Abstract
Ovarian cancer is the most fatal gynecological malignancy in women and identification of new therapeutic targets is essential for the continued development of therapy for ovarian cancer. TRRAP (transformation/transcription domain-associated protein) is an adaptor protein and a component of histone acetyltransferase complex. The present study was undertaken to investigate the roles played by TRRAP in the proliferation and tumorigenicity of ovarian cancer stem cells. TRRAP expression was found to be up-regulated in the sphere cultures of A2780 ovarian cancer cells. Knockdown of TRRAP significantly decreased cell proliferation and the number of A2780 spheroids. In addition, TRRAP knockdown induced cell cycle arrest and increased apoptotic percentages of A2780 sphere cells. Notably, the mRNA levels of stemness-associated markers, that is, OCT4, SOX2, and NANOG, were suppressed in TRRAP-silenced A2780 sphere cells. In addition, TRRAP overexpression increased the mRNA level of NANOG and the transcriptional activity of NANOG promoter in these cells. Furthermore, TRRAP knockdown significantly reduced tumor growth in a murine xenograft transplantation model. Taken together, the findings of the present study suggest that TRRAP plays an important role in the regulation of the proliferation and stemness of ovarian cancer stem cells.
Keywords: Cancer stem cells, Ovarian cancer, Spheroid, TRRAP
INTRODUCTION
Ovarian cancer occurs most frequently in older women (1) and is caused by genetic changes in several oncogenes and tumor suppressor genes (2). The majority of cases are diagnosed in the late stages of the disease because the symptoms are often barely discernable during the earlier stages of cancer development. However, a late diagnosis means that the cancer cells are likely to have disseminated into the peritoneal cavity, which imposes a clinical challenge (3). Currently, the standard treatment for advanced cancer includes cytoreductive surgery followed by chemotherapy, but the disease recurs in ~70% of patients who undergo this treatment due to the development of drug resistance (4). Thus, the identification of new diagnostic and therapeutic targets is required for the treatment of ovarian cancer.
Cancer stem cells (CSCs) are a sub-population of cancer cells with the ability to self-renew and with tumorigenic potential. CSCs are also resistant to conventional chemotherapeutic drugs and exhibit considerable metastatic potential and tumorigenic activity (5). Many studies on ovarian cancer have reported the presence of a cell population with CSC-like characteristics (6), and it has also been reported that sphere-forming ovarian cancer cells possess stem-like properties (7). CSCs share some properties with stem cells, such as the fact that both CSCs and stem cells are known to have the abilities to self-renew and differentiate (8). Accumulating evidence suggests key regulators of embryonic stem cells (ESCs), including OCT4 and SOX2, are overexpressed in poorly differentiated tumors (9), and NANOG expression has been reported to be elevated in drug resistant ovarian cancer cells with CSC-like characteristics (10). However, the molecular mechanisms underlying the transcriptional regulations of stemness-associated genes have not yet been elucidated.
TRRAP is an adaptor protein that shares homology with PIKK kinases, but lacks intrinsic kinase activity (11). TRRAP promotes histone acetylation and chromatin remodeling and also regulates gene expression and embryonic development (12). At the molecular level, TRRAP has been associated with cancer. Microarray analysis showed that TRRAP knockdown reduced the CSC-like properties of glioma (13, 14), and that TRRAP expression levels were reported to be significantly up-regulated in breast cancer (15). TRRAP has also been shown to interact with the carcinogenic gene c-Myc (16, 17), and it has been suggested that disruptions of the TRRAP/HAT complex and histone acetylation would influence chromatin-based processes and cell cycle progression, and possibly lead to abnormal proliferation (18, 19). However, although TRRAP probably plays an important role in cancer progression, its involvement in the tumorigenic potential of ovarian CSCs remains uncertain.
For these reasons, we designed the present study to investigate the roles played by TRRAP in the proliferation and tumorigenicity of ovarian cancer stem cells. A loss or gain of function approach was adopted to investigate the regulatory effects of TRRAP on the CSC-like properties of ovarian cancer cells.
