Abstract
Functional profiling from metagenomic or metatranscriptomic (“meta’omic”) sequencing provides insight into the molecular activities of microbial communities. These analyses are typically carried out using comprehensive search of sequencing reads, which is time-consuming, prone to spurious mapping, and often limited to community-level quantification. We developed a tiered meta’omic search strategy (HUMAnN2) which enables fast, accurate, and species-resolved functional profiling of host-associated and environmental communities. HUMAnN2 identifies a community’s known species, aligns reads to their pangenomes, performs translated search on unclassified reads, and finally quantifies gene families and pathways. Relative to pure translated search, HUMAnN2 is 3x faster and produces more accurate gene family profiles (89% vs. 67%). We apply HUMAnN2 to clinal variation in marine metabolism, ecological contribution patterns among human microbiome pathways, variation in species’ genomic vs. transcriptional contributions, and strain profiling. Finally, we introduce “contributional diversity” to explain patterns of ecological assembly across different microbial community types.
INTRODUCTION
Profiling microbial community function from metagenomic and metatranscriptomic (“meta’omic”) sequencing is a critically important challenge in culture-independent microbial ecology. It has the potential to characterize the extensive biochemical “dark matter” observed in many communities1, as well as to link specific molecular activities to environmental2 and health-associated3 phenotypes. In contrast with taxonomic profiling, functional profiling aims to quantify the gene and metabolic pathway content contributed by known and uncharacterized community members4. While taxonomic profiling can be performed on a maximally informative subset of meta’omic sequencing reads5, 6, comprehensive functional profiling must consider all reads and the vast space of genes from which they may derive, thus adding considerable analytical complexity.
Several previous methods exist for functional profiling of metagenomes7–9, a subset of which have been applied to metatranscriptomes10–13. These include HUMAnN14, which we developed during the Human Microbiome Project (HMP)15 for host- and environmentally-associated meta’omic functional profiling. Like later methods, HUMAnN interprets translated search of meta’omic sequencing reads to reconstruct metabolic functions. While existing methods benefit from recent methodological advances in translated search16–18, they remain considerably slower than nucleotide-level analyses. In addition, while some functional profiling methods incorporate taxonomic concepts for database refinement7 or targeted quantification9, most are limited to reporting community-level abundances (not per-organism contributions). Similarly, functional profiling lags behind efforts in strain-level analysis of microbial communities19–21, despite a growing appreciation for strain-variable functions within species.
To integrate taxonomic information with microbial community functional profiles, and to limit the bottleneck imposed by translated search, we developed HUMAnN2 as a next-generation meta’omic functional profiling method. HUMAnN2 represents both a completely new methodology and implementation: incorporating a tiered approach with accelerated nucleotide-level, translated search, and pathway reconstruction components. With these, HUMAnN2 exceeds the accuracy and performance of current pure translated search strategies. Moreover, gene and pathway abundances quantified by HUMAnN2 are automatically stratified into contributions from known and uncharacterized species. This provides previously inaccessible detail in interpreting host-associated and environmental community meta’omes.
RESULTS
Algorithm overview
HUMAnN2 implements a “tiered search” strategy to quickly and accurately profile the functional content of a meta’ome at species-level resolution (Fig. 1a, Supplementary Fig. 1, Online Methods), the results of which can also be used later for strain profiling. In the first tier, HUMAnN2 rapidly identifies known microbial species in a sample by screening DNA or RNA reads with MetaPhlAn222. HUMAnN2 then constructs a sample-specific database by merging preconstructed, functionally annotated pangenomes of the identified species23. In the second tier, HUMAnN2 performs nucleotide-level mapping of all sample reads against the sample’s pangenome database. Relative to comprehensive translated search, nucleotide-level mapping against relevant pangenomes quickly explains a large fraction of reads with fewer opportunities for spurious alignment. In the third and final tier, reads that did not align to identified species’ pangenomes are subjected to accelerated translated search against a comprehensive protein database (by default, UniRef90 or UniRef5024).
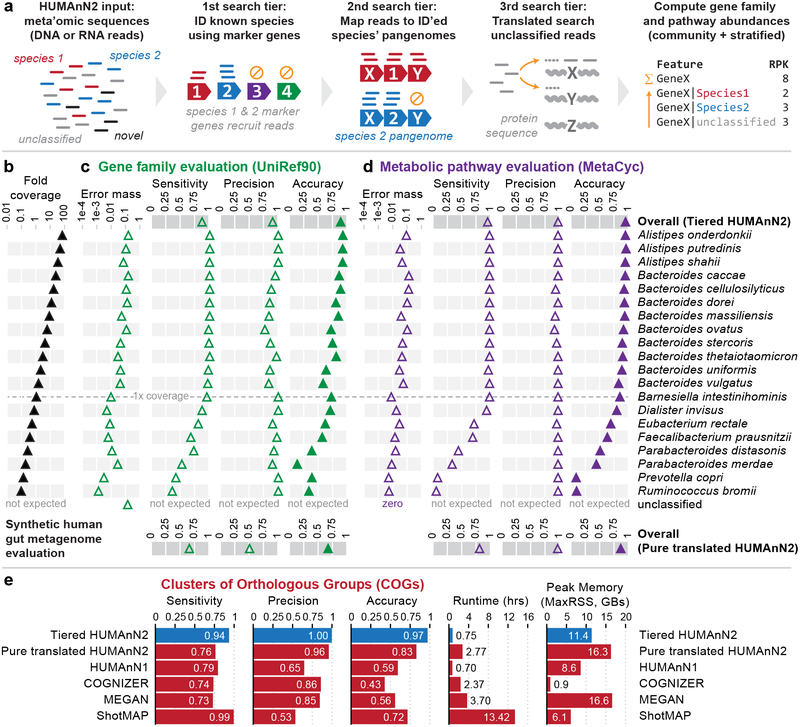
Figure 1: HUMAnN2 functionally profiles microbial communities with high accuracy using tiered search.
(a) Overview of HUMAnN2’s tiered search algorithm for meta’omic functional profiling (expanded in Supplementary Fig. 1). (b) HUMAnN2’s tiered search vs. pure translated search evaluated on a synthetic gut metagenome. Sensitivity, precision, and overall accuracy (1 - Bray-Curtis dissimilarity) were computed for (c) gene family and (d) pathway abundance profiles relative to gold standards at the whole-community level (“overall”) and for each stratification. (e) HUMAnN2 compared with other methods in the task of quantifying community-total COG abundances. Runtimes reflect multi-threaded execution on 8 CPU cores.
The tiered search generates mappings of meta’omic reads to gene sequences with known or ambiguous taxonomy. These mappings are weighted by quality and sequence length to estimate per-organism and community-total gene family abundance, which can be regrouped to other functional systems (e.g. COGs25, KOs26, Pfam domains27, and GO terms28). Finally, gene families annotated to metabolic enzymes are further analyzed to reconstruct and quantify complete metabolic pathways (by default, MetaCyc29) in the community and per-organism.
