Abstract
The administration of enzyme replacement therapies (ERTs) and other biologic therapies to patients may elicit an anti-drug immune response. The characterization of these anti-drug antibodies (ADA), especially those that may neutralize the biological activity of the drug, termed neutralizing antibodies (NAbs), is crucial in understanding the effects of these antibodies on the drug's pharmacological profile. This protocol describes a cell-based flow cytometry method to detect factors that neutralize the cellular uptake of a representative lysosomal ERT in human matrix. The protocol consists of three procedures: screening, a confirmatory step, and titer assays to detect, identify, and establish the relative level of neutralizing antibody titer in subject samples.
In this method, samples are first mixed with the fluorophore-conjugated ERT product, then incubated with cells [e.g., human T lymphocytes (Jurkat cells)] that express a cell-surface cation-independent mannose 6-phosphate receptor (CI-M6PR), and finally, analyzed with a flow cytometer. A sample without NAbs will result in the uptake of the fluorophore-conjugated ERT product via CI-M6PR, whereas, the presence of NAbs will bind to the drug and interfere with the CI-M6PR binding and uptake. The amount of the fluorophore-conjugated ERT internalized by the Jurkat cells is measured by flow cytometry and evaluated as the percentage (%) signal inhibition compared to the response obtained in the presence of a representative drug-naïve matrix. In the confirmatory step, the samples are pre-incubated with ERT-conjugated magnetic beads to deplete drug-specific factors that bind to the drug (such as NAbs) prior to an incubation with cells. Samples that screen and confirm positive for drug-specific NAbs in the assay are then serially diluted to generate an antibody titer. Semi-quantitative antibody titers may be correlated with measurements of drug safety and efficacy.
Keywords: Immunology and Infection, Issue 139, Neutralizing antibody assay, lysosomal storage disease, enzyme replacement therapy, flow cytometry, cell-based assays, assay development and validation
Introduction
Immunogenicity assessment is an important part of the safety and efficacy monitoring program for any biologic therapeutic product, including ERTs. Patients may develop an immune response that can directly impact drug safety, efficacy, and pharmacokinetic/pharmacodynamic profiles. A subset of these ADA, called NAbs, may inhibit ERT efficacy in two ways: through the inhibition of the ERT uptake into the targeted cell or by inhibiting the ERT catalytic activity. The method presented here is designed to measure NAbs that interfere with the ERT uptake into cells. To fully monitor the safety and efficacy of the therapeutic ERT, continuous monitoring of NAbs is crucial in elucidating any potential correlations with clinical outcomes or pharmacodynamic effects1.
Platforms for evaluating NAbs against protein therapeutics include cell-based, enzymatic activity and ligand-binding assays1. The optimal assay platform is selected based on a variety of criteria: the mechanism of action of the therapeutic product, the assay platform sensitivity, selectivity, precision, and importantly, its ability to mimic the inhibitory action of NAbs in vivo. Ligand-binding assays may be appropriate in certain cases (e.g., when a relevant cell line cannot be identified or if the appropriate sensitivity cannot be achieved in a cell-based assay). However, in the 2016 draft FDA Guidance for Industry document and other industry-accepted white papers, cell-based NAb assays are recommended because they may better reflect the biological mechanism of the drug in vivo1,2,3.
The critical components for developing a flow cytometry cell-based NAb assay include a suitable cell line that responds to drug stimulation, a surrogate positive-control NAb that neutralizes the ERT, a fluorophore-conjugated ERT, and the test species biological matrix4,5,6. The cell line selection is dependent on the ERT mechanism of action and multiple cell lines should be evaluated during the assay development3. In the method described here, human Jurkat T cells were selected for their endogenous CI-M6PR expression on the cell surface and the lack of antibody fragment receptors (FcRs) that indiscriminately bind the Fc region of most antibodies7,8. During the assay development, it is important to establish a negative control for the validation studies and the patient sample testing, such as serum pooled from individuals that have not been treated with the test article1. Cell lines should also tolerate relevant matrices from different species for continuity across the nonclinical, clinical, and post-marketing stages of drug development1. Another component is the selection of the assay's positive control. The positive control for the ERT cell uptake assay was chosen based on its ability to bind the therapeutic and neutralize the uptake through CI-M6PR9,1. It is often difficult to obtain useful or sustainable amounts of neutralizing antisera from human subjects for use as an assay control, especially in rare disease patient populations2. Alternatives include antisera from hyper-immunized animals, or affinity-purified polyclonal or monoclonal antibodies spiked into the assay relevant matrix1. While using a species-specific matrix, it is possible that inhibitory factors other than antibodies present in the matrix may inhibit the ERT uptake. Another critical component of the assay is the fluorophore-conjugated ERT. The selection of the fluorophore for the ERT conjugation should be evaluated for each ERT, based on the assay's need for brightness, pH stability, and potential spectral overlap into other channels on the flow cytometer.
