Abstract
It is necessary to accurately quantify the anatomical structures of maize materials based on high-throughput image analysis techniques. Here, we provide a 'sample preparation protocol' for maize materials (i.e., stem, leaf, and root) suitable for ordinary microcomputed tomography (micro-CT) scanning. Based on high-resolution CT images of maize stem, leaf, and root, we describe two protocols for the phenotypic analysis of vascular bundles: (1) based on the CT image of maize stem and leaf, we developed a specific image analysis pipeline to automatically extract 31 and 33 phenotypic traits of vascular bundles; (2) based on the CT image series of maize root, we set up an image processing scheme for the three-dimensional (3-D) segmentation of metaxylem vessels, and extracted two-dimensional (2-D) and 3-D phenotypic traits, such as volume, surface area of metaxylem vessels, etc. Compared with traditional manual measurement of vascular bundles of maize materials, the proposed protocols significantly improve the efficiency and accuracy of micron-scale phenotypic quantification.
Keywords: Biology, Issue 140, Maize, vascular bundle, micro-CT, image analysis pipeline, three-dimensional construction, microscopic phenotype
Introduction
The maize vascular system runs through the entire plant, from the root and stem to the leaves, which forms the key transportation paths for delivering water, mineral nutrients, and organic substances1. Another important function of the vascular system is to provide mechanical support for the maize plant. For example, the morphology, number, and distribution of vascular bundles in roots and stems are closely related to the lodging resistance of maize plants2,3. At present, studies on the anatomical structure of vascular bundles mainly utilize microscopic and ultramicroscopic techniques to display the anatomical structures of a certain part of the stem, leaf, or root, and then measure and count these structures of interest by manual investigation. Undoubtedly, manual measurement of various microscopic structures in large-scale microimages is a very tedious and inefficient work and severely limits the precision of microphenotypic traits, owing to its subjectivity and inconsistency4,5.
Maize has no secondary growth, and the cell content essentially consists of water in the primary meristem. Without any pretreatment, fresh samples of maize tissues can be directly scanned using a micro-CT device; however, the scanning results are probably poor and rough. The main reasons are summarized as follows: (1) low attenuation densities of plant tissues, resulting in a low contrast of atomic number and high noise in images; (2) fresh plant materials are prone to dehydrate and shrink during the normal scanning environment, as reported by Du6. The abovementioned problems have become the main constraints for the development and application of microphenotyping technology for maize, wheat, rice, and other monocotyledons. Here, we introduce the 'sample preparation protocol' to pretreat the samples of maize stem, leaf, and root. This protocol avoids the dehydration and deformation of plant materials during the CT scanning; thus, it is beneficial to increase the preservation time of plant samples with nondeformation. Moreover, the dyeing step based on solid iodine also enhances the contrast of plant materials; thus, it makes significant improvements in the imaging quality of micro-CT. Furthermore, we developed image processing software, named VesselParser, to process the CT images of maize stems and leaves. This software integrates a set of image-processing pipelines to perform high-throughput and automatic phenotyping analysis for 2-D CT images of different plant tissues. Vascular bundles in the entire cross-section of the maize stem and leaf are detected, extracted, and identified using an automatic image-processing method. As a result, we obtain 31 microscopic phenotypes of the maize stem and 33 microscopic phenotypes of the maize leaf. For the CT image series of the maize root, we developed an image-processing scheme to acquire 3-D phenotypic traits of metaxylem vessels. This scheme is superior in efficiency of image acquisition and reconstruction compared with traditional methods.
These results indicate that the image processing pipelines considering the imaging characteristics of ordinary X-ray micro-CT provide an effective method for the microscopic phenotyping of vascular bundles; this extremely widens the applications of CT techniques in plant science and improves the automatic phenotyping of plant materials at cellular resolution6,7.
Protocol
1. Sample Preparation Protocol
For the sampling, collect the stem, leaf, and root from fresh maize plants and divide them into three types of sample groups (each group with four replications). Then, cut them into small segments using a surgical blade in the following manner: (1) cut a segment of the middle stem internode 1 - 1.5 cm in length; (2) cut a segment of the maximum width of the leaf 0.5 - 3 cm in length along the vertical direction with the main vein; (3) cut a segment of the crown root 0.5 cm in length.
