Abstract
Currently there are still no early biomarkers to detect infants with risk of autism spectrum disorder (ASD), which is mainly diagnosed based on behavior observations at three or four years old. Since intervention efforts may miss a critical developmental window after 2 years old, it is significant to identify imaging-based biomarkers for early diagnosis of ASD. Although some methods using magnetic resonance imaging (MRI) for brain disease prediction have been proposed in the last decade, few of them were developed for predicting ASD in early age. Inspired by deep multi-instance learning, in this paper, we propose a patch-level data-expanding strategy for multi-channel convolutional neural networks to automatically identify infants with risk of ASD in early age. Experiments were conducted on the National Database for Autism Research (NDAR), with results showing that our proposed method can significantly improve the performance of early diagnosis of ASD.
Keywords: Autism, Convolutional neural network, Early diagnosis Deep multi-instance learning
1. Introduction
Autism, or autism spectrum disorder (ASD), refers to a range of conditions characterized by challenges with social skills, repetitive behaviors, speech and nonverbal communication, as well as by unique strengths and differences. Globally, autism is estimated to affect 24.8 million people as of 2015 [1]. The diagnosis of ASD is mainly based on behaviors. Studies demonstrate that behavioral signs can begin to emerge as early as 6–12 months [2]. However, most professionals who specialize in diagnosing the disorder won’t attempt to make a definite diagnosis until 2 or 3 years old [3]. As a result, the window of opportunity for effective intervention may have passed when the disorder is detected. Thus, it is of great importance to detect ASD earlier in life for better intervention.
Magnetic resonance (MR) examination allows researchers and clinicians to noninvasively examine brain anatomy. Structural MR examination is widely used to investigate brain morphology because of its high contrast sensitivity and spatial resolution [4]. Imaging plays an increasingly pivotal role in early diagnosis and intervention of ASD. Many neuroscience studies on children and young adults with ASD demonstrate abnormalities in the hippocampus [5], precentral gyrus [6], and anterior cingulate gyrus [7]. These researches demonstrate that it is highly feasible to detect ASD in early age.
In recent years, deep learning has demonstrated outstanding performances in a wide range of computer vision and image analysis applications. Many methods based on convolutional neural networks (CNNs) [8] are proposed for MRI analysis, e.g., classification of the Alzheimer’s disease (AD). For example, Sarraf et al. [9] proposed a DeepAD for AD diagnosis, where the existing CNN architectures, LeNet [10] and GoogleNet [11], were used for MR brain scans on the slice level and subject level, respectively. But feature representations defined at whole-brain level may not be effective in characterizing early structural changes of the brain. Several patch-level (an intermediate scale between voxel level and region of interest (ROI)) features have been proposed to represent structural MR images for brain disease diagnosis. For example, a deep multi-instance learning framework for AD classification using MRI was proposed by Liu et al. [12], achieving promising results in AD diagnosis.
Compared with adult brains, the analysis of infant brains is more difficult due to the low tissue contrast caused by the largely immature myelination. Furthermore, the number of autistic subjects is usually very limited, making the task of computer-aided diagnosis of ASD more challenging.
Motivated by deep multi-instance learning [12], in this paper, we propose a patch-level data-expanding strategy in multi-channel CNN for prediction of ASD in early age. Figure 1 shows a schematic diagram of our method. We first preprocess MR images and then identify anatomical landmarks in a data-driven manner. In the patch extraction step, we utilize a patch-level data expanding strategy based on identified landmarks. In the last step, we use the multi-channel CNNs for ASD classification. Experimental results on 276 subjects with MRIs show that our method outperforms conventional methods in ASD classification.
Fig. 1.
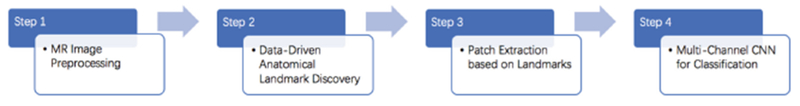
Block diagram of the proposed scheme for automatic classification of ASD.
2. Materials and Methods
Data description.
A total of 276 subjects gathered from National Database for Autism Research (NDAR) [13] were used in the study. More specifically, the dataset consists of 215 normal controls (NCs), 31 mild condition autism spectrum subjects, and 30 autistic subjects, as listed in Table 1. In the experiments, we regard the last two types as one group. All images were acquired at around 24 months of age on a Siemens 3T scanner, while subjects were naturally sleeping and fitted with ear protection, with their heads secured in a vacuum-fixation device. T1-weighted MR images were acquired with 160 sagittal slices using parameters: TR/TE = 2400/3.16 ms and voxel resolution = 1 × 1 × 1 mm3. T2-weighted MR images were obtained with 160 sagittal slices using parameters: TR/TE = 3200/499 ms and voxel resolution = 1 × 1 × 1 mm3.