HUMAnN2’s tiered search produces more accurate functional profiles 3x faster than pure translated search
We assessed HUMAnN2’s accuracy by profiling synthetic metagenomes of known taxonomic and functional composition (Online Methods). We first simulated a human gut metagenome containing 10 million 100-nt DNA reads (1 Gnt) drawn from the 20 most abundant bacterial species detected in HMP stool samples15. Species’ abundances were geometrically staggered from ~0.1x to ~70x genomic coverage (Fig. 1b) and included nine members of genus Bacteroides: both challenges for accurate per-species profiling. We analyzed this synthetic metagenome using HUMAnN2’s tiered search and a pure translated search strategy (see also Supplementary Note 1 and Supplementary Fig. 2 for a parallel analysis of a 100-member, non-human-associated community).
Focusing first on community-level gene family (UniRef90) abundances, the sensitivity, precision, and overall accuracy (1 - Bray-Curtis dissimilarity) of HUMAnN2’s tiered search were high at 86%, 90%, and 89%, respectively (Fig. 1c). Hence, HUMAnN2 i) detected most expected gene families in the community, ii) reported only a small proportion of spuriously detected families, and iii) correctly assigned the vast majority of reads to their source families. Gene families profiled by pure translated search were less accurate (overall accuracy 67%), due in part to the greater potential for spurious alignment when aligning all sample reads against a comprehensive protein database.
The per-species accuracy of HUMAnN2’s tiered search remained high for the 14 species present at 1x genomic coverage or greater, including the nine Bacteroides species. Below 1x coverage, sensitivity and overall accuracy dropped off with coverage, as greater numbers of gene families were under-sampled in that domain. However, precision remained consistently high for low-coverage species, indicating that their pangenomes did not recruit substantial unrelated reads. The small subset of reads (1.4%) that passed into the translated search tier and mapped to proteins produced an “unclassified” stratification with a minority contribution to overall error (Supplementary Note 2).
Accuracy trends for HUMAnN2’s tiered search were similar at the pathway level, with pathway precision generally exceeding gene family precision (Fig. 1d). This is due to the greater difficulty of spuriously matching a complete pathway, which requires multiple distinct reactions (gene families) to spuriously recruit reads. Simultaneously, HUMAnN2’s requirement of detecting complete (or nearly-complete) pathways causes sensitivity and overall accuracy to decay more rapidly with decreasing coverage. Notable for less-well characterized samples, gene-level error inherent to the pure translated search strategy tended to be “smoothed out” during pathway quantification, though pathway profiles from pure translated search were still less accurate than those from HUMAnN2’s tiered search (87% vs. 98%).
In addition to being more accurate, HUMAnN2’s tiered search was 3x faster than pure translated search in the synthetic evaluation (runtime <1 hr; Fig. 1e and Supplementary Notes 1 and 3). We further benchmarked the performance of tiered search on 397 HMP metagenomes spanning six body sites (Supplementary Note 4). In a typical sample, ~60% of reads mapped during the pangenome search, and an additional ~20% mapped during translated search (Supplementary Fig. 3). Thus, for well-characterized, real-world metagenomes, HUMAnN2 explains the majority of sample reads during the fast pangenome search, thus making it considerably more efficient than a pure translated search strategy.
HUMAnN2 is more sensitive, precise, and efficient than existing methods
We compared HUMAnN2 with existing functional profiling methods built upon pure translated search: HUMAnN114, COGNIZER10, MEGAN12, and ShotMAP13 (Fig. 1e). This comparison was based on estimating community-level COG (Clusters of Orthologous Groups) abundances: an output format common to all methods (Online Methods). We constructed a custom search database for ShotMAP based on UniRef90, and used the other three methods’ recommended databases. We note that these three methods may differ in their systems of COG definition relative to our UniProt-based gold standard, which could influence their accuracy relative to HUMAnN2 and ShotMAP. However, the isolate genomes sampled in these evaluations predate all methods except HUMAnN1, which limits potential bias due to database coverage.
Overall accuracy was strongest for HUMAnN2’s tiered search (97%), followed by HUMAnN2’s pure translated search (83%), ShotMAP (72%), HUMAnN1 (59%), MEGAN (56%), and COGNIZER (43%). This result further emphasizes the power of tiered search to improve accuracy. The increased accuracy of HUMAnN2’s pure translated search may be attributed to our post hoc alignment filtering and weighting aimed at maximizing specificity (Online Methods; Supplementary Figures 4 and 5). HUMAnN2’s tiered search profiled the 10M-read synthetic metagenome in 45 minutes. This was similar to HUMAnN1 using accelerated translated search16 (42 minutes), yet HUMAnN2 provides considerably more detailed output and considers a ~20x larger sequence space. HUMAnN2 was >3x faster than all other methods.
HUMAnN2 is accurate for metatranscriptomes and robust to community species lacking reference genomes
We performed extensive additional evaluations of HUMAnN2 during development (summarized here and expanded in the supplement). We demonstrated that HUMAnN2 remains accurate and efficient when profiling broadly defined gene families (UniRef50; Supplementary Note 3). We further demonstrated that HUMAnN2 continued to perform optimally among other methods in profiling a synthetic gut metatranscriptome (Supplementary Note 5 and Supplementary Fig. 6). Most critically, we demonstrated that HUMAnN2 performed ably on metagenomes containing new isolates of known species as well as novel species (with the latter profiled by the translated search tier). This was accomplished by profiling a complex (100-member) synthetic community while holding out fractions of HUMAnN2’s pangenome database to simulate novel species (Supplementary Note 1 and Supplementary Fig. 2), and by applying HUMAnN2 and other methods to communities of isolate genomes that post-date the methods’ databases (Supplementary Note 6 and Supplementary Figures 7–10).
We additionally compared HUMAnN2 with metagenomic assembly of synthetic metagenomes (Supplementary Note 7). This evaluation expands previous comparisons of the approaches on real-world human metagenomes30, where they produced very similar rankings of domain-level functional diversity. While assembly was advantageous for uncovering novel sequence diversity in deeply sequenced human metagenomes, HUMAnN2 identified more known domains in metagenomes with modest sequencing depths. This advantage follows from the known challenge of detecting low-coverage metagenomic sequences by assembly31, which was also observed in our synthetic evaluations.
Contributional diversity of core human microbiome pathways
HUMAnN2’s tiered search quantifies community-encoded functions and stratifies their abundances according to who performs them. These data can be explored in additional detail by applying traditional within-sample (alpha) and between-sample (beta) community diversity measures32 to species’ contributions to a specific function: defined here as the function’s “contributional diversity” (Online Methods). A function contributed by a single species has low within-sample (“simple”) contributional diversity, while a function with many equal contributors has high within-sample (“complex”) contributional diversity. If a function is contributed by the same assemblage of species across samples, it has low between-sample (“conserved”) contributional diversity, while a function contributed by different assemblages has high between-sample (“variable”) contributional diversity.
