Abstract
Familial hypercholesterolemia (FH) is mostly caused by low-density lipoprotein receptor (LDLR) mutations and results in an increased risk of early-onset cardiovascular disease due to marked elevation of LDL cholesterol (LDL-C) in blood. Statins are the first line of lipid-lowering drugs for treating FH and other types of hypercholesterolemia, but new approaches are emerging, in particular PCSK9 antibodies, which are now being tested in clinical trials. To explore novel therapeutic approaches for FH, either new drugs or new formulations, we need appropriate in vivo models. However, differences in the lipid metabolic profiles compared to humans are a key problem of the available animal models of FH. To address this issue, we have generated a human liver chimeric mouse model using FH induced pluripotent stem cell (iPSC)-derived hepatocytes (iHeps). We used Ldlr-/-/Rag2-/-/Il2rg-/- (LRG) mice to avoid immune rejection of transplanted human cells and to assess the effect of LDLR-deficient iHeps in an LDLR null background. Transplanted FH iHeps could repopulate 5-10% of the LRG mouse liver based on human albumin staining. Moreover, the engrafted iHeps responded to lipid-lowering drugs and recapitulated clinical observations of increased efficacy of PCSK9 antibodies compared to statins. Our human liver chimeric model could thus be useful for preclinical testing of new therapies to FH. Using the same protocol, similar human liver chimeric mice for other FH genetic variants, or mutations corresponding to other inherited liver diseases, may also be generated.
Keywords: Developmental Biology, Issue 139, Familial hypercholesterolemia, hepatocytes, human liver chimeric mice, induced pluripotent stem cells, LDL-receptor, statins, PCSK9 antibodies
Introduction
Low-density lipoprotein receptor (LDLR) captures LDL cholesterol (LDL-C) in blood to modulate cholesterol synthesis in the liver. Mutations in the LDLR gene are the most frequent cause of familial hypercholesterolemia (FH)1. Statins have traditionally been the first line of medication to treat FH and other types of hypercholesterolemia (inherited or acquired). Statins inhibit 3-hydroxy-3-methylglutaryl-coenzyme A reductase to lower cholesterol synthesis in the liver2. Additionally, statins increase LDLR levels on the hepatocyte surface to promote plasma LDL-C clearance. However, a major caveat of treatment with statins is that they simultaneously induce the expression of proprotein convertase subtilisin/hexin 9 (PCSK9), an enzyme that binds to LDLR to promote its degradation3. This effect is responsible for the insufficient or even null response to statins observed in many patients. Studying this mechanism has, unexpectedly, led to the discovery of an alternative way to treat hypercholesterolemia. PCSK9 antibodies recently approved by the FDA are currently being used in clinical trials and show higher efficacy and better tolerance than statins4. The success of PCSK9 antibodies also implies that there may be other therapeutic possibilities to modulate the LDLR degradation pathway (besides PCSK9) in patients with hypercholesterolemia. Similarly, there is interest in developing new inhibitors of PCSK9 other than antibodies, for example, siRNA oligos5.
To test new therapies for FH and in general any other type of hypercholesterolemia, appropriate in vivo models are necessary. A major problem of current in vivo models, mostly mice6 and rabbits7, are their physiological differences with humans. Crucially, these problems include a different lipid metabolic profile. The generation of human liver chimeric animals8 might help overcome this caveat. The human liver chimeric mouse is a type of "humanized" mouse with its liver repopulated with human hepatocytes, for example, primary human hepatocytes (pHH)9. A problem with pHH is that they cannot be expanded ex vivo, quickly lose their function upon isolation, and are a limited source. An alternative to pHH is the use of induced pluripotent stem cells (iPSC)-derived hepatocytes (iHeps)10. Notably, iPSCs are patient-specific and can be grown indefinitely, so iHeps can be produced on demand, which is a significant advantage over fresh pHH. Moreover, iPSCs can also be easily genetically engineered with designer nucleases to correct or introduce mutations in an isogenic background to allow more faithful comparisons11.
Human liver chimeric mouse with engrafted pHH show similarities to humans in liver metabolic profiles, drug responses, and susceptibility to hepatitis virus infection12. This makes them a good model to study hyperlipidemia in vivo. The most widely used mouse models are based on the Fah-/-/Rag2-/-/Il2rg-/- (FRG) mouse13 and the uPA transgenic mouse8, in which up to 95% of the mouse liver can be replaced by pHH. Interestingly, a recent report described a human FH liver chimeric mouse (based on the FRG mouse) with pHH from a patient carrying a homozygous LDLR mutation14. In this model, the repopulated human hepatocytes had no functional LDLR, but the residual mouse hepatocytes did, thus reducing the utility for performing in vivo testing of drugs relying on the LDLR pathway.
Here, we report a detailed protocol based on our recently published work15 for engrafting FH iHeps into the Ldlr-/-/Rag2-/-/Il2rg-/- (LRG) mouse liver. This human liver chimeric mouse is useful for modeling FH and performing drug testing in vivo.
