Abstract
Transmission of bacteria vertically through host tissues ensures offspring acquire symbionts; however, horizontal transmission is an effective strategy for many associations and plays a role in some vertically transmitted symbioses. The bivalve Solemya velum and its gammaproteobacterial chemosynthetic symbionts exhibit evolutionary evidence of both transmission modes, but the dominant strategy on an ecological time scale is unknown. To address this, a specific primer set was developed and validated for the S. velum symbiont using a novel workflow called specific marker design (SMD). Symbionts were quantified in spawned eggs and sediment and seawater samples from S. velum habitats with qPCR. Each egg was estimated to contain 50–100 symbiont genomes. By contrast, symbiont DNA was found at low abundance/occurrence in sediment and seawater, often co-occurring with host mitochondrial DNA, obscuring its origin. To ascertain when eggs become infected, histological sections of S. velum tissues were labelled for symbiont 16S rRNA via in situ hybridization. This revealed symbionts in the ovary walls and mature oocytes, suggesting association in late oogenesis. These data support the hypothesis that S. velum symbionts are vertically transmitted every host generation, thus genetic signatures of horizontal transmission are driven by ecologically infrequent events. This knowledge furthers our understanding of vertical and horizontal mode integration and provides insights across animal–bacterial chemosynthetic symbioses.
Keywords: symbiosis, transmission mode, fluorescence in situ hybridization, qPCR, transmission bottleneck, environmental DNA
1. Introduction
Numerous eukaryotes engage in mutualistic associations with bacteria, which result in the evolution of novel metabolisms and tissue adaptations, enabling the colonization of new niches. In these intimate associations, bacterial symbionts colonize hosts either extracellularly or intracellularly, and often exhibit tissue tropism, wherein they inhabit very specific tissue types [1]. Localization patterns correspond to two aspects: the functional purpose of the association (e.g. gut bacteriomes, gills, etc.) and the symbiont transmission route between host generations (e.g. the ovary). The transmission of symbionts to new host individuals directly from parent tissues is termed vertical transmission, whereas the transmission of symbionts through an environmental intermediate is termed horizontal transmission [2,3]. Thus, mechanisms facilitating host–symbiont recognition, localization and density ensure proper symbiont functioning both within host individuals [4] as well as between host generations [5,6].
Vertical transmission ensures that symbionts reach the next host generation and aligns the fitnesses of hosts and symbionts [2]. Direct inheritance also mitigates the risk of not acquiring symbionts, as timing is important if symbiont function is needed or if colonization capability has a narrow efficacy window. While common themes and mechanisms have been demonstrated in insect symbioses [2,7], much less is known about vertical transmission in the marine environment. Many marine animals reproduce by broadcast spawning or have very exposed larval stages [8], thus vertical transmission in these species occurs through symbiont association with host gametes or brooded offspring. Additionally, eggs and early developmental stages are in direct contact with the surrounding seawater, which may increase opportunities for symbionts to mix between hosts or other bacteria to be acquired.
An intermediate mode of transmission using both vertical and horizontal transmission strategies, termed mixed-mode transmission [2,3], is reported for several associations (e.g. [9,10]) and may be the dominant context in which vertical transmission occurs in the marine environment. The vast majority of marine symbioses are horizontally transmitted [2], and the ones that undergo vertical transmission experience occasional horizontal events [11,12]. Both vertical and horizontal transmissions have clear fitness benefits. Specifically, vertical transmission ensures offspring–symbiont association and horizontal transmission may mitigate the deleterious effects of continuous host restriction (i.e. small population size, transmission bottlenecks, etc.) and enable rapid host local adaptation; therefore, both could be selected for under different circumstances. While it is not yet clear whether mixed modes are an unintended artefact of bacterial dispersal and colonization abilities in the marine environment or a consequence of selection pressure to maintain both modes, they are probably more prevalent than appreciated and have major implications for symbiont evolution [10].
The symbiosis between the marine bivalve Solemya velum and its chemosynthetic gammaproteobacterial symbionts undergoes mixed-mode transmission, prompting the question of how and when each mode is accomplished in the life cycle of this species. Solemya velum is found in reducing sediments along the Atlantic coast of North America, where it digs a Y-shaped burrow to access reduced pore water. Symbionts are contained within specialized gill cells, termed bacteriocytes, giving them access to the electron acceptor oxygen, in addition to sulfide and carbon dioxide for carbon fixation, fuelling both host and symbionts [13]. Symbiont 16S rRNA sequences are identical within hosts and nearly identical within geographical localities, indicating this is a highly specific association consisting of a single bacterial lineage [10,13]. Early research concluded that symbionts are vertically transmitted based upon detection of symbiont DNA in host ovaries via PCR and bacterial cells in developing juvenile gill buds within the egg capsule [14]. Recent genomic data revealed evidence of evolutionarily frequent rates of horizontal transmission [10,15], prompting a more detailed analysis of the life history of the S. velum symbiont to compare the dynamics of these transmission modes at ecological time scales (e.g. within host tissues and environmental substrates, such as seawater and sediment).
Interrogating the life history of a bacterial symbiont is not trivial. The majority of genetic markers developed to uniquely identify bacteria in samples containing diverse genotypes are highly conserved. The most commonly employed markers include 16S ribosomal RNA, cell division contractile ring (ftsZ) and RNA polymerase subunit B (rpoB), among others [16,17]. However, none are variable enough within most bacterial strains, species, genera or even families to be useful in detecting genetic variation at these levels (see [18,19]). Similarly, the S. velum symbiont is closely related to free-living bacteria at the 16S locus [10,13] and across the genome [20], making earlier attempts to specifically detect it in the environment ineffective (S.L.R. 2012, unpublished data). Thus, a method was needed to identify markers that are sufficiently diverse to resolve strain-level differences, but conserved enough to the designed primer sets effective across the genetic diversity of the S. velum symbionts.