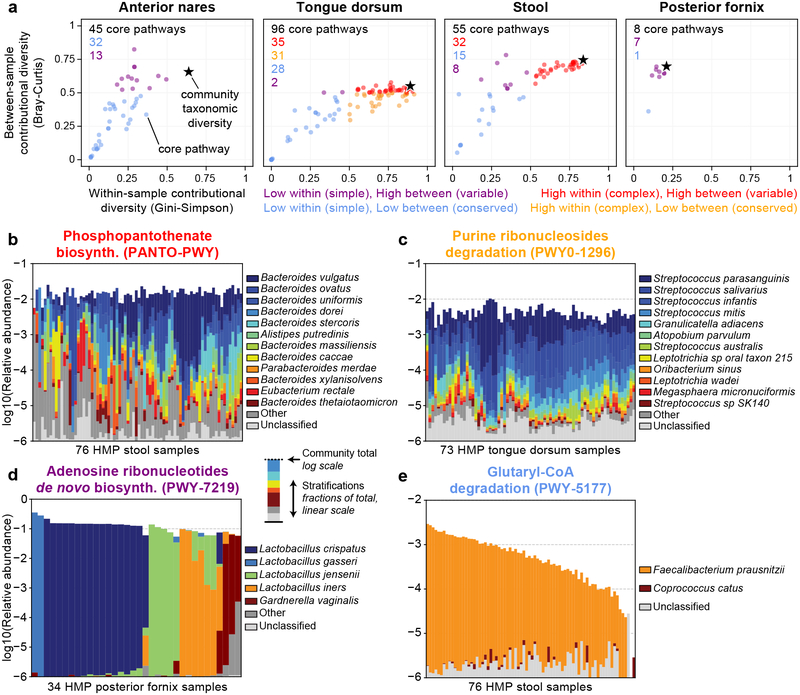
We explored the contributional diversity of human microbiome pathways that were core to a body site (non-zero in >75% of individuals) and largely explained by known species (<25% unclassified in >75% of individuals) among the 397 HMP metagenomes introduced above. (Note that functions with extensive “unclassified” abundance could be contributed by one or many different species within and across samples—hence their exclusion from this analysis.) Within- and between-sample contributional diversities were intuitively bounded above by their community-level analogs (Fig. 2a and Supplementary Fig. 11; examples in Fig. 2b–e). Contributional diversity rivals community diversity for functions that are broadly distributed in a given ecology. For example, phosphopantothenate biosynthesis in the gut had complex, variable contributors across subjects (mirroring gut ecology; Fig. 2b). Conversely, human microbiomes often contained pathways contributed by the same dominant organism across subjects, resulting in low within- and between-sample contributional diversity (Supplementary Fig. 12). For example, glutaryl-CoA biosynthesis in the gut was contributed principally by Faecalibacterium prausnitzii (Fig. 2e).
Figure 2: Contributional diversity of core human microbiome pathways.
(a) Within- and between- sample contributional diversity for core metabolic pathways (individual points) from HMP metagenomes. Stars indicate background species-level whole-community diversity. (b-e) Examples of pathways with “extreme” diversity patterns. The top of each set of stacked bars indicates the total stratified abundance of the pathway within a single sample (log-scaled). Species and “unclassified” stratifications are linearly (proportionally) scaled within the total bar height.
Oral sites were the most enriched for pathways with high within-subject but low between-subject contributional diversities, suggesting that they were encoded by complex yet similar mixtures of species across individuals (Fig. 2c). Core pathways at the vaginal site exhibited low within-sample but high between-sample contributional diversity, consistent with vaginal ecologies dominated by single, differing Lactobacillus species among subjects33 (Fig. 2d). That said, a subset of core pathways in non-vaginal sites also exhibited the same “simple but variable” contributions, which is further evidence for potential discordance between per-function and community-level diversities (Supplementary Fig. 13).
Clinal variation in marine microbial community function
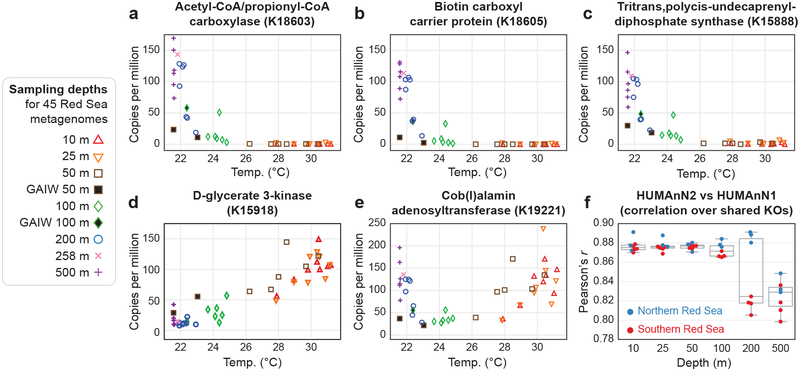
To demonstrate HUMAnN2’s applicability to environmental microbial communities, we applied the tiered search to quantify KEGG Orthogroup (KO) abundance in a dataset of 45 marine metagenomes from the epipelagic and mesopelagic zones of the Red Sea (Fig. 3 and Supplementary Note 4). We identified a number of high-variance KOs that were not detected in a previous analysis of the same samples with HUMAnN134 (examples in Fig. 3 a–e). Notably, KOs detected by both HUMAnN1 and HUMAnN2 were in the majority and their abundances were well correlated between the two methods (Fig. 3f).
Figure 3: Thermocline-associated microbial enzymes in the marine pelagic zone.
(a-e) Examples of KEGG Orthogroups (KOs) demonstrating strong temperature associations across 45 Red Sea metagenomes; all were newly quantified by HUMAnN2 relative to the samples’ initial publication. (f) Pearson correlations for 4,609 KOs that were quantified by both HUMAnN2 and HUMAnN1. “GAIW” indicates “Gulf of Aden Intermediate Water”: a cool nutrient-rich water mass within the Red Sea. The n=45 total samples in (f) are subdivided by depth layers (the sample from 258 m was grouped with the 500-m samples) and colored by latitude. From smallest to largest, box plot elements represent the lower inner fence, first quartile, median, third quartile, and upper inner fence.
Variation in KO abundance was often associated with sample temperature: the primary predictor of genetic diversity in the marine water column34, 35. Many high-variance KOs were maximally abundant in deep/cool waters and sharply less abundant at warmer temperatures. Three such KOs, among the six most variable overall, were implicated in fatty acid biosynthesis, particularly in archaea (see Fig. 3a–c). Indeed, HUMAnN2’s taxonomic stratifications revealed that the community abundances of these KOs were dominated by contributions from a single-cell genome36 of Marine Group I Thaumarchaeota (47–89% of copies).
