Abstract
Intra-arterial therapies are the standard of care for patients with hepatocellular carcinoma who cannot undergo surgical resection. The objective of this study was to develop a method to predict response to intra-arterial treatment prior to intervention.
The method provides a general framework for predicting outcomes prior to intra-arterial therapy. It involves pooling clinical, demographic and imaging data across a cohort of patients and using these data to train a machine learning model. The trained model is applied to new patients in order to predict their likelihood of response to intra-arterial therapy.
The method entails the acquisition and parsing of clinical, demographic and imaging data from N patients who have already undergone trans-arterial therapies. These data are parsed into discrete features (age, sex, cirrhosis, degree of tumor enhancement, etc.) and binarized into true/false values (e.g., age over 60, male gender, tumor enhancement beyond a set threshold, etc.). Low-variance features and features with low univariate associations with the outcome are removed. Each treated patient is labeled according to whether they responded or did not respond to treatment. Each training patient is thus represented by a set of binary features and an outcome label. Machine learning models are trained using N - 1 patients with testing on the left-out patient. This process is repeated for each of the N patients. The N models are averaged to arrive at a final model.
The technique is extensible and enables inclusion of additional features in the future. It is also a generalizable process that may be applied to clinical research questions outside of interventional radiology. The main limitation is the need to derive features manually from each patient. A popular modern form of machine learning called deep learning does not suffer from this limitation, but requires larger datasets.
Keywords: Medicine, Issue 140, Machine learning, artificial intelligence, interventional radiology, hepatocellular carcinoma, trans-arterial chemoembolization, supervised machine learning, predictive modeling, predicting outcomes, pre-procedure planning
Introduction
Patients with hepatocellular carcinoma who are not surgical candidates are offered intra-arterial therapies1,2,3. There is no single metric that determines whether a patient will respond to an intra-arterial therapy before the treatment is administered. The objective of this study was to demonstrate a method that predicts treatment response by applying methods from machine learning. Such models provide guidance to practitioners and patients when choosing whether to proceed with a treatment.
The protocol entails a reproducible process for training and updating a model starting from primary patient data (clinical notes, demographics, laboratory data, and imaging). The data is initially parsed for specific features, with each patient represented by a set of binary features and a binary outcome target label. The outcome label is determined using an established imaging-based response criterion for hepatocellular therapy4,5,6,7. The features and target labels are passed to machine learning software that learns the mapping between features and outcomes under a specific learning model (logistic regression or random forest)8,9,10. Similar techniques have been applied in radiology and other areas of cancer research for diagnosis and treatment prediction11,12,13.
The method adapts techniques from computer science to the field of interventional radiology. Traditional significance studies in interventional radiology, and medicine in general, rely upon mono- or oligo- feature analyses. For example, the Model for End-Stage Liver Disease incorporates five clinical metrics to assess the extent of liver disease. The benefit of the proposed method is the ability to add features liberally; twenty-five features are considered in the example analysis. Additional features may be added as desired.
The technique may be applied to other radiographic interventions where pre- and post- intervention imaging data are available. For example, outcomes following percutaneous treatments could be predicted in a similar manner. The main limitation of the study is the need to manual curate features for inclusion in the model. Data curation and feature extraction is time-consuming for the practitioner and may impede clinical adoption of such machine learning models.
Protocol
1 . Workstation Setup for Machine Learning
Use a system with the following: Intel Core 2 Duo or higher CPU at 2.0 GHz 4 GB or more system memory POSIX-compliant operating system (Linux or Mac OS) or Microsoft Windows 7 User permissions for executing programs and saving files
- Install the following tools: Anaconda Python3: https://www.anaconda.com/download DICOM to NIfTI converter (dcm2niix) - https://github.com/rordenlab/dcm2niix Sublime Text Editor: https://www.sublimetext.com/ itk-SNAP (optional): http://www.itksnap.org
- Install Anaconda Python3, dcm2nii and Sublime Text, visit their respective websites for operating system specific installation steps.
- Create and activate an Anaconda environment. conda create --name mlenv conda activate mlenv
- Install Anaconda packages for machine learning. conda install numpy scipy scikit-learn nltk nibabel NOTE: The nltk package is useful for parsing plaintext clinical notes, while the nibabel package provides useful functions for medical image manipulation. itk-SNAP may be installed for segmenting organs and tumors from medical images. It is useful for constraining features to specific regions.
2 . Feature Extraction from Plaintext Clinical Notes and Structured Clinical Data
Create a parent directory for the project and create a folder for each patient within the parent folder. Directory structure should resemble: Project/ Project/Patient_1/ Project/Patient_2/ Project/Patient_3/ ...
