Summary
Background
Enteropathogen detection traditionally relies on diarrhoeal stool samples, but these are inconvenient to collect if they are not immediately available, leading to suboptimum return rates of samples and delayed or missed diagnostic opportunities. We sought to compare the enteropathogen yields of rectal swabs and stool specimens in children with diarrhoea or vomiting, or both.
Methods
The Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE) did a study in three outpatient cohorts in Calgary and Edmonton (AB, Canada)—children enrolled in the Pediatric Emergency Research Canada emergency departments, children receiving routine vaccinations at a Calgary health clinic, and symptomatic children who met criteria for treatment at home. Eligible participants were children younger than 18 years, with at least three episodes of vomiting or diarrhoea in the preceding 24 h and fewer than 7 days of symptoms. After excluding those enrolled within the previous fortnight, unable to follow-up, or having psychiatric illness, neutropenia, or requiring emergent care, we attempted to collect rectal swabs and stool from all participants. Specimens were tested with the multianalyte assay Luminex xTAG Gastrointestinal Pathogen Panel, an in-house five-virus panel and bacterial culture. Primary outcomes were comparative yield (calculated as the proportion of submitted paired specimens only in which at least one pathogen was identified) and overall yield (which calculated the proportion of study participants in whom at least one pathogen was identified in all specimens, where unsubmitted specimens were analysed as negative). We used McNemar’s test to do pathogen-specific analyses, and generalised estimating equations (GEE) for the global (ie, any) pathogen analyses, with adjustments made for the presence of diarrhoea, location, and their interactions with specimen type.
Findings
Between Dec 12, 2014, and Aug 31, 2016, we studied 1519 eligible participants, 1147 (76%) of whom provided stool specimens and 1514 (>99%) provided swab specimens. 871 (76%) of 1147 stool specimens and 1024 (68%) of 1514 swabs were positive for any pathogen (p<0·0001). Comparative yield adjusted odds ratios (ORs) for stool specimens relative to swabs were 1·24 (95% CI 1·11–1·38) in children with diarrhoea at presentation and 1·76 (1·47–2·11) in children without diarrhoea. GEE analysis identified an interaction between the presence of diarrhoea and specimen type (p=0·0011) and collection location (p=0·0078). In an overall yield analysis, pathogen yield was 57% (871 of 1519 children) for stool specimens and 67% (1024 of 1519 children) for rectal swabs, with an unadjusted OR of 0·65 (95% CI 0·59–0·72) for stool relative to swab.
Interpretation
Rectal swabs should be done when enteropathogen identification and rapid detection are needed, appropriate molecular diagnostic technology is available, and a stool specimen is not immediately available. In view of their high yield, we urge that the recommendation against the use of rectal swabs as diagnostic specimens be reconsidered.
Introduction
Microbiological diagnoses in children with vomiting or diarrhoea, or both, provide clarity, guide treatment, and prompt public health responses. The pathogen-specific burden of disease estimates help to prioritise public health interventions.1 Traditionally, testing for entero pathogens has relied on analysis of diarrhoeal stool specimens. However, some laboratories will not test stool specimens if the consistency is incompatible with diarrhoea, thereby preventing enteropathogen identification in patients with vomiting in the absence of diarrhoea. Stool collection and transportation are burdensome and increase the potential for disease transmission. Additionally, waiting for stool while patients are on site is impractical and post-visit return rates are poor, even in children with diarrhoea,2 leading to delays or missed diagnostic opportunities that can adversely affect outcomes.3 Point-of-care acquisition of rectal swabs might overcome these barriers,4 but there have been few comparative analyses of stool versus swabs,4–9 and none that have included children with isolated vomiting. Currently, there is a recommendation against the use of rectal swabs as a diagnostic specimen.10
As we enter an era of sensitive, rapid, nucleic acid amplification testing, an increasingly large proportion of the time-to-result interval reflects the components of specimen collection and transportation. Although rectal swabs can be obtained expeditiously at the point of care, their diagnostic yield compared with that of stool specimens is unclear. Therefore, we compared enteropathogen identification yields from rectal swabs and stool specimens in an outpatient cohort of children with vomiting or diarrhoea, or both.
Methods
Study design and participants
Three cohorts of participants were consecutively recruited by the Alberta Provincial Pediatric Enteric Infection Team:11 (1) children with vomiting or diarrhoea in Pediatric Emergency Research Canada (PERC) emergency departments in Calgary and Edmonton (AB, Canada), and children in these departments with non-infectious illness whose caregivers agreed to submit specimens if they later developed vomiting or diarrhoea; (2) children receiving routine vaccinations at a Calgary public health clinic whose caregivers agreed to submit specimens if vomiting or diarrhoea developed later; and (3) symptomatic children identified via a province-wide nursing triage telephone resource called HealthLinkwho met triage criteria for the provision of care at home instead of seeking medical care.12 Consent in this cohort was provided by telephone. Approvals were obtained from the University of Calgary and University of Alberta research ethics boards.
Eligible children were younger than 18 years and had at least three episodes of vomiting or diarrhoea in the preceding 24 h and fewer than 7 days of symptoms.13 We excluded children enrolled in this study in emergency departments within the previous 14 days or who were unable to complete follow-up, and those with current or past psychiatric illness, neutropenia, or requiring emergent medical intervention. Informed consent was provided by caregivers; assent was obtained from the participants themselves when they were deemed to be mature enough to understand the study procedures and the potential benefits and harms.