In this paper, we investigated whether the ecological transmission route of the S. velum symbiont reflects the high rates of mixed mode transmission evident in its evolutionary genomics. First, as horizontal transmission events should be more likely if symbionts are at high abundance in the environment, we quantified symbiont DNA in sediment and water samples as a proxy for horizontal transmission frequency. Next, we quantified symbionts associated with spawned host oocytes and ovary tissues as a proxy for the reliability of vertical transmission, as failure to transmit at this stage could necessitate horizontal transmission and drive the mixed-mode signal. To accomplish these aims, first, the gross morphology of whole female S. velum was characterized to inform on possible routes of symbiont entry to/exit from host tissues. Next, we developed a workflow called specific marker design (SMD) (see electronic supplementary material, figure S1), which incorporates population-level genetic variation for the identification and design of genetic markers to specifically detect the S. velum symbionts. Third, using the new primers, symbiont abundance was quantified in spawned oocytes and environmental water and sediment samples via quantitative PCR (qPCR). Finally, in situ hybridization (ISH) was performed on adult S. velum tissues to determine the symbiont route of entry into host oocytes. Understanding the balance between mixed transmission routes in S. velum will inform on the transmission strategies available to symbionts of marine invertebrates broadly.
2. Material and methods
(a). Collections
For spawning and whole-bivalve tissue samples (e.g. figure 1), adult S. velum (7.5–14.5 mm long) were collected from an intertidal mudflat in Point Judith, Narragansett, Rhode Island with a shovel and sieve. Bivalves were either fixed in the field for microscopy (n = 23; 21 females and 2 males) or transported back to the laboratory for spawning (approx. 30 individuals; see electronic supplementary material, Material and methods for spawning protocol). Collections of female S. velum for microscopy were spread across the following months to assess ovary maturity throughout the year: March (n = 5), June (n = 4), July (n = 4), September (n = 5) and November (n = 6). Males collected for light microscopy were sampled in July and November.
Figure 1.
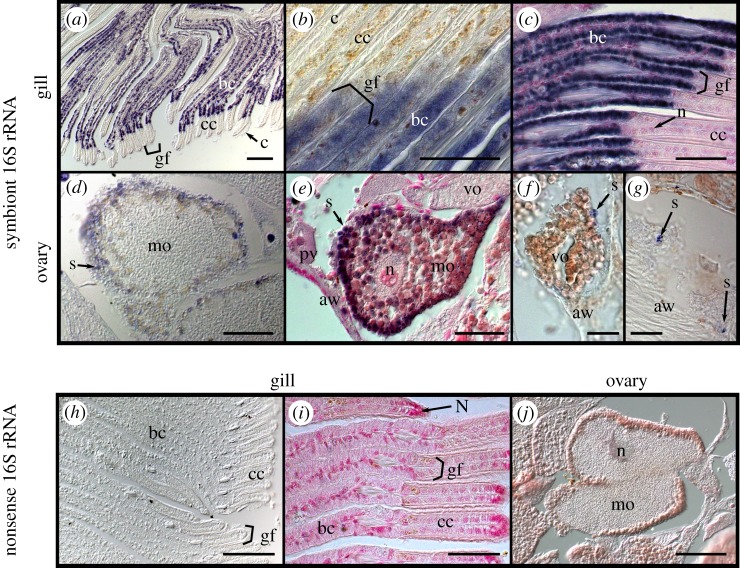
Tissue ultrastructure of female S. velum imaged with differential interference contrast (DIC) microscopy. (a) Section of whole S. velum removed from shell hybridized with the symbiont probe Svsym47 (purple) and counterstained in nuclear fast red (pink/red). (b) Close-up of gill filaments stained with nuclear fast read. (c,d) Haematoxylin eosin (HE) stained (c) gill filaments and (d) ovary within foot. Abbreviations: (a) acinus outlined by bracket, (b) bacteriocyte, (c) ciliated cell, (f) foot, (g) gill, (gf) gill filament, (h) hypobranchial gland, (mo) mature ooctye, (N) nucleus, (oo) oogonium, (ov) ovary, (pv) previtellogenic oocyte, (st) stomach, (ve) vitelline envelope, (vo) vitellogenic oocyte. Scale bars: (a) 1 mm, (b–d) 100 µm.
Sediment and seawater samples were collected from habitats of the bivalve Solemya velum along the east coast of North America (figure 2b; electronic supplementary material, table S1). Sediment samples were obtained using a shovel to dig to the depth at which S. velum occurs, near a S. velum burrow if possible, and a sterile spatula to collect approximately 0.25 g into each pre-weighed PowerSoil (MoBio, Carlsbad, CA, USA) tube. Tissues were flash frozen in cryotubes in a dry ice-ethanol slurry the field. Large volumes of sediment from RI were frozen in 15 ml tubes for extraction with the PowerMax kit (MoBio). For seawater samples, a 150 ml syringe was used to push from 130 to 400 ml of seawater, depending on the turbidity, through a 25 mm wide 0.22 µm nitrocellulose filter (Millipore). Filters were flash frozen in 15 ml tubes and the whole filter was later extracted with PowerWater kits (MoBio). Water samples of 1 l each were taken with a filter rig (100 µm glass fibre prefilter and 0.22 µm nitrocellulose filter; Millipore) and pump from Rhode Island, and approximately one-eighth of each was extracted at a time. Gill samples from two adult S. velum from Point Judith were dissected and extracted with the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) for primer testing and as positive controls in qPCR.
Figure 2.
