Abstract
Aberrant cell signaling plays a central role in cancer development and progression. Most novel targeted therapies are indeed directed at proteins and protein functions, and cell signaling aberrations may therefore serve as biomarkers to indicate personalized treatment options. As opposed to DNA and RNA analyses, changes in protein activity can more efficiently evaluate the mechanisms underlying drug sensitivity and resistance. Phospho flow cytometry is a powerful technique that measures protein phosphorylation events at the cellular level, an important feature that distinguishes this method from other antibody-based approaches. The method allows for simultaneous analysis of multiple signaling proteins. In combination with fluorescent cell barcoding, larger medium- to high-throughput data-sets can be acquired by standard cytometer hardware in short time. Phospho flow cytometry has applications both in studies of basic biology and in clinical research, including signaling analysis, biomarker discovery and assessment of pharmacodynamics. Here, a detailed experimental protocol is provided for phospho flow analysis of purified peripheral blood mononuclear cells, using chronic lymphocytic leukemia cells as an example.
Keywords: Cancer Research, Issue 140, Biomarker, Cell Signaling, Chronic Lymphocytic Leukemia (CLL), Fluorescent Cell Barcoding (FCB), Phospho Flow Cytometry, Phospho-Proteins, Single Cell Profiling
Introduction
Phospho flow cytometry is used to analyze protein phosphorylation levels at single-cell resolution. The overall goal of the method is to map cellular signaling patterns under specified conditions. By exploiting the multiparameter capacity of flow cytometry, several signaling pathways can be analyzed simultaneously in different subsets of a heterogeneous cell population such as peripheral blood. These traits offer advantages over other antibody-based technologies such as immunohistochemistry, enzyme-linked immunosorbent assay (ELISA), protein array, and reverse phase protein array (RPPA)1. Phospho flow cytometry can be combined with fluorescent cell barcoding (FCB), which means that individual cell samples are labeled with unique signatures of fluorescent dyes so that they can be mixed together, stained and analyzed as a single sample2. This reduces the antibody consumption, increases the data robustness through the combination of control and treated samples, and enhances the speed of acquisition. The combined FCB population can then be divided into smaller samples and stained with up to 35 distinct phospho-specific antibodies, depending on the amount of starting material. Large profiling experiments can, thereby, be run with standard cytometer hardware. Phospho flow cytometry has been applied to profile signaling pathways in patient samples from several hematological cancers including chronic lymphocytic leukemia (CLL)3,4,5, acute myeloid leukemia (AML)6 and non-Hodgkin lymphomas7. Phospho flow cytometry is thus a powerful approach to characterize signaling aberrations, identify and validate biomarkers, and assess pharmacodynamics.
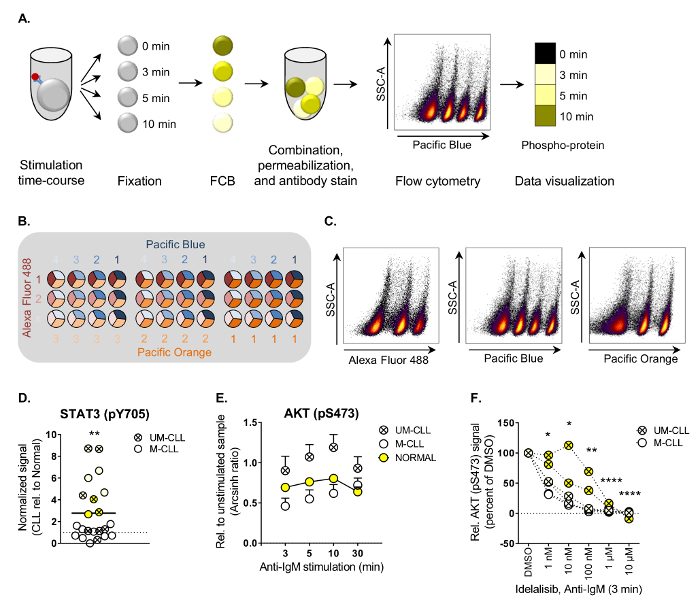
Here, the optimized protocol for analysis of CLL patient samples by phospho flow cytometry is provided (Figure 1A). Examples of basal signaling characterization, anti-IgM/B cell receptor stimulation and drug perturbation are shown. A detailed description of an FCB matrix is provided. The protocol can easily be adapted to other suspension cell types.
