Abstract
Protein-Protein Interactions (PPIs) play essential roles in diverse biological processes and their misregulations are associated with a wide range of diseases. Especially, the growing attention to PPIs as a new class of therapeutic target is increasing the need for an efficient method of cell-based PPI analysis. Thus, we newly developed a robust PPI assay (SeePPI) based on the co-translocation of interacting proteins to the discrete subcellular compartment ‘processing body’ (p-body) inside living cells, enabling a facile analysis of PPI by the enriched fluorescent signal. The feasibility and strength of SeePPI (Signal enhancement exclusively on P-body for Protein-protein Interaction) assay was firmly demonstrated with FKBP12/FRB interaction induced by rapamycin within seconds in real-time analysis of living cells, indicating its recapitulation of physiological PPI dynamics. In addition, we applied p53/MDM2 interaction and its dissociation by Nutlin-3 to SeePPI assay and further confirmed that SeePPI was quantitative and well reflected the endogenous PPI. Our SeePPI assay will provide another useful tool to achieve an efficient analysis of PPIs and their modulators in cells.
Keywords: P-body, PPI modulator, Protein-Protein interaction, Translocation-based PPI assay
INTRODUCTION
Since most proteins in living cells function as multi-protein complexes rather than as a single molecule, protein-protein interactions (PPIs) are critical for most biological processes (1). Many human diseases, such as cancers, Huntington’s disease, cystic fibrosis, Parkinson’s disease, and viral infections, are often caused by aberrant PPI, undesired PPI or pathogen-host PPI (2). Thus, targeting disease-associated PPIs has been increasingly considered for a potential therapeutic approach, especially with small molecules, which can intervene in the surfaces between proteins. In another aspect, PPIs have been generally viewed as challenging and undruggable targets because of (i) too large and flat interfaces and (ii) lack of deep grooves on their surfaces, unlike classical enzyme-ligand-binding pockets. However, recent identification of the ‘hot spots’, the specific residues or regions responsible for binding of PPI interfaces, have refueled the targeting of PPIs for drug discovery (3). Along with the existence of hot spots on PPI interfaces, several recent advances in technologies associated with structure-based design, PPI assays, and computational screening have remarkably facilitated the discovery of PPI modulators (4). Out of them, PPI assays are of particular importance, in that they are critical for evaluation of the PPI modulator throughout the drug discovery and further development processes. Despite many currently available PPI analysis methods, there is still a demand for a mammalian cell-based PPI assay that enables a robust, easy, and quantitative analysis of small-molecule drug candidates. Fluorescence resonance energy transfer (FRET) and biomolecular fluorescence complementation (BiFC) are two major platform technologies that are actively used to analyze PPIs in mammalian cells. However, both of them require inconvenient pre-optimization steps to find the optimal configuration of fusion proteins. Moreover, FRET has low sensitivity and requires a specialized instrument to detect the signals. BiFC has low dynamics because of a long fluorophore maturation time, which prevents real-time detection of rapid changes in interactions (5, 6). Alternatively, a few translocation-based cellular PPI assays with different principles were devised. These assays work by co-translocation of test proteins with bait proteins to a specific location inside cells, which is visualized by the use of fluorescent dye or protein. Translocation-based PPI assays require less effort for pre-optimization and give a relatively high signal-to-background ratio, making this assay more suitable for high-throughput screening (HTS) approaches to drug discovery. However, most currently available translocation-based assays are (i) not sufficiently sensitive, because they are based on the translocation between the large spaces inside cells (e.g., into the plasma membrane), resulting in dispersed and low translocation signals (7), or (ii) require an exogenous stimulus (e.g., phorbol 12-myristate 13-acetate, rapamycin) to induce the translocation of PPI into specific compartments in the cells (7–9), which may give unexpected or wrong readouts by affecting endogenous target protein.
