453. ( 10.1242/dev.113167) [DOI] [PubMed] [Google Scholar]
- 47.Koizumi K, Hayashi T, Wu S, Gallagher KL. 2012. The SHORT-ROOT protein acts as a mobile, dose-dependent signal in patterning the ground tissue. Proc. Natl Acad. Sci. USA 109, 13 010–13 015. ( 10.1073/pnas.1205579109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. 2000. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127, 595–603. [DOI] [PubMed] [Google Scholar]
- 49.Cui H, Benfey PN. 2009. Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J. 58, 1016–1027. ( 10.1111/j.1365-313X.2009.03839.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rizzardi K, Landberg K, Nilsson L, Ljung K, Sundas-Larsson A. 2011. TFL2/LHP1 is involved in auxin biosynthesis through positive regulation of YUCCA genes. Plant J. 65, 897–906. ( 10.1111/j.1365-313X.2010.04470.x) [DOI] [PubMed] [Google Scholar]
- 51.Chen W, Drapek C, Li D, Xu Z, Benfey P, Bai S. 2018. Histone deacetylase HDA19 affects cortical cell fate by interacting with SCARECROW in the Arabidopsis root. bioRxiv. ( 10.1101/313791) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heo JO, Chang KS, Kim IA, Lee MH, Lee SA, Song SK, Lee MM, Lim J. 2011. Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc. Natl Acad. Sci. USA 108, 2166–2171. ( 10.1073/pnas.1012215108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koizumi K, Gallagher KL. 2013. Identification of SHRUBBY, a SHORT-ROOT and SCARECROW interacting protein that controls root growth and radial patterning. Development 140, 1292–1300. ( 10.1242/dev.090761) [DOI] [PubMed] [Google Scholar]
- 54.Cui H, Kong D, Wei P, Hao Y, Torii KU, Lee JS, Li J. 2014. SPINDLY, ERECTA, and its ligand STOMAGEN have a role in redox-mediated cortex proliferation in the Arabidopsis root. Mol. Plant 7, 1727–1739. ( 10.1093/mp/ssu106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK. 2010. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197. ( 10.1104/pp.110.154773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith S, De Smet I. 2012. Root system architecture: insights from Arabidopsis and cereal crops. Proc. R. Soc. B. 367, 1441–1452. ( 10.1098/rstb.2011.0234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qin F, Kodaira KS, Maruyama K, Mizoi J, Tran LS, Fujita Y, Morimoto K, Shinozaki K, Yamaguchi-Shinozaki K. 2011. SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 157, 1900–1913. ( 10.1104/pp.111.187302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui H. 2015. Cortex proliferation in the root is a protective mechanism against abiotic stress. Plant Signal. Behav. 10, e1011949 ( 10.1080/15592324.2015.1011949) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coudert Y, Perin C, Courtois B, Khong NG, Gantet P. 2010. Genetic control of root development in rice, the model cereal. Trends Plant Sci. 15, 219–226. ( 10.1016/j.tplants.2010.01.008) [DOI] [PubMed] [Google Scholar]
- 60.Kamiya N, Itoh J, Morikami A, Nagato Y, Matsuoka M. 2003. The SCARECROW gene's role in asymmetric cell divisions in rice plants. Plant J. 36, 45–54. ( 10.1046/j.1365-313X.2003.01856.x) [DOI] [PubMed] [Google Scholar]
- 61.Wu S, Lee CM, Hayashi T, Price S, Divol F, Henry S, Pauluzzi G, Perin C, Gallagher KL. 2014. A plausible mechanism, based upon short-root movement, for regulating the number of cortex cell layers in roots. Proc. Natl Acad. Sci. USA 111, 16 184–16 189. ( 10.1073/pnas.1407371111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henry S, Dievart A, Divol F, Pauluzzi G, Meynard D, Swarup R, Wu S, Gallagher KL, Perin C. 2017. SHR overexpression induces the formation of supernumerary cell layers with cortex cell identity in rice. Dev. Biol. 425, 1–7. ( 10.1016/j.ydbio.2017.03.001) [DOI] [PubMed] [Google Scholar]
- 63.Stern DL, Orgogozo V. 2009. Is genetic evolution predictable? Science 323, 746–751. ( 10.1126/science.1158997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Romero IG, Ruvinsky I, Gilad Y. 2012. Comparative studies of gene expression and the evolution of gene regulation. Nat. Rev. Genet. 13, 505–516. ( 10.1038/nrg3229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mentink RA, Tsiantis M. 2015. From limbs to leaves: common themes in evolutionary diversification of organ form. Front. Genet. 6, 284 ( 10.3389/fgene.2015.00284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hay A, Barkoulas M, Tsiantis M. 2006. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133, 3955–3961. ( 10.1242/dev.02545) [DOI] [PubMed] [Google Scholar]
- 67.Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. 2008. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40, 1136–1141. ( 10.1038/ng.189) [DOI] [PubMed] [Google Scholar]
- 68.Vlad D, et al. 2014. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343, 780–783. ( 10.1126/science.1248384) [DOI] [PubMed] [Google Scholar]
- 69.Pieper B, Monniaux M, Hay A. 2016. The genetic architecture of petal number in Cardamine hirsuta. New Phytol. 209, 395–406. ( 10.1111/nph.13586) [DOI] [PubMed] [Google Scholar]
- 70.Kougioumoutzi E, et al. 2013. SIMPLE LEAF3 encodes a ribosome-associated protein required for leaflet development in Cardamine hirsuta. Plant J. 73, 533–545. ( 10.1111/tpj.12072) [DOI] [PubMed] [Google Scholar]
- 71.Rast-Somssich MI, et al. 2015. Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta. Genes Dev. 29, 2391–2404. ( 10.1101/gad.269050.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hofhuis H, et al. 2016. Morphomechanical innovation drives explosive seed dispersal. Cell 166, 222–233. ( 10.1016/j.cell.2016.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vuolo F, Mentink RA, Hajheidari M, Bailey CD, Filatov DA, Tsiantis M. 2017. Corrigendum: coupled enhancer and coding sequence evolution of a homeobox gene shaped leaf diversity. Genes Dev. 31, 2199 ( 10.1101/gad.309179.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hay AS, et al. 2014. Cardamine hirsuta: a versatile genetic system for comparative studies. Plant J. 78, 1–15. ( 10.1111/tpj.12447) [DOI] [PubMed] [Google Scholar]
- 75.Gan X, et al. 2016. The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat. Plants 2, 16167 ( 10.1038/nplants.2016.167) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No original data were reported.