Protocol
Blood samples were received following written informed consent from all donors. The study was approved by the Regional Committee for Medical and Health Research Ethics of South-East Norway and the research on human blood was carried out in accordance with the Declaration of Helsinki8.
NOTE: Steps 1-3 should be performed under sterile conditions in a tissue culture hood.
1. Isolation of Peripheral Blood Mononuclear Cells (PBMCs) from CLL Patient Blood Samples
CAUTION: Human blood should be handled according to regulations for Biosafety Level 2.
Dilute the blood 1:1 with phosphate-buffered saline (PBS: 136.9 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4 x 2H2O, 1.8 mM KH2PO4, pH 7.4) and transfer to 50 mL tubes (30 mL/tube).
Carefully layer 10 mL of a density gradient medium (e.g., Lymphoprep) to the bottom of the tube using a 10 mL pipette.
Centrifuge at 800 x g for 20 min at 4 °C. The PBMCs are now visible on top of the density gradient medium layer.
Use a Pasteur pipette to transfer the cells into two new 50 mL tubes. Wash twice with PBS (fill up the tubes).
Centrifuge at 350 x g for 15 min. Discard the supernatant and resuspend in 3 mL of PBS.
Count the cells using a preferred method.
Centrifuge the cells at 350 x g for 5 min. Discard the supernatant. NOTE: Steps 1.8 to 3.2 are optional. It is possible to proceed directly to step 3.3.
Resuspend the cells in fetal bovine serum (FBS) supplemented with 10% dimethyl sulfoxide (DMSO) and freeze down in suitable aliquots using cryo tubes. NOTE: DMSO is toxic to the cells. Work fast once the cells are mixed with FBS/DMSO. Cells can be stored long term in liquid nitrogen.
2. Thawing of Cells
Quickly thaw the cells in a 37 °C water bath. NOTE: DMSO is toxic to the cells. Work fast to limit the exposure to DMSO.
Wash the cells once with 10 mL of cold Roswell Park Memorial Institute medium (RPMI 1640 with supplemental GlutaMAX, see Table of Materials).
Centrifuge at 300 x g for 5 min. Discard the supernatant.
Resuspend the cells in RPMI 1640 medium supplemented with sodium pyruvate, MEM non-essential amino acids and penicillin/streptomycin (added at 1x dilution according to instructions) and 10% FBS. Transfer the cells to a small cell culture flask and leave in an incubator at 5% CO2, 37 °C for 1 hour to allow the cells to calibrate.
3. Preparation of Cells
Count the viable cells using a preferred method.
Transfer the cells to a 50 mL tube and centrifuge at 300 x g for 5 min. Discard the supernatant.
Resuspend the cells in RPMI 1640 medium (step 2.2) supplemented with 1% FBS to no more than 50 x 106 cells/mL.
Transfer the required amount of cell suspension to wells in a 96 well V-bottom plate. NOTE: Number of wells corresponds to the number of conditions to be tested. Samples for a stimulation time-course are drawn from a single well. Calculate 50 µL sample per time-point + 50 µL of dead volume. Save samples for compensation controls (one unstained sample + one sample per barcoding dye).
Transfer the 96 well plate to a pre-heated 37 °C water bath. Rest the cells for 10 min.
4. Stimulation and Fixation of Cells
NOTE: Perform steps 4-8 on the lab bench (i.e., not sterile).
CAUTION: The main ingredient of Fix Buffer I is paraformaldehyde, which is toxic (inhalation and skin contact). Handle with care.
Prepare a 96 well V-bottom plate with 60 µL of Fix Buffer I per well per sample. Leave in the 37 °C water bath. NOTE: Cells: Fix buffer should be 1:1. In order to allow for evaporation at 37 °C, the Fix buffer is initially in abundance.
Optionally, treat the cells with drugs before stimulation.
Transfer a 50 µL control sample to the fix plate. Mix by pipetting up and down.
Optionally, start the stimulation time-course by adding 10 µg/mL anti-IgM to the cells. Mix by pipetting up and down.
Transfer a 50 µL sample to the fix plate at each time-point. Mix by pipetting up and down. NOTE: Anti-IgM induced signaling is usually initiated early (minutes).
Leave the fix plate at 37 °C for 10 min after the last sample has been added.
5. Fluorescent Cell Barcoding (FCB)
NOTE: See Table 1 for a list of barcoding reagents.
Wash the fixed cells 3x with PBS (fill up the wells).