RESULTS
TRRAP knockdown abrogated the CSC-like properties of ovarian cancer cells
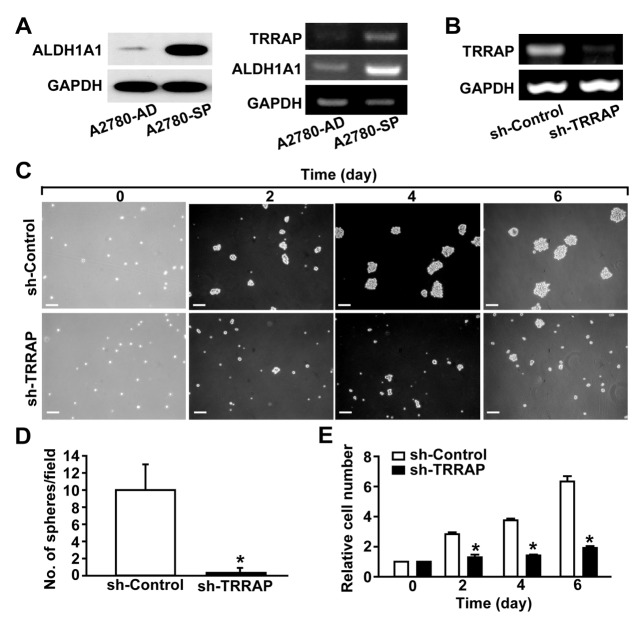
We recently reported that the sphere-forming cells of the A2780 ovarian cancer cell line exhibit CSC-like phenotypes and characteristics (20–22). The sphere-forming cells derived from suspension culture of A2780 cells (A2780-SP) exhibited increased expression of ALDH1A1, a CSC-specific marker, compared with the parental A2780 cells cultured in the adherent status (A2780-AD) (Fig. 1A). To determine whether TRRAP is associated with the generation of CSCs, we measured the mRNA levels of TRRAP in A2780-SP and A2780-AD cells. The mRNA levels of not only ALDH1A1 but also TRRAP in A2780-SP cells were higher than those in A2780-AD cells (Fig. 1A). To investigate the role of TRRAP in the sphere-forming ability of CSCs, TRRAP was silenced using TRRAP-specific shRNA (Fig. 1B). TRRAP knockdown substantially decreased the number of spheroids (Fig. 1C and 1D) and markedly reduced the number of cells in suspension cultures of A2780-SP (Fig. 1E). These results indicate that TRRAP is required for the proliferation of ovarian CSCs.
Fig. 1.
Effects of TRRAP knockdown on the proliferation of ovarian CSCs. (A) Increased expression of TRRAP in A2780-SP cells. The protein levels of ALDH1A1 and GAPDH were determined through Western blotting (Left panels). The mRNA levels of TRRAP, ALDH1A1, and GAPDH in A2780-AD and A2780-SP cells were determined by RT-PCR (Right panels). (B) shRNA-mediated knockdown of TRRAP in A2780-SP cells. A2780-SP cells were infected with lentiviruses harboring control shRNA (sh-control) or TRRAP shRNA (sh-TRRAP), and the mRNA levels of TRRAP and GAPDH were determined by RT-PCR. (C) A2780-SP cells were infected with sh-control or sh-TRRAP lentivirus, and bright field images of the spheroid cells are shown from day 0 to day 6. Scale bar = 100 μm. (D) Numbers of spheroids were quantified six days after infecting cells with lentiviruses bearing sh-control or sh-TRRAP. (E) Effects of TRRAP knockdown on the proliferation of A2780-SP cells. The cell numbers were counted from days 0 to 6 after TRRAP knockdown. Results are presented as mean ± SD. *P < 0.05 (n = 6).
Involvement of TRRAP in the regulation of cell survival of ovarian CSCs
It has been reported that TRRAP participates in the cell cycle progression of neural progenitors (23). To investigate the effect of TRRAP knockdown on the cycle progression and viability of ovarian CSCs, we measured the cell cycles of A2780-SP cells. TRRAP knockdown reduced the percentages of cells in the S and G2/M phases (Fig. 2A), whereas it increased the percentage of cells in the G0/G1 phase, suggesting cell cycle arrest at the G0/G1 phase in response to the silencing of TRRAP. Moreover, the percentage of cells at the sub-G1 phase was also augmented in TRRAP-knockdown cells, indicating that apoptotic cells were induced by TRRAP silencing (Fig. 2B). In support of these results, the expression levels of the cell cycle-associated proteins associated with the G1/S transition, such as p21, CDK4, and cyclin D3, were found to be suppressed in TRRAP-silenced A2780-SP cells (Fig. 2C). In addition, TRRAP knockdown decreased the level of PARP-1 in A2780-SP cells. These results suggest that TRRAP knockdown induces cell cycle arrest at the G0/G1 phase, and that TRRAP contributes to the acquisition of CSC-like properties of A2780 ovarian cancer cells through stimulation of cell survival and proliferation.