Table 1.
Description of 24-month subjects in the NADR datasets.
| Category | Male/female |
|---|---|
| Autism | 25/5 |
| Autism spectrum | 21/10 |
| Normal control | 133/82 |
MRI Pre-processing.
We use in-house tools to perform skull stripping and histogram matching for T1 MR images. After skull stripping, we can estimate the whole brain volume for each subject. This pre-processing step is important for subsequent analysis steps.
Data-Driven Anatomical Landmark Identification.
To extract informative patches from MRI for model training, we first identify discriminative ASD-related landmark locations using a data-driven landmark discovery algorithm [14]. The aim is to identify the landmarks with statistically significant group difference between ASD and NC subjects in local brain structures. The pipeline for defining ASD landmarks is shown in Fig. 2. Specifically, we first choose one T1-weighted image with high quality as a template. Then, a linear registration is used to locate the same location across all training images in the pre-processing step. Since the linearly-aligned images are not voxel-wisely comparable, nonlinear registration is further used for spatial normalization, thus corresponding relationship among voxels can be established. To identify local morphological patterns with statistically significant differences between groups, we use the histogram of oriented gradients (HOG) features as morphological patterns. Finally, the Hoteling’s T2 statistic [15] is adopted for group comparison. Accordingly, each voxel in the template is assigned with a p-value. Any voxels in the template with p-values smaller than 0.001 are regarded as significantly different positions. To avoid redundancy, only local minimum (whose p-values are also smaller than 0.001) are defined as ASD landmarks in the template image.
Fig. 2.
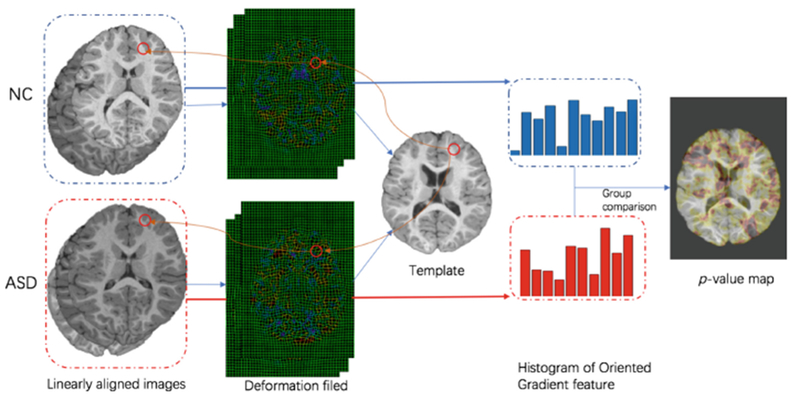
The proposed pipeline for anatomical landmark definition.
Patch Extraction from MRI.
By using the deformation field from nonlinear registration, we can get the corresponding patches in each linearly-aligned image. To address the issue of the limited number of autistic subjects, we propose a data expansion strategy on autistic subjects to have comparable number of samples as the number of NC subjects. As shown in Fig. 3(b), for each landmark, we randomly extract N (> 1) image patches as ASD training samples, but only 1 image patch as NC training sample (Fig. 3(c)). In the datasets, since the NC sample size is about 4 times larger than the ASD, we set N = 4 in the experiment.
Fig. 3.
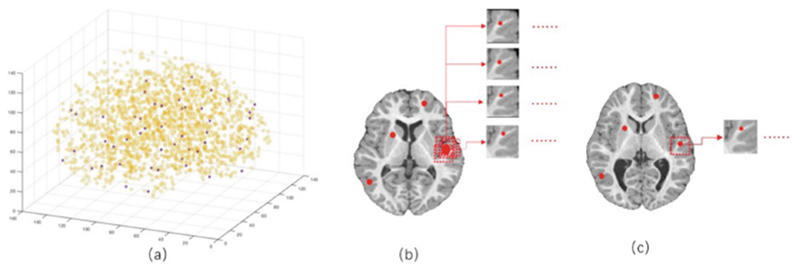
Illustration of landmarks selection and patch extraction. The yellow color in a is the illustration of all identified anatomical landmarks, and the red points show the selected 50 landmarks which are chosen by both their p-values and spatial distances from other nearby points. b shows 4 randomly extracted patches around a landmark which are as ASD training samples, and c shows 1 randomly extracted patch which is used as NC training sample.
Multi-Channel Convolutional Neural Networks.