RESULTS AND DISCUSSION
Concept of SeePPI
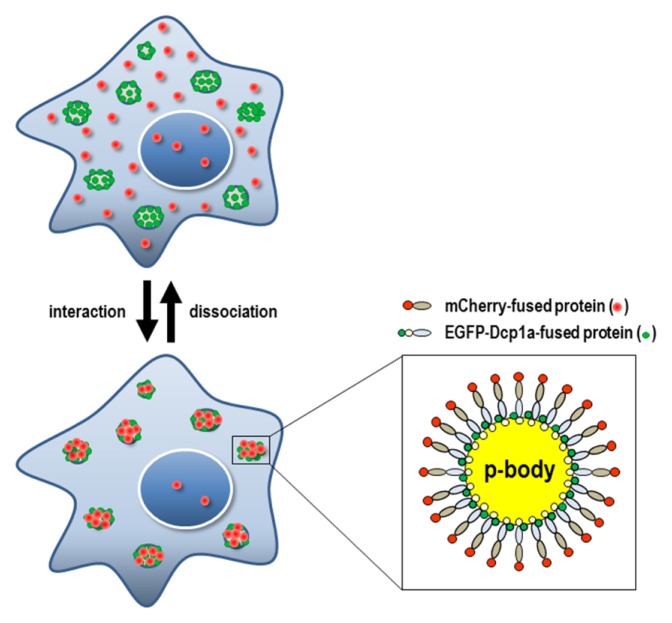
In this study, we devised a new PPI assay platform employing the translocation of interacting proteins into a discrete cytoplasmic compartment, such as the processing body (p-body), mitochondria, peroxisome, autophagosome, or stress granule. We believed that in such a way an otherwise dispersed translocation signal would be more condensed and intensified, making it much easier to analyze PPIs. Of many possible cytoplasmic compartments, we chose the p-body as the best docking system, since the formation and visualization of a p-body can be quite easily achieved by a simple overexpression of a single protein, such as Dcp1a, inducing an appropriate number (average number range per cell : 3–15) and visible size of spot signals in a cell (10). In contrast, the formation of stress granules usually requires a further process for efficient visualization, like stress conditions or the treatment with stress-inducing chemical (11). Mitochondria and peroxisome, despite being a decent number and size of cytoplasmic organelles, are physically associated with the ER and Golgi complex (12, 13), so it is harder to get a discrete spot signal than it is from a p-body. P-bodies are distinct foci within the cytoplasm of eukaryotic cells and consist of many enzymes involved in mRNA turnover. Dcp1a is a well-known component of the p-body, and the overexpression of Dcp1a itself is sufficient to get enlarged p-bodies (14, 15). Our PPI assay used the translocation of a Dcp1a-fused bait protein toward the p-body by the dominant action of Dcp1a and its visualization by GFP fusion, ending up with a GFP-Dcp1a-bait or bait-GFP-Dcp1a protein. A test protein that might interact with the bait protein was fused with mCherry (test-mCherry or mCherry-test proteins). If the bait and test fusion proteins were co-expressed in a cell and interacted with each other, both of them were supposed to be co-localized at the p-body and to show intense spot signals almost exclusively on the p-body. Therefore, we named this method SeePPI, i.e., Signal enhancement exclusively on P-body for Protein-protein Interaction; the schematic is shown in Fig. 1. Our system necessitated the localization of the bait fusion protein to the p-body.
Fig. 1.
Schematic diagram of the SeePPI concept. If mCherry-fused test proteins interact with EGFP-Dcp1a-fused bait proteins, both red and green fluorescent signals will be translocated onto p-bodies at discrete cytoplasmic spots (lower & left figure). Fluorescent fusion proteins co-localized on a p-body were enlarged (lower & right figure). Conversely, when the interaction is disrupted, the red fluorescent signals will be dispersed throughout the cells, leaving a weakened signal on the p-body (upper figure).