For the FAA fixation, soak the sample segments in an FAA solution (90:5:5 v/v/v, 70% ethanol:100% formaldehyde:100% acetic acid) for at least 3 d.
Perform the dehydration procedure in six sequential ethanol gradients (i.e., 30%, 50%, 70%, 85%, 95% and 100%) and set the processing time of each ethanol gradient as 30 min.
- Place the plant materials in the corresponding sample baskets manufactured using a 3-D printer; then, quickly transfer the sample baskets to the sample cell of a CO2 critical-point drying system. Set the drying parameters as follows: (1) CO2 in: fast speed. Holder fillers: 100%. (2) Charge: CO2 charge delay 120 s into the cycle. Exchange speed: 5. Cycle number: 12. (3) Gas out: heat, fast. Speed: slow, 50%.
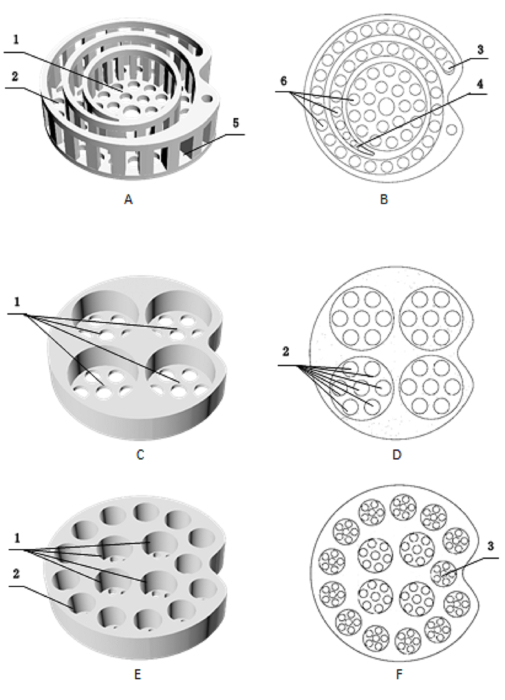
- According to the morphological differences of maize root, stem, and leaf, design and print sample baskets using a 3-D printer (e.g., Figure 1).
Place the dried plant materials (maize root, stem, or leaf) in a 50-mL centrifuge tube with 2 g of solid iodine to dye the plant materials with volatile iodine vapor and, then, place the tubes in a lightproof room for 4 - 5 h.
2. Micro-CT Scanning Protocol
To scan into the raw CT data, set the CT scanning parameters as follows: 40 kV/250 µA (for stem and leaf) or 34 kV/210 µA (for root). Set the corresponding scanning ranges for the different sizes and volumes of the plant materials used, and adjust the imaging pixel sizes as follows: 2.0 µm (for the maize root), 6.77 µm (for the maize stem), and 10.0 µm (for the maize leaf).
To reconstruct slice images, convert the raw CT data into CT slice images with a 2K resolution (2,000 x 2,000 pixels) using an image reconstruction software. More details are provided in the NRecon User Manual (http://bruker-microct.com/next/NReconUserGuide.pdf).
3. Image Analysis Protocol for a Single CT Image of a Maize Stem or Leaf
NOTE: Use automatic imaging software for vascular bundles to conduct the phenotyping analysis of vascular bundles within the CT slice images of the maize stem and leaf (Figure 2). The software usage steps are described as follows.
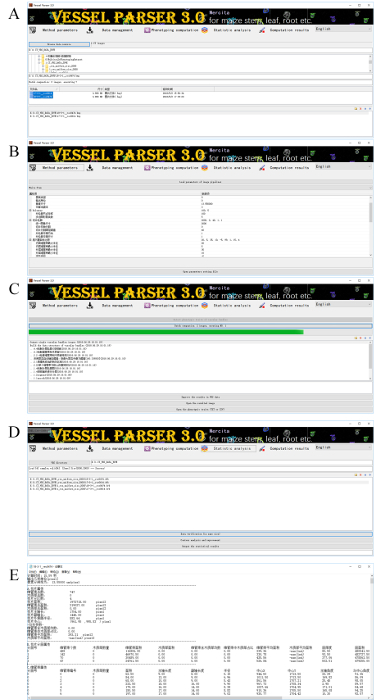
Appoint the organ type to initialize different algorithm pipelines. Click the Method parameters button and select maize stem or maize leaf in the first drop-down box.