Centrifuge at 500 x g for 5 min. Discard the supernatant.
Prepare a 96 well V-bottom plate with barcoding reagents. Pipet 5 µL of each barcoding reagent per well in the number of combinations required to stain all samples following the staining matrix, e.g., in Figure 1B. Each sample will have a unique combination of different barcoding concentrations.
Resuspend the cells in 190 µL of PBS and transfer to the barcoding plate. Mix thoroughly. NOTE: Stain one compensation sample with the highest final concentration used for each barcoding reagent and save one unstained sample.
Leave the cells for 20 min at room temperature, in the dark.
Wash the stained cells 2x with flow wash (PBS, 1% FBS, 0.09% sodium azide) (fill up the wells).
Centrifuge at 500 x g for 5 min. Discard the supernatant.
Add 190 µL of flow wash to the cells and combine the barcoded samples in one 15 mL tube. Transfer each compensation control to a separate 1.7 mL tube.
Centrifuge at 500 x g for 5 min. Discard the supernatant.
6. Cell Permeabilization for Intracellular Antigen Staining
CAUTION: The main ingredient of Perm Buffer III is methanol which is toxic (inhalation and skin contact) and flammable. Handle with care.
Transfer 2 mL of Perm Buffer III to a 15 mL tube. Leave at -20 °C so it is ice-cold upon use. NOTE: The Perm Buffer can be left at -20 °C from the start of the experiment.
Add 1.5 mL of ice-cold Perm Buffer to the barcoded cell population (in a 15 mL tube) and 100 µL to each compensation control (in 1.7 mL tubes) drop-wise while vortexing to avoid that the cells clump together.
Transfer the cells directly to -80 °C. Leave for a minimum of 30 min. NOTE: It is natural to pause the experiment at this point. Cells in Perm Buffer can be stored long term at -80 °C.
7. Antibody Staining
NOTE: See Table of Materials for a list of reported phospho-specific antibodies.
Transfer the cells from -80 °C to a box of ice.
Wash 3x with flow wash. NOTE: It is important to add flow wash in excess to see the cell pellet, e.g., add 3 mL of flow wash to the barcoded cell population and 1 mL to each compensation control.
Centrifuge at 500 x g for 5 min at 4 °C. Discard the supernatant.
Resuspend the barcoded cell population in a volume of flow wash, which allows 25 µL of cell suspension per phospho-antibody stain. Resuspend the compensation controls in 200 µL of flow wash.
Prepare antibodies for staining in a 96 well V-bottom plate. The final volume will be 50 µL/well. Per well, add phospho-specific antibody diluted in flow wash to a final volume of 10 µL, surface marker diluted in flow wash to a final volume of 15 µL, and 25 µL of cell suspension. NOTE: Antibody dilutions should be titrated prior to the experiment. Include isotype control.
Leave the cells for 30 min at room temperature, in the dark.
Wash the stained cells 2x with flow wash (fill up the wells).
Centrifuge at 500 x g for 5 min. Discard the supernatant.
Resuspend the cells in 150 µL of flow wash.
8. Preparation of Compensation Controls
Prepare compensation controls for the antibody-conjugated fluorochromes in parallel with the antibody staining. Use compensation beads according to the vendor’s instructions.
9. Flow Cytometry Analysis
NOTE: The experiment can be run on a flow cytometer with a High Throughput Sampler (HTS).
Optimize the photomultiplier tube (PMT) voltage with the unstained control.
Run compensation controls and calculate the compensation matrix.
Run samples. The event rate should be in accordance with the instrument specifications.
10. Gating Strategy and Data Analysis
Import the FCS files from the experiment to a flow cytometry analysis software like FlowJo or Cytobank (https://cellmass.cytobank.org).
- Gating strategy
- Select lymphocytes by plotting SSC-A versus FSC-A in a density dot plot.
- Display the lymphocytes and select the singlets by plotting SSC-A versus FSC -W.
- Display the single cells and gate the cell type by plotting SSC-A versus the surface marker.
- Display the cell type population in a Pacific Blue versus SSC-A density plot and select the different FCB populations based on their Pacific Blue staining intensity (see Figure 1A).
- Plot the phospho antibody channel against the FCB channel, or as a heatmap (see Figure 1A) to display the phosphorylation events.
Calculate phospho-signals using the inverse hyperbolic sine (arcsinh) of the MFI (median fluorescent intensity) of phospho-signal versus isotype control (basal phosphorylation levels, see Figure 1D), or of stimulated versus unstimulated cell populations (see Figure 1E).