Protocol
All methods described here that involve the use of animals have been approved by the Committee on the Use of Live Animals in Teaching and Research (CULATR) of the University of Hong Kong.
1. Mouse Preparation and Phenotypic Testing
- Generation of immunodeficient Ldlr knockout (KO) mice.
- Use the mice strains Ldlr-/-, Rag2-/-, and Il2rg-/- (see Table of Materials).
- Cross Rag2-/- mice with Il2rg-/- mice to generate Rag2-/-/ Il2rg-/- mice, then cross Ldlr-/- mice with Rag2-/-/ Il2rg-/- mice to generate Ldlr-/-/Rag2-/-/Il2rg-/- (LRG) mice15. At the age of 3 to 4 weeks, collect genomic DNA from the ear to determine the genotype by PCR and sequencing.
- At the age of 8 to 12 weeks, use male LRG mice as recipients for iHeps to generate human liver chimeric mice.
Feed the mice with a high-fat and high-cholesterol (HFHC) diet 7 days before iHep transplantation to help them develop hypercholesterolemia.
To induce liver injury that facilitates the proliferation of engrafted iHeps in the liver, place the mice into a sterile container and irradiate them using a gamma irradiator with γ-rays at a dose of 3 Gy 24 h prior to engraftment. Then, return the mice to their housing cages. The healthy status of these irradiated mice can be monitored by measuring their weight. Mice with 20% weight loss or greater will be euthanized.
2. iHep Differentiation and Dissociation
LDLR heterozygous KO (+/-) or homozygous KO (-/-) human iPSCs, or FH patient-iPSCs with heterozygous mutations in LDLR (FH iPSCs) are used to produce iHeps. The generation of LDLR +/- or -/- iPSCs and FH iPSCs is described in our previous report15.
- Directed differentiation of iPSCs into iHeps NOTE: The method for differentiation of human iPSCs into iHeps is modified from a previous report16.
- Three days prior to differentiation (day -3), seed the iPSCs onto extracellular matrix coated 6-well plates (see Table of Materials) at a density of 300,000 cells/well in 1.5 mL (also hereafter) of human iPSC maintenance medium (see Table of Materials) supplemented with 5 µM ROCK inhibitor (Y27632). Culture the cells at 37 °C in a 5% CO2 humidified incubator. Normally, iHeps from a 6-well plate are sufficient to engraft 5 mice.
- 24 hours later (day -2) and 48 h later (day -1), change the medium to fresh human iPSC maintenance medium without ROCK inhibitor. NOTE: iPSCs before differentiation should express high levels of OCT4 and NANOG, which can be examined by quantitative RT-PCR, immunofluorescence, or flow cytometry.
- At day 0 of differentiation, wash the cells with RPMI 1640 once and then add RPMI 1640 supplemented with 100 ng/mL Activin A and 25 ng/mL WNT3a.
- At day 1 and day 2 of differentiation, change the medium for RPMI 1640 supplemented with 100 ng/mL Activin A. NOTE: If there is too much cell death at this stage (days 0–3), up to 0.5% fetal bovine serum (FBS) can be added to the medium to improve cell viability. However, we suggest testing the minimal amount of FBS to be added for each specific iPSC line, as excess FBS can also affect the differentiation.
- At day 3, wash the cells with DMEM once and then switch to 2nd stage medium (20% serum replacement, 1x non-essential amino acids, 2 M L-glutamine, 0.1 M beta-mercaptoethanol, and 1% dimethyl sulfoxide in DMEM medium). Change the medium every other day until day 10. NOTE: On day 7, the cells should have reached confluence and display clear edges. There might be some undifferentiated areas appearing at this stage; ignore them if the percentage of these areas is small. If the percentage is high, reduce the confluence of iPSCs at day 0 and reduce the amount of FBS used in the first stage of differentiation.
- At day 10, wash the cells with hepatocyte basal medium once and then switch to hepatocyte culture medium supplemented with 20 ng/mL human hepatic growth factor and 20 ng/mL oncostatin M to promote iHep maturation. Change the medium every other day. NOTE: At this stage, >90% of the cells should be positive for HNF4A, according to immunofluorescence staining or flow cytometry.
- At day 15–17, the cells are ready for engraftment. Optionally, the supernatant of the cell culture may be collected and stored at -80 °C for quantifying the secreted albumin (ALB) by iHeps. NOTE: At this stage, >80% iHeps should be positive for HNF4A, ALB, and α1-antitrypsin (AAT) according to immunofluorescence staining. Around 60–70% of day 17 iHeps should be positive for ASGPR, as shown by flow cytometry. In our hands, iHeps at this period have optimal capacity to repopulate the mouse liver; iHeps may secrete higher levels of ALB in vitro but also become senescent if the engraftment is delayed.