To import the images, click the Data management button, set the work directory, and import automatically all slice images in this directory. Select single or multi-slice images into the image pipelines.
Determine the actual pixel size of the image. Click the Method parameters button and enter the actual pixel size of the image in the edit item of the pixel size.
For the phenotyping computation, click the Phenotyping computation button to automatically extract phenotypic traits of the vascular bundles for all selected slice images.
Click the Statistic analysis button to output the results as a TXT or CSV file format.
4. Image Analysis Protocol for CT Image Series of a Maize Root
NOTE: The CT image series of maize roots are utilized to extract the 3-D structures of metaxylem vessels using image-processing software. The main steps are as follows.
Import the reconstructed images of maize roots (in BMP file format) and determine the accurate spacing parameters (the size of a voxel [i.e., x, y, z]). Use the recursive Gaussian tool to smoothen these images to improve the image quality.
Conduct 3-D segmentation of metaxylem vessels by adjusting the threshold parameters; this generates a uniform color label for each connected metaxylem vessel.
Improve and identify the metaxylem vessels interactively using morphology, bitwise, and flood-fill operations.
Conduct volume visualization and surface reconstruction of the vessels. Use the mask statistics tool to count and measure the phenotypic traits of a vessel in the 2-D and 3-D levels.
Representative Results
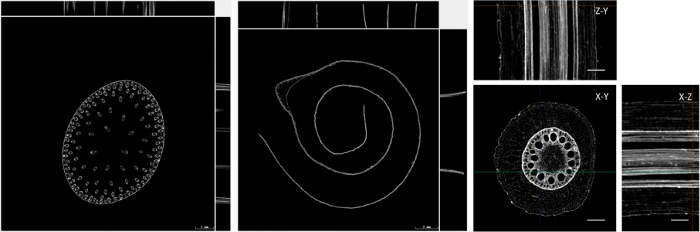
The sample preparation protocol suitable for ordinary micro-CT scanning not only prevents the deformation of plant tissues but also enhances the X-ray absorption contrast. Pretreated plant materials are scanned using a micro-CT system into high-quality slice images, and the highest resolution can reach 2 µm/pixel. Figure 4 shows the scanned micro-CT images of stem, leaf, and root, and the image contrast has a significant improvement compared with the results scanned from the fresh plant materials. In these CT slice images, significant differences in gray-level values are observable among vascular bundles, parenchyma cells, xylem vessels, epidermis, and other tissues.
Based on this imaging software for vascular bundles, the structure and distribution features of vascular bundles within the cross-section of the entire stem or leaf can be automatically analyzed and quantified. Take the image of a maize stem as an example, and the phenotyping algorithm of software consists of five steps: segment the slice image, segment the vascular bundles, analyze the spatial distribution of the vascular bundles, identify and improve the vascular bundles, and compute and output the phenotypic traits of the vascular bundles. The entire epidermis region in the image is, first, segmented based on specific or adaptive threshold values, and contour analysis techniques are performed to extract the epidermis region (Figure 3A and 3B). Next, segmentation of the vascular bundles is conducted. Vascular bundles are surrounded in the epidermis; therefore, the epidermis region with the predefined thickness is removed from the slice image. The rest of the image only consists of scattered vascular bundles with high pixel intensities and parenchyma cells with weak pixel intensities. Then, the image segmentation based on the fixed threshold value is performed to extract all candidate regions of the vascular bundles, and these regions are further determined as valid vascular bundles according to the limitations of area and shape features (Figure 3C and 3D).