Representative Results
The main steps of the phospho flow cytometry protocol are illustrated in Figure 1A. In the presented example, CLL cells were stained with the barcoding reagent Pacific Blue at four dilutions. Three-dimensional barcoding can be performed by combining three barcoding dyes, as illustrated in Figure 1B. The individual samples are then deconvoluted by subsequent gating on each barcoding reagent versus SSC-A (Figure 1C). Detailed information about the barcoding reagents are listed in Table 1.
Following the procedure described here, phospho-protein levels were characterized in B cells from CLL patients and normal controls under various conditions3. Both basal and stimulation-induced phosphorylation levels of 20 signaling molecules downstream of the B cell receptor (BCR) were analyzed (see Table of Materials for a list of reported phospho-specific antibodies). Basal phospho-protein levels were mapped in 22 CLL patient samples relative to the mean of normal controls. This analysis showed that STAT3 (pY705) is significantly upregulated in CLL cells (Figure 1D). Constitutive activation of STAT3 has been reported in other hematological malignancies and is associated with resistance to apoptosis9.
In order to identify signaling aberrations induced through the BCR pathway, cells were stimulated with anti-IgM for up to 30 min. It has been shown that CLL cells from patients with IgVH unmutated status (UM-CLL) display increased sensitivity towards anti-IgM stimulation10. This was indeed observed for the majority of the analyzed proteins, but the effect was statistically significant only for AKT (pS473) (Figure 1E, UM-CLL versus M-CLL and Normal). To test if the aberrant AKT (pS473) signal could be reversed CLL cells were exposed to the PI3Kδ inhibitor idelalisib, which is used in the clinic to treat CLL patients11. As shown in Figure 1F, AKT (pS473) levels were significantly reduced upon idelalisib treatment in a concentration-dependent manner, demonstrating that kinase inhibitors can be applied to normalize aberrant signaling in CLL cells.
These results show that phospho flow cytometry in combination with FCB is a powerful approach to perform signaling analysis studies, identify potential biomarkers, and assess pharmacodynamics.
Figure 1. Work flow and examples of applied phospho flow cytometry analysis. (A) The main steps of the phospho flow procedure are illustrated. Cells are first stimulated, then fixed and subjected to FCB before they can be combined in one tube for permeabilization and subsequent antibody staining. The cells are run on a flow cytometer and the cell populations are deconvoluted by gating during the data analysis. The results can be visualized as histograms or heatmaps, as shown. (B) Example of a three-dimensional FCB staining matrix using Alexa Fluor 488 (three dilutions), Pacific Blue (four dilutions) and Pacific Orange (three dilutions). This matrix will allow combination of up to 36 samples. (C) The FCB cell population can be deconvoluted by gating on each FCB channel versus SSC-A. Combination of the gates in the analysis software generates the correct populations for analysis. (D) Unstimulated B cells from healthy donors (n = 25) and CLL patients (n = 22) were subjected to analysis by phospho flow following the procedure in (A). The basal fluorescence intensity signals were calculated relative to IgGκ isotype control as arcsinh ratio. The signals in CLL B cells were then normalized to the signals in B cells from normal controls. **p < 0.01, calculated by an unpaired two-sample t-test. UM-CLL: IgVH unmutated CLL, M-CLL: IgVH mutated CLL. Symbols of the same color represent patient samples which grouped together in a hierarchical agglomerative cluster based on levels of 20 phospho-proteins3. (E) B cells from normal controls (n = 10, mean + SEM) or CLL patients (n = 11 [M-CLL] and n = 8 [UM-CLL], mean + SEM) were stimulated with anti-IgM for the indicated time-course and subjected to phospho flow analysis. The fluorescence intensity signals were measured relative to unstimulated samples and shown as arcsinh ratio. **p < 0.01 (Normal vs UM-CLL) and ***p < 0.001 (M-CLL vs UM-CLL), calculated by multiple comparison testing with Holm-Sidak's correction. UM-CLL: IgVH unmutated CLL, M-CLL: IgVH mutated CLL. (F) CLL cells were incubated with DMSO or idelalisib as indicated for 20 min before anti-IgM stimulation for 3 min. The cells were then processed following the phospho flow protocol. *p < 0.05, **p < 0.01, ****p < 0.0001, calculated by multiple comparison testing with Holm-Sidak's correction. UM-CLL: IgVH unmutated CLL, M-CLL: IgVH mutated CLL. See (D) for explanation of symbol color. (D-F) are modified from3. Please click here to view a larger version of this figure.