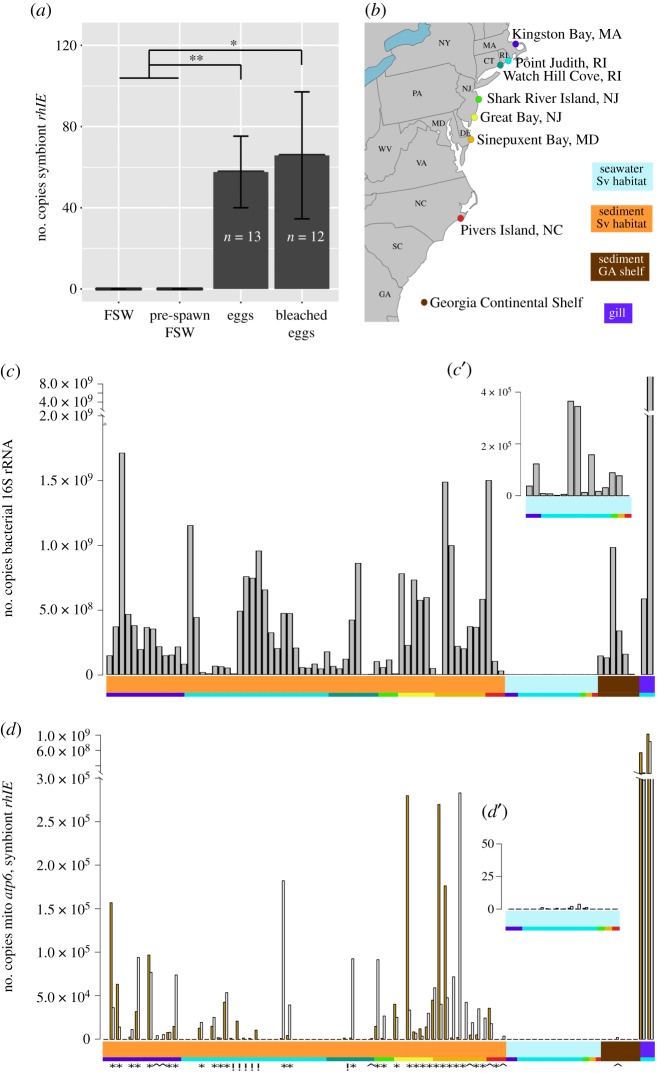
Quantification of S. velum symbionts in spawned host eggs and environmental samples with the newly designed symbiont-specific rhlE primer set. (a) Copies of symbiont rhlE detected by qPCR in untreated and bleached eggs spawned in the laboratory. Adults for spawning experiments were collected from Point Judith, RI (see (b), adapted from [10]). The number of rhlE copies per egg or per ml of filtered seawater (FSW) is reported. There is no significant difference between the mean counts for bleached and untreated eggs, whereas both contained significantly more than the seawater used for spawning (Welch's two-sample t-test; *p < 0.05, **p < 0.01). (b) Collection localities for S. velum sediment and water samples, marked with coloured dots that correspond to the bottom row of coloured blocks below plots (c,d). The coloured blocks with sample type descriptions correspond to the top row of coloured blocks. Copy numbers of qPCR-quantified (c) bacterial 16S rRNA (grey bars), (d) symbiont rhlE (yellow bars) and (d) mitochondrial atp6 (white bars) loci from individual environmental samples shown in copies per gram sediment, millilitre seawater or gram gill tissue. Inset graphs c’,d’ show the FSW samples at higher y-axis magnification. Counts are ordered along the x-axis by sample type and locality. Symbols below the coloured blocks in (d) indicate samples in which (*) both symbiont and mitochondrial sequences, (^) only mitochondria or (!) only symbionts were detected.
(b). Light microscopy
(i). Fixation and embedding
Solemya velum were fixed whole and used for the characterization of gross morphology and detection of symbionts in host tissues with ISH (n = 24). After cutting the adductor muscles, specimens were fixed in Davidson's fixative [21] for 48 h at 4°C with agitation, rinsed in 1× phosphate buffered saline (PBS; pH 7.4) and dehydrated through an ethanol series. Shells were removed from specimens while immersed in 70% ethanol. Specimens were infiltrated with xylene and embedded in paraffin, applying a vacuum during the final paraffin incubation (−15 mm Hg) to complete infiltration. Paraffin-embedded whole S. velum specimens were serially sectioned at 10 µm (Leica Rm2155 microtome), 5–10 sections were collected per Superfrost Plus slide (VWR, Pennsylvania, USA) and slides were heated at 42°C overnight to dry. On average, 50–100 sections were mounted and labelled per specimen (i.e. 10 slides of 1–2 strips with 5 sections each). These sections were used for ISH and haematoxylin and eosin staining. More females than males were examined with microscopy because symbionts were previously not detected in the testes [14], and the focus of this study was on the ovarial transmission route of the symbionts.
(c). Specific marker design workflow
We designed a novel workflow, named SMD, using established concepts for specific bacterial detection (outlined in electronic supplementary material, figures S1 and S2, Material and methods, Results, table S2). This approach BLASTs the genome of the taxon of interest, one gene at a time, against a database of bacterial sequences, and sorts and filters them according to (i) how well represented and (ii) how well conserved the sequence is among bacteria. If population-level genetic data are available for the taxon of interest, the genes can be sorted by the amount of genetic variation to find genes that have high conservation within the taxon of interest, but low conservation to other bacterial sequences. The scripts used to perform SMD are available from https://github.com/shelbirussell/SMD_workflow.
(d). Symbiont detection in spawned eggs via PCR
DNA was extracted from frozen spawned eggs using the UltraClean Tissue and Cells DNA Isolation Kit (MoBio) by bead beating and eluted in 50 µl of buffer. To control for symbiont contamination in filtered seawater (FSW) from the S. velum habitat, subsamples of the spawning seawater were filtered through 0.2 µm nitrocellulose filters (MoBio) pre- and post-spawning, and extracted with the PowerWater DNA Extraction Kit (MoBio). Two primer sets were used for symbiont detection, targeting 1457 bp of the common bacterial marker gene gyrB (DNA gyrase subunit B) with PCR and 606 bp of the newly identified marker gene from this study rhlE (ATP-dependent RNA helicase) with qPCR. A subset of amplicons was directly Sanger sequenced to confirm sequence identity.
(e). Symbiont quantification via qPCR
We quantified the number of symbiont genomes present in eggs and sediment and seawater samples from the S. velum habitat with qPCR using the newly designed primers to rhlE. Markers for host mitochondrial atp6 and bacterial 16S rRNA were used as controls for host presence and bacterial content in environmental samples, respectively. See electronic supplementary material, Material and methods for standard construction and qPCR parameters.
DNA extracts from single and pooled eggs (n = 25), gills (n = 2), sediment and seawater filters were tested (see electronic supplementary material, table S1). Standards were run in triplicate on each 96-well plate, and any replicate that differed by more than 0.5 CT from the other two replicates was not included in standard curve calculations. Each egg sample and filters from pre/post-spawning seawater were also run in triplicate. No-template controls (NTC) returned no CT on most plates, and CT values significantly lower than the most dilute standard on the remaining plates (see electronic supplementary material, table S3). Assays were required to have R2 > 0.98 and efficiencies greater than 85% for their results to be included. A subset of rhlE and atp6 qPCR products was Sanger-sequenced directly. Data were plotted and statistics were calculated in R [22].
(f). In situ hybridization
(i). In situ hybridization probes and probe design
Symbiont 16S rRNA sequences were aligned to the SILVA database (SSURef version 111) [23] and the symbiont-specific oligonucleotide probe Svsym47 was designed in ARB [24]. Control probes were designed or selected for host binding, bacterial binding and nonsense binding (see electronic supplementary material). Oligonucleotide probes were labelled at the 5′-end with FITC (Sigma Aldrich) or horseradish peroxidase (HRP) (Biomers, Ulm, Germany).