Specimen acquisition, locations, and processes
For symptomatic children with vomiting or diarrhoea in the emergency departments, two rectal swabs were collected from each participant: a flocked swab and a FecalSwab (both from Copan Italia, Brescia, Italy)—each was inserted sequentially into the rectum and rotated once through 360°. Flocked swabs were transported in a sterile tube and FecalSwabs in 2 mL modified Cary Blair transport media. Stool specimens were collected in sterile containers (V302-F, Starplex Scientific, ON, Canada). If a stool specimen was not provided before discharge from the emergency department, caregivers collected stool at home. For asymptomatic children11 in the emergency departments or vaccination clinics who developed vomiting or diarrhoea later, specimen collection kits were provided for the collection of stool and rectal swab samples when symptoms developed. For symptomatic children assessed via Health Link, care-givers collected samples using specimen collection kits consisting of two rectal swabs, a stool container, and instructions that were couriered to their homes.
14 days after enrolment, we used a standardised data collection form to obtain follow-up information using phone or electronic surveys.11 The electronic surveys were emailed daily (up to three times) until completed. If follow-up was not completed after three emails, we did a telephone follow-up. The survey included details regarding the ease of rectal swab use and acceptability of this specimen collection approach.
For stool specimens and FecalSwabs collected in the emergency departments, enteric culture was done upon receipt at the laboratory. Stools collected at home were stored at room temperature for up to 12 h, then retrieved by a study-funded courier and transported to the laboratory on ice packs. After doing enteric culture, the remaining stool samples and dry rectal swabs were stored at −80°C until analysed with nucleic acid amplification testing.
Molecular testing
Flocked dry rectal swabs were placed into 750 μL of phosphate-buffered saline (PBS; Life Technologies, Carlsbad, CA, USA). 100–150 mg of solid stool, 100 μL of liquid stool, or 300 μL of dry rectal swab suspension with the PBS was added to Bertin SK38 soil grinding lysis bead tubes with 10 μL of bacteriophage MS2 (both Luminex Molecular Diagnostics, ON, Canada) to a final volume of 1000 μL. Total nucleic acid was extracted and eluted in 70 μL using the NucliSENS easyMag extractor (bioMérieux, Marcy-l’Étoile, France) according to manufacturer’s instructions and stored at −80°C.
We used a real-time PCR in-house gastroenteritis virus panel (GVP)14 that detects norovirus GI and GII, group A rotavirus, adenovirus (all serotypes), sapovirus, and astrovirus; and a multianalyte assay (Luminex xTAG gastrointestinal pathogen panel, Luminex Molecular Diagnostics, ON, Canada). The gastroenteritis virus panel assay incorporates reverse transcription with three Taqman-probe based duplex real-time PCR reactions, modified from a previous publication:14 5 μL of nucleic acid extracts were used to generate 20 μL of complementary DNA by reverse transcription reactions.14 Each duplex real-time PCR reaction containing 3·5 μL of complementary DNA in a 10 μL reaction was done with the 7500 Fast real-time PCR System (Applied Biosystems, Foster City, CA, USA). Cycle threshold values of 38 or lower were considered positive, with values inversely proportional to viral target density. Luminex xTAG gastrointestinal pathogen panel,6,15 is a bead-based assay that incorporates multiplex real-time-PCR with a hybridisation-based universal tag sorting system, and detects group A rotavirus, norovirus GI and GII, adenovirus 40 and 41, Campylobacter spp, Clostridium difficile, Cryptosporidium, Entamoeba histolytica, Escherichia coli O157, enterotoxigenic E coli, Giardia spp, Salmonella spp, Shiga toxin-producing E coli, Shigella spp, Vibrio cholera, and Yersinia enterocolitica.16 10 μL of nucleic acids was used in gastrointestinal pathogen panel testing.
Enteric bacterial culture was done on submitted stool specimens and rectal swab specimens following routine procedures17 for isolation of Aeromonas, Campylobacter, E coli O157:H7, Salmonella, Shigella, and Yersinia (appendix p 2). Stool specimens weighing less than 1 g were considered insufficient and not cultured. For rectal swabs, tubes were vortexed and 100 μL of the modified Cary Blair medium was plated and streaked for isolation. For enrichment broths, 200 μg of solid stool or around 200 μL of liquid stool was used. All stool specimens and rectal swabs were plated on the following agars: sheep blood agar, MacConkey agar with crystal violet, Hektoen agar, Colorex O157 agar with 2·5 mg/L potassium tellurite (Alere), Yersinia (CIN) agar (Dalynn Biologicals, Calgary), Campylobacter blood free agar, and mannitolselenite broth, all supplied by Dalynn Biologicals, Calgary. After 24 h enrichment at 35°C, the broth was plated to Salmonella-Shigella and Wilson Blair agar (ProvLab). Campylobacter plates were incubated under microaerophilic conditions (42°C), and all other media were incubated at 35°C ± 2°. The duration of incubation to designate a specimen as negative ranged from 24 h (Yersinia and E coli O157), to 72 h (Shigella and Campylobacter), to 96 h (Salmonella).