Fig. 2.
Effects of TRRAP knockdown on cell cycle and cell death in A2780-SP cells. (A) Flow cytometry was used for cell cycle analysis (right) after infecting A2780-SP cells with sh-control or sh-TRRAP lentivirus. The percentages of G01/G1, S, and G2/M Phases were counted. Representative data from four independent experiments are shown. (B) Percentages of apoptotic (sub-G1 phase) A2780-SP cells. Results are presented as mean ± SD. *P < 0.05 (n = 4). (C) The effects of TRRAP knockdown on the levels of cell cycle- and apoptosis-associated genes in A2780-SP cells. The protein levels of p21, CDK4, cyclin D3, PARP-1, and GAPDH were probed through Western blot analysis. Representative data from three independent experiments are shown.
TRRAP increased the expression levels of stemness markers in ovarian CSCs
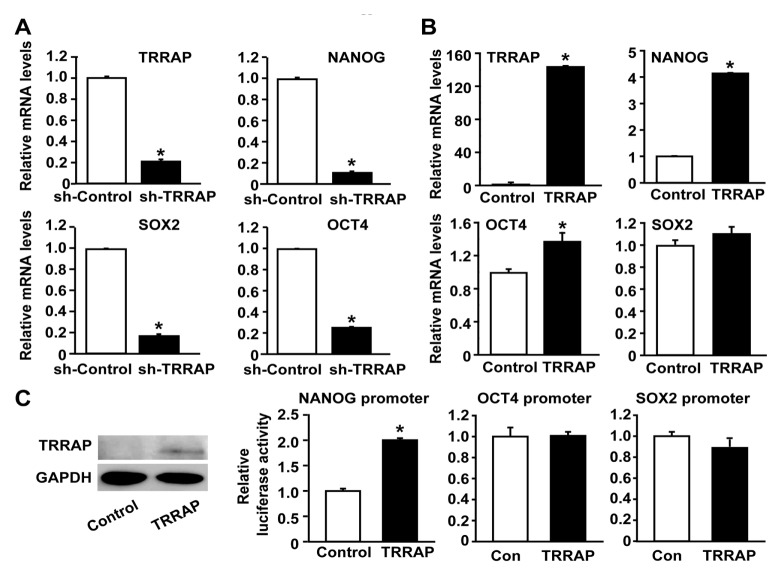
TRRAP has been reported to regulate the expression of stemness markers in ESCs (24). To investigate the role of TRRAP in the expression of stemness markers in ovarian CSCs, we measured the mRNA levels of OCT4, SOX2, and NANOG after TRRAP knockdown in A2780-SP cells. As shown in Fig. 3A, the expression levels of OCT4, SOX2, and NANOG were decreased by the silencing of TRRAP in A2780-SP cells. Interestingly, it was observed that the mRNA levels of NANOG were markedly up-regulated in A2780-AD cells in response to overexpression of TRRAP (Fig. 3B). Moreover, the mRNA level of OCT4 was slightly increased by TRRAP overexpression, whereas the mRNA level of SOX2 was not affected by TRRAP overexpression. To further investigate the effect of TRRAP on gene transcription of the stemness markers, we performed a luciferase assay of promoters for NANOG, OCT4, and SOX2. TRRAP overexpression markedly increased NANOG promoter activity in A2780-AD cells (Fig. 3C), whereas the luciferase activities of OCT4 and SOX2 promoters in A2780-AD cells were not significantly affected by TRRAP overexpression. These results suggest that TRRAP is involved in the expression of the stemness markers in ovarian CSCs through directly promoting transcription of NANOG gene.
Fig. 3.