- Dissociation and loading of iHeps into an insulin syringe
- Prepare the required reagents and materials:
- 24 h prior to engraftment, thaw the required amount (V = 40 µL * mice number) of extracellular matrix in an ice box in a cold room, and put the insulin syringe and a box of 200 µL tips into a 4 °C refrigerator.
- One hour prior to engraftment, warm the cell dissociation enzyme supplemented with 50 µg/mL DNase I to room temperature and place the RPMI 1640 medium supplemented with 20% serum replacement on ice.
- Take phase contrast images (100X and 200X) of iHeps to record their status, including cell morphology, growth, and cell density.
- Wash iHeps with 2 mL/well of room temperature Ca2+ and Mg2+-free PBS twice, and then add 1 mL of cell dissociation enzyme supplemented with 50 µg/mL DNase I to each well. Put the cells back into the incubator for 8–10 min. NOTE: To improve cell dissociation with cell dissociation enzyme, wash the cells with Ca2+ and Mg2+-free PBS. To maximize cell viability, we suggest dissociating no more than 6 wells per batch at a time. Furthermore, we suggest restricting the cell dissociation enzyme treatment to less than 10 minutes.
- Monitor cell morphology under the microscope. When most of the cells become round, add an equal volume of cold RPMI 1640 supplemented with 20% serum replacement to each well, pipette the cells gently to detach from the plate, and transfer the cell suspension to a new 15 mL tube. NOTE: This step is critical for harvested cells' viability. If the cells are difficult to detach from the plate, it does not matter if some of the cells are left over. If the cells detach as large squares of monolayer, then pipette gently after centrifugation to obtain a single cell suspension.
- Repeat step 2.2.4 for each well until nearly all attached cells are collected. Centrifuge at 200 x g for 3 min at 4 °C.
- Remove the supernatant and resuspend cells with 2 mL of cold PBS in a 15 mL tube. Pipette the cells gently to obtain a single-cell suspension and then add cold PBS to a final volume of 1 mL * number of dissociated wells. Finally, pass the cells through a 40 µm cell strainer to remove aggregates. NOTE: Normally 1-2 x 106 cells/well can be harvested after filtering.
- Aliquot 20 µL of cell suspension and add 20 µL of 0.4% trypan blue solution, then count the cells using an automated cell counter and record the concentration of cell suspension as "C". Calculate the required volume (V1 = [106 * (n+1)]/C, where n is the number of mice to be engrafted) of cell suspension (1 million/mouse) for intrasplenic injection.
- Aliquot the required volume of cell suspension into 15 mL tubes and centrifuge at 200 x g for 3 min at 4 °C.
- Remove the supernatant, resuspend the cells in the appropriate volume of cold PBS to make the volume of cell suspension (n+1) * 55/2 µL (n = number of mice), and then add an equal volume of extracellular matrix to make the final volume of cell suspension (n+1) * 55 µL.
- Place the cold insulin syringe on ice, unplug the piston, transfer 55 µL of cell suspension into the syringe and then put the piston back. Discharge bubbles carefully and put the syringe back on ice.
- Repeat 2.3.10 until all syringes have been loaded with the cells. The syringes are now ready for injection. NOTE: To maximize cell viability, we recommend keeping the cells on ice after dissociation. For the above steps 2.2.4–2.2.11, make sure all the reagents are kept at 2–8 °C before use.
3. Intrasplenic Injection of iHeps
Anesthetize the mice with ketamine (100 mg/kg) and xylazine (10 mg/kg) injected intraperitoneally. Put vet ointment on mice eyes to prevent dryness during the anesthesia procedure, and monitor their muscle reflexes to observe the anesthesia's effect and assess pain.
Once mice have lost muscle reflex to stimulation, place them in the right lateral decubitus position. Apply a thick layer of depilatory cream to the incision area in the left flank for 5–8 min. Then, remove the depilatory cream and hair by wiping the area with a water-moistened gauze pad.
Scrub the left flank with povidone-iodine or, alternatively, with 70% ethanol 3 times followed by a final soaking with povidone-iodine for disinfection.
In a level 2 biosafety cabinet, locate the position of the spleen, which can be visualized transparently in the left flank, and then incise the skin and abdominal wall for 0.5–1 cm. Exteriorize the spleen by pulling out the adipose tissue gently near it using pointy forceps. Stabilize the spleen using a swab gently.
Insert the needle of the insulin syringe 3–4 mm into the parenchyma of the spleen and inject approximately 50 µL of cell suspension gently. Retract the needle and place a cotton swab over the injection site for 1 minute to prevent bleeding and spillage of material.
Return the spleen to the peritoneum and close the wound with 5–0 nylon sutures. Then, keep mice in a warm chamber/incubator for 3–16 h to return them to their normal body temperature and to revive them. Mice that have undergone surgery are not returned to the company of other animals until fully recovered.
Add meloxicam (26 μg/mL) to the drinking water and inject buprenorphine (50 μg/kg) intramuscularly as anti-inflammatory drug and analgesic, respectively. Monitor surgical wounds daily to prevent any inflammation. NOTE: Make sure all the instruments and supplies used in steps 4-7 are sterile.