The assay described here is an example for measuring a NAb to a therapeutic protein, such as an ERT, that enters the cell via CI-M6PR. Several ERTs, intended to treat lysosomal storage disorders (LSDs), utilize this pathway for cell uptake and lysosomal targeting, including elosulfase alfa for Morquio A syndrome, cerliponase alfa for CLN2 Batten disease, agalsidase alfa for Fabry disease, and alglucosidase alfa for Pompe disease10,11. The purpose of this method is to measure the relative levels of NAbs that interfere with drug binding and internalization via CI-M6PR. This is performed in tiered screening, confirmatory, and titer steps3. Samples are first screened for NAb positivity and then confirmed positive in the confirmatory step. Finally, samples that screen and confirm positive may be serially diluted to generate an antibody titer1. This cell-based flow cytometry drug uptake assay provides a sensitive and mechanistically relevant in vitro method of measuring drug-specific NAbs that may affect a drug's pharmacological profile. We previously validated the method and tested clinical samples using this platform for the drug elosulfase alfa8. Here we describe the detailed step-by-step protocol that may be applied to other therapeutic proteins or ERTs.
Protocol
Human matrices were purchased from commercial sources with approval from their Institutional Review Board (IRB) but should be treated as potentially infectious. Ensure that the laboratory environment used maintains a culture of safety12.
1. Before Starting the Assay
Prepare ERT-conjugated streptavidin magnetic beads according to the manufacturer’s instructions13.
- Prepare quality control samples (QCs): spike a positive control antibody (PC) (e.g., an NAb) in the species-specific matrix (e.g., pooled CSF or serum). NOTE: FDA and EMA guidance recommends preparing a high and a low QC for assay validation and routine testing1,14.
- For example, to obtain a QC at 10 µg/mL, add 10 µL of 1 mg/mL stock positive control into 990 µL of the relevant matrix (e.g., CSF) for a total volume of 1,000 µL.
- Aliquot the QC samples in a volume suitable for a single use (e.g., 50 µL) and freeze the aliquots at -60 to -80 °C1.
- Aliquot a cut-point-control (CC), species-specific matrix not treated with the test article (e.g., pooled CSF or serum) for a single use (e.g., 50 µL) and freeze the aliquots at -60 to -80 °C1.
Perform a conjugation reaction between the fluorophore and the ERT using a protein-labeling kit according to the manufacturer’s protocol15. Aliquot the sample in a volume suitable for a single use (e.g., 50 µL) and freeze the aliquots at -60 to -80 °C.
2. Day 1: Cell Plating and Sample Preparation
- Cell plate preparation
- Warm the cell growth medium (e.g., RPMI-1640 with 10% fetal bovine serum and 1% penicillin-streptomycin) in a water or bead bath at 37 °C19.
- Count the human T lymphocytes Jurkat cells and plate 100 µL per well at 7.5 x 105 cells/mL in a 96-well round-bottom cell culture plate appropriate for use on a flow cytometer equipped with a plate loader.
- Ensure that the cells have at least 70% viability before proceeding with the experiment (e.g., assess the viability with trypan blue staining)17.
- Incubate the cells in a 37 °C incubator with 5% CO2 and 95% humidity overnight for 14–20 h.
- Screening sample preparation
- Dilute the subject or assay control samples using serum-free media (e.g., RPMI-1640). In the example method here, dilute samples 1:2.5 in RPMI-1640 by adding 60 µL of the sample to 90 µL of serum-free media. (e.g., 8-strip tubes). Proceed to step 3.1 for the assay procedure to prepare a fluorophore-labeled ERT. Note: The dilution of 1:2.5 was experimentally determined and is optimal for the method presented here. Optimal sample dilutions should be determined for each method that is developed.
- Confirmatory bead and sample preparation
- Calculate the number of ERT-conjugated beads needed for confirmatory samples.
- For example, for 10 samples, use 10 samples x 100 µL of ERT-conjugated beads per sample + 30% extra to compensate for any losses during the washing step = 1,300 µL of ERT-conjugated beads.
- Vortex the beads thoroughly. Add the calculated number of beads to a 15 mL conical tube for washing.
- Wash the ERT-conjugated magnetic beads by adding 1,300 µL of coupling buffer or as suggested by the manufacturer (e.g., with phosphate-buffered saline with 0.1% polysorbate 20) following the steps below13.
- Vortex the tube thoroughly. Place the tube in a magnetic tube rack for 2 min. Without disturbing the beads, carefully aspirate and discard the supernatant. NOTE: The solution will turn from dark brown to clear when beads are pulled to the magnet.
- Resuspend the beads with 1,300 µL of coupling buffer. Repeat the wash steps for a total of four washes.
- After the final wash, suspend the beads in 1,300 µL of coupling buffer (previously calculated in step 2.3.1.1). Add 100 µL per well to a 96-well, white, round-bottom, non-binding polypropylene plate according to the plate map (e.g., one confirmatory well per sample or QC; the sample will be split into duplicates when incubated with the fluorophore-labeled ERT).
- Place the plate on a 96-well side-skirted magnet and allow the beads to form a pellet. Carefully aspirate the clear supernatant and discard it. NOTE: The solution will turn from dark brown to clear after approximately 1–2 min on the side-skirted magnet. The obtained beads are called the “dry” beads.
- If the samples screened positive and are being confirmed, add 100 µL of each sample with and without ERT-conjugated beads into the appropriate/assigned well. Seal the plate with a plastic film and shake it for at least 60 min on a rotary shaker at approximately 800 rpm at room temperature (RT). NOTE: Previously prepared and frozen positive and negative QCs and the CC should be included on every plate.