Role of TRRAP in the expression of NANOG in CSCs. (A) Effects of TRRAP knockdown on the expressions of stemness factors in A2780-SP cells. The mRNA levels of OCT4, SOX2, and NANOG were assessed after silencing TRRAP in A2780-SP cells. (B) RT-PCR analysis results for OCT4, SOX2, and NANOG mRNA levels after TRRAP overexpression in A2780-AD cells. (C) Luciferase reporter assay of human NANOG, OCT4, and SOX2 promoters after overexpression of control or FLAG-tagged TRRAP in A2780-AD cells (right panel). The expression levels of FLAG-TRRAP and GAPDH were determined through Western blotting analysis with anti-FLAG and anti-GADPH antibodies (left panel). Results are presented as mean ± SD. *P < 0.05 (n = 4).
Knockdown of TRRAP inhibited tumor growth in vivo
To investigate the involvement of TRRAP in tumor growth, we evaluated the effects of TRRAP silencing on the tumorigenicity of ovarian CSCs using an in vivo murine xenograft transplantation model. A2780-SP cells infected with control or sh-TRRAP lentivirus were cultured under puromycin selection pressure, and then selected cells were subcutaneously injected into nude mice. In mice injected with control shRNA-transfected A2780-SP cells, tumor volumes increased time-dependently, whereas in vivo growth and tumor weights were markedly attenuated in mice injected with TRRAP shRNA-transfected cells (Fig. 4). These results suggest that TRRAP is required for the tumorigenesis of ovarian CSC in vivo.
Fig. 4.
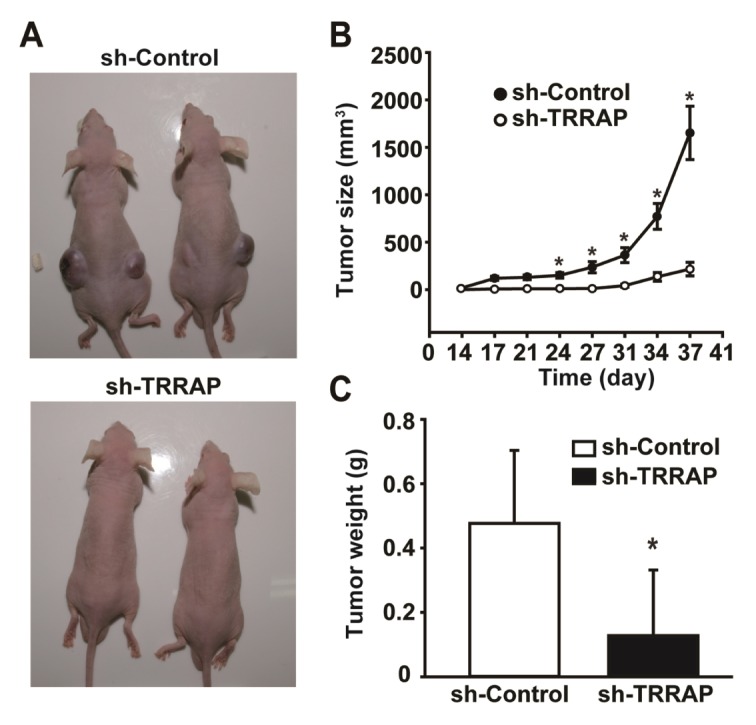
Role of TRRAP on in vivo tumor growth of A2780 sphere cells. (A) Effects of TRRAP knockdown on the in vivo growth of xenograft transplanted A2780-SP cells. A2780-SP cells were infected with lentiviruses expressing sh-control or sh-TRRAP and then transplanted into nude mice. Representative images are shown of xenograft tumors 38 days after transplanting A2780-SP cells infected with sh-control or sh-TRRAP lentiviruses. (B) Tumor volumes were measured daily from days 14 to 38 after injecting A2780-SP cells. (C) Tumor weights were measured 38 days after transplanting A2780-SP cells. Results are presented as mean ± SD. *P < 0.05 (n = 8).
DISCUSSION
The present study shows that TRRAP is essentially required for the proliferation of ovarian CSCs in vitro. TRRAP knockdown attenuated the proliferation and sphere-forming ability of A2780-SP cells and also caused cell cycle arrest and subsequent cell death. In addition, the in vivo growth of transplanted A2780-SP cells was greatly attenuated by the silencing of TRRAP expression. TRRAP depletion has been reported to cause early embryonic lethality in mice as well as defects in cell cycle progression in normal cells (19). TRRAP knockdown has been shown to significantly suppress tumor formation through intracranially implanted brain tumor-initiating cells in mice (11). Knockdown of TRRAP increased the differentiation of cultured brain tumor-initiating cells, sensitized these cells to apoptotic stimuli, and inhibited the cell cycle progression of a glioblastoma multiforme cell line. (13). These results suggest that TRRAP plays a key role in the proliferation and in vivo tumor growth of ovarian CSCs.