Visualization of rapamycin-induced interaction of FKBP12/FRB on the p-body
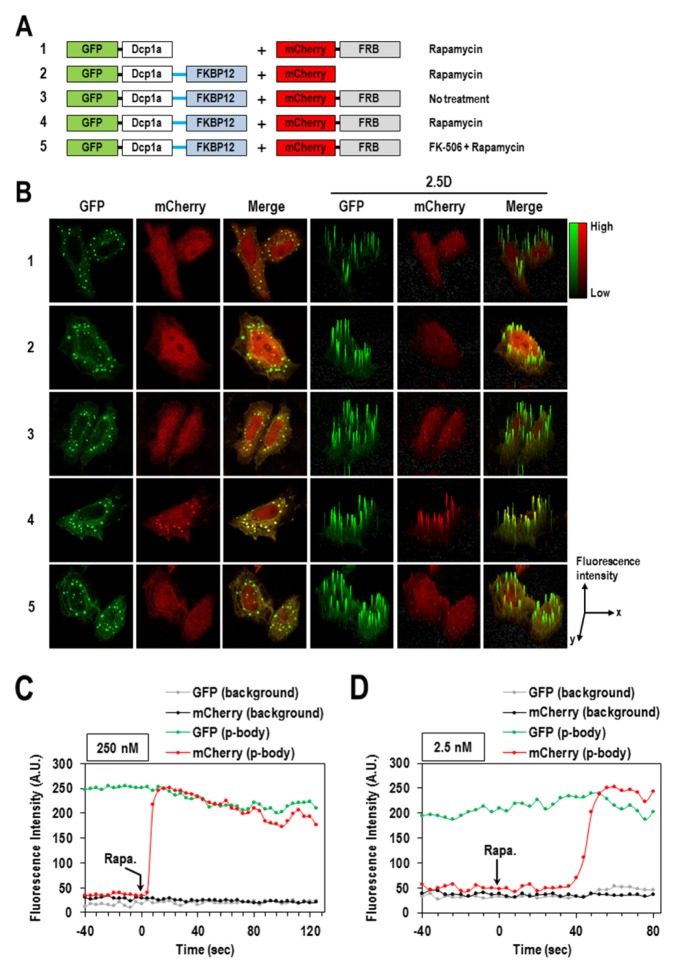
In order to verify whether our concept is working per se, we first employed the well-studied FKBP12/FRB interaction (16) that is induced by the treatment with rapamycin (Fig. 2A). From the preliminary tests with the exogenous expression of four different fusion combinations in HeLa cells, the configuration of EGFP-Dcp1a-FKBP12 and mCherry-FRB were shown the highest green and red fluorescent signals, respectively; so further analysis was done with that fusion pair (data not shown). As expected, the green signal of EGFP-Dcp1a-FKBP12 was mainly detected at discrete spots of the p-body. Then, rapamycin-induced translocation of mCherry-FRB into the p-body was observed only when it was co-expressed with EGFP-Dcp1a-FKBP12 (Fig. 2B-4). EGFP-Dcp1a did not lead the red fluorescent signal of mCherry-FRB to the p-body, and EGFP-Dcp1a-FKBP12 with mCherry was not able to do so (Fig. 2B-1 and -2). Moreover, when pre-treated with FK-506, a drug that disrupts FKBP12/FRB interaction, there was little p-body translocation of mCherry-FRB observed in spite of rapamycin treatment (Fig. 2B-5). Collectively, these results firmly demonstrated that our newly developed SeePPI assay enabled an easy and solid evaluation of PPI with intensified fluorescent signals on the p-body.
Fig. 2.
Visualizing rapamycin-induced interaction of FKBP12/FRB on the p-body. (A) Schematic diagrams of pairs of bait and test fusion proteins. (B) HeLa cells were transfected with pairs of plasmids, as shown in panel (A). Twenty-four hours after co-transfection, cells were treated with DMSO or 250 nM rapamycin for 10 min, then fixed and visualized under confocal microscopy. FK-506 was treated 10 min prior to rapamycin addition to disrupt FKBP12/FRB interaction. The 2.5D surface plots represent the level of intensity of FKBP12/FRB interaction. EGFP-Dcp1a-FKBP12 and mCherry-FRB were co-expressed in HeLa cells for 24 hours, treated with 250 nM (C) or 2.5 nM (D) of rapamycin, and then quantitatively analyzed for their real-time translocation onto the p-body. Rapamycin was added at time = 0 second.