- After the incubation, place the confirmatory plate on the 96-well side-skirted magnet and allow the beads to form a pellet.
- Titer dilution series preparation
- To prepare a titer series, serially dilute the sample in the pooled matrix a sufficient number of times to cross the predetermined titer cut point1.
- For example, mix 30 µL of the sample to 60 µL of pooled matrix in a 1:3 dilution series for a total of 8 dilutions (e.g., 8-strip tubes). Proceed to step 3.1 for the assay procedure to prepare fluorophore-labeled ERT.
3. Day 1: Assay Procedure
- Prepare the fluorophore-labeled ERT in the serum-free medium (e.g., 60 µL per well, or approximately 6 mL/plate).
- For example, to prepare 10 mL of 1 µg/mL fluorophore-labeled ERT, add 10 µL of stock 1 mg/mL fluorophore-labeled ERT to 9.99 mL of serum-free media (e.g., RPMI-1640).
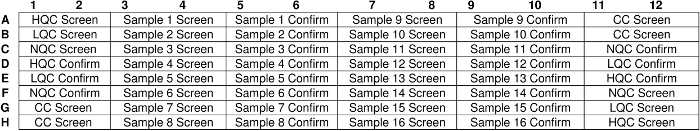
In a new incubation plate (e.g., 96-well, white, round-bottom, non-binding polypropylene plate), add the prepared samples from step 2.2, 2.3, or 2.4 and a fluorophore-labeled ERT in a 1:1 mixture in duplicate wells (e.g., transfer 60 µL of each prepared sample and add 60 µL of the fluorophore-labeled ERT per well). NOTE: An example experiment plate map is provided in Figure 1.
Figure 1: Example of an experiment plate layout. Samples and a fluorophore-labeled ERT were incubated overnight at 2–8 °C. Controls such as high-quality control (HQC), low-quality control (LQC), and negative quality control (NQC) were screened and confirmed on opposite corners of the plate to assess plate uniformity. Please click here to view a larger version of this figure.
Mix the samples and the fluorophore-labeled ERT by pipetting up and down several times with a multichannel pipette. Wrap the plates in the foil and incubate them overnight at 2–8 °C for 14–20 h.
4. Day 2: Adding the Prepared Samples to the Cells
Remove the sample incubation plate(s) from the incubation at 2–8 °C and warm the plates by placing them in a bead bath at 37 °C for 10–15 min.
Remove the cells from the CO2 incubator. Perform a visual check of the plated cells using an inverted microscope to ensure that the cells appear healthy and evenly distributed across the wells17,19.
Add 100 µL of the previously mixed samples and fluorophore-labeled ERT to a cell plate according to the experimental plate map.
Place the cell plate with the added samples back into the CO2 incubator at 37 °C. Incubate the plate for 3 h and ± 15 min. NOTE: This time was established in the lab as optimal for the signal-to-noise response in the assay.
Centrifuge the cell plate for 6 min at 320 x g in a tabletop centrifuge at 14–18 °C. Confirm the presence of a cell pellet at the bottom of the plate wells.
Hold the plate at a 30–45° angle. Carefully remove the supernatant from each well without disturbing the cell pellet. Add 200 µL of 1x DPBS onto the cell pellet in each well to resuspend the cells.
Repeat the cell washing steps for a total of 3 washes. Proceed immediately to step 5 for the cell viability staining.
5. Day 2: Cell Viability Staining
Using a multichannel pipette, add 100 µL of a working solution of 1x Live/Dead stain to each well, selected for an emission wavelength different from that of the fluorophore-labeled ERT and prepared according to the manufacturer’s instructions22.
Incubate the plate for 15 min at RT in the dark. Centrifuge the plate for 6 min at 320 x g in a tabletop centrifuge at 14–18 °C.
Carefully remove the supernatant from each well without disturbing the cell pellet. Wash the cells 1x with 1x DPBS.
6. Day 2: Fixing the Cells with 1% Paraformaldehyde
Using a multichannel pipette, dispense 100 µL of chilled (at 2–8 °C) 1% paraformaldehyde (PFA) into each well.
Seal the plates and gently pulse vortex to mix. Wrap the plate in the foil and place it at 2–8 °C for at least 10 min to allow for fixation. NOTE: Be careful to avoid splashing onto the plate seal.
Add 50 µL of 1x DPBS into each well using a multichannel pipette up and down prior to the analysis.
7. Flow Cytometry
Place the plates on the flow cytometer with the plate loader and run them.