In mouse ESCs, TRRAP has been reported to be involved in maintenance of self-renewal, and TRRAP loss resulted in downregulation of stemness marker genes NANOG, OCT4, and SOX2 (24). We demonstrated here that the silencing of TRRAP expression led to reduced expression levels of NANOG, OCT4, and SOX2 in A2780-SP cells. Whereas, in A2780-AD cells, overexpression of TRRAP significantly increased the expression levels of NANOG and OCT4 but not SOX2. Moreover, TRRAP overexpression stimulated transcription of NANOG promoter, but not that of OCT4 or SOX2. These results suggest that TRRAP directly regulates NANOG gene transcription, whereas TRRAP may indirectly regulate the expression of OCT4 and SOX2. However, the molecular mechanism involved in the TRRAP-dependent regulation of NANOG gene expression is still unclear. When Tip60-p400 complex was knocked down in ESCs, the changes in gene expression profile overlapped with those observed after NANOG knockdown, which was observed to be associated with decreased binding between p400 and target promoters (25, 26). It has been established that TRRAP in Tip60-p400 complex binds to the promoters of stem cell markers and recruits other transcription initiation complexes (26). An increasing body of evidence suggests that NANOG plays a key role in the regulation of the stemness-like characteristics of CSCs (27–29). These observations suggest that TRRAP in Tip60-p400 complex plays a key role in the gene transcription of NANOG, followed by NANOG-dependent increased transcription of stemness markers.
Our study provides the finding that the TRRAP gene is critically required for the regulation of the tumorigenic potential of ovarian CSCs. We also found that the expression of the stemness factor NANOG was regulated by TRRAP in CSCs. Collectively, these results suggest TRRAP as a potential target for the eradication of CSCs in ovarian cancer.
MATERIALS AND METHODS
Materials
Fetal bovine serum, trypsin, and Hank’s balanced salt solution were purchased from Invitrogen (Carlsbad, CA). RPMI 1640 medium was obtained from Welgene (Gyeongsan-si, Republic of Korea), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was obtained from EMD Millipore (Billerica, MA), and M2 anti-FLAG antibody was obtained from Sigma-Aldrich (Saint Louis, MO). Basic fibroblast growth factor and epidermal growth factor were purchased from R&D Systems (Minneapolis, MN). Cell culture plates with ultra-low attachment surfaces were purchased from Corning Life Sciences (Tewksbury, MA). Neurobasal medium, RPMI 1640, FBS, B-27 Supplement, penicillin, streptomycin, and Accutase cell detachment solution were purchased from Life Technologies (Grand Island, NY). The epithelial human ovarian cancer cell line A2780 was purchased from the American Type Culture Collection (Manassas, VA). Cell cycle regulation antibody sampler kit and anti-PARP-1 antibody were purchased from Cell Signaling Technology, Inc. (Danvers, MA).
Cell culture
A2780 cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) at 37°C in a 5% CO2 atmosphere. Cell culture plates for adherent cells were purchased from Thermo Fisher Scientific Inc. (Waltham, MA). Sphere-forming ovarian CSCs were generated from A2780 cells as previously described (20). Briefly, A2780 cells were seeded in an ultra-low attachment dish at a density of 2 × 103 cells/10 cm dish and cultured in sphere culture medium (Neurobasal medium supplemented with B-27, 10 ng/ml basic fibroblast growth factor, 20 ng/ml epidermal growth factor, 2.5 μg/ml amphotericin B, 100 U/ml penicillin, and 100 μg/ml streptomycin). Fresh sphere culture medium was added every two or three days. Spheroids were dissociated into single cells through treatment with Accutase cell detachment solution, followed by filtering through a 40-μm cell strainer and plating at 1 × 104 cells/ml. Spheroids were subcultured in sphere culture medium for at least four passages before analysis.
Overexpression and knockdown of TRRAP
For the overexpression of TRRAP, A2780 cells were transfected with CbS-FLAG-TRRAP vector (Addgene, www.addgene.org) using Lipofectamine (Thermo Fisher Scientific, Waltham, MA). To confirm TRRAP overexpression, mRNA levels were determined through real time RT-PCR and protein levels were detected through Western blotting with anti-FLAG antibody.