4. Test of Plasma LDL-C Level
At days 0, 7, 14, 21, and 28 post-engraftment, restrain the LRG mice tightly to minimize side-to-side movement of the head using two-handed restrain method, then puncture the facial vein using the lancet. Blood will begin to flow after removal of the lancet. Collect around 50 µL of blood into 1.5 mL tubes containing 1 µL of EDTA. NOTE: If the grip is too tight, the collected blood volume may be reduced and mice may die because of breathing difficulty.
Mix the blood gently by inverting the tubes several times and then centrifuge at 950 x g for 15 min. Collect the supernatants and store them at -80 °C immediately.
Once all the samples are collected, thaw the plasma at room temperature and test the LDL-C level using a LDL-C detection kit according to the manufacturer's manual.
5. In Vivo Drug Testing in Chimeric Mice Engrafted with LDLR +/- and FH iHeps
To induce hypercholesterolemia, feed the mice a HFHC diet 7 days prior to engraftment.
At 7 days post-engraftment, treat each group of mice with vehicle (PBS, injected subcutaneously), 10 mg/kg/week PCSK9 antibodies (a clinical-grade formulation of PCSK9 monoclonal antibodies, injected subcutaneously too), 10 mg/kg/day simvastatin (40 mg/L mg/mL in drinking water), or combined PCSK9 antibodies and simvastatin.
Collect mouse plasma at days 0 (the day of engraftment), 14, 21, and 28 post-engraftment. Store the samples at -80 °C immediately.
Once all the samples are available, test the plasma LDL level using a LDL-C detection kit, according to the manufacturer's manual.
6. Endothelial Function Test
Endothelial function is affected early in FH and can be tested in our mouse model as an indicator of the severity of the disease or to evaluate the improvement with different treatments. A stereomicrocope, dissection forceps, scissors, a wire myograph, acquisition hardware (see Table of Materials), and a computer are needed for this.
Sacrifice the mice by intraperitoneal injection of 100 mg/kg phenobarbital. Remove internal organs so that the descending aorta parallel to the spine is visible and can be dissected out by scissors together with adjacent tissue and heart. Dissect the aortae using fine scissors and place them in cold oxygenated Krebs solution (mM: 119 NaCl, 4.7 KCl, 2.5 CaCl2, 1 MgCl2, 25 NaHCO3, 1.2 KH2PO4, and 11 D-glucose).
Transfer the aortae in Krebs solution to a silicone-coated petri dish. Pin the connective tissue to fix the aorta position without stretching it. Under a stereomicroscope, use fine forceps to spring the scissors and dissect the aorta free from the surrounding fat and adventitial tissue without damaging the vessel wall, then cut it into 1.5 to 2 mm length segments.
Cut an approximately 2 cm long and 40 μm thick stainless wire and gently put through the aorta lumen. Transfer the segment to the wire myograph chamber filled with oxygenated Krebs solution by holding the wire.
To measure the length of the aortae segments when studying contractility, place each segment perpendicularly in between the jaws and record the reading (D1) on the micrometer. Remove the segment and move the jaws together and record the reading (D0). The length of the segment would be L = D1-D0.
Follow the user guide to clamp the wire and secure it with the screwdriver whilst placing the segment in between the jaws, but leave it unstretched.
For normalization before the experiment, set the myograph to zero in the unstretched position. Then, slowly move the jaw apart and observe the aorta tension change until reaching 3 mN. After 15 minutes, drain the solution from the myograph chamber, and replace with fresh Krebs solution, wait for 15 min and adjust the tension to 3 mN again.
Change the standard Krebs solution to 60 mM KCl-containing Krebs solution to induce a contraction for at least 15 min. Rinse with fresh Krebs solution 3 times.
Add increasing concentrations of phenylephrine (Phe; e.g., from 10 nM to 100 μM). Wash out with standard Krebs solution and add a single concentration of Phe at ~70% of maximal contraction from the previous contraction. When the contraction is stable, add increasing concentrations of acetylcholine (ACh; e.g., from 1 or 3 nM to 10 or 30 µM) to induce vasodilation. ACh is added at approximately 2 min interval. NOTE: In some aortic segments with small Phe-induced contraction, for example smaller than 30% of KCl induced contraction, U46619 (another vasoconstrictor) at a concentration from 1 nM to 30 nM can be used to induce a stable contraction that is bigger than 70% of KCl-induced contraction.
Clean the myograph according to the manufacturer's manual.
Using data analysis software (see Table of Materials), record the force (F) after every addition of drugs. Draw the marker to basal tension (Fbasal) for relative measurement, move the arrow to the highest point after Phe as FPhe, and then move to the lowest point after every addition of ACh as FACh. Calculate the ACh-induced endothelium-dependent vasodilation (EDV) by % relaxation = (FACh-Fbasal)/(FPhe-Fbasal) on the Y-axis. Prepare a concentration response curve on statistical software using log10 value of concentration in M on the X-axis.