Conversely, D-glycerate 3-kinase was more abundant in warmer, surface waters (see Fig. 3d), and largely attributed to Prochlorococcus marinus (25%) and Candidatus Pelagibacter ubique (21%): the two most abundant bacterial species in the surface ocean. These two species may use this enzyme to salvage glycerate in different aspects of central carbon metabolism (Prochlorococcus in photorespiration and Candidatus Pelagibacter as an entry point to glycolysis). Cob(I)alamin adenosyltransferase was notable for being enriched at low and high depths and depleted at intermediate depths (Fig. 3e). Cobalamin is a required cofactor for ribonucleotide reductase in certain marine bacteria, including Prochlorococcus37. Indeed, Prochlorococcus was the enzyme’s dominant contributor in surface samples (71–96%), whereas Verrucomicrobia was dominant in the deepest samples (36–41%).
Profiling strain-level functional variation
HUMAnN2’s accurate gene presence/absence calls (see Fig. 1c) can be applied to track strain-level20 functional variation in well-covered community species (Supplementary Note 4). While HUMAnN2 cannot assign new functions to a species, it identifies (potentially novel) subspecies-level clades from metagenomes based on presence/absence of functions observed across the species’ sequenced isolate genomes. For example, HUMAnN2’s gene family profiles of the HMP metagenomes introduced above revealed putative subspecies-level clades of Lactobacillus jensenii and Eubacterium eligens in the posterior fornix and gut, respectively (Supplementary Fig. 14).
Critically, HUMAnN2’s strain profiles provide a means of functionally explaining subspecies-level variation based on enrichments in “variable” gene families20. For example, strain-variable genes in HMP species were intuitively enriched for mobile-element processes such as DNA-mediated transposition (Wilcoxon enrichment test; FDR-corrected q<0.2 in 42 species) and DNA integration (q<0.2 in 105 species). In some cases, gene presence/absence was strongly correlated with body site, indicative of possible niche-adapted subspecies. For example, Haemophilus haemolyticus strains from tongue metagenomes were enriched for genes involved in outer cell membrane assembly relative to plaque and buccal strains (q=0.03; Supplementary Fig. 15).
Analyzing paired metatranscriptomes and metagenomes
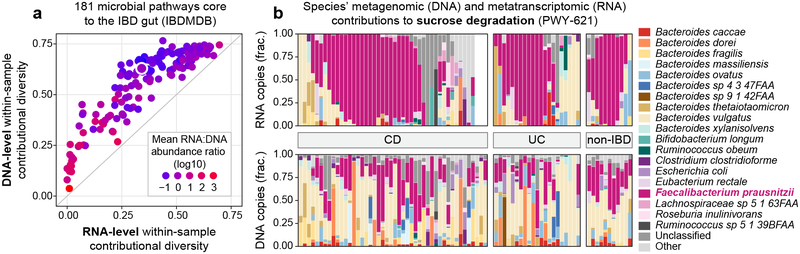
HUMAnN2 can profile paired metagenomes (DNA reads) and metatranscriptomes (RNA reads) to compare and contrast microbial community functional potential and activity4, as well as their respective contributional diversities. To illustrate this, we profiled core pathways (as defined above) from 78 paired meta’omes from the Inflammatory Bowel Disease Multi’omics Database (IBDMDB)38 (Supplementary Note 4). Within-sample contributional diversity at the DNA and RNA levels were well-correlated across 181 pathways, suggesting that more diverse pathway encoding tends to result in more diverse transcription (Spearman’s r=0.91; Fig. 4a). Simultaneously, DNA diversity tended to exceed RNA diversity, suggesting that pathways are not proportionally transcribed by the community species that encode them. Sucrose degradation was one such striking example: while encoded by many species, the pathway’s transcript pool was dominated by Faecalibacterium prausnitzii (Fig. 4b).
Figure 4: Metatranscriptomic functional profiling and multi’omic data integration with HUMAnN2.
(a) Average within-sample metagenomic (DNA) versus metatranscriptomic (RNA) contributional diversities for n=181 core pathways profiled from 78 paired inflammatory bowel disease (IBD) meta’omes from the IBDMDB cohort. Pathways are colored by “relative expression” (RNA:DNA ratio). (b) Sucrose degradation (outlined in ‘a’) is a prevalent pathway with high within-subject contributional diversity at the DNA level but low within-subject contributional diversity at the RNA level. This pattern was conserved across three IBD phenotypes: Crohn’s disease (CD), ulcerative colitis (UC), and non-IBD controls. Species’ contributions were rescaled to sum to 1 within each sample (set of stacked bars).
To differentiate changes in community gene expression from changes in gene copy number, it is critical to normalize functions’ RNA abundances against their DNA abundances. For example, within these profiles of the IBD gut, 71% of pathways’ RNA abundances fell within an order of magnitude of their DNA abundances. Methanogenesis pathways were among the largest outliers, with RNA:DNA ratios indicative of strong expression39. HUMAnN2’s stratified profiles confirmed Methanobrevibacter smithii as a consistent, dominant contributor to these pathways, resulting in low within- and between-subject contributional diversity.
DISCUSSION
HUMAnN2 is a new approach for functional profiling of meta’omically sequenced microbial communities. The method introduces a novel tiered search algorithm that provides exceptionally accurate profiles for characterized members of microbial communities, with fallback to translated search for uncharacterized members. These tiers operate jointly in far less time than traditional pure translated search. Moreover, tiered search provides taxonomic stratification of microbial functions at the species-level, thus quantifying the community abundance of functions while simultaneously assigning them to specific contributors. The utility of tiered search will only improve as reference catalogs continue to rapidly expand. In addition, tiered search facilitates this expansion by identifying unclassified meta’omic sequencing reads for external assembly of novel genes.
HUMAnN2’s functional stratifications enable discussion of “contributional diversity”: an analog of community-level diversity for individual microbial functions. Community-level function is often more conserved than community composition15, 39–41, consistent with a functional repertoire “defining” a niche and satisfied by different microbial assemblages. Contributional diversity adds another means by which this feature of functional ecology may be understood1, in that, while some functions do appear to be distributed evenly across community members, others are more restricted. Similarly, modern “multi’omic” analyses of microbial communities distinguish between community functional potential (encoding by genomes) and functional activity (gene or protein expression)39, 42, 43. Contributional diversity reveals another way in which these measurements can differ: for example, broadly encoded functions that are expressed dominantly by one or a few species.
Functional meta-analysis44 of diverse, new and existing meta’omic profiles are among the future biological areas opened up by the HUMAnN2 methodology, with the potential to reveal i) novel microbial community biochemistry and signaling, ii) determination of these functions’ source species and contributional diversity patterns and, in multi’omic datasets, iii) species-resolved deviations between functional potential and activity. In the human microbiome, HUMAnN2 provides the opportunity to generate testable hypotheses regarding specific species- (or strain-) level functions associated with health-related differences in community-level function. To support these future discoveries, the method is implemented as open source, fully documented software, packaged with demonstration data and training materials, and supports an active user community, accessible via http://huttenhower.sph.harvard.edu/humann2.
