Abstract
Cell reprogramming has been considered a powerful technique in the regenerative medicine field. In addition to diverse its strengths, cell reprogramming technology also has several drawbacks generated during the process of reprogramming. Telomere shortening caused by the cell reprogramming process impedes the efficiency of cell reprogramming. Transcription factors used for reprogramming alter genomic contents and result in genetic mutations. Additionally, defective mitochondria functioning such as excessive mitochondrial fission leads to the limitation of pluripotency and ultimately reduces the efficiency of reprogramming. These problems including genomic instability and impaired mitochondrial dynamics should be resolved to apply cell reprograming in clinical research and to address efficiency and safety concerns. Sirtuin (NAD+-dependent histone deacetylase) has been known to control the chromatin state of the telomere and influence mitochondria function in cells. Recently, several studies reported that Sirtuins could control for genomic instability in cell reprogramming. Here, we review recent findings regarding the role of Sirtuins in cell reprogramming. And we propose that the manipulation of Sirtuins may improve defects that result from the steps of cell reprogramming.
Keywords: Cell reprogramming, Genome stability, Induced pluripotent stem cells (iPSCs), Mytochondria dynamics, Sirtuins (Sirts)
INTRODUCTION
Cell reprogramming techniques have emerged with novel techniques to treat a variety of human diseases in the regenerative medicine field (1). In the reprogramming process, ‘immortality’ is regarded as a key to develop rejuvenation strategies (2). Takahashi et al. stated that cell reprogramming using four transcription factors such as Oct4, Sox2, Klf4, and c-Myc could convert terminally differentiated cells into induced pluripotent stem cells (iPSCs) (1). The pluripotency of iPSCs has opened up numerous possibilities for regenerative medicine to treat many diseases (3). Despite the powerful ability of iPSCs to treat numerous diseases, major concerns in recent iPSCs research include enhancing reprogramming efficiency and genomic stability. Genomic instability in iPSCs is generated in several steps of the cell reprogramming process (4). Cellular reprogramming goes through an intricate process that is similar to biological pathways of tumorigenesis (5). The essential factors for cell reprogramming are associated with tumorigenesis. For example, c-Myc and Klf4 play central roles in tumorigenesis, and Oct4 acts as an important initiator for germ cell tumors (5). In addition, to inducing changes in the original cell identity, cell reprogramming needs reactivation of the telomerase to continue to survive (6). Maintenance of telomere as an enzyme for telomere elongation is important for genomic stability during reprogramming (7). Telomerase is reactivated during reprogramming and the length and epigenetic state of the telomere contributes to rejuvenation in iPSCs. Shortening of the telomeres influences the reprogramming efficiency and the quality of the iPSCs (8). The strategy to solve the genome instability in cell reprogramming research for application in disease modeling and clinical cell therapy (9). During cell reprogramming, cells experience a metabolic shift into the glycolytic state (10). Oxidative stress and DNA damage from the cell reprogramming process results in a metabolic imbalance (11). Because of these metabolic shifts, mitochondrial activity is hampered and cannot react when energy is demanded due to cellular respiration. The reduction of mitochondrial activity during cell reprogramming is a matter that should be resolved for increasing iPSCs efficiency. Sirtuins known as histone deacetylases are relevant to the control of longevity, energy metabolism, and cell development in mammals (12). It was reported that sirtuins can affect the fate of stem cells through deacetylation of histone and non-histone proteins involved in gene expression (13). Recent studies demonstrated that the deficiency of Sirtuins influences reprogramming efficiency (14) and contributes to genomic instability, which as we noted, is an important issue in the cell reprogramming process (15). Here, we review evidence on the significant role of Sirtuins in the cell reprogramming process.