- Obtain plaintext clinical notes from the electronic medical record (EMR). Retrieve notes manually through the EMR or by means of the hospital information technology (IT) office through a data-dump. Store each patient's notes in their respective folders. Project/Patient_1/History_and_Physical.txt Project/Patient_1/Procedure_Note.txt
- Decided which clinical features to include in the model. Parse the plaintext clinic notes for these features. The Python Natural Language Toolkit (nltk) library provides useful commands for splitting documents into sentences. Each sentence may be searched for appropriate terms such as jaundice. Store each patient's features in a file with one feature per line. Project/Patient_1/Features.txt: age 67 sex male albumin 3.1 cirrhotic no hepatitis_c no ...
- For non-binary features, take the median value of each feature across all patients. Binarize each feature as a true(1) or false(0) value based on the median value. Project/Patient_1/Binary_Features.txt: age_over_60 0 male_sex 1 albumin_less_than_3.5 1 presence_of_cirrhosis 0 hepatitis_c 0 ...
3 . Feature Extraction from Medical Images
NOTE: See Step 3 Supplementary Materials for Code Examples.
Download pre- and post- therapy magnetic resonance DICOM images from the hospital PACS. Store images in the corresponding patient folders. Project/ Project/Patient_1/Pre_TACE_MRI_Pre-Contrast.dcm Project/Patient_1/Pre_TACE_MRI_Arterial.dcm Project/Patient_1/Post_TACE_MRI_Pre-Constrast.dcm Project/Patient_1/Post_TACE_MRI_Arterial.dcm
Convert DICOM images into NIfTI format using the dcm2niix program. The following commands converts all .dcm images in specified folder. Repeat for all patients. dcm2niix Project/Patient_1/ dcm2niix Project/Patient_2/
- Load each NIfTI file into Python. import nibabel image = nibabel.load('Project/Patient_1/Pre_TACE_MRI_Pre-Contrast.dcm')
- Canonicalize the orientation of each image. This ensures that the x, y, and z axes are identical, irrespective of the machine used to acquire the images. cImage = nibabel.as_closest_canonical(image)
Use itk-SNAP (or an equivalent software package) to segment binary liver and tumor masks for each image. Project/Patient_1/Pre_TACE_MRI_Pre-Contrast_Liver_Mask.bin Project/Patient_1/Pre_TACE_MRI_Pre-Contrast_Tumor_Mask.bin
- Read the liver and tumor masks into Python. The code below demonstrates how to correct orientation issues in order to orient the masks along the same canonical axes as the MR images. import numpy as np with open(liver_mask_file, 'rb') as f: liver_mask = f.read() liver_mask = np.fromstring(liver_mask, dtype='uint8') liver_mask = np.reshape(liver_mask, diff.shape, order='F') liver_mask = liver_mask[:,::-1,:] liver_mask[liver_mask > 0] = 1
- Use the liver and tumor masks to isolate voxels containing liver and tumor. liver = np.copy(cImage) liver[liver_mask <= 0] = 0
- Calculate mean liver enhancement feature. mean_liver_enhancement = mean(liver)
- Calculate liver volume feature. pixdim = cImage.header['pixdim'] units = pre.header['xyzt_units'] dx, dy, dz = pre_pixdim[1:4] liver_volume = length(liver) * dx * dx * dz
- (Optional) Calculate additional features as desired.
- Update patient-specific features file with the image features. Project/Patient_1/Features.txt: age 67 sex male albumin 3.1 cirrhotic no hepatitis_c no pre_tace_mean_liver_enhancement 78 pre_tace_liver_volume 10000
- Calculate median values for each imaging feature and binarize as in Step 2.2.2. Project/Patient_1/Binary_Features.txt: age_over_60 0 male_sex 1 albumin_less_than_3.5 1 presence_of_cirrhosis 0 hepatitis_c 0 pre_tace_mean_liver_enhancement 1 pre_tace_liver_volume 0
4. Feature Aggregation and Reduction
NOTE: See Step 4 Supplementary Materials for Code Examples.
- Combine the Binary_Features.txt files for each patient into a spreadsheet with patients on the y-axis and features on the x-axis.
Patient Age > 60 Male Sex Albumin < 3.5 Presence of Cirrhosis Hepatitis C Present mean liver enhancement > 50 liver volume > 20000 1 0 1 1 0 0 1 0 2 1 1 1 0 0 0 0 3 0 1 1 0 1 0 0 - Add qEASL outcome response labels as the final column.