To generate lentiviruses bearing shRNA targeting TRRAP, plat GP cells were co-transfected with 6.67 μg of pLKO.1-puro lentiviral expression vector bearing TRRAP-specific shRNA (GCCCTGTTCTTTCGCTTTGTA), 5 μg VSV-G envelope plasmid, and 3.33 μg Δ 8.9 packaging plasmid using 15 μl Lipofectamine and Lipofectamine PLUS reagent. Culture supernatants containing lentivirus particles were harvested at 48 h after transfection and concentrated using a Lenti-X Concentrator at 4°C (Clontech Laboratories, Inc., Mountain View, CA). For viral infection, A2780-SP cells were infected by treatment with the concentrated viral supernatants in the presence of 5 μg/ml Polybrene (Sigma-Aldrich) for 24 h. The lentivirus-infected cells were then selected by maintaining them in RPMI1640 supplemented with 10% fetal bovine serum and 1 μg/ml of puromycin.
Cell proliferation
To measure cell proliferation, A2780-SP cells were seeded in a 24-well culture plate at a density of 1 × 104 cells/well. After being cultured under various experimental conditions, the cells were washed twice with HBSS and incubated with 200 μl of MTT solution (0.5 mg/ml) for 2 h at 37°C. After this incubation, the MTT solution was removed, the cells were treated with dimethyl sulfoxide (200 μl/well), and the absorbance of the solution at 570 nm was determined using a micro-plate spectrophotometer (TECAN) after dilution to a linear range.
Luciferase assay
A2780 cells were plated in 12-well culture plates at a density of 3 × 105 cells/well, and transfected with luciferase reporter plasmids using Lipofectamine/Lipofectamine Plus according to the manufacturer’s instructions. Transfection mixtures contained 400 ng of NANOG reporter construct (pNANOG-Luc) and 10 ng of an internal control plasmid (pCMV-RL vector containing Renilla luciferase, Promega). The luciferase assay vectors containing OCT4 and SOX2 promoters were prepared as previously reported (30). One day following transfection with plasmids, the cells were harvested, lysed, and centrifuged at 2000×g for 3 min at 4°C. Luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) using a VICTOR3 (Perkin Elmer, Inc., Waltham, MA).
Cell cycle analysis
Cell cycle was determined through propidium iodide staining and FACS analysis. Briefly, the shRNA-transfected A2780-SP cells were harvested by trypsinization, fixed in 70% cold ethanol for 15 min at −20°C, and then rehydrated in PBS for another 15 min. The cells were stained with 1 ml of 3 μM propidium iodide solution in incubation buffer (100 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM CaCl2, 0.5 mM MgCl2, and 0.1% Nonidet P-40). The cell suspension was then subjected to fluorescence-activated cell sorting (FACS CANTO II (BD Biosciences, San Jose, CA)), and each cell was assigned to the G0/G1, S, or G2/M phases of the cell cycle, based on their propidium iodide intensities.
Western blotting
Cells were lysed in a lysis buffer (pH 7.4; 20 mM Tris-HCl, 1 mM EGTA, 1 mM EDTA, 10 mM NaCl, 0.1 mM phenylmethyl sulfonyl fluoride, 1 mM Na3VO4, 30 mM sodium pyrophosphate, 25 mM β-glycerol phosphate, and 1% Triton X-100). Lysates were resolved by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and the proteins were transferred to nitrocellulose membranes. The proteins were stained with 0.1% Ponceau S solution (Sigma-Aldrich), blocked with 5% non-fat milk, and immunoblotted with anti-FLAG or anti-GAPDH antibodies overnight. Bound antibodies were visualized using horseradish peroxidase-conjugated secondary antibodies using the enhanced Chemiluminescence Western blotting system (GE Healthcare Life Sciences, Pittsburgh, PA, USA).