(ii). In situ hybridization (CARD-FISH and CISH)
Paraffin sections were processed according to [25] with optimization modifications for S. velum tissues, including extended permeabilization treatments and endogenous enzyme inactivation. HRP oligonucleotides were detected by precipitating fluorescently labelled tyramides (as described in [25]) and alkaline phosphatase (AP)-labelled antibodies bound to FITC were detected with NBT-BCIP precipitation. See electronic supplementary material for full protocols.
(iii). Imaging
Tissue sections prepared with H&E staining or CISH were imaged with DIC microscopy on a Zeiss AxioImager. Sections prepared with CARD-FISH were imaged on a Zeiss LSM700 confocal microscope. Confocal laser intensity and gain settings were calibrated to minimize background autofluorescence for each laser line. See electronic supplementary material for image analysis.
3. Results
(a). Morphology and anatomy of S. velum
The gross morphology of adult S. velum was characterized by differential interference contrast (DIC) microscopy of whole bivalve histological sections to orient subsequent investigations of symbiont location in host tissues (figure 1; originally described in [26]). Adult S. velum shell lengths were an average of 10 ± 1.5 mm (n = 24). The large protobranch gill filled the majority of the posterior end of the mantle cavity and consisted of two ctenidia surrounding a hypobranchial gland (figure 1a–c). The large, muscular foot encompassed the majority of the anterior and ventral ends of the mantle cavity. The gonad filled the dorsal half of the interior of the foot and extended to fill the dorsal mantle cavity in mature specimens (e.g. figure 1a,d).
Dissected S. velum ovaries consisted of clusters of acini (a in figure 1d), which resemble stalks of grapes in structure and are common in bivalve ovaries (e.g. [27]). Oogenesis was asynchronous, with each acinus containing oocytes at different stages of development: oogonia, previtellogenic oocytes, vitellogenic oocytes and mature oocytes (figure 1a,d). Average oocyte sizes are described in the electronic supplementary material, table S4 (total n = 14 females), and micrographs shown in figure 1 are representative of the other samples. While the sizes of the different stages of oocytes were found to be consistent throughout the year, there were more oocytes in mid-oogenesis during the summer months than in the fall (previtellogenic and vitellogenic oocytes; chi-squared test p = 0.0336 and 0.0212, respectively). Interestingly, there were similar numbers of oogonia and mature oocytes among months (chi-squared test p = 0.360 and 0.884, respectively). For oogonia, consistent numbers across seasons are likely related to the stem cell division rate. Unspawned mature oocytes are often resorbed during winter months [28], so those remaining in November may be in the process of resorption.
(b). SMD marker design and PCR detection of S. velum symbionts
Using SMD, we successfully identified and designed three specific S. velum symbiont primer sets and selected the single copy rhlE gene to use in subsequent qPCR assays (see electronic supplementary material, Results). With this primer set and one designed to the symbiont gyrB sequence, we verified that symbionts are contained within spawned S. velum eggs with PCR. Directly sequenced gyrB and rhlE PCR products amplified from eggs were identical to the sequences for these genes from the Rhode Island symbiont subpopulation (Sanger read data not shown).
(c). qPCR quantification of symbionts in spawned S. velum eggs
Symbiont rhlE sequences amplified from both bleached and untreated eggs with qPCR (figure 2a). Symbionts were not detected in the FSW used for spawning prior to introducing S. velum adults (FSW in figure 2a). PCR product was barely detected after 16 S. velum were added (pre-spawn in figure 2a), but this amount (0.024 rhlE copies per ml of seawater) was significantly lower than the number detected per egg for both bleached and untreated eggs (Welch's two-sample t-test p = 0.0297 and 0.00335, respectively). While there was no significant difference in means between the bleached and unbleached eggs (p = 0.8224), more of the bleached eggs failed to amplify (5/12 bleached versus 1/13 unbleached reactions). In total, bleached and untreated eggs were found to contain 64.1 ± 17.4 s.e. (n = 79) copies of symbiont rhlE per egg by qPCR. As rhlE is a single copy gene, this equals the number of symbiont genomes per egg and is a lower bound given inherent variation in DNA extraction efficiencies (e.g. DNA binding to tube walls).
(d). qPCR quantification of bacteria and S. velum symbionts in environmental samples
We sought to determine the prevalence of S. velum symbionts in their mudflat habitat (figure 2b) as a proxy for the probability that a host acquires a symbiont through the environment. These data allowed us to infer that ecological opportunities for horizontal symbiont transmission between S. velum hosts are likely infrequent, and thus the genetic patterns of mixed transmission previously reported [10,15] are driven by rare events.
Using the symbiont-specific rhlE primer set, we found that S. velum symbionts are at low abundance (figure 2c) in bacteria-rich environmental samples (figure 2d) and are rarely detected independently of host DNA (6/72 samples; gold bars marked with exclamation marks in figure 2d; electronic supplementary material, Results). Amplifying with the universal bacterial primer set to the V4 region of the 16S rRNA (EUB16SV4), revealed that sediment samples from the S. velum habitats contained on average 3.57 ± 3.91 × 108 copies of bacterial 16S rRNA per gram (n = 64). By contrast, these sediment samples contained on average 3.65 ± 9.38 × 103 copies of symbiont rhlE and 4.17 ± 7.80 × 103 copies of mitochondrial atp6 per gram. The seawater samples from S. velum habitats (n = 15) contained an average of 8.54 ± 11.9 × 104 copies of bacterial 16S rRNA per ml. However, very little symbiont or mitochondrial sequence was detected, with an average 1.44 ± 2.40 × 10−1 copies of symbiont rhlE and 0.61 ± 1.07 copies mitochondrial atp6 quantified per ml. Sanger sequences of the selected rhlE and atp6 qPCR confirmed that the genotypes detected in these samples are identical to the known symbiont genotypes from each subpopulation (see electronic supplementary material, Results).
(e). Symbiont detection in host tissues by in situ hybridization
We sought to determine the distribution of symbionts in host tissues to inform on the route symbionts take between hosts. As symbionts probably colonize eggs through the ovary prior to spawning [14], we specifically sought to identify them in this tissue. Following the design of an oligonucleotide probe to the S. velum symbiont 16S rRNA (Svsym47), symbionts were detected in whole-bivalve histological sections, containing the main symbiont-containing tissue, the gill, as well as the ovary (figures 1, 3 and 4). A chromogenic detection strategy was employed for sensitive detection, to control for tissue autofluorescence and to examine samples/tissues rapidly with DIC microscopy. Positive and negative control probes confirmed tissue permeability and probe specificity. See electronic supplementary material, Results and figures S3–S5 for these data.