In this study, both local and global features are used for classification. We use a multi-channel CNN model as shown in Fig. 4 for diagnosis. Given a subject, the input data are L patches extracted from L landmarks. To learn representations of individual image patch in one group, we first run multiple sub-CNN architectures. More specifically, we embed L parallel sub-CNN architectures with a series of 5 convolutional layers with stride size 1 and 0-padding (i.e., Conv1, Conv2, Conv3, Conv4, and Conv5), and a fully-connected (FC) layer (FC7). The rectified linear unit (ReLU) activation function is used after convolutional layers, while Conv2 and Conv4 are followed by max-pooling procedures to conduct the down-sampling operation for their outputs, respectively. To model global structural information of MRI, we further concatenate the outputs of L FC7 layers and add two additional FC layers (i.e., FC9, and FC10) to capture global structural information of MRI. Moreover, we use a concatenated representation comprising the output of FC8, personal information (gender) and the whole-brain volume into FC9 layer. Finally, FC11 layer is used to predict class probability (via soft-max).
Fig. 4.
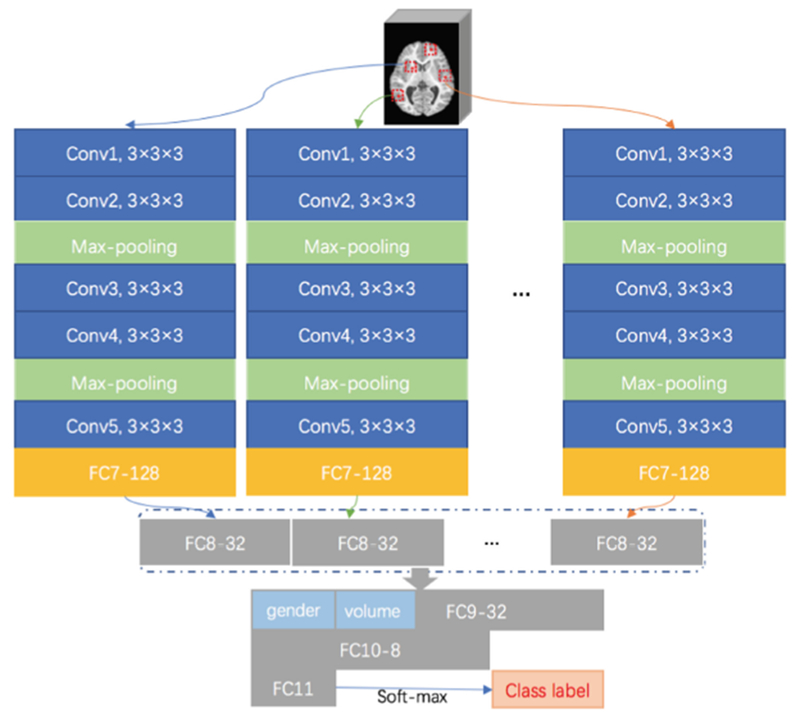
Architecture of the proposed multi-channel CNN for ASD diagnosis. In each channel CNN, there is a sequence of convolutional (Conv) layers and fully connected (FC) layers.
3. Experiments and Results
In this section, we present our experimental results using the proposed data-expanding multi-channel CNNs (DE-MC) scheme. We also evaluate performance with respect to landmark number and learning rate. Ten-fold cross-validation was used in the experiment. In each fold, we selected 264 subjects, with 55 ASD training samples and 209 NC training samples for training, and used 6 ASD subjects and 6 NC subjects as testing samples. The patch size was set as 24 × 24 × 24. Since both T1w and T2w images are available, we concatenated the extracted T1w and T2w image patches as input to the multi-channel framework. The cross-entropy loss was used to train the network.
First, we compared our DE-MC method with 3D convolutional neural networks (3D CNN). The input of 3D CNN is the whole linearly aligned MR image and the network has 10 convolutional layers and 3 fully connection layers, and we have optimized all parameters for fair comparison. The performance of classification is evaluated by the overall classification accuracy (ACC), as shown in Table 2. We can see that, compared with 3D CNN, our method can get a much higher accuracy. In terms of overall accuracy, DE-MC achieves 24% improvement over 3D CNN. To evaluate the contributions of the proposed strategies, we further compare the DE-MC with its two variants: (1) multi-channel learning without patch-level data expansion (MC), and (2) multi-channel learning using data expansion without additional information (i.e., gender information and whole-brain volume) (DE-MC2). They all share the same L = 50 landmarks and learning rate lr = 0.0001. Compared multi-channel learning without patch-level data expansion (MC), the proposed method can get better result. It proves that the data expanding strategy can help get more useful information for classification and improve the performance. Compared with the proposed methods with and without additional information (i.e., gender information and whole-brain volume), we can see this additional information is useful for ASD diagnosis.