GENOMIC INSTABILITY IN CELL REPROGRAMMING
Genomic instability occurs during the cell reprogramming process (16). A number of studies report that after reprogramming iPSCs exhibit the genomic abnormalities such as chromosomal aberrations (17). Because of the transcription factors used in cell reprogramming cells have an increased risk of both tumor formation and genetic mutation (18). Telomerase is significantly upregulated during cell programming (8). Pluripotent cells show high activity of telomerase responsible for synthesizing telomeres in the reprogramming process (19). The iPSCs generation process showed that telomerase reverse transcriptase was upregulated in cells during cellular reprogramming (1). Telomerase activity and telomere length affect the state of pluripotency (20). In cell reprogramming, reactivation of telomerase has been shown to promote efficiency of iPSC reprogramming by maintaining telomere length and self-renewal potential for a relatively long time (21). Upon reprogramming, telomere lengthening is affected by a decrease of DNA methylation (22) and a reduction of methylation in histone H3 at lysine 9 (H3K9) m3 and histone H4 at lysine 20 (H4K20) m3 (8). Some studies investigated the differences in the telomere dynamics during reprogramming (21). Telomere shortening is a crucial issue in reprogramming process in that it hampers sufficient iPSCs generation. During the cell reprogramming process the proliferation rate increases causing replication stress and genomic structural variation (23). Additionally, recent studies show that pluripotent stem cells have an abnormal cell-cycle regulation such as a shorter G1 phase. The ataxia telangiectasia mutated Rad3 (ATR)-mediated checkpoint pathway is an essential replication stress response that generates genomic instability during reprogramming (24). Other studies report that Checkpoint kinase 1 (CHK1) overexpression could enhance both the reprogramming efficiency and the iPSCs quality (25). Abnormal cell cycle regulation is a distinct feature and the control over it is considered important to current reprogramming research. Accordingly, to realize the application of iPSCs in clinical research, we need a comprehensive understanding of genetic instability and should find an appropriate solution for it in cell reprogramming.
MITOCHONDRIAL DYNAMICS IN CELL REPROGRAMMING
Mitochondria is a multifunctional organelle and plays a crucial role in many cellular mechanisms such as energy production, apoptosis, reactive oxygen species (ROS) production, senescence, and metabolism (26). Mitochondrial homeostasis has been shown to be essential for maintenance of a pluripotent state. Ji et al. report that a decrease of ROS production in the mitochondria could improve iPSCs quality (27). Also excessive mitochondrial fission and knockdown of the mitochondrial DNA polymerase could trigger a lack of pluripotency (28). Tricyclic antidepressant (TCA)-derived cytosolic acetyl-CoA is essential for maintaining histone acetylation and an open chromatin state during cell reprogramming (29). Reprogramming somatic cells into iPSCs triggers impairment of the mitochondrial network during the reprograming process (30). Besides, during cell reprogramming, cells show particular characteristics including immature and globular mitochondria (31), and poorly developed cristae (32). Reduced expression 1 (REX1) known as a pluripotency marker regulates cell fate through its effect on mitochondrial dynamics (33). The knockdown of Dynamin-related GTPases-1 (DRP1) triggers the elongation of the mitochondrial network (34) and regulates membrane dynamics in a variety of cellular mechanisms and in mitochondrial fusion (35). One study demonstrated that the DRP1-GTPase inhibitor impedes cell reprogramming of human fibroblasts to iPSCs. The mechanistic target of rapamycin (mTOR) promotes cellular homeostasis and multiple signaling events that affect reprogramming (1). Inhibition of mTOR leads to an immediate decrease in mitochondrial respiration (36) and subsequently influences the generation of iPSCs (37). Taken together, cell reprogramming influences abnormal mitochondria function and homeostasis, and mitochondrial dynamics should be a focus for future cell reprogramming research.
SIRTUINS IN GENOMIC INSTABILITY DERIVED FROM CELL REPROGRAMMING