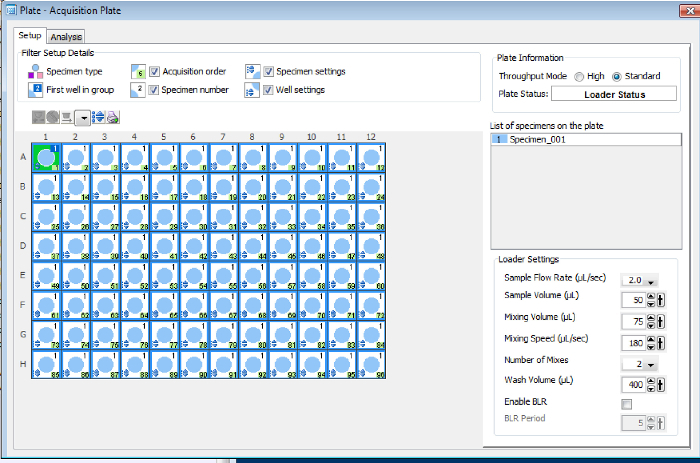
Acquire and record the measured median fluorescence intensity (MFI) using the flow cytometry software (see Table of Materials). NOTE: See Figure 2 as an example for the loader settings. The sample flow rate, sample volume, mixing volume, mixing speeds, and number of mixes are optimized for this assay. These criteria may be adjusted using the "arrows up" or "down" buttons next to the numbers for each criterion, depending on the flow cytometry acquisition software23.
Figure 2: Example of the flow cytometer loader settings. The loader settings should be optimized for each assay that is developed. Please click here to view a larger version of this figure.
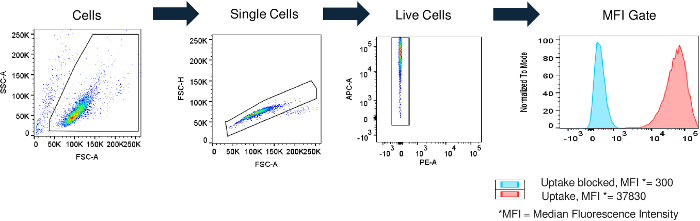
Create a cytometer analysis/gates/plots as shown in Figure 3. NOTE: The gate creation and application may differ for each flow cytometry software23.
Figure 3: Flow cytometry gating strategy. Jurkat cells were separated from the total events and collected by plotting the forward-scatter (FSC) vs. side-scatter (SSC) channels and drawing a gate around the target population. Singlets (single cells) were separated from doublets or larger cell aggregates using the FSC area (FSC-A) and FSC height (FSC-H) channels. Live cells were chosen by gating on singlet cells negative for a viability stain. The median fluorescence intensity (MFI) was measured in single, live Jurkat cells and is plotted as a histogram. The MFI values of cells with the drug uptake blocked (e.g., HQC) and of cells with control amounts of uptake are shown (red and blue, respectively). Please click here to view a larger version of this figure.
Exclude dead cells from the analysis and record approximately 10,000 events23.
8. Data Analysis
Export the data (raw MFI) to the analysis software (see Table of Materials). NOTE: Depending on the analysis needed, the following data analysis may be performed using analysis software.
Calculate the mean MFI of duplicate wells (QCs and samples) and the coefficient of variation (%CV) to evaluate the spread variability of the duplicates from the mean.
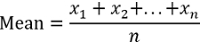
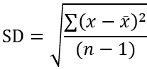
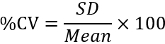
For samples and QCs that were screened in the assay, calculate the percent signal inhibition (%SI) relative to the mean MFI of the for the QCs pooled matrix CC.
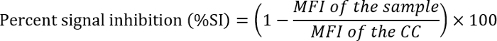
If needed, calculate the recovery ratios (RRs) for the confirmed QCs and samples relative to the corresponding screen values.
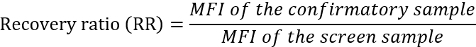
Representative Results
On the first day of the method, a frozen aliquot of Jurkat cells was thawed and plated, and the test samples were prepared. Figure 1 shows an example plate map. On the second day, the samples were mixed with the Jurkat cells and incubated at 37 °C for approximately 3 h and 15 min. The cells were then washed, fixed with PFA, and analyzed on a flow cytometer. Figure 2 shows example flow cytometer settings.
The flow cytometry gating strategy was designed to measure the amount of fluorophore-conjugated drug in live single Jurkat cells24 (Figure 3). The cells were separated from the debris by drawing a gate that excludes cellular debris [usually, low forward scatter (FSC)] and dead cells [usually, high side scatter (SSC)], leaving only live cells for analysis25. Single cells were then separated from cell clusters by drawing a gate that excludes the high FSC area and the low FSC height26. The cells were treated with a viability stain that labeled any dead or unhealthy cells without an intact membrane, and an additional gate was created to exclude the dead cells22. The MFI of the live single cells was used for the analysis of the fluorophore-conjugated ERT uptake. As an example of potential screening assay results, the matrix spiked with a high amount of surrogate anti-drug antibody positive control [e.g., high-quality control (HQC)] inhibited fluorophore-conjugated ERT uptake, resulting in a low MFI of approximately 300 (Figure 3). In contrast, cells incubated with the matrix in the absence of the positive control antibody should have a higher MFI (MFI = 37,830 in Figure 3), demonstrating the uptake of the fluorophore-conjugated ERT.
The neutralizing positive control antibody for the example here was selected based on the mechanism of action of the drug (i.e., the ERT uptake through CI-M6PR). A panel of fluorophores was also tested and compared for optimal assay sensitivity and dynamic range1. CypHer 5e was tested due to its increased fluorescence at an acidic pH, which might be relevant to some ERTs as an additional measure of lysosomal targeting of the drug27. A green fluorescent dye (e.g., Alexa Fluor 488), and a far-red fluorescent dye (e.g., Alexa Fluor 647) were also examined. An example of how different fluorophores might perform is presented in Figure 4a and 4b. In the example, the Alexa Fluor 647 ERT had the best performance due to its superior sensitivity (e.g., the highest %SI at the lowest PC concentration) and wide dynamic range (~3 orders of magnitude).