ONLINE METHODS
These methods detail the HUMAnN2 algorithm, the construction of its databases, our evaluations on synthetic metagenomes, and contributional diversity calculations. Methods related to our HUMAnN2 applications (i.e. the analyses of HMP metagenomes, Red Sea metagenomes, and paired IBDMDB meta’omes) are provided in Supplementary Note 4. Methods related to our evaluations on synthetic metatranscriptomes, novel isolate genomes, and assembled metagenomes are provided in Supplementary Notes 5, 6, and 7, respectively. Methods details can also be found in the online Life Sciences Reporting Summary.
Algorithm overview
HUMAnN2 is a system for accelerated functional profiling of shotgun metagenomic and metatranscriptomic (“meta’omic”) sequencing from host- and environmentally-associated microbial communities. HUMAnN2 implements a tiered search strategy comprised of three search phases (tiers). In the first search tier, the meta’omic sample is rapidly screened to identify known species in the underlying community. This information is then used to construct a custom gene sequence database for the sample by concatenating precomputied, functionally annotated pangenomes of detected species. In the second search tier, the entire sample is aligned against this database, yielding i) per-species, per-gene alignment statistics and ii) a collection of unmapped reads. In the final search tier, unmapped reads are aligned against a user-specified (typically comprehensive and nonredundant) protein database by translated search, yielding i) taxonomically unclassified per-gene alignment statistics and ii) a collection of novel reads. Per-gene alignment statistics are weighted based on alignment quality, coverage, and sequence length to yield gene abundance values i) for the community and ii) stratified according to per-species and “unclassified” contributions. Gene abundance values are finally applied to metabolic network reconstruction to identity and quantify pathways in the community (also stratified according to per-species and “unclassified” contributions). These processes, including the underlying databases and search parameters, are expanded in detail below.
Gene and pathway reference data as fixed inputs to HUMAnN2
Comprehensive protein databases
HUMAnN2 uses UniRef90 and UniRef5024 as comprehensive, non-redundant protein sequence databases. Briefly, UniRef90 represents a clustering of all non-redundant protein sequences in UniProt45 such that each sequence in a cluster aligns with 90% identity and 80% coverage of the longest sequence in the cluster (the cluster seed). Each resulting cluster is represented by a single sequence (usually the best-annotated member of the cluster, which is not necessarily the seed). UniRef50 is constructed by clustering all UniRef90 representative sequences to make clusters aligning with 50% amino acid sequence identity and 80% coverage of the cluster seeds. We use UniRef90 and UniRef50 clusters i) as a basis for describing gene family structure in microbial genomes and ii) as a comprehensive database for translated meta’omic search (see below). Protein annotations used by HUMAnN2 [e.g. Enzyme Commission (EC) number, COG25, KO26, Pfam domain27, and GO term28 assignments] are inferred from the annotations of representative UniProt sequences.
ChocoPhlAn pangenomes
Nucleotide-level search in HUMAnN2 is performed using collections of species pangenomes. We refer to this collection in HUMAnN2 as “ChocoPhlAn.” (An earlier version of ChocoPhlAn was published as MetaRef46; the version of ChocoPhlAn incorporated in HUMAnN2 is identical to that underlying MetaPhlAn2 and its marker database22.) A species’ pangenome is a nonredundant representation of the species’ protein-coding potential. To construct a pangenome for a given species, we download all available isolate genomes for that species from NCBI GenBank and/or RefSeq, along with associated coding sequence (CDS) annotations. Each isolate genome is analyzed with PhyloPhlAn47 to confirm correct taxonomic placement. Using UCLUST48, we then cluster all CDSs from high-quality isolate genomes of a given species at 97% nucleotide identity. One representative (centroid) sequence from each cluster is saved. These centroid sequences constitute the species’ pangenome. These steps were conducted in the course of MetaPhlAn2 development.
To use ChocoPhlAn for functional profiling, we annotated each pangenome centroid sequence to UniRef90 and UniRef50 by i) translating the centroid to produce an amino acid sequence and then ii) performing protein-level search against UniRef90. If the centroid’s best hit in UniRef90 met the criteria for inclusion in the corresponding UniRef90 cluster (>90% amino acid identity and >80% coverage), then the centroid was annotated to the UniRef90 cluster and inherited its corresponding UniRef50 annotation. If not, the centroid was labeled as “UniRef90_unknown” and a similar search was carried out against UniRef50 (requiring >50% identity to a UniRef50 sequence). If this search also failed, then the centroid was labeled as “UniRef50_unknown.” ChocoPhlAn includes pangenomes for >4K cellular microbes (bacteria, archaea, and fungi) which include >18M gene clusters. HUMAnN2 v0.9.6 adds support for >3K viral pangenomes which include >100K gene clusters.
Associating UniRef90/50 gene families with MetaCyc reactions
All alignments generated by HUMAnN2 are collapsed to UniRef90 or UniRef50 gene families, which constitute the method’s most highly-resolved main output. Gene families must be further collapsed to enzyme/reaction abundances prior to metabolic pathway reconstruction. This required generating a map linking UniRef90 and UniRef50 identifiers to MetaCyc reactions. These links were established in two ways. First, MetaCyc reactions are associated with a subset of proteins in UniProt, which are identified by UniProt accession numbers (ACs). As each protein in UniProt is associated with a UniRef90 cluster (and by extension, a UniRef50 cluster), Reaction-AC associations were converted to Reaction-UniRef90 and Reaction-UniRef50 associations for use in HUMAnN2. Second, MetaCyc reactions are associated with entries in the Enzyme Commission (EC) catalog: a four-level hierarchical description of enzymatic activities. UniProt entries (and, by extension, UniRef entries) are also associated with EC numbers. This relationship enabled additional transitive association of MetaCyc reactions and UniRef90/50 identifiers using EC annotations as a bridge. To maintain specificity, only EC annotations of the highest level of specificity were used in this process (for example, a UniRef90 entry associated with EC 1.1.1 would not be linked to a MetaCyc RXN associated with EC 1.1.1.1, nor would the reverse mapping be allowed). MetaCyc RXNs with at least one UniRef90 (or UniRef50) association are said to be “quantifiable” in HUMAnN2.
MetaCyc reaction to pathway mapping
HUMAnN114 incorporated KEGG’s structured pathway syntax26 to improve the accuracy of pathway reconstruction and quantification. This syntax i) specifies the reactions that must be satisfied to complete a pathway, as well as ii) possible alternative paths through the pathway (satisfiable by different combinations of reactions). We generated a corresponding structure for MetaCyc pathways by parsing MetaCyc’s pathway definition files. More specifically, each pathway was resolved to a directed acyclic graph connecting initial reactants with final products. (MetaCyc’s “superpathways” were resolved to their respective sub-pathways and recursive paths were removed.)
Each reaction node in a pathway was annotated to describe whether it connects with other nodes via “AND” or “OR” relationships (indicating, for example, that reactions 1 and 2 are both required to convert A to B, or that either 1 or 2 can perform the conversion). A pathway is said to be “satisfied” when there exists a path from initial reactants to final products that only passes through reaction nodes that were detected (non-zero abundance) in a given meta’omic sample (see below). Pathways were excluded if i) they contained less than four quantifiable reactions (i.e. reactions associated with level-4 EC numbers, which are in turn associated with UniRef90 and UniRef50 families) or ii) if they included >10% unquantifiable reactions. (Unquantifiable reactions in otherwise acceptable pathways were flagged as “optional” in the structured pathway syntax.)