Outcomes
This study had two primary outcomes: comparative yield, calculated as the proportion of paired specimens in which at least one pathogen was identified, and overall yield, calculated as the proportion of study participants in whom at least one pathogen was identified. Comparative yield included only paired stool specimens and rectal swabs (ie, from participants who submitted both specimen types). Overall yield included all eligible study participants as the denominator, with unsubmitted specimens scored as negative. Secondary outcomes included agreement between diagnoses using stool specimens versus rectal swabs, and real-time PCR cycle threshold values between paired specimens.
Statistical analysis
We did not do any formal sample size calculations. All specimens, regardless of location of collection, underwent identical testing. Although FecalSwabs and dry swabs were entered into different testing pathways, their combined testing protocol was identical to that of stool specimens so they were analysed as a single unit. All specimens were tested for 18 unique targets (the five viruses in the gastroenteritis virus panel; and the three viruses, nine bacteria, and three parasites in the gastrointestinal pathogen panel; and six bacteria based on cultures). In the absence of a reference standard, or an adequate test to resolve discrepant analyses, sensitivity and specificity values could not be calculated.18
McNemar’s test was used in the pathogen-specific analyses for comparative and overall yields. Because multiple specimens (ie, repeated measures) were collected from the same participant, we used generalised estimating equations (GEEs) with exchangeable correlation structures in the global pathogen analyses (ie, of any pathogen identified). In the subgroups of children with diarrhoea and isolated vomiting, the proportions of specimens positive for any pathogen were compared using GEE without adjustment. In the global pathogen analyses for comparative and overall yields, GEEs were adjusted for the presence of diarrhoea, location, and their interactions with specimen types. GEE models accounted for location of specimen collection as a proxy for the individual who did the rectal swab (ie, health-care professional or caregiver) and the presence of diarrhoea. Pairwise interactions between specimen (swab or stool), location, and diarrhoea at presentation—all three pairings with all permutations—were included in the models. Calculations were repeated as an exploratory analysis with C difficile-positive specimens classified as negative in children younger than 2 years, and restricted to paired specimens obtained within 24 h of each other.19 Other exploratory analyses examined details from the follow-up survey about the ease of rectal swab use and the acceptability of collecting rectal swabs compared with collecting stool specimens.
Agreement was assessed for paired specimens with result concordance computed by Cohen’s κ and interpreted as slight (0·00–0·20), fair (0·21–0·40), moderate (0·41–0·60), substantial (0·61–0·80), or almost perfect (0·81–1·00).20 We did the calculation for all pathogens identified in more than 25 cases. We measured correlations between cycle threshold values of positive GVP tests using Pearson correlation coefficients, and compared between paired specimens with the Wilcoxon signed rank test.
We did not use multiple imputations in our calculations because only 14 participants had incomplete clinical data.21 Analyses were done using SPSS version 22.0. We calculated two-tailed p values and set the significance level α at 0·05. To control for false discovery, we corrected p values using the Benjamini-Hochberg method within sets of tests.22
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
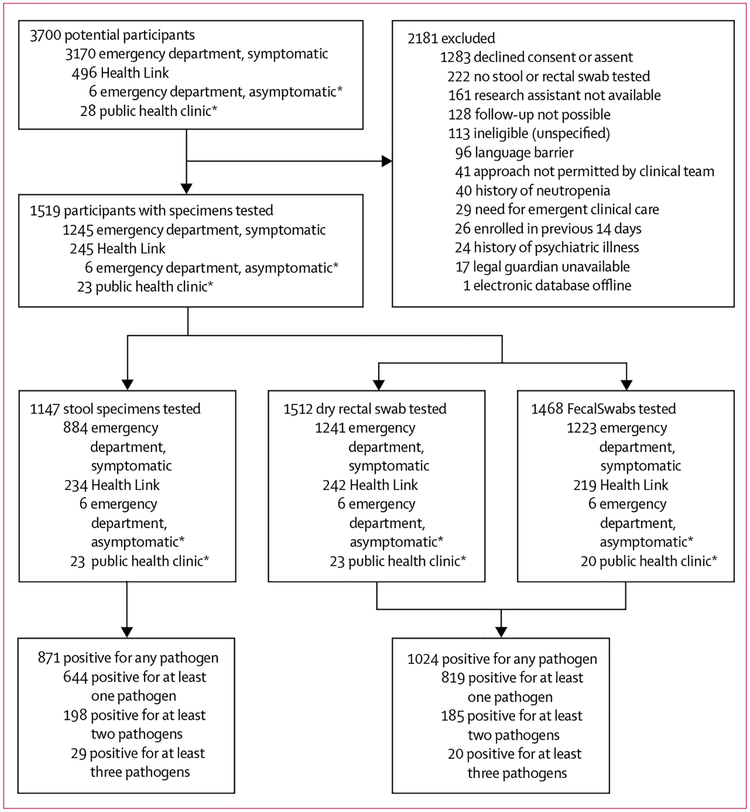
Between Dec 12, 2014, and Aug 31, 2016, 1519 eligible participants were included (figure, table 1), who submitted a rectal swab, a stool specimen, or both for testing. Of these 1519 participants, 1147 (76%) of 1519 provided stool specimens, 1514 (>99%) provided swab specimens: either a dry rectal swab (n=1512) or FecalSwab (n=1468), and 1511 (>99%) provided information about clinical symptoms (eg, vomiting and diarrhoea history). Median age was 1·6 years (IQR 0·94–3·3); at enrolment, 89% (1342/1511) reported vomiting and 67% (1015/1511) reported diarrhoea (table 1).
Figure: Trial profile.