To analyze the statistical difference, use the area under curve of each segment from one mouse, and analyze the difference of the area under curve amongst groups using one-way ANOVA. Individual points can also be analyzed if needed.
7. Evidence of iHep Repopulation in the Mouse Liver
At the endpoint, sacrifice the mice by injecting intraperitoneally 100 mg/kg phenobarbital.
Collect plasma by cardiac puncture. Dissect lobes of liver using ophthalmic scissors, then put livers into a 10-cm peri-dish and wash twice with saline. Remove saline from the liver surface with absorbent paper, then cut the lobes into around 0.6-cm-length pieces. Fix the lobes of the liver in 10% formalin.
Embed the fixed livers in paraffin and section using a tissue processing system and a sliding microtome, respectively, according to manufacturer's instructions.
Heat the slides to 60 °C for 1 h to melt the wax. Then, immerse the slides in xylene for 5 min twice, and rehydrate the tissue with 100%, 90%, and 70% ethanol successively.
Immerse the slides into the slide-chamber containing around 50 mL of antigen retrieval solution, and then put the chamber into the pressure cooker, 120 °C for 1.5 min. Wash the slides gently with pipe water for 5 minutes.
Stain the sections with primary antibodies (around 100 µL/section) targeting human ALB (hALB) and human nuclei antigen (hNA). Then, stain with secondary antibodies conjugated with a fluorescent label or horseradish peroxidase (which reacts with the DAB detection kit).
To calculate the percentage of hALB+ areas and hNA+ cells, scan the slides stained with anti-hALB and anti-hNA and reacted with DAB using an automated slide scanning system to get whole section images. NOTE: Whole section-scanned images are helpful to calculate repopulated efficiency in an unbiased manner based on hALB+ areas or hNA+ cells in the immunohistochemistry-stained livers. As an alternative method to monitor iHep repopulation efficiency, plasma human albumin level can be tested using a human specific ALB ELISA kit.
Take snapshot images to cover the whole slide using the associated digital slide viewing software under 5X view. To quantify the percentage of hALB+ areas, for each snapshot image, use the microscope imaging software to qualify the positive area (P) and the total area (T) of the image, and use Image J to qualify the blank area (B). The percentage of hALB+ areas is expressed as P/(T-B) *100%. For each group, at least 3 mice should be selected. And for each mouse, at least 4 sections from different positions of the liver should be selected.
To quantify the percentage of hNA+ cells, for each snapshot image, randomly select 1/16 of the area with an image processing software, and then manually count the number of total nuclei (T) and hNA+ nuclei (N). The percentage of hNA+ is expressed as N/T *100%. For each group, at least 3 mice should be selected. For each mouse, at least 4 sections from different positions of the liver should be selected.
Representative Results
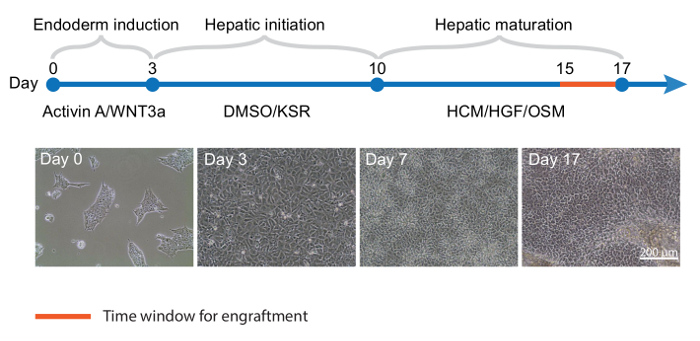
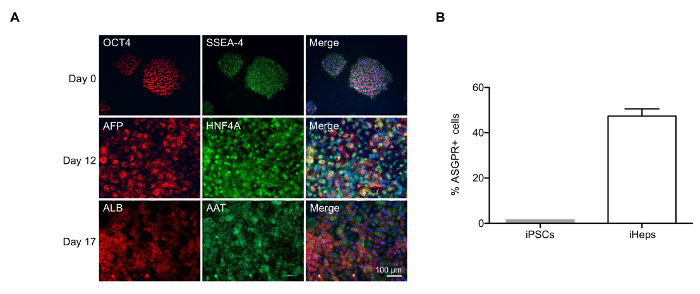
Directed Differentiation of Human iPSCs into iHeps When reaching 70% confluence, human iPSCs are differentiated into iHeps with a 3-step protocol16 (Figure 1 upper panel). After 3 days of endoderm differentiation, iPSC colonies become loosened and spread to full confluence (Figure 1 lower panel). Then, with 2nd stage medium, hepatoblasts appear and proliferate. These cells are crowded but show clear edges at this stage (day 7, Figure 1 lower panel). After 17 days of differentiation, polarized iHeps with typical hexagon morphology appear (Figure 1 lower panel). These iHeps express pHH markers, including AAT and ALB (Figure 2A). Moreover, the ratio of ASGPR+ iHeps should be relatively high, as measured by flow cytometry (Figure 2B)15.