Quantifying gene families by tiered search
Taxonomic Prescreen
HUMAnN2 takes as input a quality-controlled (including host-read-depleted) metagenome or metatranscriptome (“meta’ome”) provided as a FASTA or FASTQ file (with optional GZIP compression). DNA/RNA reads are initially screened using MetaPhlAn2 with default parameters. (The resulting MetaPhlAn2 outputs are saved as temp output in HUMAnN2.) Microbial species detected by MetaPhlAn2 above a target relative abundance threshold are passed to the next search tier (pangenome search). A lenient detection threshold of 0.0001 (0.01%) relative abundance is used as a default, which is equivalent to 0.1x fold-coverage of a 5 Mbp microbial genome in a 10 Gnt metagenome in which 50% of reads map to sequenced isolate genomes.
Pangenome search
HUMAnN2 next concatenates the pangenomes of species detected in the prescreen as a single FASTA file, which it then provides as input for building a Bowtie 2 index49. All sample reads (as introduced above) are then profiled against this index using Bowtie 2 in “very sensitive” mode. Because HUMAnN2 is aligning to isolated coding sequences, it does not consider read end-pairing relationships when evaluating Bowtie 2 alignment quality.
Translated search
Reads that failed to align against the pangenome database are mapped by translated search against a user-specified protein database. Four options are available: full versions of UniRef90 and UniRef50, as well as reduced versions of UniRef90 and UniRef50 containing only proteins associated with a MetaCyc reaction (discussed further in Supplementary Note 3). HUMAnN2 can call three translated search binaries to complete this task: DIAMOND16, RAPSearch217, and USEARCH48. DIAMOND is the recommended default. HUMAnN2 tunes the parameters of the translated search depending on whether the user is mapping against UniRef90 clusters versus the broader (more inclusive) UniRef50 clusters. For example, when using DIAMOND for translated search against UniRef50, the “--sensitive” search flag is invoked. The final output of the translated search is a tabular report of read-vs-protein alignment statistics (tabular BLAST format).
Alignment post-processing
Alignments in HUMAnN2 are post-processed to account for mapping quality and database sequence length. If a read has two or more high-quality alignments to distinct database sequences, the read’s single count is divided across the corresponding sequences in proportion to squared alignment identity. This serves as a more generic version of the default alignment weighting procedure implemented in HUMAnN1, which was based on alignment E-value (a statistic that lacks strict equivalents in some alignment software, e.g. Bowtie 2). Notably, a variety of similar weighting schemes were found to be equivalently good during HUMAnN1 evaluation, and all markedly better than naïve best-hit mapping14.
A weighted count to a sequence is further normalized by the alignable length of the database sequence (in kilobases) to produce a count in reads per kilobase (RPK) units. (Alignable length is the total length of the database sequence minus the aligned length of the read plus 1: the number of positions where an equivalent alignment to the database sequence could have begun.) These procedures are applied to nucleotide-level alignments against ChocoPhlAn pangenomes and to translated alignments against UniRef90/UniRef50. Weighted hits to sequences in the ChocoPhlAn pangenomes are summed within-species according to UniRef90/UniRef50 annotations (or UniRef90_unknown/UniRef50_unknown if no annotation exists). Weighted direct hits to UniRef90/UniRef50 families during translated search are summed and assigned to an “unclassified” species bin. These gene family abundances, along with a community total abundance (all species totals plus “unclassified”), are reported as HUMAnN2’s stratified gene family abundance table.
HUMAnN2’s translated search uses a comprehensive (rather than sample-specific) sequence database, which results in more opportunities for spurious alignments to occur. To compensate for this, HUMAnN2 filters translated alignment results in two additional ways before applying the general weighting procedures outlined above. First, we say that a read is “well aligned” to a protein if the majority of the read is used in the alignment (tunable default: 90% query coverage). This forces translated alignment of reads to more closely resemble the non-local alignment modes of Bowtie 2 (as used in pangenome search). Next, a read’s weight is only distributed over proteins whose sequences were “well covered” by well-aligned reads (tunable default: 50% of positions covered). Without such a filter, it is possible for small, frequently occurring peptide motifs to spuriously recruit compatible reads across a wide range of database proteins (most of which are not present in the underlying community; Supplementary Fig. 5). Reads that were never “well aligned” or which only aligned to poorly-covered proteins are exported alongside unaligned reads for downstream analyses (e.g. assembly of novel gene sequences) external to HUMAnN2.
Quantifying pathway abundance and coverage
Using the UniRef50/UniRef90 to MetaCyc reaction mapping described above, a reaction’s abundance is computed as the sum of the abundances for all gene families that map to the reaction. These sums are computed for each species, the “unclassified” stratum, and the community as a whole, consistent with HUMAnN2’s gene-level abundance reporting.
HUMAnN2’s procedures for computing pathway abundance (copy number) and coverage (detection confidence) are computed largely as described and benchmarked in HUMAnN114, with modifications added to account for i) the move from KEGG- to MetaCyc-based pathway definitions and ii) the need to compute the values in a stratified (per-species) manner as well as community-wide. Starting from a set of reaction abundances, HUMAnN2 first performs an (optional) gap-filling step to account for conspicuously depleted reactions or under-annotation. The default gap-filling in HUMAnN2 replaces the least-abundant reaction in the pathway with the abundance of the next-least-abundant reaction. Optional reactions are not considered in the gap-filling computations. Next, MinPath50 is applied to identify a parsimonious set of pathways to explain the observed reactions. Abundance and coverage are then computed for each pathway following HUMAnN1’s methods for structured (default) or unstructured pathway definitions. For structured pathways, abundance is computed as the harmonic mean of reaction abundances (after optimizing over alternative subpathways and optional reactions); for unstructured pathways, abundance is computed as the average of the top 50% most-abundant reactions in the pathway. Coverage is calculated similarly after converting reaction abundances to measures of reaction detection confidence. These procedures are carried out for the reactions detected in each species, “unclassified” reaction abundance values, and community total abundance values.
Evaluation details
Simulating metagenomes
We defined synthetic metagenome “templates” consisting of lists of species and target relative abundance values. For each species in a template, we selected a random isolate genome of that species from among those represented in ChocoPhlAn. We induced 3% artificial nucleotide sequence mutations in the isolate genomes to approximate the properties of previously unseen isolate genomes of the same species; genomic loci and nucleotide states were sampled randomly during the mutation process. Next, we randomly pulled 5 million 250-nucleotide fragments (substrings) from among those genomes. To guarantee that genome copies in the synthetic metagenome followed the target relative abundance distribution, fragments were pulled from each genome with probability proportional to the product of the genome’s size and corresponding species’ target relative abundance. We converted each fragment to a pair of 100-nucleotide sequencing reads in FASTQ format using ART51 with its Illumina HiSeq 2500 error model (resulting in 10 million total synthetic reads or 1 Gnt).