*Children enrolled while in the emergency department or public health clinic without infectious symptoms who submitted specimens at a later time when they met study eligibility criteria.
Table 1:
Demographic characteristics of the cohort at the time of enrolment
| All patients (N=1519) | Rectal swab and stool specimen (N=1142) | Rectal swab only (N=372) | Stool specimen only (N=5) | |||||
|---|---|---|---|---|---|---|---|---|
| n | Value | n | Value | n | Value | n | Value | |
| Age (years) | 1519 | 1·6 (0·94–3·30) | 1142 | 1·5 (0·87–2·88) | 372 | 2·1 (1·10–4·50) | 5 | 1·4 (0·86–4·20) |
| Enrolled in emergency department | 1519 | 1245 (82%) | 1142 | 882 (77%) | 372 | 361 (97%) | 5 | 2 (40%) |
| Vomiting | 1511 | 1342 (89%) | 1136 | 998 (88%) | 371 | 341 (92%) | 4 | 3 (75%) |
| Number of vomiting episodes in previous 24 h* | 1338 | 5 (3–8) | 994 | 4 (2–8) | 341 | 6 (3–10) | 3 | 4 (range 1–5) |
| Vomiting duration at time of enrolment (h)* | 1341 | 38·0 (13·6–76·9) | 998 | 39·9 (14·7–78·6) | 340 | 32·1 (11·9–68·7) | 3 | 33·7 (range 11·8–36·3) |
| Diarrhoea | 1511 | 1015 (67%) | 1136 | 812 (72%) | 371 | 199 (54%) | 4 | 4 (100%) |
| Number of diarrhoea episodes in previous 24 h† | 1014 | 4 (2–7) | 811 | 4 (2–7) | 199 | 4 (2–7) | 4 | 7 (6–32) |
| Diarrhoea duration at time of enrolment (h)† | 1013 | 52·5 (22·9–93·0) | 810 | 53·7 (24·3–95·2) | 199 | 45·6 (18·1–88·6) | 4 | 34·0 (31·9–42·1) |
| Received rotavirus vaccine | 1511 | 425 (28%) | 1136 | 322 (28%) | 371 | 101 (27%) | 4 | 2 (50%) |
| Rectal swab dry | 1519 | 1512 (>99%) | NA | NA | NA | NA | NA | NA |
| Rectal fecal swabs | 1519 | 1468 (97%) | NA | NA | NA | NA | NA | NA |
| Stool specimen | 1519 | 1147 (76%) | NA | NA | NA | NA | NA | NA |
Data are median (IQR) or n (%), except where indicated. The cohort included all eligible children who submitted a rectal swab or a stool specimen, or both. NA=not applicable.
Among children who indicated presence of vomiting.
Among children who indicated presence of diarrhoea.
871 (76%) of 1147 stool specimens and 1024 (68%) of 1514 swabs were positive for any pathogen (p<0·0001; appendix p 3). 1015 (67%) of 1511 children presented with diarrhoea, and pathogen detection was achieved with 657 (81%) of 816 stool specimens and 778 (77%) of 1011 rectal swabs; the unadjusted odds ratio (OR) was 1·24 (95% CI 1·11–1·38, p=0·0001). Among the 497 participants with isolated vomiting (ie, vomiting in the absence of diarrhoea), a pathogen was detected in 209 (65%) of 324 stool specimens and 243 (49%) of 496 rectal swabs; the unadjusted OR was 1·77 (1·50–2·10; p<0·0001).
Among paired samples (appendix p 4), the comparative yield from stool specimens was higher than that from rectal swabs (866 [76%] of 1142 paired swab samples tested positive vs 793 [69%] of 1142 paired rectal swabs; unadjusted OR 1·38, 95% CI 1·26–1·51; table 2). GEE analysis identified an interaction between presence of diarrhoea and specimen type (p=0·0011), and collection location (p=0·0078) on pathogen detection.
Table 2:
Pathogen yields in relation to specimen type
| Odds ratio (95% CI)* | |
|---|---|
| Unadjusted comparative yield of at least one pathogen† | 1·38 (1·26–1·51) |
| Comparative yield adjusted for interaction | |
| With diarrhoea | 1·24 (1·11–1·38) |
| Without diarrhoea | 1·76 (1·47–2·11) |
| Unadjusted overall yield of at least one pathogen‡ | 0·65 (0·59–0·72) |
| Overall yield adjusted for interaction | |
| Emergency department with diarrhoea | 0·50 (0·43–0·58) |
| Emergency department without diarrhoea | 0·72 (0·60–0·87) |
| Home with diarrhoea | 0·94 (0·75–1·17) |
| Home without diarrhoea | 1·37 (1·03–1·92) |
Specimens were rectal swabs or stool specimens.
Odds ratios represent stool relative to rectal swab modelled to account for clustering by child, using the outcome of a positive test for at least one pathogen from either specimen as the dependent variable with generalised estimating equations containing an exchangeable correlation structure.
Although 1147 children submitted stool specimens (table 1), only 1142 had paired rectal swabs.
Proportion of study participants in whom at least one pathogen was identified by specimen type using the number of eligible study participants as the denominator. Missing specimens were defined as negative in this analysis to enable a pragmatic assessment of the specimens.