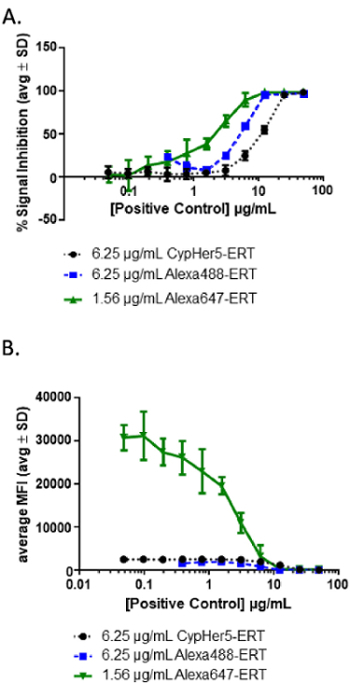
Figure 4: Example results from different fluorophore-ERT conjugates. (A) During the assay development, a positive control (PC) dilution curve should be evaluated using different fluorophore-conjugated ERTs. In the example shown here, two fluorophore-conjugated ERTs (Alexa Fluor 488 and CypHer 5e) at 6.25 µg/mL and a brighter fluorophore-conjugated ERT (Alexa Fluor 647) at 1.56 µg/mL were tested in the presence of increasing concentrations of positive-control NAbs. (B) This panel shows the curves from panel A graphed, using MFI to show the significant increase in the dynamic range using an Alexa Fluor 647-conjugated ERT. Please click here to view a larger version of this figure.
The method described is a tiered approach for detecting, confirming, and interpolating a quasi-quantitative level of NAb titer3. To establish the assay is ready for validation, the parameters including assay sensitivity, precision, selectivity, specificity, drug tolerance, robustness, and cut points should be evaluated according to established guidances1,3.
Samples were considered potentially positive in this assay when %SI values greater than the screening cut point (SCP) were obtained. The SCP was determined statistically by testing drug-naïve samples from a representative population and measuring a relative decrease in signal intensity (SI)3. Guidance's (for instance of the FDA) recommend that the SCP is established at the 95th percentile of the normally distributed data set, and methods for calculating the SCP have been described in detail elsewhere1. Any sample that decreased the assay signal (measured as an increase in the %SI) at or above the SCP was determined to be potentially positive, and samples with results higher than the SCP (with no change or decrease in the %SI) are considered negative.
Samples that screened positive (%SI above the SCP) were tested in the confirmatory assay to determine the specificity of NAbs to the ERT. The confirmatory assay was performed by pre-incubating samples with ERT-conjugated magnetic beads to remove drug-specific antibodies or inhibitory factors. The RR (the ratio of confirmatory MFI to screening MFI) was evaluated to determine the number of NAbs removed from the sample. An RR higher than the calculated threshold of positivity [i.e., the confirmatory cut point (CCP)] indicated the presence of anti-drug NAbs. As in the screening assay, the CCP should first be established by evaluating treatment-naïve samples from a representative population in the confirmatory assay. CCP was based on a statistically determined 1% false-positive rate and was the threshold designating a positively confirmed sample (Figure 5)3.
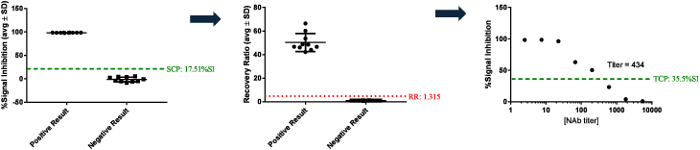
Samples that screened and confirmed positive were serially diluted and tested in the titer assay to determine the relative level or titer of NAbs in each sample. The highest dilution at which a sample tests positive when it crossed a designated threshold [e.g., the titer cut point (TCP)] is the sample titer (Figure 5)1,3.
Figure 5: Example sample testing results. These panels show examples of samples that tested positive or negative by screening either above or below an SCP of 17.51 %SI. Positive samples were then tested in the confirmatory assay using drug-conjugated magnetic beads to deplete the drug-specific antibody from the samples prior to testing in the assay. Samples with a recovery ratio (RR) greater than the confirmatory cut point (i.e., RR = 1.315) were considered to be confirmed positive. Samples that confirmed positive were diluted until the signal crossed the titer cut point (TCP) to establish the titer (dilution factor) at which the sample result equals the titer cut point. Please click here to view a larger version of this figure.
Assay repeatability and variability were tested over several days and with more than one analyst. The QCs were initially prepared in one batch and sub-aliquoted for 1x use. Over the course of 3 d, two analysts performed the assay with several sets of QCs to demonstrate the precision of the assay. In the example data shown, the %CV of the inter- and intra-assay precision for the QCs are less than the FDA guidance's recommendation of %CV < 20% (Figure 6)1.