Reverse transcription-polymerase chain reaction (RT-PCR)
After cells were harvested, total cellular RNA was extracted using TRIzol (Sigma). mRNA levels were measured by RT-PCR using a Reverse Transcription cDNA Kit (Nano helix. co). cDNA in 1 μl of reaction mixture was amplified using HelixAmp™ Ready-2×-Go PCR kit (Nano helix, Daejeon, Republic of Korea, www.nanohelix.co.kr) and 10 pmol of sense and antisense primers (as detailed below). The thermal profile was as follows: denaturation at 95°C for 30 s, annealing at 51–55°C for 30 s depending on the primers used, and extension at 72°C for 30 s. Each PCR reaction was carried out for 25–30 cycles. PCR products were then analyzed through 1% agarose gel electrophoresis. GAPDH, 5′-TCCATGACAA CTTTGGTATCG-3′, 5′-TGTAGCCAAATTCGTTGTCA-3′; TRRAP, 5′-ATGATGCAAGAAGTTAGTGAAAA-3′, 5′-GAAGATGTTCG TTGGTTGGTA-3′; NANOG, 5′-CGAGATAAACATGGCAATC AAAAT-3′, 5′-AATTCAGCAAGAAGCCTCTCCTT-3′; ALDH1, 5′-CCGTGGCGTACTATGGATGC-3′, 5′-CGCAATGTTTTGATG CAGCCT-3′.
Real time RT-PCR assay
Real time RT-PCR was performed using an ABI 7500 real time PCR system (Applied Biosystems, Foster City, CA) using the primer detailed below and the SYBR Green PCR Master Mix (ABS-4309155, Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Data were analyzed using the Δ (Δ CT) method and normalized versus GAPDH. TRRAP, 5′-CAACGATTGCTGTGAAAGTCAAC-3′, 5′-TGGAATGTGTTC GAGTCTCACT-3′; OCT4, 5′-GATACTGGTTCGCTTTCTCT-3′, 5′-GGGATATACACAGGCCGAT-3′; SOX2, 5′-CGAGATAAAC ATGGCAATCAAAAT-3′, 5′-AATTCAGCAAGAAGCCTCTCCTT-3′; NANOG, 5′-CCATCCTTGCAAATGTCTTCTG-3′, 5′-CTTTG GGACTGGTGGAAGAATC-3′.
Xenograft transplantation and in vivo monitoring
For xenograft transplantation, A2780-SP cells were infected with control lentivirus or sh-TRRAP-expressing lentivirus. The lentivirus-infected cells (1 × 105 cells/200 μl PBS) were subcutaneously injected into the right and left flanks of BALB/c-nu/nu mice. Mice were sacrificed at 5 weeks after xenograft transplantation, at which point their tumor weights and volumes were measured.
Statistical analysis
Quantitative data are presented as the mean ± SD of each representative experiment. Significance was calculated using the unpaired Student’s t-test.
ACKNOWLEDGEMENTS
The authors thank Il Ho Jang for his support and comments on the manuscript. This research was supported by the MRC program (NRF-2015R1A5A2009656), the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (NRF-2016R1D1A1B03935769), and the Korea Health Technology R&D Project, Ministry of Health and Welfare (HI17C1635).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Slotman BJ, Rao BR. Ovarian cancer (review). Etiology, diagnosis, prognosis, surgery, radiotherapy, chemotherapy and endocrine therapy. Anticancer Res. 1988;8:417–434. [PubMed] [Google Scholar]
- 2.Aunoble B, Sanches R, Didier E, Bignon YJ. Major oncogenes and tumor suppressor genes involved in epithelial ovarian cancer (review) Int J Oncol. 2000;16:567–576. doi: 10.3892/ijo.16.3.567. [DOI] [PubMed] [Google Scholar]
- 3.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351:2519–2529. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 4.Martin LP, Schilder RJ. Management of recurrent ovarian carcinoma: current status and future directions. Semin Oncol. 2009;36:112–125. doi: 10.1053/j.seminoncol.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Zhan Q, Wang C, Ngai S. Ovarian cancer stem cells: a new target for cancer therapy. Biomed Res Int. 2013;2013;916819 doi: 10.1155/2013/916819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgos-Ojeda D, Rueda BR, Buckanovich RJ. Ovarian cancer stem cell markers: prognostic and therapeutic implications. Cancer Lett. 2012;322:1–7. doi: 10.1016/j.canlet.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosca E, Cocola C, Sabour D, et al. Overlapping genes may control reprogramming of mouse somatic cells into induced pluripotent stem cells (iPSCs) and breast cancer stem cells. In Silico Biol. 2010;10:207–221. doi: 10.3233/ISB-2010-0437. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee M, Nam EJ, Kim SW, Kim S, Kim JH, Kim YT. Prognostic impact of the cancer stem cell-related marker NANOG in ovarian serous carcinoma. Int J Gynecol Cancer. 2012;22:1489–1496. doi: 10.1097/IGJ.0b013e3182738307. [DOI] [PubMed] [Google Scholar]