Sirtuins as an NAD+-dependent histone deacetylase have been involved in the improvement of longevity and metabolism in mammals (38). Given that histone acetylation is associated with gene activation (39), Sirtuins act as an epigenetic regulator of gene expression by histone deacetylation (40). Sirtuins have been shown to be essential for the silent chromatin state of the ribosomal RNA genes and telomeres. In mammals, Sirt6 has been reported to maintain telomeric chromatin and to enhance replicative capacity (41). According to cell reprogramming research the activation of Sirtuins considerably enhances the efficiency of cell reprogramming (42). Several studies demonstrate that the inhibition of histone deacetylases leads to increases of histone acetylation levels, chromatin opening, and ultimately could enhance efficiency of cell reprogramming (43). Sirtuins could possibly control the chromatin state by modulating the activation of enzymes such as H4K16Ac (44) and H3K4me3 that can upregulate cell reprogramming. Sirt1 is intimately linked to the maintenance of human embryonic stem cells pluripotency by inactivating p53 (45). Besides stem cells derived from Sirt6, knockout mice cells exhibit expression of Oct4, Sox2 and Nanog and present Sirt6’s function in balance between pluripotency and differentiation (46). Sirt1 could lead to the deacetylatization of Sox2 (14) and Sirt1’s overexpression induces the demethylation of the Oct4 promoter (47) and also affects reprogramming efficiency. Myc stability, important in cell reprogramming, could also be regulated by Sirt2 (48). Sirt1 deacetylates c-Myc by interacting physically with the C-terminus of c-Myc (49). Sirt1 induces p53 translocation into the mitochondria (50) and modulates Nanog expression (51) and is an important reprogramming factor. Judging by the metabolic state of the cell, Sirt1 can affect the epigenome change and the activity of chromatin-modifying enzymes (52). Sirt1 histone deacetylase regulates the epigenetical change and gene expressions in cells by translating a metabolic shift in the reprogramming process (53). A recent study showed that Sirt6 inhibits the transcription of Hypoxia-inducible factors (HIF1)-alpha and Myc (54). Sirt6 is essential for the maintenance of the telomere position in cells (55) and the deficiency of it leads to DNA damage and genomic instability (15). In addition, Sirt6 protects cells against stress by repairing DNA damage and preserving telomere integrity and controlling metabolic homeostasis (56). Sirt6 can deacetylate lysine 9 on histone H3 (H3K9Ac) (41) and lysine 56 on histone H3 (H3K56Ac) (57). And Sirt6 can recruit the chromatin remodeler Sucrose Nonfermenting Protein 2 Homolog (SNF2H) (58). As we have seen, Sirtuins influence cell reprogramming efficiency by regulating the activities of histone deacetylases, by controlling the chromatin state of telomere, and by being involved in metabolic shifts during cell reprogramming (Fig. 1).
Fig. 1.
The function of sirtuins on genome stability. Sirt 1, 2, and 6 control the chromatin state by regulating the activation of enzymes during chromatic remodeling. Sirt1 removes acetylates in Sox2 and Myc removes methylates in Oct4. Also, Sirt2 modulates the stability of the Myc protein. SIRT6 can deacetylate H3K9Ac and H3K56Ac and is involved in the transcription of c-Myc. AC: Acetylation, ME: Methylation, SIRT: Surtuin.
ACTIVATORS AND INHIBITORS OF SIRTUINS
Several compounds are known to be activators of Sirtuin. Resveratrol (3,5,4′-trihydroxy-trans-stilbene), SRT1720, Oxazolo [4,5-b] pyridines derivative, imidazole [1,2-b] thiazole derivative, and 1,4-dihydropyridine (DHP) derivatives are typical compounds that are known activators of Sirtuins (59). The exact mechanisms of Sirt1 activation by these activators is still unclear, but many of them seem to activate Sirt1 through allosteric activation, particularly, the resveratrol mediate activation of Protein Kinase AMP-Activated Catalytic Subunit Alpha 2 (AMPK), which is an initial sensor that increases NAD+ levels leading to activation of Sirt1 (60).
The metabolic effects of Resveratrol, the most common Sirtuin activator, relate to the cAMP level elevation in muscles (61). Also the general health in mice fed with a high caloric diet improved and they showed a marked reduction in sings of aging (62). These results open the possibility of clinical use of commercial micronized Respiratory Syncytial Virus (RSV) formulation, SRT501, for lowering blood glucose and improving insulin sensitivity in patients with type 2 diabetes (63). Moreover, SRT1720 has been shown to induce cell death in multiple myeloma cells (64) and significantly decrease tumor growth in a preclinical evaluation for cancer treatment (65). Also, as a new activators of Sirt1 unrelated to Resveratrol, a series of oxazolo pyridines was identified for potential therapeutic targets to treat different diseases (66). For example, compound 29 showed antidiabetic activity in types 2 diabetes (67) and SRT2104 was tested in a clinical trial of patients with metabolic inflammatory (68) and cardiovascular diseases (69).