Next, in order to know how well our SeePPI assay can reflect the real dynamics of PPIs inside cells, we analyzed the real-time translocation of mCherry-FRB into the p-body in response to the treatment with rapamycin. When HeLa cells co-expressing EGFP-Dcp1a-FKBP12 and mCherry-FRB were treated with a high concentration of rapamycin (250 nM), the red fluorescent signal took less than 5 seconds to initiate the formation of the p-body and took just 12 seconds to reach a maximal intensity and size comparable to those of the green fluorescent signal (Fig. 2C and Supplementary video 1A). The similar translocation of the red fluorescent signal was also seen even at a much lower concentration (2.5 nM), but with a slower response, within 56 sec (Fig. 2D and Supplementary video 1B). This ultra-fast response of FKBP12/FRB interaction to rapamycin in our SeePPI assay is extraordinary when compared to the responses in other previously reported PPI assays (~1 hour of BiFC at 100 nM (17) and ~10 minutes of CUPID at 20 nM (7)). These results demonstrated that our SeePPI assay can be suitable for the detection of dynamic PPIs and may even be more advantageous than other assays.
Visualization of p53/MDM2 interaction dissociated by Nutlin-3 on the p-body
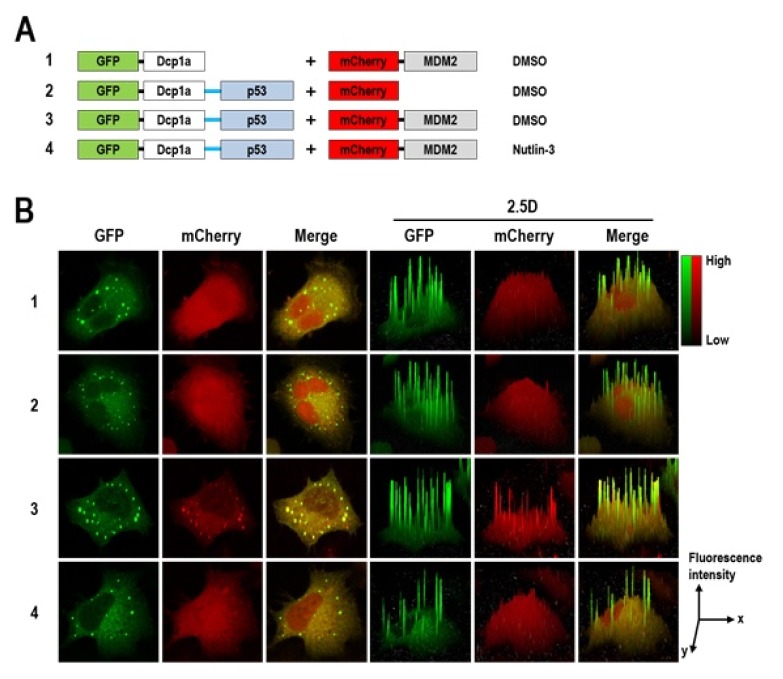
To further verify our SeePPI assay, we applied p53/MDM2 interaction, a well-studied anticancer target PPI, into our system and also examined the response to Nutlin-3, a prototype inhibitor of p53/MDM2. The p53 is a tumor suppressor, and MDM2 post-translationally regulates the protein stability of p53 by ubiquitination and subsequent proteasomal degradation (18). In many cancer cells, the level of the MDM2 protein tends to be higher than in normal cells and that of the p53 protein is inversely decreased. Thus, disrupting p53/MDM2 interaction with small molecules has long been considered to be a therapeutic strategy for several cancers (19), and Nutlin-3 is one of the leading candidates discovered from HTS and developed for a clinical trial (20). Considering that the N-terminal parts of each protein (p53N and MDM2N) are sufficient for interaction, and C-terminal fusions have been used frequently in other studies (7, 21), we also constructed plasmids encoding EGFP-Dcp1a-p53N and mCherry-MDM2N (Fig. 3A) and transfected them into HeLa cells. One day after transfection, red fluorescent signals were observed in the p-body where green fluorescent signals appeared (Fig. 3B-3). Neither EGFP-Dcp1a with mCherry-MDM2N nor EGFP-Dcp1a-p53 with mCherry was shown this p-body co-localization (Fig. 3B-1 and -2). Moreover, the red fluorescent signal of mCherry-MDM2N in the p-body disappeared almost completely when treated with Nutlin-3 (Fig. 3B-4). Collectively, these results obviously demonstrated that p53/MDM2 interaction was recapitulated in our SeePPI assay, and also suggested its feasibility for the screening of small molecules that modulate PPIs.