Figure 3.
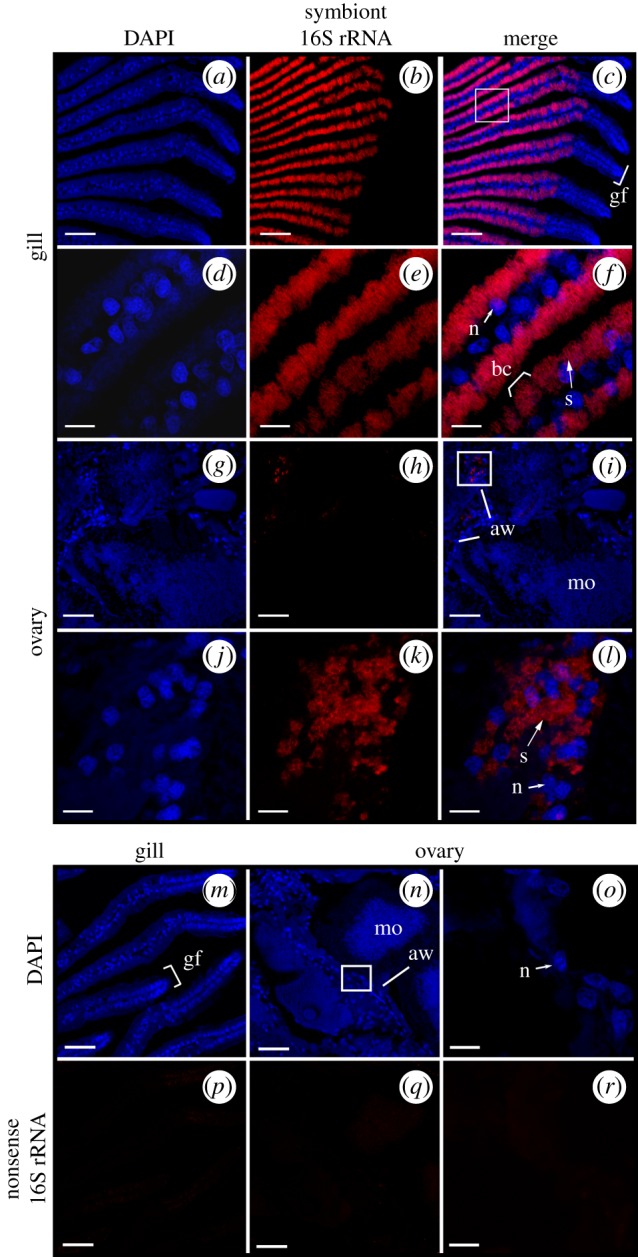
CARD-FISH localization of symbiont 16S rRNA in S. velum gill and ovary transverse sections imaged with confocal microscopy. Symbiont-containing S. velum (a–f,m,p) gill filaments and (g–l,n,o,q,r) ovary. (a–l) Columns left to right: DAPI (blue), Svsym47 signal (red, Alexa 647), merged images. Boxes in (a–c) and (g–i) show enlargement of regions featured in (d–f) and (j–l), respectively. (m–r) Hybridization of S. velum (m,p) gill and (n,o,q,r) ovary sections with the negative control probe NON338. (m–o) DAPI (blue). (p–r) NON338 signal under laser intensities and gain settings for Svsym47-hybridized gill (red, Alexa 647). (o,r) are enlargements of (n,q). Abbreviations: aw: acinal wall; bc: bacteriocyte; gf: gill filament; mo: mature oocyte; n: nucleus; s: symbiont cells. Scale bars: (a,b,m,n,o,p) 100 µm, (c–f) 50 µm, (g,h) 10 µm.
Figure 4.
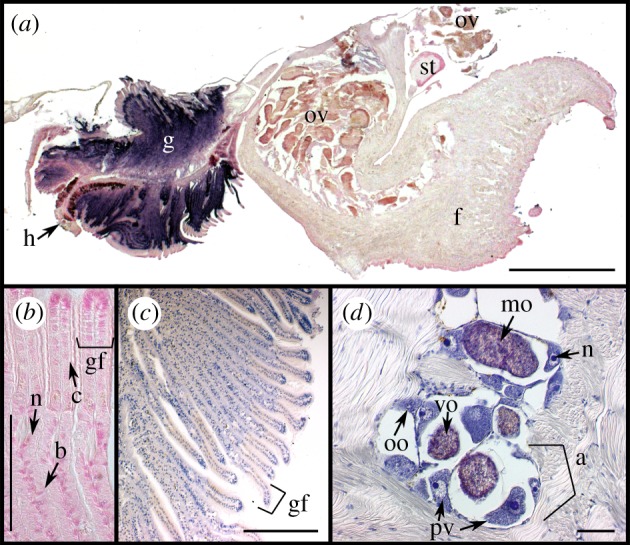
Chromogenic in situ hybridization (CISH) localization of symbiont 16S rRNA in S. velum gill and ovary imaged with DIC microscopy. (a–c,h,i) S. velum gill and (d–g,j) ovary labelled with the (a–g) symbiont probe Svsym47 or (h–j) negative control probe NONSvsym47. Probe signal in purple and DNA counterstained in nuclear fast red (pink). Note: (d,f,g,h) were not counterstained to more clearly see the probe signal. Abbreviations: (aw) acinal wall, (bc) bacteriocytes, (c) cilia, (cc) ciliated cells, (gf), gill filament shown between brackets, (mo) mature oocyte, (pv) previtellogenic oocyte, (n) nucleus, (s) symbiont cells, (vo) vitellogenic oocyte. Scale bars: (a,h,j) 100 µm, (b–e,i) 50 µm, (f,g) 20 µm.
Unambiguous positive signals were obtained from gill sections of all S. velum specimens (n = 24) labelled with the universal Bacteria probe EUB338 [29] and the symbiont-specific 16S probe Svsym47 (Material and methods and table 1) via both CARD-FISH (figure 3) and CISH (figure 4) protocols. The images in figures 3 and 4 are representative of the gill and ovary tissues in all 24 samples. In double-hybridized sections, the symbiont probe signal co-localized with the bacteria signal (electronic supplementary material, figure S4C–E).
Table 1.