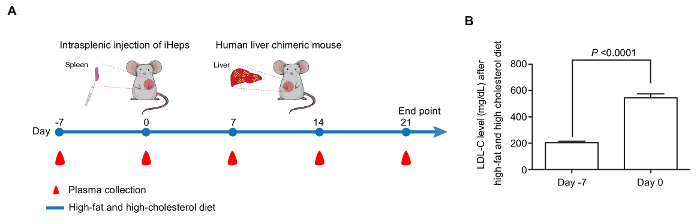
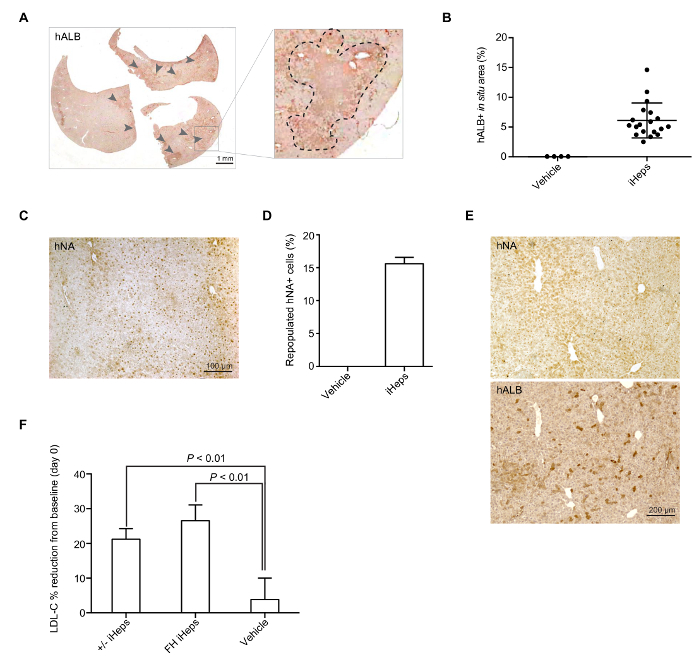
Generation of a FH In Vivo Disease Model Using FH iHeps To help LRG mice develop hypercholesterolemia, we feed them with a HFHC diet 7 days prior to engraftment (day -7). On the day of engraftment (day 0), LRG mice display around 3-fold plasma LDL-C level (around 600 mg/dL, Figure 3B). Day 15–17 iHeps are engrafted into LRG mice via intrasplenic injection; these iHeps soon reside in the liver parenchyma and proliferate there (Figure 3A). At the end-point, livers of these chimeric mice are collected and fixed, then stained with hALB and hNA, both of which should show clear proof of iHep-mediated liver repopulation in LRG mouse liver based on staining for hALB and hNA (Figure 4A-E). In our hands, both LDLR +/- and FH iHeps could reduce plasma LDL-C level significantly 21 days post-engraftment (Figure 4F). At this point, FH human liver chimeric mice can be used to test therapies for FH.
Validation of FH Human Liver Chimeric Mouse Model Using Available Drugs To validate our model, we used 2 well-known LDL-C lowering drugs, simvastatin and PCSK9 antibodies (Figure 5A). Our data demonstrate that 21 days after treatment, PCSK9 antibodies have a stronger ability for LDL-lowering and EDV than simvastatin in FH chimeric mice (Figure 5B-D). Notably, the observed percentage reduction of plasma LDL-C with PCSK9 antibodies in FH chimeric mice is similar to that reported in clinical trials4. These results demonstrate the potential utility of LRG chimeric mice engrafted with FH iHeps for preclinical testing of novel drugs for FH.
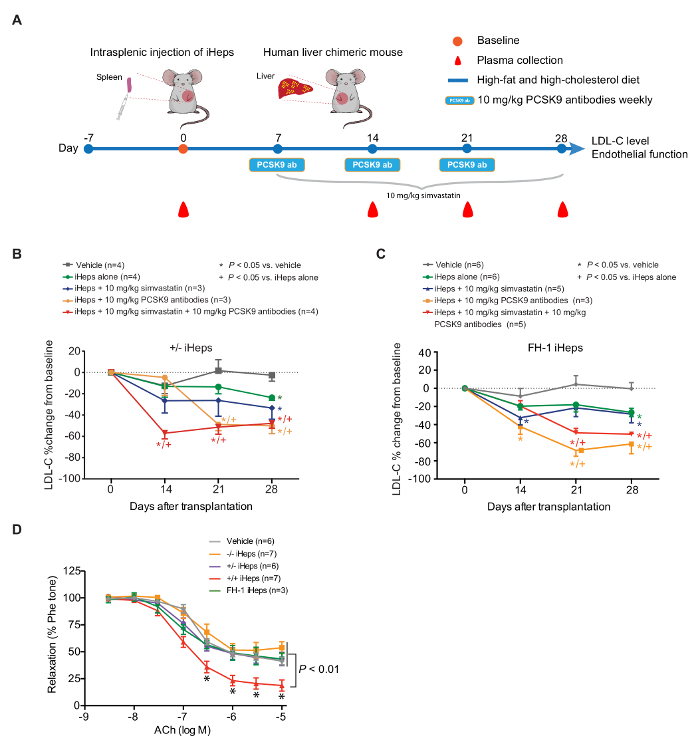
Figure 1: Directed Differentiation of Human iPSCs into iHeps.Top panel, Timeline for iPSC differentiation into iHeps; key cytokines and media are shown for each stage. (Lower panel) Representative phase contrast images of different time points of iHep differentiation. KSR: knockout serum replacement; HCM: hepatocytes culture medium; HGF: hepatocyte growth factor; OSM: oncostatin M. Please click here to view a larger version of this figure.