In contrast, Splitomicin, HR73, Sirtinol, AGK2, Cambinol, Salermide, Tenovin, and Suramin are inhibitors of Sirtuin (70). The reaction mechanism of Sirtuins is the cleavage of nicotinamide (NIC) from NAD+ whereas ADP-ribose binds its acetyl-peptide with the formation of an o-alkylamidate intermediate. Sirtuin inhibitors hamper cleavage of NIC from NAD+. Suramin is an especially potent inhibitor of Sirt1, Sirt2 and Sirt5 (71). It inhibits NAD+-dependent deacetylase activity with an IC50 value of 22uM leading to mitochondrial dynamics disruptions (72). Several studies revealed that pharmacological inhibition of Sirtuin1 by Sirtinol inhibits prostate cancer cell proliferation in which Sirtuin1 is highly enriched (73). Moreover, Salermide, a sirtinol derivative, induces cell death via inhibiting MAP kinases erk1/2, p38 and JNK paring Sirtuin1 and Sirtuin2 in various human cancer cell lines derived from leukemia, lymphoma, colon, and breast primary malignancies (74). 6-Chloro-2,3,4,9-tetrahydro-1 H-Carbazole-1-carboxamide (EX527) is also known as a Sirtuin1 inhibitor and EX-527/SEN0014196 reduced neuronal death caused by mutant Huntington proteins in cell-based assays in preclinical studies of Huntington’s disease (75). More importantly, activation and inhibition of Sirtuin by small molecules is a complicated process and the effects of activation and inhibition of Sirtuin occasionally depend on the physiological state of the specific cells for its activity. For instance, increased activity of SIRT1 after treatment with resveratrol in the immediate immune response reduced the NFkB activation in the NFkB-dependent inflammatory genes in microglia and neuronal loss (76). This suggests that Sirt1 is working as anti-inflammatory mediator, whereas decreasing Sirt1 activity by sirtinol potentiates inflammatory responses, presumably occur via Sirt1-mediated deacetylation of p65 (77). Moreover, unlike Sirt1 effects on the inflammation, Sirt6 activity is positive for a given pro-inflammatory gene expression upregulating Tumor necrosis factor alpha (TNFa) and Interferon Production Regulator (IFNr) synthesis on both innate and adaptive immune cells (78). Such complexity of sirtuin activity in the various physiological states of cells lead to the difficulties of Sirtuin activators or inhibitors in determining the desired outcome of cell reprogramming efforts.
SIRTUINS IN MITOCHONDRIA DYNAMICS DURING CELL REPROGRAMMING
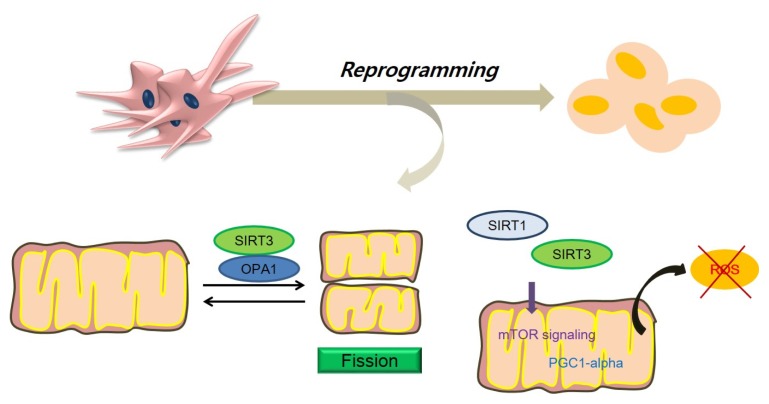
Mitochondrial dynamics are controlled by many cellular proteins such as fission proteins DRP1 (79), fusion proteins Mitofusins 1 and 2 (Mfn1/2) (80), and optic atrophy 1 (OPA1) proteins (81). The mitochondrial network was reported to have changed during the cell-cycle progression and mitosis processes (82). Mitochondrial distribution during mitosis acts in a critical role during asymmetric cell division in stem cells. Mitochondrial fusion, fission, and biogenesis are linked with mitochondrial dynamics as well (83). One study showed that mitochondrial fission and fusion contributes to the maintenance of pluripotency (84). Loss of mitochondrial fusion proteins such as Mfn1/2 (85) leads to a metabolic transition by activating HIF-1alpha signaling in iPSC reprogramming (86). Sirt1 can exert nuclear localization signals and nuclear export signals and can come and go between the cytoplasm and the nucleus (87). Sirt3 regulates the activity of both mitochondrial enzymes (88) and mitochondrial biogenesis through activation of the Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1-alpha) (89). OPA1 is a GTPase anchored to the mitochondria’s inner membrane and is linked to the maintenance of mitochondria crista structure and protection of cells against stimuli (90). Sirt3 has been known to bind directly to OPA1 and subsequently modulates mitochondrial dynamics (91). Sirt1 can enhance mitochondrial function by involving PI3K/Beclin 1 and mTOR signaling (92). Additionally, Sirt1 can destruct damaged mitochondria through a mitophagy process (93). Mitophagy is dependent on the activities of specific factors such as PTEN-induced putative kinase 1 (PINK1) and E3 ubiquitin ligase Parkin (94). According to genetic research, Sirtuins affect mitophagy by inhibiting mitochondrial defects in PINK1-null mutants (95). In addition, Sirt1 suppresses the activity of the HIF1-alpha (96) that inhibits mitochondrial function. Sirt3 is known as a powerful regulator of the ROS detoxification via deacetylation of Mangano-Superoxide Dismutase (MnSOD) in mitochondria (97). Sirt3 eliminates excessive ROS production through activation of a Forkhead box O3 FOXO3-alpha (98) and then regulates mitochondrial dynamics (99). Proceeding from what has been said above, Sirtuins may affect cell reprogramming efficiency through the regulation of mitochondrial dynamics including the regulation of fission proteins, the regulation of mitophagy, the modulation of mTOR signaling, and the control of ROS production (Fig. 2).