We produced a gene family abundance gold standard by incrementing the abundance of each gene family found in a genome by the product of the genome’s coverage [in reads per kilobase (RPK) units] and the gene family’s copy number. Note that this procedure does not account for random per-gene variation in fragment sampling, which will thus contribute to deviations from the gold standard (and be more marked for low-coverage species). This issue is discussed further in Supplementary Note 1. Gold standards for other functional categories (e.g. COGs) were generated by regrouping (summing) the gene family gold standard according to gene family functional annotations in UniProt. Gold standards for pathway coverage and abundance were generated by providing the gene family gold standard as an input file for HUMAnN2. Hence, our pathway-level accuracy assessment measures the influence of gene-level error on pathway quantification, and not the accuracy of assigning pathways to isolate genomes based on their annotated genes.
Comparing expected and observed profiles
We compared expected and observed gene and pathway abundance profiles at the community level as well as for each contributing species. Comparisons were made after sum-normalizing expected and observed profiles to relative abundance units. Four statistics were used for comparison: sensitivity, i.e. the fraction of expected features that were detected by HUMAnN2 (with “detected” defined as non-zero measured abundance); precision, the fraction of features detected by HUMAnN2 that were in the expected (gold standard) profile; overall accuracy, the fraction of feature abundance that was shared between the expected and observed datasets (1 - Bray-Curtis dissimilarity); and error mass, the proportion of total absolute error between the observed and expected profiles attributable to a particular stratification (individual species or “unclassified”).
Comparing HUMAnN2 with other methods
We profiled the 20-species, synthetic human gut metagenome with HUMAnN2, HUMAnN114, COGNIZER10, ShotMAP13, and MEGAN12 to generate profiles of COG abundance. HUMAnN2 was run in the default (tiered) mode and also in pure translated search mode against the full UniRef90 protein database. The resulting UniRef90 abundance profiles were converted to COG abundance profiles using the “humann2_regroup_table” script with the UniRef90-to-eggNOG option (which is inclusive of COGs).
To analyze the synthetic gut metagenome with HUMAnN1 (updated to use DIAMOND16 for translated search) we constructed a database from HUMAnN1’s default protein sequence collection: the last public release of KEGG (v56)26. We then aligned the synthetic reads against this database using HUMAnN1’s recommended search parameters (top-20 hits with E-value<1.0) while invoking DIAMOND’s “sensitive” mode. The resulting tabular alignment output was provided as input to HUMAnN1. HUMAnN1’s default KEGG Orthogroup (KO) output was converted to COG abundance using a KO-to-COG mapping derived from KEGG v56 (“data/cogc” in the HUMAnN1 installation).
We analyzed the synthetic metagenome in COGNIZER using the “-p 4” option, which defines a workflow in which RAPSearch217 profiles the metagenome against a reduced (non-redundant) COG sequence collection. This workflow was selected to be maximally time-efficient based on evaluations from the COGNIZER publication10. COGNIZER directly output a COG abundance table for downstream analysis.
We created a custom COG database for ShotMAP by supplying “build_shotmap_searchdb.pl” with individual FASTA files containing all UniRef90 sequences annotated to each COG. We used the option “--searchdb-split-size 30000” to split the database into subsets to improve memory efficiency. We then ran ShotMAP with the option “--class-score 31.3” which sets the minimum bit score for an alignment to be included in a family.
A DAA file was created for MEGAN by running DIAMOND to align the synthetic metagenome against the full NCBI NR database (downloaded Nov 2, 2016). Using the MEGAN GUI, the DAA file was “meganized” to COG abundance based on MEGAN’s included EggNOG mapping file (June 2016 version). Using the MEGAN GUI EggNOG viewer, we exported COG abundances to a text file for downstream analysis.
All runs were carried out in Google Cloud instances of machine type n1-standard-8 (which have 8 cores and 30 GB of memory). To benchmark the runs we captured the elapsed time along with the maximum RSS (resident set size) memory for the main process and all of its subprocesses, including all subprocesses in the process tree which have the main process as the top-most parent. These values were captured and recorded with the “humann2_benchmark” script. For workflows with separate mapping and post-processing steps (HUMAnN1 and MEGAN), elapsed time values encapsulate both steps, while maximum RSS values reflect the maximum across the two steps. Community-level COG abundances were sum-normalized and compared to the synthetic gold standard using the statistics described above.
Contributional diversity
We calculated contributional diversity for functions by applying traditional ecological similarity measures to the functions’ stratified abundance values. Here, the stratified values were renormalized after excluding “unclassified” abundance prior to computing diversity statistics. Functions with a non-trivial proportion of “unclassified” (>25%) in a non-trivial fraction of samples (>25%) were completely excluded from analysis. We used Gini-Simpson alpha diversity to measure within-sample contributional diversity of a function. This measure can be interpreted as the probability of selecting two “copies” of a function derived from different species, and varies from 0 (single contributor) to 1 (infinite contributors). We used Bray-Curtis beta diversity to measure between-subject contributional diversity of a function. This measure can be interpreted as the fraction of shared contributions between two samples, and varies from 0 (identical contributions) to 1 (no contributors in common). Diversity values for a pathway computed over samples (or sample pairs) were summarized by averaging.
Code availability
HUMAnN2 is a Python2/3 compatible package. The latest version can be installed via pip or HomeBrew (or installed from source via http://huttenhower.sph.harvard.edu/humann2). HUMAnN2 is also bundled as part of the bioBakery virtual machine, which is available as a Vagrant Box, a Google Cloud image, and an Amazon Web Services AMI (via http://huttenhower.sph.harvard.edu/biobakery). An archive of HUMAnN2 version 0.11.0 of the software (used in the evaluations reported here) is bundled with the publication.
The HUMAnN2 package includes 223 unit and functional tests, which run in ~20 minutes to verify successful installation and operation. Once installed, the complete HUMAnN2 workflow can be run with a single command by providing i) an input meta’omic sequencing dataset (fasta/fastq format) and ii) output folder. Four protein databases are available for use with HUMAnN2 (UniRef50 full, UniRef90 full, UniRef50 EC-filtered, UniRef90 EC-filtered). These databases, along with ChocoPhlAn and a collection of useful “utility” mapping files, are downloaded independently of the HUMAnN2 installation using the included “humann2_databases” script. Alternatively, the user can build and run HUMAnN2 with their own custom databases.