The overall concordance analysis yielded a κ of 0·76 (95% CI 0·71–0·80; appendix p 5). κ values were greater for viruses than with bacteria (0·82, 0·79–0·86 vs 0·74, 0·68–0·80). Pathogen-specific analysis showed that rotavirus had the highest κ value (0·95, 0·93–0·97) and C difficile the lowest (0·76, 0·70–0·82; appendix p 6).
Paired positive viral specimens had lower median cycle threshold values (ie, higher viral loads; p<0·0001) in stool specimens compared with swabs for all viruses (appendix p 7). The overall correlation between cycle threshold values was r=0·66 (appendix p 11). When cycle threshold values were compared between the presence or absence of diarrhoea, higher values were present for rotavirus (in rectal swabs and stool specimens) and astrovirus (in rectal swabs) when diarrhoea was absent (appendix p 8).
Overall pathogen yield was 57% (871/1519) and 67% (1024/1519) for stool samples and rectal swabs respectively (unadjusted OR 0·65; 95% CI 0·59–0·72; tables 2, 3). GEE analysis identified significant inter action between specimen type and presence of diarrhoea (p=0·0019), specimen type and collection location (p<0·0001), and presence of diarrhea and collection of location (p=0·023) on pathogen detection. Adjusted OR for identifying a pathogen in stool samples relative to rectal swabs ranged from 0·50 (95% CI 0·43–0·58; emergency department with diarrhoea) to 0·72 (0·60–0·87; emergency department without diarrhoea) to 0·94 (0·75–1·17; home with diarrhoea) to 1·37 (1·03–1·92; home without diarrhoea; table 2).
Table 3:
Overall yields in relation to specimen type, for any pathogen, for the entire cohort (n=1519)
| Either stool or swab positive | Rectal swab positive | Stool positive | p value* | |
|---|---|---|---|---|
| Any enteropathogen | 1121 (74%) | 1024 (67%) | 871 (57%) | <0·0001† |
| Any viral enteropathogen | 1025 (67%) | 940 (62%) | 802 (53%) | <0·0001† |
| Adenovirus | 241 (16%) | 180 (12%) | 195 (13%) | 0·176 |
| Astrovirus | 40 (3%) | 34 (2%) | 34 (2%) | >0·999 |
| Norovirus GI/GII | 374 (25%) | 325 (21%) | 289 (19%) | 0·0024† |
| Rotavirus | 400 (26%) | 380 (25%) | 320 (21%) | <0·0001† |
| Sapovirus | 126 (8%) | 116 (8%) | 96 (6%) | 0·0022† |
| Any bacterial enteropathogen | 252 (17%) | 206 (14%) | 180 (12%) | 0·021 |
| Aeromonas spp | 16 (1%) | 9 (1%) | 7 (<1%) | 0·804 |
| Campylobacter spp | 11 (1%) | 10 (1%) | 6 (<1%) | 0·219 |
| Clostridium difficile tcdA/B | 174 (11%) | 148 (10%) | 121 (8%) | 0·0032† |
| Escherichia coli O157:H7 | 7 (<1%) | 6 (<1%) | 6 (<1%) | >0·999 |
| Escherichia coli O26:H11 | 1 (<1%) | 0 | 1 (<1%) | NA |
| Enterotoxigenic Escherichia coli LT/ST | 4 (<1%) | 2 (<1%) | 3 (<1%) | >0·999 |
| Salmonella | 27 (2%) | 20 (1%) | 22 (1%) | 0·774 |
| Shigella | 5 (<1%) | 5 (<1%) | 4 (<1%) | >0·999 |
| stx 1/stx 2 | 19 (1%) | 13 (1%) | 16 (1%) | 0·508 |
| Vibrio cholerae | 0 | 0 | 0 | NA |
| Yersinia enterocolitica | 4 (<1%) | 2 (<1%) | 3 (<1%) | >0·999 |
| Any parasite | 7 (<1%) | 1 (<1%) | 6 (<1%) | 0·125 |
| Cryptosporidium | 0 | 0 | 0 | NA |
| Entamoeba | 3 (<1%) | 0 | 3 (<1%) | NA |
| Giardia | 4 (<1%) | 1 (<1%) | 3 (<1%) | 0·625 |
Data are n (%). Overall yields are for all specimens, with unsubmitted specimens analysed as negative. The analysis assumed that missing stool specimens (n=372) or rectal swabs (n=5) tested negative for enteropathogens. NA=not applicable.
p value for McNemar test. p value for summary measures (any pathogen, virus, bacteria, and parasite) adjusted using Benjamini-Hochberg procedure (n=4) and significance was determined separately from those of the individual pathogen targets (n=20).
Significant after correction via Benjamini-Hochberg procedure for multiple comparisons.
Comparative and overall yields were unchanged when repeated with C difficile considered as negative (appendix p 9) and when restricted to paired specimens collected within 24 h of each other (appendix p 10). Rectal swabs were reported as easy to do by 1386 (93%) of the 1494 individuals who did the collection; however, emergency department clinicians reported that they were easy to do more often than caregivers (1176 [95%] of 1237 vs 210 [82%] of 257; p<0·0001). Of 1363 caregivers who responded to a question comparing the entire process (ie, not the actual performance) of rectal swabs versus the collection of stool specimens, 54 (4%) reported it as not acceptable, 79 (6%) as slightly not acceptable, 246 (18%) as neutral, 112 (8%) as slightly acceptable, and 872 (64%) as acceptable.