| Serial dilute as follows (starting with the stock solution) | ||||||
| Barcoding reagent | Stock concentration | #1 | #2 | #3 | #4 | unstained |
| Alexa Fluor 488 | 10 mg/mL | 1:500 | 1:5 | x | ||
| Pacific Blue | 10 mg/mL | 1:2500 | 1:4 | 1:4 | 1:10 | |
| Pacific Orange | 2 mg/mL | 1:50 | 1:12 | 1:24 |
Table 1. Barcoding reagents.
Discussion
Phospho flow cytometry is a powerful technique to measure protein phosphorylation levels in single cells. Since the method relies on staining with antibodies, phospho flow cytometry is limited by antibody availability. Furthermore, in order to obtain reliable results, all antibodies should be titrated and verified before use. A detailed protocol for titration of phospho-specific antibodies has been described elsewhere12. During panel design, consideration of the signal-to-noise ratio is critical. In the presented example, all phospho-antibodies were conjugated to Alexa Fluor 647. This fluorophore often provides the optimal differential between samples with low versus high levels of phospho-protein. Furthermore, by using only one color for the phospho-proteins the other channels will be left free for FCB and surface marker staining. This panel design reduces spillover into the phospho channel. By having all phospho-antibodies conjugated to the same fluorophore, the data analysis will also be simplified.
In the presented protocol, all antibody stainings were performed after fixation and permeabilization of the cells. However, it is important to keep in mind that surface marker staining can be adversely affected by the fixation and permeabilization steps due to denaturation of the surface antigen or increased nonspecific staining13. The user should therefore test the reactivity of the antibodies on a case to case basis. Resources on compatible clones may also be helpful, such as the overview of different fixation/permeabilization procedures and their compatibility with various antibodies at https://www.cytobank.org/facselect/.
Protein phosphorylation or de-phosphorylation is a transient modification that occurs in response to both extrinsic and intrinsic cues. When comparing phosphorylation patterns, it is therefore crucial that the experiments are carried out under similar conditions. When studying signaling in primary cells from blood, factors that could impact the result include time elapsed after drawing the blood, storage conditions and for how long the isolated cells are rested before initiation of the experiment. When comparing signaling patterns in cryo preserved cells and freshly isolated cells from blood, only very minor significant differences could be observed (Skånland, unpublished). However, it is still advisable to use cryo preserved normal cells as a control when studying biobanked patient samples, for example. The optimal conditions for performing the phospho flow cytometry experiments and the impact of external factors should be tested by the individual user.
Here, a protocol is presented for phospho flow analysis of suspension cells. The protocol can be adapted to other cell types, but it is a prerequisite that the cells are in suspension as single cells for the analysis by flow cytometry. The procedure to achieve this must be delicate to preserve, and not affect, phosphorylation patterns. Examples exist where adherent cells are detached from the culturing dish by cold trypsination12,14, or are rather grown on microspheres15. When it comes to phospho flow cytometry on solid tissue, one report exists on lung tumors where single cells were obtained by passing the cells through a tube with a cell strainer16. Recently, phospho flow cytometry was combined with a novel approach termed Disaggregation for Intracellular Signaling in Single Epithelial Cells from Tissue (DISSECT) in order to study phospho-proteins in epithelial tissues17 and colorectal cancer18.
The FCB is a critical step in the protocol since deconvolution of the samples at the end of the experiment relies on distinct FCB populations. In order to obtain this, the cells need to be homogeneously stained. It is therefore important to prepare a barcoding plate that the cells can be added to. Adding the reagents to the cells will result in uneven staining and mixed populations that cannot be deconvoluted by gating. It is highly recommended to run a test of the barcoding dilutions before the experiment is performed as the staining intensity is cell-type dependent.
Additional antibody-based techniques such as protein array and reverse phase protein array (RPPA) can be applied for quantification of phospho-protein levels in a medium to high-throughput manner. However, some qualities of phospho flow cytometry distinguish this method from the others. An important advantage of phospho flow cytometry is that it allows for single cell profiling. By including surface markers for different cellular subsets, inter-cellular heterogeneity can be detected. Combination with FCB furthermore allows for analysis of several conditions in the same experimental run. These features make phospho flow cytometry an attractive method for future applications in biomarker discovery and precision medicine19.