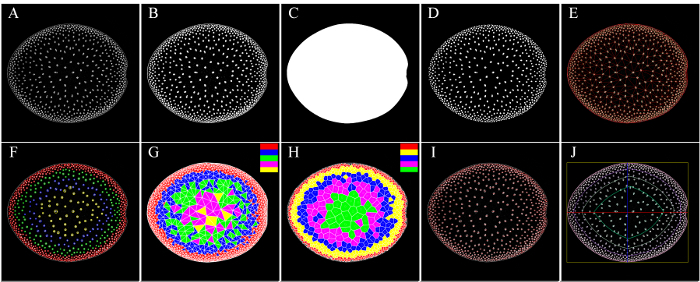
After the segmentation of vascular bundles, spatial distribution characteristics of the vascular bundles are extracted. Take the geometric centers of scattered vascular bundles as nodes to generate triangle meshes for all vascular bundles in the slice image, and these meshes are clustered into five types according to their areas. The areas of the vascular bundles manifest a significant decreasing trend from the center to the edge of the maize stem. The triangular and Voronoi meshes describe the spatial distribution and topological connections of vascular bundles, and each mesh is drawn with a specific color according to the clustered results of vascular bundles (Figure 3E - 3H). Vascular bundles that meet the constraints of spatial distribution (mesh area and shape are the important indexes to determine the availability of vascular bundle) are reserved and used to generate the final segmentation results (Figure 3I).
At last, phenotypic traits of vascular bundles, such as geometric, shape, and distribution information, can be computed according to the above analysis, which results in the output of a TXT or CSV file (Figure 3J). Based on the imaging software for vascular bundles, 31 phenotypic traits of the stem can be automatically analyzed; the average computation time for each CT image is ~30 s. The relative phenotypic parameters of the stem are shown in Table 1. Similarly, 33 phenotypic traits of the leaf can be extracted, the average computation time is ~50 s, and these parameter classifications are shown in Table 2. For a list of slice images, the above image analysis pipelines are integrated into a batch processing for automatic execution. This workflow is efficient to analyze the phenotypic traits of all the vascular bundles within an entire slice image of a maize stem and leaf. Notably, most phenotypic traits of vascular bundles, such as the total area, average area, and area ratio of vascular bundles, are significantly difficult to be measured by manual measurements.
Because the metaxylem vessels of maize roots show obvious morphological changes along the direction of root growth, it is more valuable to extract the 3-D structures of metaxylem vessels for phenotypic analysis. Based on the CT image series of maize root, 3-D segmentation, surface reconstruction, and volume visualization are performed. Based on the segmented results, the 3-D structural parameters of metaxylem vessels can be automatically calculated, including the volume, surface area, cross-sectional area (basal), and cross-sectional area (distal) of the total metaxylem vessels and each single metaxylem vessel. This workflow significantly improves the efficiency of 3-D phenotypic trait analysis. The results of segmentation, reconstruction, and volume visualization can directly manifest the spatial structures of metaxylem vessels of maize root, as shown in Figure 5.
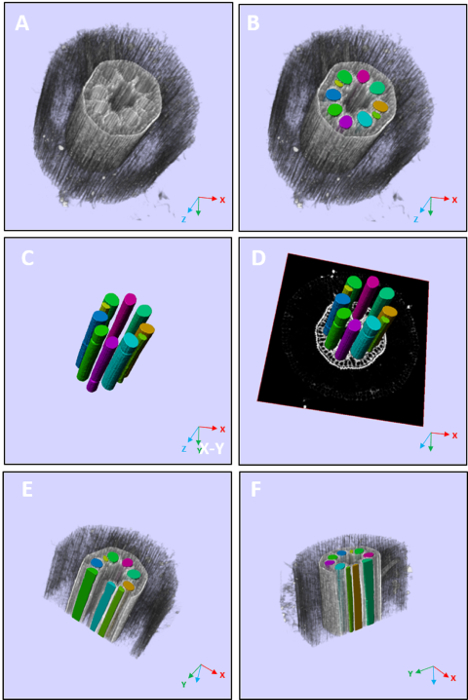
Figure 1: Different types of sample baskets printed with a 3-D printer. (A and B) The spiral sample basket for the leaf is composed with (A1) a central oval groove and (A2) the surrounding spiral groove. The width of the spiral groove is set at about 4 mm to accommodate the leaf with the main vein. The sidewall of the spiral groove is created with (A5) square drainage holes and the bottom of the basket with (B6) circular drain holes. (C and D) These two panels show a four-well sample basket suitable for the stem, with (C1) sample holes with a diameter of 25 mm, and the bottom of the basket with (D2) seven drain holes. (E and F) These two panels show a multiwell sample basket suitable for the root, with (E1) four circular holes with a diameter of 10 mm in the central area, and (E2) 13 circular holes with a diameter of 8 mm arranged near the edge of the basket. The bottom of the sample basket has drain holes with a diameter of 1 mm to ensure that the tiny root tissue does not leak out. Please click here to view a larger version of this figure.