Discussion
In this large cohort study, we identified a slightly higher comparative yield of at least one pathogen with stool specimens than with rectal swabs, particularly in children with isolated vomiting. However, when con sidering the entire cohort, the overall pathogen yield (unsubmitted specimens analysed as negative) with rectal swabs was 10 percentage points higher than with stool specimens because fewer bulk (or cup) stools were submitted. Rectal swabs are easy to do, generally well accepted, have high diagnostic utility, and should be considered when enteropathogen identification is needed and a stool specimen is unavailable or unlikely to be submitted.
We anticipated contradictory findings (ie, ORs in opposite directions) for the primary outcomes of comparative and overall yield because we hypothesised that rectal swabs would have similar diagnostic test characteristics as stool specimens but that potentially many more swabs than stool specimens would be submitted. Despite stool specimens being submitted for 76% of participants, which greatly exceeds submissions in previously reported studies2,23,24 likely due to use of a study-funded courier, rectal swabs still had a higher overall pathogen yield. Similarly high specimen sub mission rates have been shown with courier use to identify infectious agents in outbreaks.25
Although previous studies assessing rectal swab yields to detect enteropathogens have included children,5,6,8,9 most of these focused on patients admitted to hospital5–8 and were done in low-income and middle-income countries.5,6,8,9 Two similar studies have been done in an emergency department setting,24,26 but to our knowledge ours is the first to include children with isolated vomiting. Although our findings are consistent with most previous reports,5–8,26 they differ from the only North American study based in the emergency department setting24 that assessed unpaired specimens in 364 adults and identified an enteropathogen in 49% of stool specimens and in only 9% of rectal swabs. Similarly, a paediatric study of unpaired samples reported a lower pathogen detection rate in rectal swabs.27 In addition to using paired specimens and including children with isolated vomiting, our pathogen detection values might have been higher because we tested for two additional viruses than did the other studies, and used nucleic acid amplification test technologies to identify bacteria.
Professional organisations have recommended test ing diarrhoeal stool specimens for enteropathogens in lieu of formed stools or swab samples.10 Indeed, when compared head-to-head, stool specimens are superior to swab samples, probably because of the smaller amount of faecal material collected with rectal swabs. The higher rectal swab cycle-threshold values, particularly among discordant samples,4 probably reflect a smaller amount of faecal material and the dilution with buffers to elute material for nucleic acid extraction.14,16 This finding is highlighted by the higher cycle threshold values in children with isolated vomiting (appendix p 8). If the lower sensitivity of rectal swabs is due to the lower amount of stool, perhaps modified extraction methods can remedy this deficiency to improve their comparative yield.7,9,28 Moreover, with highly automated,29 1-h run time syndromic panels now available,30 challenges of specimen collection, handling, and transportation are increasingly the rate-limiting steps. In view of the high yield and agreement of rectal swabs, ease and acceptability of sample collection, lower biohazard exposure, timeliness, and ability to obtain specimens from individuals with isolated vomiting, we urge that the recommendation against the use of rectal swabs as a diagnostic specimen be reconsidered.
For many enteropathogens the strength of association with disease increases with greater pathogen loads.4 Moreover, swab specimens are more likely to collect mucosal adherent microorganisms (suggesting a pathogenic role), while stool specimens contain those that exist freely within the lumen. Thus in children with discordant, stool positive-swab negative results and relatively low pathogen abundance (high cycle threshold counts), the detected pathogens might represent non-disease states.29
The limitation of using stool specimens is likely underestimated in this study because real-world stool submission rates, in the absence of a courier system, are significantly lower. Additionally, the collection of many stool specimens would be delayed compared with collection of rectal swabs. Future research, incorporating a cost-benefit analysis, should assess the added benefit of diagnosis based on rectal swabs when a stool specimen is unavailable. A key aspect of such work should include a focus on actions taken, treatment decisions, and outcomes that were changed as a result of the micro biological analyses. Additionally, future research should assess whether the presence of visible faecal material on rectal swabs is associated with specimen adequacy and yield, because evidence addressing this issue is not available.
There were few bacterial and parasitic pathogens identified in our cohort, and thus more evidence is required regarding the use of rectal swabs for such enteropathogens. Additionally, although a broad range of enteropathogens was sought, some of the detected organisms are not always the cause of the disease, most particularly C difficile. Additionally, the lack of control data limits conclusions that can be drawn regarding the pathogenicity of individual organisms. Lastly, our overall yield analysis, in which unsubmitted specimens were analysed as negative, needs to be interpreted for clinical applicability in context.
In conclusion, in children with vomiting or diarrhoea, or both, rectal swabs have an approximately 10 percentage point greater chance of enteropathogen identification compared with stool specimens, despite stool specimens having a higher comparative yield when compared within the same individual. Because rectal swab specimens are easy to obtain and are more likely to be submitted, they can be used to expedite diagnosis, and minimise the burden on families when enteropathogen identification is needed, the appropriate technology is available, and stool specimens are unavailable or unlikely to be submitted. Given the importance of patient preferences and of the cost of diagnostics to health systems, it will be important in future work to understand how to optimally acquire specimens and maximise patient and family satis faction, and to establish the role of syndromic molecular panels in the diagnosis of gastrointestinal disease.