Disclosures
The author has nothing to disclose.
Acknowledgments
This work was conducted in the lab of Professor Kjetil Taskén, and was supported by the Norwegian Cancer Society and Stiftelsen Kristian Gerhard Jebsen. Johannes Landskron and Marianne Enger are acknowledged for critical reading of the manuscript.
References
- Lu Y, et al. Using reverse-phase protein arrays as pharmacodynamic assays for functional proteomics, biomarker discovery, and drug development in cancer. Seminars in Oncology. 2016;43(4):476–483. doi: 10.1053/j.seminoncol.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nature Methods. 2006;3(5):361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- Myhrvold IK, et al. Single cell profiling of phospho-protein levels in chronic lymphocytic leukemia. Oncotarget. 2018;9(10):9273–9284. doi: 10.18632/oncotarget.23949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente-Ribes A, et al. Spleen tyrosine kinase inhibitors reduce CD40L-induced proliferation of chronic lymphocytic leukemia cells but not normal B cells. Haematologica. 2016;101(2):e59–e62. doi: 10.3324/haematol.2015.135590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blix ES, et al. Phospho-specific flow cytometry identifies aberrant signaling in indolent B-cell lymphoma. BMC Cancer. 2012;12:478. doi: 10.1186/1471-2407-12-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish JM, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118(2):217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Myklebust JH, et al. Distinct patterns of B-cell receptor signaling in non-Hodgkin lymphomas identified by single-cell profiling. Blood. 2017;129(6):759–770. doi: 10.1182/blood-2016-05-718494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. THE JOURNAL OF THE AMERICAN MEDICAL ASSOCIATION. 2013;310(20):2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- Siveen KS, et al. Targeting the STAT3 signaling pathway in cancer: role of synthetic and natural inhibitors. Biochimica et Biophysica Acta. 2014;1845(2):136–154. doi: 10.1016/j.bbcan.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nature Reviews Cancer. 2016;16(3):145–162. doi: 10.1038/nrc.2016.8. [DOI] [PubMed] [Google Scholar]
- Arnason JE, Brown JR. Targeting B Cell Signaling in Chronic Lymphocytic Leukemia. Current Oncology Reports. 2017;19(9):61. doi: 10.1007/s11912-017-0620-7. [DOI] [PubMed] [Google Scholar]
- Landskron J, Tasken K. Phosphoprotein Detection by High-Throughput Flow Cytometry. Methods in Molecular Biology. 2016;1355:275–290. doi: 10.1007/978-1-4939-3049-4_19. [DOI] [PubMed] [Google Scholar]
- Krutzik PO, Clutter MR, Nolan GP. Coordinate analysis of murine immune cell surface markers and intracellular phosphoproteins by flow cytometry. Journal of Immunology. 2005;175(4):2357–2365. doi: 10.4049/jimmunol.175.4.2357. [DOI] [PubMed] [Google Scholar]
- Pollheimer J, et al. Interleukin-33 drives a proinflammatory endothelial activation that selectively targets nonquiescent cells. Arteriosclerosis, Thrombosis, and Vascular Biology. 2013;33(2):e47–e55. doi: 10.1161/ATVBAHA.112.253427. [DOI] [PubMed] [Google Scholar]
- Ertsås HC, Nolan GP, LaBarge MA, Lorens JB. Microsphere cytometry to interrogate microenvironment-dependent cell signaling. Integrative biology: quantitative biosciences from nano to macro. 2017;9(2):123–134. doi: 10.1039/c6ib00207b. [DOI] [PubMed] [Google Scholar]
- Lin CC, et al. Single cell phospho-specific flow cytometry can detect dynamic changes of phospho-Stat1 level in lung cancer cells. Cytometry A. 2010;77(11):1008–1019. doi: 10.1002/cyto.a.20965. [DOI] [PubMed] [Google Scholar]
- Simmons AJ, et al. Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-alpha-induced apoptosis in vivo. Molecular Systems Biology. 2015;11(10):835. doi: 10.15252/msb.20156282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons AJ, et al. Impaired coordination between signaling pathways is revealed in human colorectal cancer using single-cell mass cytometry of archival tissue blocks. Science Signaling. 2016;9(449):rs11. doi: 10.1126/scisignal.aah4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nature Reviews Cancer. 2015;15(12):747–756. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]