Figure 2: Characterization of iHeps Produced with Modified Differentiation Protocol. (A) Immunofluorescence of iHeps at various stages of differentiation. Nuclei are stained in blue in the merged compositions. (B) Bar graph shows the percentage of ASGPR+ iHeps derived obtained at day 17, as measured by flow cytometry. Samples were measured in 3 independent experiments; mean values are shown and error bars indicate standard deviation (SD). Please click here to view a larger version of this figure.
Figure 3: Generation of a FH In Vivo Disease Model with iHeps. (A) Time line for the generation of human liver chimeric mice by intrasplenic injection of FH iHeps into LRG mice. (B) Bar graph shows that feeding LRG mice with HFHC diet leads to significantly increased LDL-C level (n = 10). P values are indicated on the figure and were obtained using an unpaired t-test; mean values are shown and error bars indicate standard error of the mean (SEM). Panel B is modified from Figure 3I of our previous report15. Please click here to view a larger version of this figure.
Figure 4: iHep Mediated Repopulation of LRG Mice Liver. (A) Representative whole section-scanned image of hALB staining in a mouse liver repopulated with LDLR +/- iHeps. Arrows indicate clusters of human iHeps engrafted into the mouse liver; zoomed sections are shown in the right panel. (B) Scatter plot graph shows the percentage of repopulated hALB+ corresponding to iHep-containing areas in a mouse liver (from different donor iPSCs), calculation was based on whole section-scanned images (n = 19). Mean values are shown and error bars indicate SD. (C) Representative images of immunohistochemical staining for hNA in a mouse liver with engrafted FH iHeps. (D) Bar graph shows the percentage of repopulated hNA+ iHeps (from different donor iPSCs) in LRG mouse livers (n = 3). Mean values are shown and error bars indicate SD. (E) hALB and hNA staining on two consecutive sections of a mouse liver repopulated with wild type iHeps. (F) Bar graph shows the percentage of plasma LDL-C reduction from baseline at day 21 post-engraftment; n = 5 for LDLR +/- iHeps and n = 6 for FH iHeps and the vehicle. P values are indicated on the figure and were obtained using an unpaired t-test; mean values are shown error bars indicate SEM. Panels B, D, and E are modified from Figure 3G-3I of our previous report15. Please click here to view a larger version of this figure.
Figure 5: PCSK9 Antibodies Show Stronger LDL Lowering Ability than Simvastatin in Human Liver Chimeric LRG Mice. (A) schematic view of the in vivo drug testing approach using FH human liver chimeric mice. (B and C) Percentage change of plasma LDL-C from baseline at days 14, 21, and 28 in FH chimeric mice fed with HFHC diet and treated with the indicated drugs; n indicates number of mice. P values were obtained using a Kruskal-Wallis test; mean values are shown and error bars indicate SEM. (D) EDV in response to increasing concentrations of ACh. P values are indicated on the figure for indicated concentration. P values were obtained using two-way ANOVA adjusted with Dunnett's multiple comparison; error bars indicate SEM. Figure 5 is modified from Figure 4A-4C and Figure 5A of our previous report15. Please click here to view a larger version of this figure.
Discussion
Previous studies using iHeps in rodents have confirmed that they are an effective way to study inherited liver diseases17. To further expand the use of this technology and because current FH animal models are suboptimal, we engrafted FH iHeps into LRG mice and showed that the engrafted LDLR +/- or heterozygous LDLR-mutated FH iHeps can reduce mice plasma LDL-C level and respond to lipid-lowering drugs in vivo.
There are 3 critical steps in our protocol for generating human FH liver chimeric mice using iHeps: 1) Production of high-quality iHeps through directed differentiation. Given the clonal variability between iPSC lines18, it is important to use isogenic iPSCs for proper comparisons and to test the iHep differentiation efficiency of the mother cell line before performing the engineering and the engraftment.
2) Correct iHep upload into the syringe. Differently from other protocols, we use 50% extracellular matrix (final concentration, v/v) to resuspend iHeps, and then upload them into the insulin syringe. We believe that the extracellular matrix protects cells and provides a microenvironment that facilitates iHep migration into the liver from the spleen. Bubbles are a lethal factor for surgery and should be avoided completely in the syringe.