Supplementary Material
Research in context
Evidence before this study
We did a PubMed search for studies assessing rectal swabs’ diagnostic utility using the terms “accuracy”, “yield”, or “diagnosis”, in combination with “rectal swab” and “gastroenteritis” on Feb 5, 2017, without date or language restrictions. From this search we identified 44 publications. Several reports described the use of rectal swabs, but none were large cohort studies of outpatient children with vomiting or diarrhoea, or both, and none were done in high-income countries comparing paired stool and swab specimens using broad diagnostic syndromic panels. Previous studies described small cohorts of individuals admitted to hospital with diarrhoea (ie, excluding those with vomiting in the absence of diarrhoea) from low-income and middle-income countries, in whom only a limited range of pathogens were sought. Although these early studies reported that rectal swabs had comparatively lower sensitivity than stool specimens, the recent advent of flocked swabs and the introduction of molecular diagnostic approaches necessitate a re-evaluation of rectal swab diagnostic test characteristics. Thus, we sought to compare the diagnostic sensitivity of rectal swabs with that of stool specimens for enteropathogen identification in an outpatient cohort of children with diarrhoea or vomiting, or both.
Added value of this study
Our findings from this large cohort of outpatient children showed that although pathogens were identified in a greater proportion of stool specimens among participants submitting both stool specimens and rectal swabs, use of rectal swabs increased the overall yield by 10 percentage points. This analysis incorporated the ability of a patient to submit a specimen for analysis, which, despite the use of a study-funded courier service to maximise the submissions of stool specimens, was significantly greater for rectal swabs.
Implications of all the available evidence
When paired with stool specimens from the same participants, rectal swabs had lower diagnostic yields of pathogens, but had greater absolute yields when the pragmatic consideration of lower stool specimen submission rates was taken into account. Thus, when stool is not immediately available and enteropathogen identification is needed, rectal swabs are a suitable diagnostic alternative.
Acknowledgments
This research was supported by APPETITE, which is funded by a grant from the Alberta Innovates–Health Solutions Team Collaborative Innovation Opportunity. PIT is also supported by grant number NIH P30DK052574 (Digestive Diseases Research Core Center). APPETITE is also supported by the Alberta Children’s Hospital Research Institute (Calgary, AB, Canada) and the Women and Children’s Health Research Institute (Edmonton, AB, Canada), through a Partnership Award. SBF is supported by the Alberta Children’s Hospital Foundation Professorship in Child Health and Wellness. The Pediatric Emergency Medicine Research Associate Program (PEMRAP) is supported by a grant from the Alberta Children’s Hospital Foundation. We thank the patients and their families for cooperating with our study, Bryanne Crago, Christina Ferrato from Calgary Laboratory Services, and Judy Qui from the Department of Laboratory Medicine and Pathology, University of Alberta, DynaLIFE Dx Diagnostic Laboratory Services, community laboratories as well as Provincial Laboratory for Public Health (ProvLab), Edmonton and Calgary, especially the bacteriology staff for their assistance with receiving, handling, and processing specimens; the emergency department research nurses and PEMRAP at the Alberta Children’s Hospital for recruiting study participants; the emergency department bedside nurses for assisting with rectal swab performance; Nadia Dow and Manasi Rajagopal, as well as the research assistants, research nurses, and the Little Bit of Help research volunteer programme for their assistance with participant recruitment at the Stollery Children’s Hospital; the nurses at Health Link who responded to calls from across the province for their assistance with participant recruitment; and Laurel Ryan for her role as a patient adviser. We would like to extend special thanks to Marie Louie for building the connections that have made our endeavours possible, and Carey-Ann Burnham for her guidance and review of our manuscript. No compensation for the assistance of any aforementioned individuals was provided.
Funding Alberta Innovates—Health Solutions Team Collaborative Research Innovation Opportunity.
Footnotes
Declaration of interests
SBF reports grants from Alberta Innovates–Health Solutions, the Alberta Children’s Hospital Research Institute, the Women and Children’s Partnership Award Health Research Institute, and the Alberta Children’s Hospital Foundation, and non-financial support from Calgary Laboratory Services, ProvLab Alberta, Luminex Corporation, and Copan Italia. PIT reports personal fees and other funding from MediBeacon Inc. All other authors declare no competing interests.