New marker genes, primers and probes for the S. velum symbiont used in the PCR, qPCR and ISH experiments.
| target | gene | name |
sequence (5'–3′) | references | ||
|---|---|---|---|---|---|---|
| PCR primers | S. velum symbionts | rhlE (JV46_09660)a | SvrhlE | 112F | GGACGGGATATCATGGGTGG | this study |
| 717R | CAGCCAGGAGAGGAGTTCAC | |||||
| S. velum symbionts | gyrB (JV46_17100) | SvgyrB | 856F | GCCCTGACAAGGACGCTCAACC | this study | |
| 2312R | GCATCAGCAGCAACAGCATCTTCG | |||||
| S. velum mitochondria | atp6 | Svatp6 | 70F | TGAGCAGGAACTTTATGGGTT | this study | |
| 522R | AAGAACCACATGCCCAGCTC | |||||
| bacteria (universal) | 16S rRNA V4 region | EUB | 16SV4F | GTGCCAGCMGCCGCGGTAA | Caporaso et al. [30] | |
| 16SV4R | GGACTACHVGGGTWTCTAAT | |||||
| ISH probes | S. velum symbionts | 16S rRNA (47–76 bp) | Svsym47 | CCCGAAGGTATTTTCGCGTTCGACTTGCAT | this study | |
| none; reverse complement of Svsym47 | NONSvsym47 | ATGCAAGTCGAACGCGAAAATACCTTCGGG | this study | |||
| eubacteria (most) | 16S rRNA (338–355 bp) | EUB338 | GCTGCCTCCCGTAGGAGT | Amann et al. [29] | ||
| none; reverse complement of EUB338 | NON338 | ACTCCTACGGGAGGCAGC | ||||
| eukaryotes (most) | 18S rRNA gene (516–531 bp) | EUK516 | ACCAGACTTGCCCTCC | Amann et al. [29] | ||
aMarker gene identified with SMD.
Consistent with TEM evidence [31], the symbiont 16S rRNA signal was strong in the epithelium of the gill filaments and occurred in non-ciliated cells, termed bacteriocytes (bc), interspersed between intercalary cells (figure 3a–f and figure 4a–c). Symbiont signal was strongest along the outer half of each bacteriocyte on both sides of each gill filament, opposite the bacteriocyte nuclei (n in figure 3f).
Punctate signals consistent with groups of bacteria or single bacteria were localized in the ovary with CARD-FISH and CISH (figure 3g–l and figure 4d–g). This signal was found associated with the cells of the acinal wall with both methods and localized to areas of approximately 1 by 2–3 µm (s in figures 3 and 4), consistent with the known locations of the bacteria [31]. The opposite binding intensities of the Svsym47 and EUB338 probes relative to the host EUK516 probe underscore the difficulty of detecting low-copy targets in S. velum tissue (electronic supplementary material, Results and figure S3). Owing to the high autofluorescence of oocytes, bacteria were only imaged in direct association with oocytes via CISH (figure 4d–g). With CISH, symbiont signal was detected in the periphery of many vitellogenic and mature oocytes (figure 4d,e), although the limited resolution of the 10 µm sections in DIC made it unclear as to whether symbionts were positioned immediately inside or outside of the acinal and oocyte membranes (figure 4d–g).
4. Discussion
Transmission of microbial symbionts can occur via horizontal or vertical mechanisms depending on the evolutionary and life history of the host and symbiont. Often horizontally transmitted symbionts are present at detectable abundances in the environment [18,31], whereas vertically transmitted symbionts become associated with hosts prior to reproduction, generally through association with host oocytes. However, examples of later timing do exist for brooded host species. In the case of hosts with broadcast-spawned gametes, such as S. velum, transmission of symbionts within or attached to gametes exposed to the extracellular environment introduces opportunities for horizontal transmission. For example, novel symbionts could be taken up by embryos and these would either replace or mix with the endogenous symbiont population. Given that even low rates of horizontal transmission can have significant effects on symbiont genome evolution [10,32], knowledge of how symbionts are vertically transmitted between host species is important to understanding the evolutionary pressures shaping these associations.
Detecting symbiotic bacteria in complex samples can be challenging. SMD helps to resolve this issue by leveraging the information in a bacterial genome against known bacterial sequences. This method is effective, as only the top (eight) candidate genes were selected for primer design, and 38% of these yielded specific primer sets in the first round of testing (electronic supplementary material, table S2), although we selected one for qPCR quantification (table 1).
The qPCR assays suggest that the S. velum symbionts are associated with spawned host eggs, are in low abundance in sediment and are undetectable in seawater (figure 2a,c,d). Bacterial 16S sequences were present at expected abundances in sediment and seawater samples—an average of 108–109 bacteria per gram of soil and 106 per ml of water [33–35]—indicating that PCR inhibitors or extraction efficiency did not affect quantification. Many sediment samples failed to amplify with symbiont primers and those that did often also amplified with mitochondrial primers. These results do not preclude a low abundance of free-living (and dividing) symbionts, as symbionts may have been incidentally sampled along with host cells or larvae. Detecting more copies of the symbiont genome in the sediment samples than the seawater samples is consistent with observations of the burrowing Codakia clam symbioses [36]. However, the S. velum symbionts are present at lower abundances in the environment (4 × 103 cells g−1 and 1.4 cells ml−1) than symbionts from strictly horizontally transmitted associations, e.g. Vibrio fischeri, the bioluminescent symbionts of bobtail squid, which require 10–105 cells ml−1 for colonization [6] and rhizobia symbionts of legume plants, which require 104–106 cells g−1 [37]. Therefore, horizontal transmission is probably a rare event on ecological time scales for the S. velum symbionts (i.e. it does not occur every generation).
S. velum symbionts were detected in the acinal walls and some mature oocytes in the S. velum ovary via ISH with specific 16S probes (figures 1, 3 and 4). Despite the lack of symbionts in many seemingly mature oocytes in the ovary, symbionts were detected in 76% of PCR amplifications from egg DNA (92% of untreated and 58% of bleached; figure 2a). These results suggest that symbionts colonize oocytes prior to or during spawning via the acinal wall. The acinal walls contain supportive cells that supply nutrients or cellular factors during oogenesis [38], making this a candidate entry point to the eggs. Two additional lines of evidence suggest that symbionts enter oocytes near the time of spawning: (1) oogonia are likely too small to contain symbionts without interfering with oogenesis (average 22.8 µm in diameter versus 1–10 µm in length, for oogonia and symbiont cells, respectively); and (2) symbionts were detected in fewer than 10% of all mature oocytes examined fully or partially in serial sections. Furthermore, incorporation of symbionts late in oogenesis, specifically post-vitellogenesis, has been reported in a number of other associations (e.g. [39,40]).