- 11.Murr R, Vaissiere T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene. 2007;26:5358–5372. doi: 10.1038/sj.onc.1210605. [DOI] [PubMed] [Google Scholar]
- 12.Murr R, Loizou JI, Yang YG, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 13.Wurdak H, Zhu S, Romero A, et al. An RNAi screen identifies TRRAP as a regulator of brain tumor-initiating cell differentiation. Cell Stem Cell. 2010;6:37–47. doi: 10.1016/j.stem.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Charles NA, Holland EC. TRRAP and the maintenance of stemness in gliomas. Cell Stem Cell. 2010;6:6–7. doi: 10.1016/j.stem.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Shan M, Liu T, et al. Analysis of TRRAP as a Potential Molecular Marker and Therapeutic Target for Breast Cancer. J Breast Cancer. 2016;19:61–67. doi: 10.4048/jbc.2016.19.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/MCB.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikiforov MA, Chandriani S, Park J, et al. TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol Cell Biol. 2002;22:5054–5063. doi: 10.1128/MCB.22.14.5054-5063.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichim G, Mola M, Finkbeiner MG, Cros MP, Herceg Z, Hernandez-Vargas H. The histone acetyltransferase component TRRAP is targeted for destruction during the cell cycle. Oncogene. 2014;33:181–192. doi: 10.1038/onc.2012.570. [DOI] [PubMed] [Google Scholar]
- 19.Herceg Z, Hulla W, Gell D, et al. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat Genet. 2001;29:206–211. doi: 10.1038/ng725. [DOI] [PubMed] [Google Scholar]
- 20.Seo EJ, Kwon YW, Jang IH, et al. Autotaxin Regulates Maintenance of Ovarian Cancer Stem Cells through Lysophosphatidic Acid-Mediated Autocrine Mechanism. Stem Cells. 2016;34:551–564. doi: 10.1002/stem.2279. [DOI] [PubMed] [Google Scholar]
- 21.Kim DK, Seo EJ, Choi EJ, et al. Crucial role of HMGA1 in the self-renewal and drug resistance of ovarian cancer stem cells. Exp Mol Med. 2016;48:e255. doi: 10.1038/emm.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi EJ, Seo EJ, Kim DK, et al. FOXP1 functions as an oncogene in promoting cancer stem cell-like characteristics in ovarian cancer cells. Oncotarget. 2016;7:3506–3519. doi: 10.18632/oncotarget.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tapias A, Zhou ZW, Shi Y, et al. Trrap-dependent histone acetylation specifically regulates cell-cycle gene transcription to control neural progenitor fate decisions. Cell Stem Cell. 2014;14:632–643. doi: 10.1016/j.stem.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 24.Sawan C, Hernandez-Vargas H, Murr R, et al. Histone acetyltransferase cofactor Trrap maintains self-renewal and restricts differentiation of embryonic stem cells. Stem Cells. 2013;31:979–991. doi: 10.1002/stem.1341. [DOI] [PubMed] [Google Scholar]
- 25.Chen PB, Hung JH, Hickman TL, et al. Hdac6 regulates Tip60-p400 function in stem cells. Elife. 2013;2:e01557. doi: 10.7554/eLife.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeter CR, Liu B, Liu X, et al. NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Oncogene. 2011;30:3833–3845. doi: 10.1038/onc.2011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang ML, Chiou SH, Wu CW. Targeting cancer stem cells: emerging role of Nanog transcription factor. Onco Targets Ther. 2013;6:1207–1220. doi: 10.2147/OTT.S38114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeter CR, Yang T, Wang J, Chao HP, Tang DG. Concise Review: NANOG in Cancer Stem Cells and Tumor Development: An Update and Outstanding Questions. Stem Cells. 2015;33:2381–2390. doi: 10.1002/stem.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo EJ, Kim DK, Jang IH, et al. Hypoxia-Notch1-Sox2 signaling is important for maintaining cancer stem cells in ovarian cancer. Oncotarget. 2016;7:55624–55638. doi: 10.18632/oncotarget.10954. [DOI] [PMC free article] [PubMed] [Google Scholar]