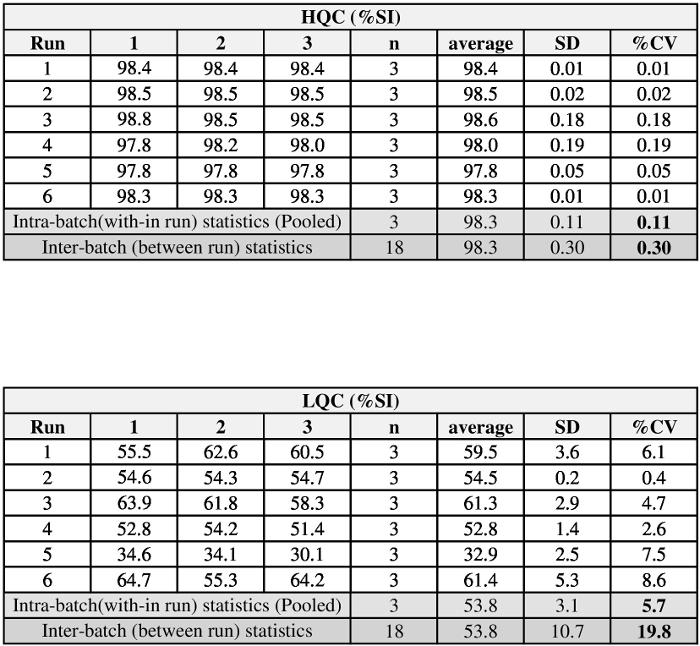
Figure 6: Example of QC precision data generated over three days with two analysts. The precision data were calculated by an analysis of variance (ANOVA) using the formula presented in DeSilva et al.28. The intra-batch (within runs) and inter-batch (between runs) statistics are reported as %CV. Please click here to view a larger version of this figure.
Discussion
Neutralizing antibodies or other factors that prevent the ERT uptake through CI-M6PR have the potential to impact ERT safety or efficacy1. Therefore, it is important to evaluate NAbs in subject samples using a robust model relevant to the drug's mechanism of action. We and others have found that the performance of some bioassay formats (e.g., receptor dimerization, luciferase expression, etc.) does not align with health authority recommendations for assay precision, reproducibility, and sensitivity29. In the bioassay method described here, Jurkat cells that express endogenous CI-M6PR are employed to monitor NAbs specific to the lysosomal ERT uptake. Combined with a flow cytometry readout, this assay uses a physiological cell model with suitable assay precision, sensitivity, reproducibility, and high sample throughput. NAb detection with a flow cytometry readout has also been applied to other indications. For example, methods have been developed to detect pre-existing antibodies against hepatitis E and adeno-associated virus (AAV)30,31.
Critical steps in the protocol include selecting a suitable fluorophore, incorporating an LQC that monitors assay sensitivity, ensuring a sufficient level of cell viability, and maintaining strict adherence to the incubation times. In the example presented here, fluorophores (e.g., Alexa Fluor 488, Alexa Fluor 647, and CypHer 5e), conjugated to a lysosomal ERT, varied widely in their performance. Alexa Fluor 647, which was also the brightest fluorophore tested32, was selected for further assay development. We recommend that several fluorophores be evaluated early on in the development of the assay to provide the best sensitivity and dynamic range. Assay sensitivity during sample testing should be monitored by including an LQC that is close enough to the sensitivity limit that it will fail in 1% of the testing runs1. Together with the HQC, the LQC also acts as a system suitability QC to monitor assay drift over time3. Optimal cell viability and performance is achieved by preparing a cell bank of single-use aliquots and qualifying in new cell banks based on comparable QC results3,18. Cell and fluorophore-conjugated drug incubation times are also central to a consistent assay performance since drug uptake increases with the amount of time the drug is incubated with cells. To minimize day-to-day variability in drug uptake, sample data may be normalized to pooled matrix control samples on each plate (e.g., the cut point control samples in the method above). Another important factor in producing consistent data with this method is to monitor plate uniformity and minimize possible edge effects. An initial experiment may include performing the assay using a single QC across the whole plate, while more robust experiments may be performed during assay validation (e.g., precision and accuracy)1. This demonstrates the importance of the plate map layout33. As shown in the plate map example, quality control samples placed on both sides of the plate monitor a uniformity that can be tracked over time with Levey-Jennings charts34.
While this assay monitors NAbs and other factors that may inhibit lysosomal ERT uptake, additional experiments may be conducted to confirm an uptake inhibition due to antibodies. One option is to treat samples with the protein A/G/L, which non-specifically binds immunoglobulin35. If samples continue to test positive after protein A/G/L depletion, a non-antibody inhibitory factor may be responsible for blocking the drug uptake. The method described here was designed to measure antibody-mediated uptake inhibition, and the positive control and other assay parameters should be reconsidered if a high percentage of subject samples with non-antibody inhibitory factors are found.