Detection of symbionts in surface-sterilized eggs suggests that the symbionts are intracellular. However, if the symbionts are contained within the vitelline envelope, but outside the cell membrane, they may not have been degraded by the bleach treatment. Consistent with this idea, more bleach-treated eggs samples failed to amplify than untreated. Furthermore, symbiont localization underneath the vitelline envelope has been reported for the chemosynthetic symbionts of vesicomyid clams [41]. Thus, an intracellular location cannot be concluded until symbionts are imaged at a higher resolution within the eggs (e.g. via TEM).
Finding the symbiont rhlE sequence in eggs indicates a reliable cellular mechanism for the vertical transmission of symbionts to offspring. Indeed, other mixed-mode associations have found that the vertical route to be more faithful than the horizontal route [42]. This is likely to be the situation with S. velum given the exceedingly low abundance of symbionts found in their habitat (figure 2d). Future efforts will continue to address the need to image the passage of symbiont cells to oocytes from other ovary tissues in order to time and place their route to the oocytes. Collectively, these data support that symbionts are vertically transmitted to S. velum eggs at high fidelity.
Accurate measurement of the distribution of symbionts in hosts and the environment is important to understanding how symbioses are maintained. Association with the germline, in addition to the functional tissue type, indicates the reproductive interests of hosts and symbionts are linked. However, the detection of S. velum symbionts in the environment indicates that environmental pressures may also influence symbiont populations. Further research into precise routes of transmission into host oocytes, and more general investigations into the roles environmental symbionts play in this mixed-mode association, will build on the work presented here to inform on how symbiont transmission in extant populations influences the association over evolutionary time.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Russ Corbett-Detig, Gonzalo Giribet, Peter Girguis, Cassandra Extavour, members of the Cavanaugh Lab and two reviewers for valuable suggestions and comments that improved the manuscript. We also thank the Extavour laboratory for the use of their Zeiss AxioImager widefield microscope and the Harvard Arboretum for the use of their Zeiss LSM700 confocal microscope.
Data accessibility
This article has no additional data.
Authors' contributions
S.L.R. and C.M.C. designed the study, interpreted the data and wrote the original manuscript. S.L.R. and E.M. performed the experiments, S.L.R. analysed the data. All authors contributed to the final draft.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by Harvard University's William F. Milton Fund, Department of Organismic and Evolutionary Biology, and Microbial Sciences Initiative.
References
- 1.Hentschel U, Steinert M, Hacker J. 2000. Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. 8, 226–231. ( 10.1016/S0966-842X(00)01758-3) [DOI] [PubMed] [Google Scholar]
- 2.Bright M, Bulgheresi S. 2010. A complex journey: transmission of microbial symbionts. Nat. Rev. Microbiol. 8, 218–230. ( 10.1038/nrmicro2262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebert D. 2013. The epidemiology and evolution of symbionts with mixed-mode transmission. Annual Reviews Ecology , Evol. Syst. 44, 623–643. ( 10.1146/annurev-ecolsys-032513-100555) [DOI] [Google Scholar]
- 4.Login FH, Balmand S, Vallier A, Vincent-Monegat C, Vigneron A, Weiss-Gayet M, Rochat D, Heddi A. 2011. Antimicrobial peptides keep insect endosymbionts under control. Science 334, 362–365. ( 10.1126/science.1209728) [DOI] [PubMed] [Google Scholar]
- 5.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. 2007. How rhizobial symbionts invade plants: the Sinorhizobium–Medicago model. Nat. Rev. Microbiol. 5, 619–633. ( 10.1038/nrmicro1705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nyholm SV, McFall-Ngai M. 2004. The winnowing: establishing the squid–vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642. ( 10.1038/nrmicro957) [DOI] [PubMed] [Google Scholar]
- 7.Stoll S, Feldhaar H, Fraunholz MJ, Gross R. 2010. Bacteriocyte dynamics during development of a holometabolous insect, the carpenter ant Camponotus floridanus. BMC Microbiol. 10, 308 ( 10.1186/1471-2180-10-308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strathmann RR, Staver JM, Hoffman JR. 2002. Risk and the evolution of cell-cycle durations of embryos. Evolution 56, 708–720. ( 10.1111/j.0014-3820.2002.tb01382.x) [DOI] [PubMed] [Google Scholar]
- 9.Itoh H, et al. 2014. Evidence of environmental and vertical transmission of Burkholderia symbionts in the oriental chinch bug, Cavelerius saccharivorus (Heteroptera: Blissidae). Appl. Environ. Microbiol. 80, 5974–5983. ( 10.1128/AEM.01087-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell SL, Corbett-Detig R, Cavanaugh CM. 2017. Mixed transmission modes and dynamic genome evolution in an obligate animal-bacterial symbiosis. ISME J. 11, 1359–1371. ( 10.1038/ismej.2017.10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sipkema D, de Caralt S, Morillo JA, Al-Soud WA, Sørensen SJ, Smidt H, Uriz MJ. 2015. Similar sponge-associated bacteria can be acquired via both vertical and horizontal transmission: microbial transmission in Petrosia ficiformis. Environ. Microbiol. 17, 3807–3821. ( 10.1111/1462-2920.12827) [DOI] [PubMed] [Google Scholar]
- 12.Ozawa G, et al. 2017. Ancient occasional host switching of maternally transmitted bacterial symbionts of chemosynthetic vesicomyid clams. Genome Biol. Evol. 9, 2226–2236. ( 10.1093/gbe/evx166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stewart FJ, Baik A, Cavanaugh CM. 2009. Genetic subdivision of chemosynthetic endosymbionts of Solemya velum along the southern New England coast. Appl. Environ. Microbiol. 75, 6005–6007. ( 10.1128/AEM.00689-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krueger DM, Gustafson RG, Cavanaugh CM. 1996. Vertical transmission of chemoautotrophic symbionts in the bivalve Solemya velum (Bivalvia: Protobranchia). Biol. Bull. 190, 195–202. ( 10.2307/1542539) [DOI] [PubMed] [Google Scholar]