References
- 1.Caliendo AM, Gilbert DN, Ginocchio CC, et al. Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis 2013; 57 (suppl 3): S139–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein EJ, Boster DR, Stapp JR, et al. Diarrhea etiology in a children’s hospital emergency department: a prospective cohort study. Clin Infect Dis 2006; 43: 807–13. [DOI] [PubMed] [Google Scholar]
- 3.Freedman SB, Eltorki M, Chui L, et al. Province-wide review of pediatric Shiga toxin-producing Escherichia coli case management. J Pediatr 2017; 180: 184–90. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Gratz J, Amour C, et al. Optimization of quantitative PCR methods for enteropathogen detection. PLoS One 2016; 11: e0158199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arvelo W, Hall AJ, Estevez A, et al. Diagnostic performance of rectal swab versus bulk stool specimens for the detection of rotavirus and norovirus: implications for outbreak investigations. J Clin Virol 2013; 58: 678–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldfarb DM, Steenhoff AP, Pernica JM, et al. Evaluation of anatomically designed flocked rectal swabs for molecular detection of enteric pathogens in children admitted to hospital with severe gastroenteritis in Botswana. J Clin Microbiol 2014; 52: 3922–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gustavsson L, Westin J, Andersson LM, Lindh M. Rectal swabs can be used for diagnosis of viral gastroenteritis with a multiple real-time PCR assay. J Clin Virol 2011; 51: 279–82. [DOI] [PubMed] [Google Scholar]
- 8.Rodrigues A, de Carvalho M, Monteiro S, et al. Hospital surveillance of rotavirus infection and nosocomial transmission of rotavirus disease among children in Guinea-Bissau. Pediatr Infect Dis J 2007; 26: 233–37. [DOI] [PubMed] [Google Scholar]
- 9.Kabayiza JC, Andersson ME, Welinder-Olsson C, Bergstrom T, Muhirwa G, Lindh M. Comparison of rectal swabs and faeces for real-time PCR detection of enteric agents in Rwandan children with gastroenteritis. BMC Infect Dis 2013; 13: 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baron EJ, Miller JM, Weinstein MP, et al. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2013 recommendations by the Infectious Diseases Society of America (IDSA) and the American Society for Microbiology (ASM) (a). Clin Infect Dis 2013; 57: e22–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freedman SB, Lee BE, Louie M, et al. Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE): epidemiology, emerging organisms, and economics. BMC Pediatr 2015; 15: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letourneau S Health Link Alberta: a model for successful health service integration. Health Q 2009; 13: 56–60. [DOI] [PubMed] [Google Scholar]
- 13.Guarino A, Albano F, Ashkenazi S, et al. European Society for Paediatric Gastroenterology, Hepatology, and Nutrition/European Society for Paediatric Infectious Diseases: evidence-based guidelines for the management of acute gastroenteritis in children in Europe. J Pediatr Gastroenterol Nutr 2008; 46 (suppl 2): S81–122. [DOI] [PubMed] [Google Scholar]
- 14.Pang X-L, Preiksaitis JK, Lee BE. Enhanced enteric virus detection in sporadic gastroenteritis using a multi-target real-time PCR panel: a one-year study. J Med Virol 2014; 86: 1594–601. [DOI] [PubMed] [Google Scholar]
- 15.Claas EC, Burnham CA, Mazzulli T, Templeton K, Topin F. Performance of the xTAG(R) gastrointestinal pathogen panel, a multiplex molecular assay for simultaneous detection of bacterial, viral, and parasitic causes of infectious gastroenteritis. J Microbiol Biotechnol 2013; 23: 1041–45. [DOI] [PubMed] [Google Scholar]
- 16.Perry MD, Corden SA, Howe RA. Evaluation of the Luminex xTAG gastrointestinal pathogen panel and the Savyon Diagnostics gastrointestinal infection panel for the detection of enteric pathogens in clinical samples. J Med Microbiol 2014; 63: 1419–26. [DOI] [PubMed] [Google Scholar]
- 17.Leber AL. Clinical Microbiology Procedures Handbook, 4th edn. ASM Press, Washington DC, 2016. [Google Scholar]
- 18.Glasziou P, Irwig L, Deeks JJ. When should a new test become the current reference standard? Ann Intern Med 2008; 149: 816–22. [DOI] [PubMed] [Google Scholar]
- 19.Sammons JS, Toltzis P. Pitfalls in diagnosis of pediatric Clostridium difficile infection. Infect Dis Clin North Am 2015; 29: 465–76. [DOI] [PubMed] [Google Scholar]
- 20.Feuerman M, Miller AR. Relationships between statistical measures of agreement: sensitivity, specificity and kappa. J Eval Clin Pract 2008; 14: 930–33. [DOI] [PubMed] [Google Scholar]
- 21.Bennett DA. How can I deal with missing data in my study? Aust N Z J Public Health 2001; 25: 464–69. [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 1995; 57: 289–300. [Google Scholar]
- 23.Hall AJ, Rosenthal M, Gregoricus N, et al. Incidence of acute gastroenteritis and role of norovirus, Georgia, USA, 2004–2005. Emerg Infect Dis 2011; 17: 1381–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bresee JS, Marcus R, Venezia RA, et al. The etiology of severe acute gastroenteritis among adults visiting emergency departments in the United States. J Infect Dis 2012; 205: 1374–81. [DOI] [PubMed] [Google Scholar]
- 25.Jones TF, Bulens SN, Gettner S, et al. Use of stool collection kits delivered to patients can improve confirmation of etiology in foodborne disease outbreaks. Clin Infect Dis 2004; 39: 1454–59. [DOI] [PubMed] [Google Scholar]
- 26.Sidler JA, Kach R, Noppen C, et al. Rectal swab for detection of norovirus by real-time PCR: similar sensitivity compared to faecal specimens. Clin Microbiol Infect 2014; 20: O1017–19. [DOI] [PubMed] [Google Scholar]
- 27.Stockman LJ, Staat MA, Holloway M, et al. Optimum diagnostic assay and clinical specimen for routine rotavirus surveillance. J Clin Microbiol. 2008; 46: 1842–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips G, Lopman B, Tam CC, Iturriza-Gomara M, Brown D, Gray J. Diagnosing norovirus-associated infectious intestinal disease using viral load. BMC Infect Dis 2009; 9: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Kabir F, Manneh J, et al. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis 2014; 14: 716–24. [DOI] [PubMed] [Google Scholar]
- 30.Spina A, Kerr KG, Cormican M, et al. Spectrum of enteropathogens detected by the FilmArray GI Panel in a multicentre study of community-acquired gastroenteritis. Clin Microbiol Infect 2015; 21: 719–28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.