Figure 2: Screenshots of automatic imaging software for vascular bundles. (A) This panel shows the data management to import CT images from any file directory and select CT slice images for the subsequent processing. (B) This panel shows the method parameters to identify the organ type of CT slice images and configure the corresponding method parameters. (C) This panel shows the phenotyping computation to perform the batch computation of CT slice images and show the execution progress. (D) This panel shows the statistical analysis to verify the computational results and generate phenotypic traits for all the CT images. (E) This panel shows the computational results to output the analysis results as a TXT or CVS file. Please click here to view a larger version of this figure.
Figure 3: Workflow of image-processing and phenotyping computation based on CT slice images. (A) Import the CT slice image of the maize stem. (B) Segment the slice image with a fixed threshold value. (C) Extract the entire region of the maize stem. (D) Delete the epidermis region of the maize stem. (E) Triangular meshes of vascular bundles. (F) This panel shows a clustering analysis according to the areas of the vascular bundles. (G) This panel shows a clustering analysis according to the areas of the triangular meshes. (H) This panel shows a clustering analysis according to the areas of Voronoi meshes. (I) This panel shows the final segmented results of the vascular bundles. (J) This panel shows the spatial distribution of the vascular bundles. Please click here to view a larger version of this figure.
Figure 4: Multiplanar reconstruction (MPR) images of the maize stem, leaf, and root. The left panel shows an MPR image of a stem cross-section. The middle panel shows an MPR image of a leaf cross-section. The right panel shows an MPR image of a root cross-section. Please click here to view a larger version of this figure.
Figure 5: 3-D visualization of root metaxylem vessels. The bars = 0.2 mm. (A and B) These panels show a 3-D visualization of a root. (C - F) These panels show a 3-D visualization of metaxylem vessels. Please click here to view a larger version of this figure.
 Table 1: Microscopic phenotypic traits of the maize stem.
Table 1: Microscopic phenotypic traits of the maize stem.
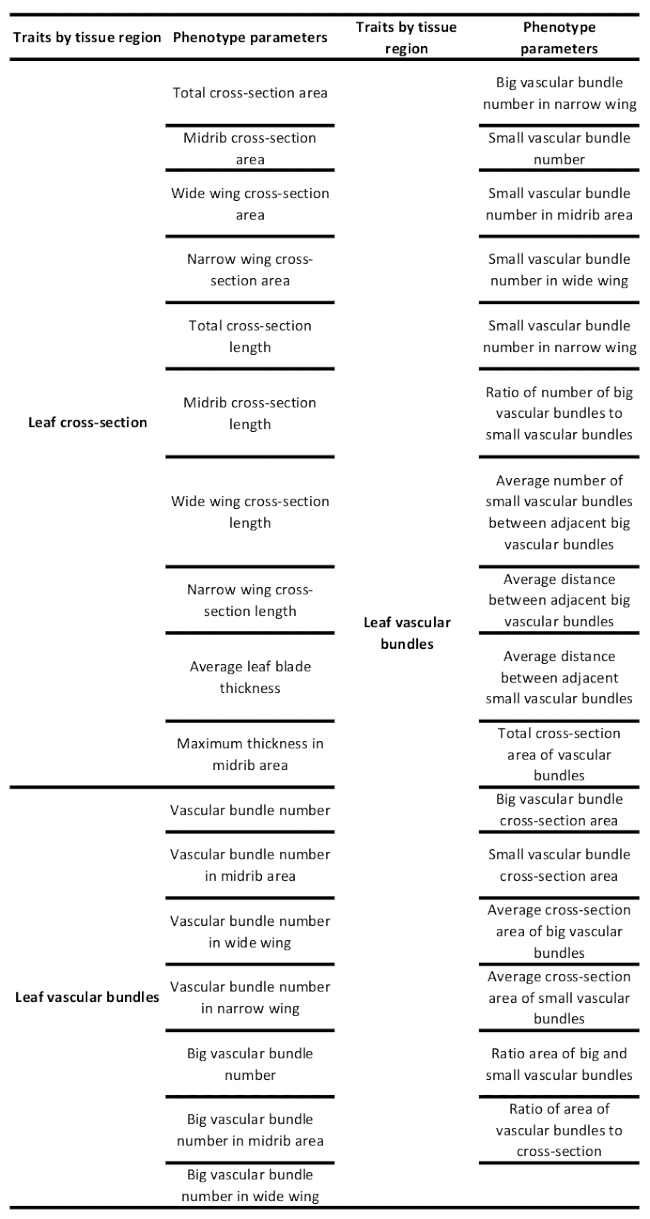 Table 2: Microscopic phenotypic traits of the maize leaf.
Table 2: Microscopic phenotypic traits of the maize leaf.
Discussion