Fig. 2.
The relationship between sirtuins and mitochondrial dynamics caused by cell reprogramming. Cell reprogramming leads to mitochondrial dynamics such as changes in fission and fusion. The mitochondrial dynamics are linked with the maintenance of the pluripotent state. Sirtuins regulates mitochondria fission by binding with fission proteins such as OPA1 proteins. Also, sirtuins promote mTOR signaling, the activity of PGC1-alpha, and ultimately eliminate ROS production during cell reprogramming. mTOR: The mechanistic target of rapamycin, OPA1: Optic atrophy 1, PGC1-alpha: Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ROS: reactive oxygen species.
PERSPECTIVES AND CONCLUSIONS
In conclusion, cell reprogramming has limitations including genomic instability and impaired mitochondrial dynamics. Until now, the appropriate solution to overcome these limitations was not fully investigated. Sirtuins contribute to genomic stability and mitochondrial dynamics through several signaling reactions and the activation of enzymes. After examining the roles of Sirtuins, we propose further research should look at the multiple other functions of Sirtuins in cell reprogramming. We suggest investigating more advanced manipulation of Sirtuins in cell reprogramming and ultimately expect to promote more efficient and safe cell reprogramming processes and technology.
ACKNOWLEDGEMENTS
This work was supported by the Korea Health Technology R&D Project, Ministry of Health & Welfare (HI16C1176).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Rando TA, Chang HY. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 2012;148:46–57. doi: 10.1016/j.cell.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Blasco MA, Serrano M, Fernandez-Capetillo O. Genomic instability in iPS: time for a break. EMBO J. 2011;30:991–993. doi: 10.1038/emboj.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz S, Panopoulos AD, Herrerias A, et al. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marion RM, Strati K, Li H, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 9.Weissbein U, Benvenisty N, Ben-David U. Quality control: Genome maintenance in pluripotent stem cells. J Cell Biol. 2014;204:153–163. doi: 10.1083/jcb.201310135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folmes CD, Nelson TJ, Martinez-Fernandez A, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banito A, Rashid ST, Acosta JC, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura H, Hayashi-Takanaka Y, Stasevich TJ, Sato Y. Visualizing posttranslational and epigenetic modifications of endogenous proteins in vivo. Histochem Cell Biol. 2015;144:101–109. doi: 10.1007/s00418-015-1344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao Y, Chen J. Proteomics approaches in the identification of molecular signatures of mesenchymal stem cells. Adv Biochem Eng Biotechnol. 2013;129:153–176. doi: 10.1007/10_2012_143. [DOI] [PubMed] [Google Scholar]
- 14.Mu WL, Wang YJ, Xu P, et al. Sox2 Deacetylation by Sirt1 Is Involved in Mouse Somatic Reprogramming. Stem Cells. 2015;33:2135–2147. doi: 10.1002/stem.2012. [DOI] [PubMed] [Google Scholar]
- 15.Mostoslavsky R, Chua KF, Lombard DB, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 16.Laurent LC, Ulitsky I, Slavin I, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taapken SM, Nisler BS, Newton MA, et al. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol. 2011;29:313–314. doi: 10.1038/nbt.1835. [DOI] [PubMed] [Google Scholar]
- 18.Araten DJ, Golde DW, Zhang RH, et al. A quantitative measurement of the human somatic mutation rate. Cancer Res. 2005;65:8111–8117. doi: 10.1158/0008-5472.CAN-04-1198. [DOI] [PubMed] [Google Scholar]
- 19.Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 20.Vaziri H, Chapman KB, Guigova A, et al. Spontaneous reversal of the developmental aging of normal human cells following transcriptional reprogramming. Regen Med. 2010;5:345–363. doi: 10.2217/rme.10.21. [DOI] [PubMed] [Google Scholar]