Patient Age > 60 Male Sex Albumin < 3.5 Presence of Cirrhosis Hepatitis C Present mean liver enhancement > 50 liver volume > 20000 qEASL Responder 1 0 1 1 0 0 1 0 1 2 1 1 1 0 0 0 0 1 3 0 1 1 0 1 0 0 0 - Export the spreadsheet as a tab-delimited file. Project/ML_Matrix.tsv: PatientAge > 60 Male Sex Albumin < 3.5 Presence of Cirrhosis Hepatitis C Present mean liver enhancement > 50 liver volume > 20000 qEASL Responder 1 0 1 1 0 0 1 0 1 2 1 1 1 0 0 0 0 1 3 0 1 1 0 1 0 0 0
- Remove low-variance features from consideration. import numpy as np from sklearn.feature_selection import VarianceThreshold # Read in the binary matrix. features = [] labels = [] for i, L in enumerate(sys.stdin): if i == 0 continue n_fs_L = L.strip().split('\t') features.append([float(_) for _ in n_fs_L[1:-1]]) labels.append(n_fs_L[-1]) X = np.array(features) y = np.array(labels) # Compute features appearing in at least 20% of both responders and non-respnders. model = VarianceThreshold(threshold=0.8 * (1 - 0.8)) X_new = model.fit_transform(X, y) The male sex, albumin < 3.5, presence of cirrhosis, and liver volume > 2000 features have been removed.
Patient Age > 60 Hepatitis C Present mean liver enhancement > 50 qEASL Responder 1 0 0 1 1 2 1 0 0 1 3 0 1 0 0 Remove features with low univariate-association with the outcome. Filter only those features that passed 4.2. Retain ceil(log2(N)) features, where N is the number patients. Ceil(Log2(3)) = 2. import math from sklearn.feature_selection import SelectKBest from sklearn.feature_selection import chi2 # Read in the binary matrix as in 4.2.1 ... # Compute top ceil(log2(N)) features by univariate association. k = math.ceil(log2(length(y))) model = SelectKBest(chi2, k=k) X_new = model.fit_transform(X, y) The male sex age > 60 feature has been removed from the remaining features from 4.2.1.
| Patient | Hepatitis C Present | mean liver enhancement > 50 | qEASL Responder |
| 1 | 0 | 1 | 1 |
| 2 | 0 | 0 | 1 |
| 3 | 1 | 0 | 0 |
5 . Model Training and Testing
See Step 5 Supplementary Materials for Code Examples
Train a logistic regression model using the binary features matrix from 4.3. import math from sklearn.linear_model import LogisticRegression # Read in the binary matrix as in 4.2 and 4.3. ... # For each patient, train a model on all other patients. score = 0.0 models = [] for patient in len(X): # Train model on all but one of the patients. train_x = np.array([_ for i, _ in enumerate(X) if i != patient]) train_y = np.array([_ for i, _ in enumerate(y) if i != patient]) model = LogisticRegression(C=1e15) model.fit(train_x, train_y) # Test on the left-out patient. y_prediction = model.predict(X[patient]) if y_prediction == y[patient]: score += 1 models.append(model)
Train a random forest model using the binary features matrix from 4.2.2. Steps are identical to 5.2.1, except the model instantiation should be updated as follows: from sklearn.ensemble import RandomForestClassifier ... model = RandomForestClassifier(n_estimators=100) ...
Print out score / len(X)for 5.1 and 5.2. This represents the average accuracy of all logistic regression models and all random forest models, respectively. All N models should be applied to new patients with the average classification taken as the prediction outcome
Representative Results
The proposed method was applied to 36 patients who had undergone trans-arterial therapies for hepatocellular carcinoma. Twenty-five features were identified and binarized using steps 1-5. Five features satisfied both the variance and univariate association filters (see steps 5.1 and 5.2) and were used for model training. Each patient was labeled as either a responder or non-responder under the qEASL response criteria. The features matrix was thus a 36 x 5 array while the target labels vector was 36 x 1.
Logistic regression and random forest classifiers were used for model fitting. Leave-one-out cross-validation was used to assess the performance of the resulting models. Two additional models were trained using just the top-two features (presence of cirrhosis and pre-TACE tumor signal intensity greater than 27.0). Figure 1 illustrates the performance of the models as features were added. Both logistic regression and random forest models predicted trans-arterial chemoembolization treatment response with an overall accuracy of 78% (sensitivity 62.5%, specificity 82.1%, positive predictive value 50.0%, negative predictive value 88.5%).