With the successful application of CT technology in the fields of biomedicine and materials science, this technology has been gradually introduced into the fields of botany and agriculture, promoting researches in plant life sciences as a promising technical tool. In the late 1990s, CT technology was first used to study the morphological structures and development of plant root systems. In the past decade, synchrotron HRCT has become a powerful, nondestructive tool for plant biologists, and has been successfully used to identify the tissue structures of grape vascular system8, tissue structure of Arabidopsis leaf9,10, and seed structure of rape11. Through the synchrotron HRCT, great progress has been made in the study of structure and function of vascular bundles in woody plants12,13,14. However, little research of HRCT technology for maize, wheat, rice, and other crops has been done15. Maize has no secondary growth, and the cell essentially consists of water in the primary meristem. Although fresh samples can be scanned by micro-CT without any pretreatment, the scanning results are very poor. The main reasons are as follows: (1) low attenuation densities of plant tissues, resulting in a low contrast in atomic number and high noise in images; (2) fresh material tends to dehydrate and shrink during the scanning period, as reported by Du6. The above-mentioned reasons have become the main factors limiting the application of this technology in maize, wheat, rice, and other monocotyledons.
Here, we introduce a simple and practical sample preparation protocol that not only prevents the deformation of plant tissues but also enhances the X-ray absorption contrast. High-quality and high-resolution CT images of root, stem, and leaf were obtained based on the sample preparation protocol and micro-CT imaging system, and the highest resolution was up to 2 µm/pixel. Thus, the sample preparation protocol is suitable for ordinary micro-CT scanning and provides a great opportunity for broader applications in monocot and other plant sciences. This protocol can be easily modified to accommodate other plant materials such as dehydration or drying procedure, and its parameter setting can also be adjusted according to specific plant materials for the best results. Notably, this approach is limited by the size and volume of the plant sample. A too thick sample segment may lead to incomplete drying or deformation of the sample. Therefore, this sample preparation protocol is applicable for small plant materials with a thickness of less than 3 cm and not for much larger maize materials such as maize ear or cob.