- 21.Marion RM, Blasco MA. Telomere rejuvenation during nuclear reprogramming. Curr Opin Genet Dev. 2010;20:190–196. doi: 10.1016/j.gde.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 22.Yehezkel S, Rebibo-Sabbah A, Segev Y, et al. Reprogramming of telomeric regions during the generation of human induced pluripotent stem cells and subsequent differentiation into fibroblast-like derivatives. Epigenetics. 2011;6:63–75. doi: 10.4161/epi.6.1.13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasi CE, Dereli-Oz A, Negrini S, et al. Genomic instability in induced stem cells. Cell Death Differ. 2011;18:745–753. doi: 10.1038/cdd.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez-Contreras AJ, Gutierrez-Martinez P, Specks J, Rodrigo-Perez S, Fernandez-Capetillo O. An extra allele of Chk1 limits oncogene-induced replicative stress and promotes transformation. J Exp Med. 2012;209:455–461. doi: 10.1084/jem.20112147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz S, Lopez-Contreras AJ, Gabut M, et al. Limiting replication stress during somatic cell reprogramming reduces genomic instability in induced pluripotent stem cells. Nat Commun. 2015;6:8036. doi: 10.1038/ncomms9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji J, Sharma V, Qi S, et al. Antioxidant supplementation reduces genomic aberrations in human induced pluripotent stem cells. Stem Cell Reports. 2014;2:44–51. doi: 10.1016/j.stemcr.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci. 2007;120:4025–4034. doi: 10.1242/jcs.016972. [DOI] [PubMed] [Google Scholar]
- 29.Moussaieff A, Rouleau M, Kitsberg D, et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402. doi: 10.1016/j.cmet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- 31.Mandal S, Lindgren AG, Srivastava AS, Clark AT, Banerjee U. Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cells. 2011;29:486–495. doi: 10.1002/stem.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sathananthan H, Pera M, Trounson A. The fine structure of human embryonic stem cells. Reprod Biomed Online. 2002;4:56–61. doi: 10.1016/S1472-6483(10)61916-5. [DOI] [PubMed] [Google Scholar]
- 33.Han H, Irimia M, Ross PJ, et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Ye X, Zhao Q, et al. Drp1 is dispensable for mitochondria biogenesis in induction to pluripotency but required for differentiation of embryonic stem cells. Stem Cells Dev. 2014;23:2422–2434. doi: 10.1089/scd.2014.0059. [DOI] [PubMed] [Google Scholar]
- 35.Otera H, Mihara K. Molecular mechanisms and physiologic functions of mitochondrial dynamics. J Biochem. 2011;149:241–251. doi: 10.1093/jb/mvr002. [DOI] [PubMed] [Google Scholar]
- 36.Ramanathan A, Schreiber SL. Direct control of mitochondrial function by mTOR. Proc Natl Acad Sci U S A. 2009;106:22229–22232. doi: 10.1073/pnas.0912074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Xia P, Ye B, Huang G, Liu J, Fan Z. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell. 2013;13:617–625. doi: 10.1016/j.stem.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Greer EL, Dowlatshahi D, Banko MR, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S, Park JR, Seo MS, et al. Histone deacetylase inhibitors decrease proliferation potential and multilineage differentiation capability of human mesenchymal stem cells. Cell Prolif. 2009;42:711–720. doi: 10.1111/j.1365-2184.2009.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michishita E, McCord RA, Berber E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen T, Shen L, Yu J, et al. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell. 2011;10:908–911. doi: 10.1111/j.1474-9726.2011.00722.x. [DOI] [PubMed] [Google Scholar]
- 43.Huangfu D, Maehr R, Guo W, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 45.Zhang ZN, Chung SK, Xu Z, Xu Y. Oct4 maintains the pluripotency of human embryonic stem cells by inactivating p53 through Sirt1-mediated deacetylation. Stem Cells. 2014;32:157–165. doi: 10.1002/stem.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Etchegaray JP, Chavez L, Huang Y, et al. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol. 2015;17:545–557. doi: 10.1038/ncb3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng CH, Cherng JY, Chiou GY, et al. Delivery of Oct4 and SirT1 with cationic polyurethanes-short branch PEI to aged retinal pigment epithelium. Biomaterials. 2011;32:9077–9088. doi: 10.1016/j.biomaterials.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 48.Liu PY, Xu N, Malyukova A, et al. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ. 2013;20:503–514. doi: 10.1038/cdd.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mao B, Zhao G, Lv X, et al. Sirt1 deacetylates c-Myc and promotes c-Myc/Max association. Int J Biochem Cell Biol. 2011;43:1573–1581. doi: 10.1016/j.biocel.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 50.Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/S0092-8674(01)00527-X. [DOI] [PubMed] [Google Scholar]