- 15.Russell SL, Cavanaugh CM. 2017. Intrahost genetic diversity of bacterial symbionts exhibits evidence of mixed infections and recombinant haplotypes. Mol. Biol. Evol. 34, 2747–2761. ( 10.1093/molbev/msx188) [DOI] [PubMed] [Google Scholar]
- 16.Kim M, Morrison M, Yu Z. 2011. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J. Microbiol. Methods 84, 81–87. ( 10.1016/j.mimet.2010.10.020) [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Li L, Khan MA, Zhu F. 2012. Popular molecular markers in bacteria. Molecular Genetics , Microbiol. Virol. 27, 103–107. ( 10.3103/S0891416812030056) [DOI] [PubMed] [Google Scholar]
- 18.Klose J, Polz MF, Wagner M, Schimak MP, Gollner S, Bright M. 2015. Endosymbionts escape dead hydrothermal vent tubeworms to enrich the free-living population. Proc. Natl Acad. Sci. USA 112, 11 300–11 305. ( 10.1073/pnas.1501160112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hosokawa T, Kikuchi Y, Nikoh N, Meng X-Y, Hironaka M, Fukatsu T. 2010. Phylogenetic position and peculiar genetic traits of a midgut bacterial symbiont of the stinkbug Parastrachia japonensis. Appl. Environ. Microbiol. 76, 4130–4135. ( 10.1128/AEM.00616-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cúcio C, Overmars L, Engelen AH, Muyzer G.. 2018. Metagenomic analysis shows the presence of bacteria related to free-living forms of sulfur-oxidizing chemolithoautotrophic symbionts in the rhizosphere of the seagrass Zostera marina. Front. Marine Sci. 5, 171 ( 10.3389/fmars.2018.00171) [DOI] [Google Scholar]
- 21.Kim Y, Ashton-Alcox KA, Powell EN. 2006. Histological techniques for marine bivalve molluscs: update. Silver Spring, MD: NOAA Technical Memorandum NOS NCCOS. [Google Scholar]
- 22.R Core Development Team. 2012. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 23.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. ( 10.1093/nar/gks1219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig W. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371. ( 10.1093/nar/gkh293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wendeberg A. 2010. Fluorescence in situ hybridization for the identification of environmental microbes. Cold Spring Harb. Protoc. 2010, pdb.prot5366 ( 10.1101/pdb.prot5366) [DOI] [PubMed] [Google Scholar]
- 26.Yonge CM. 1939. The protobranchiate mollusca; a functional interpretation of their structure and evolution. Phil. Trans. R. Soc. Lond. B 230, 79–148. ( 10.1098/rstb.1939.0005) [DOI] [Google Scholar]
- 27.Fabioux C, Pouvreau S, Roux FL, Huvet A. 2004. The oyster vasa-like gene: a specific marker of the germline in Crassostrea gigas. Biochem. Biophys. Res. Commun. 315, 897–904. ( 10.1016/j.bbrc.2004.01.145) [DOI] [PubMed] [Google Scholar]
- 28.Camacho-Mondragón MA, Ceballos-Vázquez BP, Uría-Galicia E, López-Villegas EO, Pipe R, Arellano-Martínez M. 2015. Ultrastructural and histological study of oogenesis and oocyte degeneration in the penshell Atrina maura (Bivalvia: Pinnidae). Malacologia 59, 1–12. ( 10.4002/040.059.0102) [DOI] [Google Scholar]
- 29.Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS 108, 4516–4522. ( 10.1073/pnas.1000080107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavanaugh CM. 1983. Symbiotic chemoautotrophic bacteria in marine invertebrates from sulphide-rich habitats. Nature 302, 58–61. ( 10.1038/302058a0) [DOI] [Google Scholar]
- 32.Brandvain Y, Goodnight C, Wade MJ. 2011. Horizontal transmission rapidly erodes disequilibria between organelle and symbiont genomes. Genetics 189, 397–404. ( 10.1534/genetics.111.130906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuwae T, Hosokawa Y. 1999. Determination of abundance and biovolume of bacteria in sediments by dual staining with 4Ј,6-diamidino-2-phenylindole and acridine orange: relationship to dispersion treatment and sediment characteristics. Appl. Environ. Microbiol. 65, 3407–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hobbie JE, Daley RJ, Jasper S. 1977. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33, 1225–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor JP, Wilson B, Mills MS, Burns RG. 2002. Comparison of microbial numbers and enzymatic activities in surface soils and subsoils using various techniques. Soil Biol. Biochem. 34, 387–401. ( 10.1016/S0038-0717(01)00199-7) [DOI] [Google Scholar]
- 36.Gros O, Frenkiel L, Mouëza M. 1998. Gill filament differentiation and experimental colonization by symbiotic bacteria in aposymbiotic juveniles of Codakia orbicularis (Bivalvia: Lucinidae). Invert. Reprod. Dev. 34, 219–231. ( 10.1080/07924259.1998.9652656) [DOI] [Google Scholar]
- 37.Meade J, Higgins P, O'Gara F. 1985. Studies on the inoculation and competitiveness of a Rhizobium leguminosarum strain in soils containing indigenous Rhizobia. Appl. Environ. Microbiol. 49, 899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferreira MAP, et al. 2006. Morphological and morphometric aspects of Crassostrea rhizophorae (Guilding, 1828) oocytes in three stages of the gonadal cycle. Int. J. Morphol. 24, 437–442. [Google Scholar]
- 39.Nan G-H, Xu Y-P, Yu Y-W, Zhao C-X, Zhang C-X, Yu X-P. 2016. Oocyte vitellogenesis triggers the entry of yeast-like symbionts into the oocyte of brown planthopper (Hemiptera: Delphacidae). Ann. Entomol. Soc. Am. 109, 753–758. ( 10.1093/aesa/saw025) [DOI] [Google Scholar]
- 40.Herren JK, Paredes JC, Schupfer F, Lemaitre B. 2013. Vertical transmission of a Drosophila endosymbiont via cooption of the yolk transport and internalization machinery. mBio 4, e00532-12 ( 10.1128/mBio.00532-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikuta T, et al. 2016. Surfing the vegetal pole in a small population: extracellular vertical transmission of an ‘intracellular’ deep-sea clam symbiont. R. Soc. open sci. 3, 160130 ( 10.1098/rsos.160130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vorburger C, Siegrist G, Rhyner N. 2017. Faithful vertical transmission but ineffective horizontal transmission of bacterial endosymbionts during sexual reproduction of the black bean aphid, Aphis fabae: endosymbiont transmission in aphids. Ecol. Entomol. 42, 202–209. ( 10.1111/een.12379) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.