The microscopic phenotyping technology of plant materials is one of the hot topics of plant phenotypic studies in recent years, and it is gradually becoming one of the basic support technologies for genetic breeding and plant physiology. Traditional microscopic phenotypic analysis of plants requires a large number of complex sample preparations and tedious manual operations. It is very labor-intensive and time-consuming to count and measure the microscopic traits; the results are also prone to subjective errors. For example, to quantify the phenotypic characteristics of vascular bundles in a maize stem, the stem sample needs to be embedded in paraffin and then sliced, stained, and imaged. For a stained slice image, it is difficult to perform an automated image processing due to the ambiguous definition of cell boundaries; thus, manual identification and segmentation are indispensable16. To satisfy the requirements of large-scale measurements for anatomical traits of the maize stem, Legland and Heckwolf introduced different image-processing methods; however, the structures of vascular bundles in the rind are still a challenge17,18. Hence, high-throughput image analysis and accurate quantification of anatomical traits of maize tissues are necessary. Here, we provide automatic imaging software for vascular bundles, that can automatically extract 31 phenotypic traits of vascular bundles in ~30 s for each CT image of the maize stem and 33 phenotypic traits in ~50 s for each CT image of the maize leaf. The structure and distribution features of vascular bundles within the cross-section of entire stems or leaves can be automatically analyzed and quantified. This software has the following advantages: (1) it automatically processes the CT slice images of the maize stem and leaf and extracts the phenotypic traits of vascular bundles; (2) it has a higher recognition rate of vascular bundles in the CT image, especially for small vascular bundles on the edge; (3) a novel graphic analysis method is used to reveal the distribution characteristics of vascular bundles.
Further, the X-ray micro-CT scanning technology has obvious advantages in the efficiencies of image acquisition and reconstruction compared with traditional reconstruction techniques based on paraffin section images19,20,21. Based on the CT image series of maize root, an image-processing scheme is developed to extract the spatial structures of metaxylem vessels and successfully used for the 3-D measurement of microscopic traits. The main limitation of this scheme is that the 3-D segmented results depend on slight manual interaction. In the future, we aim to develop a set of automated 3-D image analysis software for the CT dataset of maize roots to improve the efficiency of 3-D segmentation and reconstruction.
In conclusion, an ordinary micro-CT scanning based on a practical sample preparation protocol for the maize stem, leaf, and root is constructed to produce high-resolution CT images. The sample preparation protocol provided here not only prevents the deformation of plant tissues but also enhances the X-ray absorption contrast. This protocol is also suitable for other CT scanning applications in wheat, rice, and other monocotyledons. So far, we have developed automatic imaging software for vascular bundles, that is capable of automatically and rapidly extracting the phenotypic traits of vascular bundles from a single CT image of the maize stem and leaf. Based on the CT image series of the maize root, an image-processing scheme is successfully set up to extract the 3-D phenotypic traits of metaxylem vessels. Microscopic phenotyping techniques of plant materials based on X-ray micro-CT provide a new prospect for the accurate and rapid quantification and identification of maize vascular bundles.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This research was supported by the National Nature Science Foundation of China (No.31671577), the Science and Technology Innovation Special Construction Funded Program of Beijing Academy of Agriculture and Forestry Sciences(KJCX20180423), the Research Development Program of China (2016YFD0300605-01), the Beijing Natural Science Foundation (5174033), the Beijing Postdoctoral Research Foundation (2016 ZZ-66), and the Beijing Academy of Agricultural and Forestry Sciences Grant (KJCX20170404), (JNKYT201604).