HUMAnN2 features four “bypass” modes to allow the user to tailor his or her workflow, e.g. including/excluding tiers in the tiered search. A “resume” feature allows the user to bypass compute-intensive sections of the workflow that have already completed while fine-tuning downstream analyses. HUMAnN2 includes 43 command-line arguments to customize runs for a user’s compute environment and to allow for parameter tuning (though a typical user will only interact with the two required “input” and “output” parameters). HUMAnN2 is bundled with a (growing) library of support scripts to facilitate downstream analyses, such as merging and normalizing profiles, regrouping default gene family abundances to other functional categories, combining RNA and DNA profiles to generate “relative expression” measurements, inferring approximate taxonomic assignment for proteins in the “unclassified” stratum, generating strain profiles, and plotting stratified abundances. These and other topics are expanded in detail in HUMAnN2’s user manual: http://huttenhower.sph.harvard.edu/humann2/manual.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank M. Wong, T. Sharpton, and the members of the HUMAnN user group for their feedback on the development and evaluation of HUMAnN2. Funding for this work was provided by NSF 1565100 (to JGC); People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007–2013) under REA grant agreement PCIG13-GA-2013–618833 and by MIUR “Futuro in Ricerca” RBFR13EWWI_001 (to NS); and NIH NIDDK U54DE023798, NSF MCB-1453942, NIH NIDDK P30DK043351, and NSF DBI-1053486 (to CH).
Footnotes
COMPETING FINANCIAL INTERESTS
None declared.
DATA AVAILABILITY STATEMENT
The Human Microbiome Project (HMP) metagenomes analyzed in this work are available via http://hmpdacc.org. The IBDMDB metagenomes and metatranscriptomes analyzed in this work are available via http://ibdmdb.org. The Red Sea metagenomes analyzed in this work were previously deposited as NCBI BioProject PRJNA289734. The synthetic metagenomes and metatranscriptomes used in the evaluation of HUMAnN2 and other methods are available from the authors and at http://huttenhower.sph.harvard.edu/humann2.
REFERENCES
- 1.Shafquat A, Joice R, Simmons SL & Huttenhower C Functional and phylogenetic assembly of microbial communities in the human microbiome. Trends in microbiology 22, 261–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuhrman JA Microbial community structure and its functional implications. Nature 459, 193–199 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Lloyd-Price J, Abu-Ali G & Huttenhower C The healthy human microbiome. Genome medicine 8, 51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franzosa EA et al. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nature reviews. Microbiology 13, 360–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segata N et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nature methods 9, 811–814 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sunagawa S et al. Metagenomic species profiling using universal phylogenetic marker genes. Nature methods 10, 1196–1199 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Silva GG, Green KT, Dutilh BE & Edwards RA SUPER-FOCUS: a tool for agile functional analysis of shotgun metagenomic data. Bioinformatics (Oxford, England) 32, 354–361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma AK, Gupta A, Kumar S, Dhakan DB & Sharma VK Woods: A fast and accurate functional annotator and classifier of genomic and metagenomic sequences. Genomics 106, 1–6 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Petrenko P, Lobb B, Kurtz DA, Neufeld JD & Doxey AC MetAnnotate: function-specific taxonomic profiling and comparison of metagenomes. BMC biology 13, 92 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bose T, Haque MM, Reddy C & Mande SS COGNIZER: A Framework for Functional Annotation of Metagenomic Datasets. PloS one 10, e0142102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Kim MS, Koh AY, Xie Y & Zhan X FMAP: Functional Mapping and Analysis Pipeline for metagenomics and metatranscriptomics studies. BMC bioinformatics 17, 420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huson DH et al. MEGAN Community Edition - Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS computational biology 12, e1004957 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nayfach S et al. Automated and Accurate Estimation of Gene Family Abundance from Shotgun Metagenomes. PLoS computational biology 11, e1004573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abubucker S et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS computational biology 8, e1002358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchfink B, Xie C & Huson DH Fast and sensitive protein alignment using DIAMOND. Nature methods 12, 59–60 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Tang H & Ye Y RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics (Oxford, England) 28, 125–126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauswedell H, Singer J & Reinert K Lambda: the local aligner for massive biological data. Bioinformatics (Oxford, England) 30, i349–355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Truong DT, Tett A, Pasolli E, Huttenhower C & Segata N Microbial strain-level population structure and genetic diversity from metagenomes. Genome research. 27, 626–638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholz M et al. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nature methods 13, 435–438 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Luo C et al. ConStrains identifies microbial strains in metagenomic datasets. Nature biotechnology 33, 1045–1052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Truong DT et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nature methods 12, 902–903 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Medini D, Donati C, Tettelin H, Masignani V & Rappuoli R The microbial pan-genome. Current opinion in genetics & development 15, 589–594 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Suzek BE, Wang Y, Huang H, McGarvey PB & Wu CH UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics (Oxford, England) 31, 926–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galperin MY, Makarova KS, Wolf YI & Koonin EV Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic acids research 43, D261–269 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Sato Y, Kawashima M, Furumichi M & Tanabe M KEGG as a reference resource for gene and protein annotation. Nucleic acids research 44, D457–462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finn RD et al. The Pfam protein families database: towards a more sustainable future. Nucleic acids research 44, D279–285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic acids research 43, D1049–1056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caspi R et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic acids research 44, D471–480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloyd-Price J et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sczyrba A et al. Critical Assessment of Metagenome Interpretation-a benchmark of metagenomics software. Nature methods 14, 1063–1071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamady M & Knight R Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome research 19, 1141–1152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravel J et al. Vaginal microbiome of reproductive-age women. Proceedings of the National Academy of Sciences of the United States of America 108 Suppl 1, 4680–4687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson LR et al. Metagenomic covariation along densely sampled environmental gradients in the Red Sea. The ISME journal (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunagawa S et al. Ocean plankton. Structure and function of the global ocean microbiome. Science (New York, N.Y.) 348, 1261359 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Swan BK et al. Genomic and metabolic diversity of Marine Group I Thaumarchaeota in the mesopelagic of two subtropical gyres. PloS one 9, e95380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson LR et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proceedings of the National Academy of Sciences of the United States of America 108, E757–764 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell host & microbe 16, 276–289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Franzosa EA et al. Relating the metatranscriptome and metagenome of the human gut. Proceedings of the National Academy of Sciences of the United States of America 111, E2329–2338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turnbaugh PJ et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burke C, Steinberg P, Rusch D, Kjelleberg S & Thomas T Bacterial community assembly based on functional genes rather than species. Proceedings of the National Academy of Sciences of the United States of America 108, 14288–14293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duran-Pinedo AE et al. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. The ISME journal 8, 1659–1672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mason OU et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. The ISME journal 6, 1715–1727 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasolli E, Truong DT, Malik F, Waldron L & Segata N Machine Learning Meta-analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLoS computational biology 12, e1004977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Consortium UniProt. UniProt: a hub for protein information. Nucleic acids research 43, D204–212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang K et al. MetaRef: a pan-genomic database for comparative and community microbial genomics. Nucleic acids research 42, D617–624 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Segata N, Bornigen D, Morgan XC & Huttenhower C PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nature communications 4, 2304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC Search and clustering orders of magnitude faster than BLAST. Bioinformatics (Oxford, England) 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye Y & Doak TG A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS computational biology 5, e1000465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang W, Li L, Myers JR & Marth GT ART: a next-generation sequencing read simulator. Bioinformatics (Oxford, England) 28, 593–594 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.