- 51.Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241–251. doi: 10.1016/j.stem.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 53.Ryall JG, Dell’Orso S, Derfoul A, et al. The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell. 2015;16:171–183. doi: 10.1016/j.stem.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zwaans BM, Lombard DB. Interplay between sirtuins, MYC and hypoxia-inducible factor in cancer-associated metabolic reprogramming. Dis Model Mech. 2014;7:1023–1032. doi: 10.1242/dmm.016287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tennen RI, Bua DJ, Wright WE, Chua KF. SIRT6 is required for maintenance of telomere position effect in human cells. Nat Commun. 2011;2:433. doi: 10.1038/ncomms1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kugel S, Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem Sci. 2014;39:72–81. doi: 10.1016/j.tibs.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michishita E, McCord RA, Boxer LD, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Toiber D, Erdel F, Bouazoune K, et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell. 2013;51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galleano I, Schiedel M, Jung M, Madsen AS, Olsen CA. A Continuous, Fluorogenic Sirtuin 2 Deacylase Assay: Substrate Screening and Inhibitor Evaluation. J Med Chem. 2016;59:1021–1031. doi: 10.1021/acs.jmedchem.5b01532. [DOI] [PubMed] [Google Scholar]
- 60.Baur JA. Biochemical effects of SIRT1 activators. Biochim Biophys Acta. 20101804:1626–1634. doi: 10.1016/j.bbapap.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gu XS, Wang ZB, Ye Z, et al. Resveratrol, an activator of SIRT1, upregulates AMPK and improves cardiac function in heart failure. Genet Mol Res. 2014;13:323–335. doi: 10.4238/2014.January.17.17. [DOI] [PubMed] [Google Scholar]
- 62.Chaudhary N, Pfluger PT. Metabolic benefits from Sirt1 and Sirt1 activators. Curr Opin Clin Nutr Metab Care. 2009;12:431–437. doi: 10.1097/MCO.0b013e32832cdaae. [DOI] [PubMed] [Google Scholar]
- 63.Bruckbauer A, Zemel MB, Thorpe T, et al. Synergistic effects of leucine and resveratrol on insulin sensitivity and fat metabolism in adipocytes and mice. Nutr Metab (Lond) 2012;9:77. doi: 10.1186/1743-7075-9-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chauhan D, Bandi M, Singh AV, et al. Preclinical evaluation of a novel SIRT1 modulator SRT1720 in multiple myeloma cells. Br J Haematol. 2011;155:588–598. doi: 10.1111/j.1365-2141.2011.08888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lahusen TJ, Deng CX. SRT1720 induces lysosomal-dependent cell death of breast cancer cells. Mol Cancer Ther. 2015;14:183–192. doi: 10.1158/1535-7163.MCT-14-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maya JD, Morello A, Repetto Y, et al. Trypanosoma cruzi: inhibition of parasite growth and respiration by oxazolo(thiazolo)pyridine derivatives and its relationship to redox potential and lipophilicity. Exp Parasitol. 2001;99:1–6. doi: 10.1006/expr.2001.4638. [DOI] [PubMed] [Google Scholar]
- 67.Vu CB, Bemis JE, Disch JS, et al. Discovery of imidazo[1,2-b]thiazole derivatives as novel SIRT1 activators. J Med Chem. 2009;52:1275–1283. doi: 10.1021/jm8012954. [DOI] [PubMed] [Google Scholar]
- 68.Bonkowski MS, Sinclair DA. Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat Rev Mol Cell Biol. 2016;17:679–690. doi: 10.1038/nrm.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Camins A, Sureda FX, Junyent F, et al. Sirtuin activators: designing molecules to extend life span. Biochim Biophys Acta. 2010;1799:740–749. doi: 10.1016/j.bbagrm.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 70.Botta G, De Santis LP, Saladino R. Current advances in the synthesis and antitumoral activity of SIRT1-2 inhibitors by modulation of p53 and pro-apoptotic proteins. Curr Med Chem. 2012;19:5871–5884. doi: 10.2174/092986712804143303. [DOI] [PubMed] [Google Scholar]
- 71.Villalba JM, Alcain FJ. Sirtuin activators and inhibitors. Biofactors. 2012;38:349–359. doi: 10.1002/biof.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang TT, Schoene NW, Kim EK, Kim YS. Pleiotropic effects of the sirtuin inhibitor sirtinol involves concentration-dependent modulation of multiple nuclear receptor-mediated pathways in androgen-responsive prostate cancer cell LNCaP. Mol Carcinog. 2013;52:676–685. doi: 10.1002/mc.21906. [DOI] [PubMed] [Google Scholar]