The assay may also be further characterized to demonstrate fluorophore-labeled drug traffics to the appropriate cellular compartment based on the drug's mechanism of action. In the example presented here, an ERT is expected to bind CI-M6PR on the cell surface and traffic it to the lysosome. We previously reported the results of several experiments indicating that nearly all of the fluorescent signal observed in the method results from fluorophore-labeled ERT in the lysosome8. We found that the fluorophore-labeled ERT signal was eliminated following a treatment of the cells with cytochalasin B, which disrupts internalization through the inhibition of actin reorganization. Also, experiments that quenched external fluorophore with trypan blue, or slowed internalization by placing cells at 4 °C, indicate that the fluorophore-labeled ERT is rapidly internalized and very little fluorescence is due to an ERT bound at the cell surface. Lysosomal targeting was confirmed by visualizing the co-localization of pH-sensitive lysotracker dye with the fluorophore-conjugated drug using confocal microscopy. A specificity for uptake through CI-M6PR may also be verified using exogenous M6P to compete with labeled drugs for receptor binding. Similar experiments should be performed to verify the uptake kinetics and cellular localization of fluorophore-conjugated drugs in other assays.
This cell-based assay platform has been used to study NAbs for therapeutic drugs that utilize CI-M6PR receptor-mediated endocytosis. We recently reported results using this assay platform that demonstrated no correlation between the development of an NAb and drug efficacy for elosulfase alfa36,37. To validate the assay for clinical sample testing, the parameters, including assay sensitivity, precision, selectivity, specificity, drug tolerance, robustness, and cut points, should be evaluated according to established guidances3,4. It should also be noted, for lysosomal ERTs, that NAbs may develop with the potential to interfere with drug activity through binding near the enzyme catalytic site. In general, we consider monitoring this type of NAb to be of a lower priority since the harsh acidic and proteolytic environment of the lysosome is not favorable to antibody-ERT interactions2,39,40. However, it is possible that proteolytic-resistant NAbs exist and may inhibit the catalytic portion of a drug40. This assay monitors fluorophore-labeled ERT uptake to the lysosome, and a limitation of the assay is the inability to monitor proteolytic-resistant NAbs. If this type of NAb is suspected based on safety or efficacy data, an assay that monitors ERT activity should be developed and used to test samples.
The assessment of immunogenicity is important in understanding the effects of NAbs on drug safety and efficacy. The identification of NAbs capable of inhibiting in vitro drug uptake via CI-M6PR provides an opportunity for understanding NAb activity in vivo. The method presented here utilizes a human cell line that expresses CI-M6PR to measure the interference of fluorophore-conjugated lysosomal ERT cellular uptake. This method has already been used to monitor NAbs for several ERTs intended to treat lysosomal storage diseases. This assay platform may apply to other methods for studying the effects of NAbs on biologic therapeutics that require a cellular internalization for their proper function.
Disclosures
The authors are employees and shareholders of BioMarin Pharmaceutical Inc.
Acknowledgments
The authors have no acknowledgments.
References
- CDER. Assay Development and Validation for Immunogenicity Testing of Therapeutic Protein Products (Draft) FDA Guidance for Industry; 2016. [Google Scholar]
- Gupta S, et al. Recommendations for the design, optimization, and qualification of cell-based assays used for the detection of neutralizing antibody responses elicited to biological therapeutics. Journal of Immunological Methods. 2007;321:1–18. doi: 10.1016/j.jim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Gupta S, et al. Recommendations for the validation of cell-based assays used for the detection of neutralizing antibody immune responses elicited against biological therapeutics. Journal of Pharmaceutical and Biomedical Analysis. 2011;55:878–888. doi: 10.1016/j.jpba.2011.03.038. [DOI] [PubMed] [Google Scholar]
- Pedras A. NAb me well- FDA Regulatory Perspectives on Neutralizing Antibody Assays. BEBPA Forum. 2015.
- Stovold C. NAB Assay Development and Validation Immunogencity Workflow. Annual BEBPA Bioassay Meeting. 2013.
- Kromminga A. Neutralizing anti-drug antibodies Emerging Trends and Clinical Impact. Symposium. IPM Biotech; 2013. [Google Scholar]
- Davis RS, Wang YH, Kubagawa H, Cooper MD. Identification of a family of Fc receptor homologs with preferential B cell expression. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9772–9777. doi: 10.1073/pnas.171308498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton AC, et al. Antibodies that neutralize cellular uptake of elosulfase alfa are not associated with reduced efficacy or pharmacodynamic effect in individuals with Morquio A syndrome. Journal of Immunological Methods. 2017;440:41–51. doi: 10.1016/j.jim.2016.10.006. [DOI] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services. Food and Drug Administration. Center for Drug Evaluation and Research (CDER). Center for Biologics Evaluation and Research (CBER) Guidance for Industry. S6 Addendum to Preclinical Safety Evaluation of Biotechnology-Derived Pharmaceuticals. ICH; 2012. [Google Scholar]
- Lee K, et al. A biochemical and pharmacological comparison of enzyme replacement therapies for the glycolipid storage disorder Fabry disease. Glycobiology. 2003;13:305–313. doi: 10.1093/glycob/cwg034. [DOI] [PubMed] [Google Scholar]
- Kirkegaard T. Emerging therapies and therapeutic concepts for lysosomal storage diseases. Expert Opinion on Orphan Drugs. 2013;1:385–404. [Google Scholar]
- Miller JM, et al. Guidelines for Safe Work Practices in Human and Animal Medical Diagnostic Laboratories. Morbidity and Mortality Weekly Report. 2012. pp. 1–101. Available from: https://www.cdc.gov/mmwr/preview/mmwrhtml/su6101a1.htm. [PubMed]
- Invitrogen by ThermoFisher Scientific. DynabeadsTMM-270 Streptavidin. 2015.