References
- Lucas WJ, et al. The plant vascular system: evolution, development and functions. Journal of Integrative Plant Biology. 2013;55:294–388. doi: 10.1111/jipb.12041. [DOI] [PubMed] [Google Scholar]
- Gou L, et al. Effect of population density on stalk lodging resistant mechanism and agronomic characteristics of maize. Acta Agronomica Sinia. 2007;33:1688–1695. [Google Scholar]
- Hu H, et al. QTL mapping of stalk bending strength in a recombinant inbred line maize population. Theoretical and Applied Genetics. 2013;126:2257–2266. doi: 10.1007/s00122-013-2132-7. [DOI] [PubMed] [Google Scholar]
- Wilson JR, Mertens DR, Hatfield RD. Isolates of cell types from sorghum stems: Digestion, cell wall and anatomical characteristics. Journal of the Science of Food and Agriculture. 1993;63:407–417. [Google Scholar]
- Hatfield R, Wilson J, Mertens D. Composition of cell walls isolated from cell types of grain sorghum stems. Journal of the Science of Food and Agriculture. 1999;79:891–899. [Google Scholar]
- Du J, et al. Micron-scale phenotyping quantification and three-dimensional microstructure reconstruction of vascular bundles within maize stems based on micro-CT scanning. Functional Plant Biology. 2016;44(1):10–22. doi: 10.1071/FP16117. [DOI] [PubMed] [Google Scholar]
- Pan X, et al. Reconstruction of Maize Roots and Quantitative Analysis of Metaxylem Vessels based on X-ray Micro-Computed Tomography. Canadian Journal of Plant Science. 2018;98(2):457–466. [Google Scholar]
- McElrone AJ, Choat B, Parkinson DY, MacDowell AA, Brodersen CR. Using high resolution computed tomography to visualize the three dimensional structure and function of plant vasculature. Journal of Visualized Experiments. 2013. p. e50162. [DOI] [PMC free article] [PubMed]
- Cloetens P, Mache R, Schlenker M, Lerbs-Mache S. Quantitative phase tomography of Arabidopsis seeds reveals intercellular void network. Proceedings of the National Academy of Sciences of the Unites States of America. 2006;103:14626–14630. doi: 10.1073/pnas.0603490103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorca-Fornell C, et al. Increased leaf mesophyll porosity following transient retinoblastoma-related protein silencing is revealed by microcomputed tomography imaging and leads to a system-level physiological response to the altered cell division pattern. Plant Journal. 2013;76(6):914–929. doi: 10.1111/tpj.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboven P, et al. Void space inside the developing seed of Brassica napus and the modelling of its function. New Phytologist. 2013;199:936–947. doi: 10.1111/nph.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen CR, Roark LC, Pittermann J. The physiological implications of primary xylem organization in two ferns. Plant, Cell & Environment. 2012;35:1898–1911. doi: 10.1111/j.1365-3040.2012.02524.x. [DOI] [PubMed] [Google Scholar]
- Choat B, Brodersen CR, McElrone AJ. Synchrotron X-ray microtomography of xylem embolism in Sequoia sempervirens saplings during cycles of drought and recovery. New Phytologist. 2015;205:1095–1105. doi: 10.1111/nph.13110. [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz JM, et al. Direct x-ray microtomography observation confirms the induction of embolism upon xylem cutting under tension. Plant Physiology. 2015;167:40–43. doi: 10.1104/pp.114.249706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staedler YM, Masson D, Schönenberger J. Plant tissues in 3D via. x-ray tomography: simple contrasting methods allow high resolution imaging. PLoS One. 2013;8:75295. doi: 10.1371/journal.pone.0075295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Legay S, Barrière Y, Méchin V, Legland D. Color quantification of stained maize stem section describes lignin spatial distribution within the whole stem. Journal of the Science of Food and Agriculture. 2013;61:3186–3192. doi: 10.1021/jf400912s. [DOI] [PubMed] [Google Scholar]
- Legland D, Devaux MF, Guillon F. Statistical mapping of maize bundle intensity at the stem scale using spatial normalisation of replicated images. PLoS One. 2014;9(3):90673. doi: 10.1371/journal.pone.0090673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckwolf S, Heckwolf M, Kaeppler SM, de Leon N, Spalding EP. Image analysis of anatomical traits in stem transections of maize and other grasses. Plant Methods. 2015;11:26. doi: 10.1186/s13007-015-0070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Jaeger M, Wang M, Li B, Zhang BG. Three-dimensional distribution of vessels, passage cells and lateral roots along the root axis of winter wheat (Triticum aestivum) Annals of Botany. 2011;107:843–853. doi: 10.1093/aob/mcr005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin J, Laga H, Huang CY, Heuer S, Miklavcic SJ. RootAnalyzer: A Cross-Section Image Analysis Tool for Automated Characterization of Root Cells and Tissues. PLoS One. 2015;10:0137655. doi: 10.1371/journal.pone.0137655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passot S, et al. Characterization of pearl millet root architecture and anatomy reveals three types of lateral roots. Frontiers in Plant Science. 2016;7:829. doi: 10.3389/fpls.2016.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]