3) Correct number of engrafted cells. Overload of cells can also lead to high lethality rate. We recommend engrafting 1–1.5 million iHeps per 25–30 g mouse.
Our protocol also has some limitations: the liver injury induced by irradiation is moderate and single-dose, and the maturation state of our iHeps is not comparable to pHH. Related to both considerations, the degree of chimerism of our model is similar to recent reports describing NOD/Lt-SCID/IL-2Rγ−/− mice or Gunn rats engrafted with iPSC-derived iHeps17,19, but significantly lower than FRG mice or uPA transgenic mice engrafted with pHH. To overcome this caveat, on one hand, one could further optimize the hepatic differentiation protocol to improve the maturation of iHeps. On the other hand, LRG mice could be crossed with FRG mice to generate Ldlr-/-/Fah-/-/Rag2-/-/Il2rg-/- mice.
In summary, here we have described a detailed protocol for generating human liver chimeric animals with FH iHeps, and for testing the functionality of the engrafted iHeps. Importantly, these chimeric mice can be used for in vivo drug testing. Our model can likely be optimized by improving the functionality of iHeps or knocking out additional genes (e.g., Fah) in the recipient LRG mice, and it will be useful to investigate pathological mechanisms of the disease and perform preclinical studies.
Disclosures
H.-F.T. is the National Coordinator and Investigator of the ODYSSEY OUTCOMES study sponsored by Sanofi and Regeneron Pharmaceuticals.
Acknowledgments
This work was supported by the Shenzhen Science and Technology Council Basic Research Program (JCYJ20150331142757383), Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16030502), Hong Kong Research Grant Council Theme Based Research Scheme (T12-705/11), Cooperation Program of the Research Grants Council of the Hong Kong Special Administrative Region and the National Natural Science Foundation of China (N-HKU730/12 and 81261160506), Research Team Project of Guangdong Natural Science Foundation (2014A030312001), Guangzhou Science and Technology Program (201607010086), and Guangdong Province Science and Technology Program (2016B030229007 and 2017B050506007).
References
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232(4746):34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- Endo A. The discovery and development of HMG-CoA reductase inhibitors. J Lipid Res. 1992;33(11):1569–1582. [PubMed] [Google Scholar]
- Dubuc G, et al. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler Thrombo Vasc Biol. 2004;24(8):1454–1459. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- Robinson JG, et al. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med. 2015;372(16):1489–1499. doi: 10.1056/NEJMoa1501031. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K, et al. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet. 2014;383(9911):60–68. doi: 10.1016/S0140-6736(13)61914-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi S, et al. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J Clin Invest. 1993;92(2):883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. Serial inbreeding of rabbits with hereditary hyperlipidemia (WHHL-rabbit) Atherosclerosis. 1980;36(2):261–268. doi: 10.1016/0021-9150(80)90234-8. [DOI] [PubMed] [Google Scholar]
- Carpentier A, et al. Engrafted human stem cell-derived hepatocytes establish an infectious HCV murine model. J Clin Invest. 2014;124(11):4953–4964. doi: 10.1172/JCI75456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno C, et al. Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol. 2004;165(3):901–912. doi: 10.1016/S0002-9440(10)63352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basma H, et al. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology. 2009;136(3):990–999. doi: 10.1053/j.gastro.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146(2):318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig KD, et al. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest. 2010;120(3):924–930. doi: 10.1172/JCI40094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma H, et al. Robust expansion of human hepatocytes in Fah(-/-)/Rag2(-/-)/Il2rg(-/-) mice. Nat Biotechnol. 2007;25(8):903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig-Choisat B, et al. Development and rescue of human familial hypercholesterolaemia in a xenograft mouse model. Nat Commun. 2015;6:7339. doi: 10.1038/ncomms8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, et al. Generation of human liver chimeric mice with hepatocytes from familial hypercholesterolemia induced pluripotent stem cells. Stem Cell Rep. 2017;8(3):605–618. doi: 10.1016/j.stemcr.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara M, et al. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109(31):12538–12543. doi: 10.1073/pnas.1209979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, et al. Amelioration of hyperbilirubinemia in gunn rats after transplantation of human induced pluripotent stem cell-derived hepatocytes. Stem Cell Rep. 2015;5(1):22–30. doi: 10.1016/j.stemcr.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann D, Vallier L. Variability of human pluripotent stem cell lines. Curr Opin Genet Dev. 2017;46:179–185. doi: 10.1016/j.gde.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Liu H, Kim Y, Sharkis S, Marchionni L, Jang YY. In vivo liver regeneration potential of human induced pluripotent stem cells from diverse origins. Sci Transl Med. 2011;3(82):82ra39. doi: 10.1126/scitranslmed.3002376. [DOI] [PMC free article] [PubMed] [Google Scholar]