- European Medicines Agency. Guideline on Immunogenicity assessment of biotechnology-derived therapeutic proteins. European Medicines Agency; 2017. [Google Scholar]
- ThermoFisher Scientific. Molecular ProbesTMAlexa FluorTM647 Protein Labeling Kit (A20173) 2004.
- JoVE Science Education Database. An Introduction to Working in the Hood. Journal of Visualized Experiments. 2018. General Laboratory Techniques.
- Invitrogen and Gibco. Cell Culture Basics Handbook. ThermoFisher Scientific Inc; 2010. [Google Scholar]
- Coecke S, et al. Guidance on Good Cell Culture Practice. Alternatives to Laboratory Animals. 2005;33:261–287. doi: 10.1177/026119290503300313. [DOI] [PubMed] [Google Scholar]
- ThermoFisher Scientific. Introduction to Cell Culture. 2018. Available from: https://www.thermofisher.com/us/en/home/references/gibco-cell-culture-basics/introduction-to-cell-culture.html.
- JoVE Science Education Database. Using a Hemacytometer to Count Cells. Journal of Visualized Experiments. 2018. Basic Methods in Cellular and Molecular Biology.
- Bio-Rad Laboratories. Counting Cells with Bio-Rad's TC20(TM) Automated Cell Counter. 2012. Available from: https://www.youtube.com/watch?v=sDHOL7UEL1M.
- ThermoFisher Scientific. User Guide LIVE / DEADTMFixable Dead Cell Stain Kits. 2016.
- Biosciences. Getting Started with BD FACSDiva Software. BD Biosciences; 2007. [Google Scholar]
- Biosciences. BD FACS Canto II Instructions For Use. 2007.
- Quirke P. Introduction to Flow Cytometry: A Learning Guide. Journal of Clinical Pathology. 2002;45(3) [Google Scholar]
- Bio-Rad Laboratoratories. A guide to gating in flow cytometry. 2016. Available from: https://www.bio-rad-antibodies.com/blog/a-guide-to-gating-in-flow-cytometry.html.
- Minor LK. Handbook of Assay Development in Drug Discovery. Boca Raton, FL: CRC Press; 2006. [Google Scholar]
- Desilva B, et al. Recommendations for the Bioanalytical Method Validation of Ligand-binding Assays to Support Pharmacokinetic Assessments of Macromolecules. Pharmaceutical Research. 2003;20 doi: 10.1023/b:pham.0000003390.51761.3d. [DOI] [PubMed] [Google Scholar]
- Hu J, et al. Comparison of cell-based and non-cell-based assay platforms for the detection of clinically relevant anti-drug neutralizing antibodies for immunogenicity assessment of therapeutic proteins. Journal of Immunological Methods. 2015;419:1–8. doi: 10.1016/j.jim.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Cai W, et al. A high-throughput neutralizing assay for antibodies and sera against hepatitis E virus. Scientific Reports. 2016;6 doi: 10.1038/srep25141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles River. A Flow Cytometric Method to Assess Neutralizing Antibodies to AAV Gene-Therapy Vectors. Charles River Researcher. 2015.
- Biosciences. BD Biosciences Relative Fluorochrome Brightness. BD Biosciences. 2014. p. 16181. Available from: http://www.bdbiosciences.com/us/applications/research/multicolor-flow/m/745795/overview.
- Waritani T, Chang J, McKinney B, Terato K. An ELISA protocol to improve the accuracy and reliability of serological antibody assays. MethodsX. 2017;4:153–165. doi: 10.1016/j.mex.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen PW, et al. HTS Assay Validation. Assay Guidance Manual. 2004. pp. 1–30.
- Thermo Fisher Scientific. Tech Tip #34. Binding characteristics of antibody-binding proteins: Protein A, Protein G, Protein A/G and Protein L. 2013.
- Long B, et al. Long-term Immunogenicity of Elosulfase Alfa in the Treatment of Morquio A Syndrome: Results From MOR-005, a Phase III Extension Study. Clinical Therapeutics. 2017;39 doi: 10.1016/j.clinthera.2016.11.017. [DOI] [PubMed] [Google Scholar]
- Schweighardt B, et al. Immunogenicity of Elosulfase Alfa, an Enzyme Replacement Therapy in Patients with Morquio A Syndrome: Results from MOR-004, a Phase III Trial. Clinical Therapeutics. 2015;37 doi: 10.1016/j.clinthera.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Schneider Z. Importance of isoelectric point (pI) of antibodies. Antibody Society. 2017. Available from: http://www.antibodysociety.org/importance-isoelectric-point-pi-antibodies/
- Mellman IRA, Plutner H. Internalization and Degradation of Macrophage Fc Receptors Bound to Polyvalent Immune Complexes. The Journal of Cell Biology. 1984;98:1170–1177. doi: 10.1083/jcb.98.4.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. Neutralizing antibodies to the therapeutic enzymes: considerations for testing, prevention and treatment. Nature Biotechnology. 2008;26:901–908. doi: 10.1038/nbt.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]