Table 2.
Classification results achieved by different methods.
| Method | 3D CNN | DE-MC | MC | DE-MC2 |
|---|---|---|---|---|
| ACC | 0.5234 | 0.7624 | 0.6231 | 0.7432 |
Hyper parameters, e.g., number of landmarks and learning rate, in the deep learning may significantly impact the performance, and hence they should be carefully chosen. Table 3 shows the results with respect to the use of different combination of learning rate and landmark number.
Table 3.
Classification accuracy of our method with different hyperparameters.
| L | lr | 0.0001 | 0.001 | 0.01 |
|---|---|---|---|---|
| 30 | 0.6132 | 0.5244 | 0.4243 | |
| 50 | 0.7624 | 0.6138 | 0.5471 | |
| 100 | 0.7620 | 0.6249 | 0.5731 | |
| 200 | 0.7431 | 0.6169 | 0.5708 | |
4. Conclusion
In this paper, we proposed a patch-level data-expanding strategy to multi-channel CNN for early ASD diagnosis. In the patch extraction step, we utilized a patch-level data expanding strategy to balance the samples from NC and autistic groups, and the experiments show that this strategy is effective. The experiment results also showed that our proposed networks can achieve reasonable diagnosis accuracy. In our future work, we will further validate the proposed model on 6-month-old infant subjects with risk of ASD.
Acknowledgments.
Data used in the preparation of this manuscript were obtained from the NIH-supported National Database for Autism Research (NDAR). NDAR is a collaborative informatics system created by the National Institutes of Health to provide a national resource to support and accelerate research in autism. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or of the Submitters submitting original data to NDAR.
This work was supported in part by National Institutes of Health grants MH109773, MH100217, MH070890, EB006733, EB008374, EB009634, AG041721, AG042599, MH088520, MH108914, MH107815, and MH113255.
References
- 1.Newschaffer CJ, et al. : The epidemiology of autism spectrum disorders. Dev. Disab. Res. Rev 8, 151–161 (2002) [Google Scholar]
- 2.Filipek PA, et al. : The screening and diagnosis of autistic spectrum disorders. J. Autism Dev. Disorders 29, 439–484 (1999) [DOI] [PubMed] [Google Scholar]
- 3.Baird G, Cass H, Slonims V: Diagnosis of autism. BMJ Brit. Med. J 327, 488–493 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen R, Jiao Y, Herskovits EH: Structural MRI in autism spectrum disorder. Pediatr. Res 69, 63R (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schumann CM, et al. : The amygdala is enlarged in children but not adolescents with autism; the hippocampus is enlarged at all ages. J. Neurosci 24, 6392 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greimel E, et al. : Changes in grey matter development in autism spectrum disorder. Brain Struct. Funct 218, 929–942 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thakkar KN, et al. : Response monitoring, repetitive behaviour and anterior cingulate abnormalities in autism spectrum disorders (ASD). Brain 131, 2464–2478 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciresan DC, Meier U, Masci J, Maria Gambardella L, Schmidhuber J: Flexible, high performance convolutional neural networks for image classification In: IJCAI Proceedings-International Joint Conference on Artificial Intelligence, pp. 1237 Barcelona, Spain, (2011) [Google Scholar]
- 9.Sarraf S, Tofighi G: DeepAD: Alzheimer’s Disease Classification via Deep Convolutional Neural Networks using MRI and fMRI. bioRxiv 070441 (2016) [Google Scholar]
- 10.LeCun Y: LeNet-5, convolutional neural networks (2015). http://yann/.lecun.com/exdb/lenet20
- 11.Zhong Z, Jin L, Xie Z: High performance offline handwritten chinese character recognition using googlenet and directional feature maps. In: 13th International Conference on Document Analysis and Recognition (ICDAR), pp. 846–850. IEEE; (2015) [Google Scholar]
- 12.Liu M, Zhang J, Adeli E, Shen D: Landmark-based deep multi-instance learning for brain disease diagnosis. Med. Image Anal 43, 157–168 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Payakachat N, Tilford JM, Ungar WJ: National Database for Autism Research (NDAR): Big data opportunities for health services research and health technology assessment. PharmacoEconomics 34, 127–138 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Gao Y, Gao Y, Munsell BC, Shen D: Detecting anatomical landmarks for fast Alzheimer’s disease diagnosis. IEEE Trans. Med. Imaging 35, 2524 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mardia K: Assessment of multinormality and the robustness of Hotelling’s T2 test. Appl. Stat, 163–171 (1975) [Google Scholar]