Fig. 3.
Monitoring the dissociation of p53/MDM2 interaction by Nutlin-3. (A) Schematic diagrams of pairs of bait and test fusion proteins. (B) HeLa cells were transfected with pairs of plasmids as shown in panel (A). Twenty-four hours after transfection, cells were fixed and visualized under confocal microscopy. Before cell fixation, 1 μM of Nutlin-3 was treated for 30 min to disrupt p53/MDM2 interaction. The 2.5D surface plots represent the level of intensity of p53/MDM2 interaction.
Since our assay made it possible to visualize the inhibition of p53/MDM2 interaction by Nutlin-3, we sought to quantify the level of inhibitory effect of both Nutlin-3 and cmpd 7, an ineffective fragment of the Nutlin family (22) (Fig. 4A), by measuring the mean intensity of the red fluorescent signal on the p-body. Twenty-four hours after co-transfection of EGFP-Dcp1a-p53N and mCherry-MDM2N into U2OS cells, the cells were treated with 1 μM of each compound for 30 min and measured by the mean intensity of green and red fluorescent signals on the p-body. As shown in Fig. 4B, Nutlin-3 exhibited a 68% decrease of the mean intensity of red fluorescent signals on the p-body, whereas cmpd 7 did only 18% decrease of it, indicating that a SeePPI assay demonstrates the effect of small molecule inhibitors for PPI. To further confirm whether their inhibitory effects were correlated with the amount of endogenous p53 proteins, we performed Western blotting with an anti-p53 antibody. The increase of the amount of p53 protein represents the inhibitory effect of p53/MDM2 interaction, which would be inversely correlated with the decrease of the mean intensity of red fluorescent signals on the p-body from SeePPI. As agreeing well with our expectation, after the treatment with 10 μM of each compound for 10 hrs in U2OS cells, the amount of p53 proteins was remarkably increased by Nutlin-3 (7.4-fold), but weakly by cmpd 7 (1.8-fold) (Fig. 4C and D), indicating that the SeePPI of p53/MDM2 well reflected the endogenous p53/MDM2 interaction.
Fig. 4.
Quantitative analysis of the inhibitory effect of Nutlin-3 and cmpd 7 for p53/MDM2 interaction. (A) Chemical structure of Nutlin-3 and cmpd 7 (an inactive fragment of Nutlin family). (B) Indicated pair of fusion proteins were expressed in U2OS cells for 24 hours and then treated with Nutlin-3 and cmpd 7 for 30 min prior to cell fixation and subsequent visualization. The mean intensity of green and red fluorescent signals on the p-body was quantitatively measured, and the relative intensities were presented (n of p-body spots = 41, 52, 34 for DMSO, Nutlin-3, and cmpd 7, respectively). (C) Ten hours after the treatment with Nutlin-3 and cmpd 7 on U2OS cells, the amount of endogenous p53 protein was analyzed by Western blotting. The asterisk indicates a nonspecific band. HnRNP A1 was monitored as a loading control. (D) The p53 and hnRNP A1 proteins from Western blotting were quantified, and the amount of protein relative to that in the DMSO-treated samples are presented. The mean ± s.d. was calculated from three independent experiments.
In summary, we provide a clear proof of the concept of our new translocation-based cellular SeePPI assay that employs the p-body as a natural docking place inside cells. Our SeePPI assay would be useful for research on PPIs and their modulators with the following advantages:
easier to obtain acceptable readouts regardless of fusion configuration,
highly sensitive and confers a high signal-to-noise ratio,
sufficiently quantitative to be suitable for high-throughput screening,
stimulus-free to translocate the PPI into a discrete spot of the p-body in the cells,
recapitulates the dynamic PPIs as close as to intact ones.