- 74.Peck B, Chen CY, Ho KK, et al. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther. 2010;9:844–855. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- 75.Naia L, Rego AC. Sirtuins: double players in Huntington’s disease. Biochim Biophys Acta. 20151852:2183–2194. doi: 10.1016/j.bbadis.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 76.Chen J, Zhou Y, Mueller-Steiner S, et al. SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-kappaB signaling. J Biol Chem. 2005;280:40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- 77.Risitano R, Curro M, Cirmi S, et al. Flavonoid fraction of Bergamot juice reduces LPS-induced inflammatory response through SIRT1-mediated NF-kappaB inhibition in THP-1 monocytes. PLoS One. 2014;9:e107431. doi: 10.1371/journal.pone.0107431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Polyakova O, Borman S, Grimley R, Vamathevan J, Hayes B, Solari R. Identification of novel interacting partners of Sirtuin6. PLoS One. 2012;7:e51555. doi: 10.1371/journal.pone.0051555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mopert K, Hajek P, Frank S, Chen C, Kaufmann J, Santel A. Loss of Drp1 function alters OPA1 processing and changes mitochondrial membrane organization. Exp Cell Res. 2009;315:2165–2180. doi: 10.1016/j.yexcr.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 80.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 82.Christiansen EG. Orientation of the mitochondria during mitosis. Nature. 1949;163:361. doi: 10.1038/163361a0. [DOI] [PubMed] [Google Scholar]
- 83.Cho DH, Nakamura T, Lipton SA. Mitochondrial dynamics in cell death and neurodegeneration. Cell Mol Life Sci. 2010;67:3435–3447. doi: 10.1007/s00018-010-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilkerson DC, Sankar U. Mitochondria: a sulfhydryl oxidase and fission GTPase connect mitochondrial dynamics with pluripotency in embryonic stem cells. Int J Biochem Cell Biol. 2011;43:1252–1256. doi: 10.1016/j.biocel.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 85.Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Son MJ, Kwon Y, Son MY, et al. Mitofusins deficiency elicits mitochondrial metabolic reprogramming to pluripotency. Cell Death Differ. 2015;22:1957–1969. doi: 10.1038/cdd.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 88.Bell EL, Guarente L. The SirT3 divining rod points to oxidative stress. Mol Cell. 2011;42:561–568. doi: 10.1016/j.molcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giralt A, Hondares E, Villena JA, et al. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. J Biol Chem. 2011;286:16958–16966. doi: 10.1074/jbc.M110.202390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Olichon A, Baricault L, Gas N, et al. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 91.Park SH, Ozden O, Jiang H, et al. Sirt3, mitochondrial ROS, ageing, and carcinogenesis. Int J Mol Sci. 2011;12:6226–6239. doi: 10.3390/ijms12096226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ou X, Lee MR, Huang X, Messina-Graham S, Broxmeyer HE. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells. 2014;32:1183–1194. doi: 10.1002/stem.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yoshii SR, Mizushima N. Autophagy machinery in the context of mammalian mitophagy. Biochim Biophys Acta. 20151853:2797–2801. doi: 10.1016/j.bbamcr.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 94.Eiyama A, Okamoto K. PINK1/Parkin-mediated mitophagy in mammalian cells. Curr Opin Cell Biol. 2015;33:95–101. doi: 10.1016/j.ceb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 95.Koh H, Kim H, Kim MJ, Park J, Lee HJ, Chung J. Silent information regulator 2 (Sir2) and Forkhead box O (FOXO) complement mitochondrial dysfunction and dopaminergic neuron loss in Drosophila PTEN-induced kinase 1 (PINK1) null mutant. J Biol Chem. 2012;287:12750–12758. doi: 10.1074/jbc.M111.337907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 97.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 98.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morigi M, Perico L, Rota C, et al. Sirtuin 3-dependent mitochondrial dynamic improvements protect against acute kidney injury. J Clin Invest. 2015;125:715–726. doi: 10.1172/JCI77632. [DOI] [PMC free article] [PubMed] [Google Scholar]