MATERIALS AND METHODS
Construction of plasmids
The plasmid of pEGFP-Dcp1a was constructed and kindly provided by Dr. Jong Heon Kim (National Cancer Center, South Korea) and the plasmid of pΔEGFP-mCherry was a kind gift from Dr. Nak-Kyun Soung (Korea Research Institute of Bioscience and Biotechnology, South Korea). To construct the pEGFP-Dcp1a-bait (FKBP12:1-108aa and p53N:1-72aa) expression plasmids, each of the PCR-amplified FKBP12 and p53N cDNA fragments was cloned into the plasmid pEGFP-Dcp1a at the KpnI site. To generate the pmCherry-target (FRB:2019-2114aa and MDM2N:1-119aa) expression plasmids, each of the PCR-amplified FRB and MDM2N cDNA fragments was inserted into the plasmid pΔEGFP-mCherry at the XhoI site.
Cells, transfection and reagents
HeLa and U2OS cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) and RPMI-1640 medium containing 10% fetal bovine serum (FBS; Hyclone and Gibco) supplemented with 1% penicillin and streptomycin. HeLa cells were grown on round coverslips (Marlenfeld GmbH & Co.) in a 24-well culture plate to 50–70% confluency. Transient co-transfection of plasmids expressing the desired protein pairs (FKBP12/FRB and p53N/MDM2N) was conducted using the XtremeGene Transfection Reagent (Roche) and Lipofectamine 3000 Reagent (ThermoFisher Scientific) according to the manufacturer’s instructions. Rapamycin (Calbiochem), FK-506 (Cayman), and Nutlin-3 (Sigma-Aldrich) were dissolved in dimethyl sulfoxide (DMSO) prior to treatment. Cmpd 7 was kindly provided by Dr. Peppi Prasit (USA).
Confocal imaging
Confocal microscopy was performed on the LSM 700 with a Plan-Apochromat 63X/1.40 Oil DIC M27 objective lens along with ZEN 2010 software according to the manufacturer’s instructions. For rapamycin-induced FKBP12/FRB interaction, transiently co-transfected HeLa cells were treated with 250 nM rapamycin for 10 min, and 25 μM FK-506 was pre-incubated for 10 min before the treatment with rapamycin for the competition on FKBP12/FRB interaction. For the inhibition of p53/MDM2 interaction by Nutlin-3, transiently co-transfected HeLa cells were treated with 1 μM Nutlin-3 for 30 min. Briefly, cells were washed with 1X PBS twice, fixed in 4% paraformaldehyde at room temperature for 5 min, and then washed with 1X PBS twice again, and mounted on the microscope slides.
Real-time imaging
For live cell imaging, transiently co-transfected Hela cells were prepared on the four wells of a Nunc Lab Teck-II Chamber Slide, which was connected to a temperature controller set at 37°C. Sequential images of the same cell were collected at 6.2 s (for 2.5 nM rapamycin) and at 3.8 s (for 250 nM rapamycin) intervals using a laser-scanning confocal microscope (LSM 710, Carl Zeiss, Germany) with a Plan-Apochromat 20X/0.8 M27 objective lens. During imaging, rapamycin was treated in the chamber.
Quantitative western blot analysis
Quantitative western blot analysis was performed as described previously (23). The following antibodies were used: anti-p53 antibody (catalog # sc-98, Santa Cruz Biotechnology, 1:1000 dilution), Anti-hnRNP-A1 antibody (catalog # CSB-PA002942, Cusabio, 1:1000 dilution).
Quantitative image analysis
For measuring the mean intensity of both green and red fluorescent signals on the same p-body spot, the circular area of the p-body in the merged image was specified and then measured for its intensity using the “Histo” tool for “Mean Intensity” in the ZEN 2010 software. PhotoScape software was used to convert the videos in .Ism file format to GIF file format.
Supplementary Information
ACKNOWLEDGEMENTS
This research was supported by grants funded by the ministry of Health & Welfare (HI14C2124), the National Research Foundation of Korea (NRF-2016K1A1A8A01938649 & NRF-2015M3A9C7030128), and the KRIBB Research Initiative Programs.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Fletcher S, Hamilton AD. Protein-protein interaction inhibitors: small molecules from screening techniques. Curr Top Med Chem. 2007;7:922–927. doi: 10.2174/156802607780906735. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez MW, Kann MG. Chapter 4: Protein interactions and disease. PLoS Comput Biol. 2012;8:e1002819. doi: 10.1371/journal.pcbi.1002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 4.Scott DE, Bayly AR, Abell C, Skidmore J. Small molecules, big targets: drug discovery faces the protein-protein interaction challenge. Nat Rev Drug Discov. 2016;15:533–550. doi: 10.1038/nrd.2016.29. [DOI] [PubMed] [Google Scholar]
- 5.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 6.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee KB, Hwang JM, Choi IS, et al. Direct monitoring of the inhibition of protein-protein interactions in cells by translocation of PKCdelta fusion proteins. Angew Chem Int Ed Engl. 2011;50:1314–1317. doi: 10.1002/anie.201005333. [DOI] [PubMed] [Google Scholar]
- 8.Lee KH, Lee S, Lee WY, Yang HW, Heo WD. Visualizing dynamic interaction between calmodulin and calmodulin-related kinases via a monitoring method in live mammalian cells. Proc Natl Acad Sci U S A. 2010;107:3412–3417. doi: 10.1073/pnas.0911262107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, Ha JS, Lee SG, Kim TK. Inducible biosynthetic nanoscaffolds as recruitment platforms for detecting molecular target interactions inside living cells. J Am Chem Soc. 2012;134:11346–11349. doi: 10.1021/ja303518d. [DOI] [PubMed] [Google Scholar]
- 10.Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell. 2008;19:4154–4166. doi: 10.1091/mbc.e08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchi S, Patergnani S, Pinton P. The endoplasmic reticulum-mitochondria connection: one touch, multiple functions. Biochim Biophys Acta. 20141837:461–469. doi: 10.1016/j.bbabio.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 13.Smith JJ, Aitchison JD. Peroxisomes take shape. Nat Rev Mol Cell Biol. 2013;14:803–817. doi: 10.1038/nrm3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeRose R, Miyamoto T, Inoue T. Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 2013;465:409–417. doi: 10.1007/s00424-012-1208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robida AM, Kerppola TK. Bimolecular fluorescence complementation analysis of inducible protein interactions: effects of factors affecting protein folding on fluorescent protein fragment association. J Mol Biol. 2009;394:391–409. doi: 10.1016/j.jmb.2009.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res. 2003;1:993–1000. [PubMed] [Google Scholar]
- 19.Shangary S, Wang S. Targeting the MDM2-p53 interaction for cancer therapy. Clin Cancer Res. 2008;14:5318–5324. doi: 10.1158/1078-0432.CCR-07-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgess A, Chia KM, Haupt S, Thomas D, Haupt Y, Lim E. Clinical Overview of MDM2/X-Targeted Therapies. Front Oncol. 2016;6:7. doi: 10.3389/fonc.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulikov R, Winter M, Blattner C. Binding of p53 to the central domain of Mdm2 is regulated by phosphorylation. J Biol Chem. 2006;281:28575–28583. doi: 10.1074/jbc.M513311200. [DOI] [PubMed] [Google Scholar]
- 22.Fry DC, Wartchow C, Graves B, et al. Deconstruction of a nutlin: dissecting the binding determinants of a potent protein-protein interaction inhibitor. ACS Med Chem Lett. 2013;4:660–665. doi: 10.1021/ml400062c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung S, Choi M, Choi K, et al. Inactivation of human DGAT2 by oxidative stress on cysteine residues. PLoS One. 2017;12:e0181076. doi: 10.1371/journal.pone.0181076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.