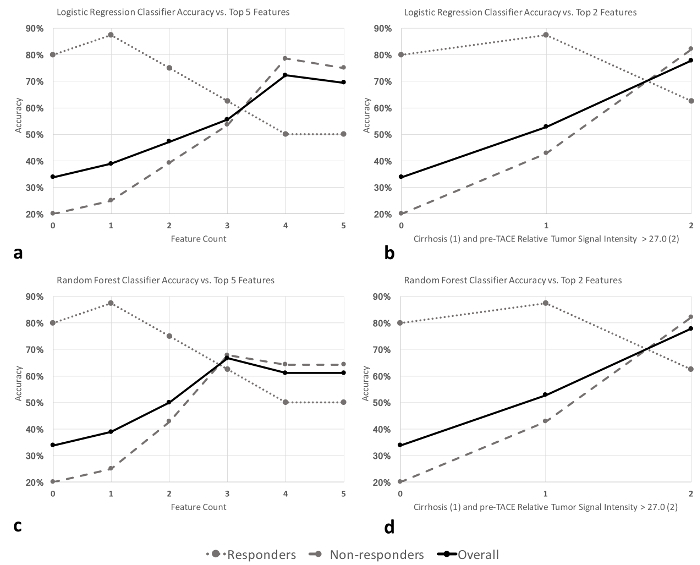
Figure 1: Performance of machine learning algorithms. (a,b) Logistic regression and (c,d) random forest classifier accuracies as features are added. Features were added in the following order: 1) ethiodized oil, 2) sorafenib, 3) cirrhosis, 4) pre-transarterial chemo- embolization relative tumor signal intensity >27.0, and 5) number of tumors >2. Figure used with permission in unmodified form from the Journal of Vascular and Interventional Radiology14. Please click here to view a larger version of this figure.
Discussion
Patients with hepatocellular carcinoma who are not candidates for surgical resection are offered intra-arterial therapies. Few methods exist to determine if a patient will respond pre-treatment. Post-treatment evaluation techniques rely upon changes in tumor size or tumor contrast uptake. These are called response criteria, with the most accurate being the Quantitative European Association for the Study of the Liver (qEASL) criterion. qEASL relies upon both volumetric and enhancement changes following therapy to predict a likelihood of response. Despite the strengths of qEASL, it is nonetheless a post-treatment evaluation criteria, and cannot aid in treatment planning.
There is a need to assess which patients are likely to respond to intra-arterial therapies before performing the intervention. The method demonstrated in this protocol incorporates clinical, laboratory and imaging features into a predictive model using techniques from the fields of computer science and statistics. A machine learning model is trained that maps these features from patients who have undergone intra-arterial therapies to their qEASL outcomes. The model may then be applied to new patients who will undergo treatment to predict their qEASL response using only their pre-treatment features.
Step 1 describes workstation setup for machine learning. It provides brief instructions on how to setup a workstation with required tooling. Steps 2-4 go into technical detail about how clinical and imaging data might be parsed to obtain features of interest. These steps are critical as selection of appropriate features will determine the effectiveness of the model. Certain substeps were chosen to ease feature extraction. For example, radiographic images are typically stored in DICOM format which is not ideal for image analysis. The National Institutes of Health (NIH) Neuroimaging Branch developed the Neuroimaging Informatics Technology Initiative (NIfTI) standard to facilitate image manipulation in research environments. Step 3.2 entails the conversion from DICOM to NIfTI format to ease the process of feature extraction. Liver and tumor masks may be extracted from NIfTI format using a program such as itk-SNAP or similar segmentation software.
Each feature, whether acquired from clinical notes or imaging data, should be binarized as true-false values. For example, a continuous image enhancement gradient ranging from 0 to 10 may be binarized to a single feature representing enhancement greater than or less than 5. Alternatively, the feature could be split into multiple binary features: x < 3; 3 <= x < 7; 7 <= x. Machine learning models that operate on binary features are easier to train.
The end-result of Steps 2-4 is a binary matrix with patients on the y-axis, features on the x-axis, and a final column representing the outcome (responder or non-responder) as determined for that patient under the qEASL response criterion. Certain features may be under-represented or over-represented in an outcome population. For example, if all treatment responders were male, it may incorrectly be concluded that male gender implies response. One way to deal with this fallacy is to remove all features that are not found in both responders and non-responders above some threshold such as 20%.
Other features may have limited importance in determining the outcome under study. For example, eye color is likely irrelevant to outcomes following intra-arterial therapies. Such features will have a low univariate association with the outcome. Although no single feature is expected to have a significant (p < 0.05) association with the outcome, an effective strategy is to require that features have some minimal univariate correlation above a defined threshold.
Step 5 covers the process of training and applying a machine learning model. It is not strictly necessary to proceed in the fashion described, as long as the end-result is the same. The training process uses leave-one-out cross-validation whereby N models are trained for each of the N patients. The N resulting models generated from leave-one-out cross-validation may be averaged to produce a final model.
The technique is also applicable to other procedures done under imaging. Machine learning takes features from a set of training patients and weights them according to their relative contribution to a target outcome. The weighting process depends on the chosen model; logistic regression models calculate an exponentiated linear combination while random forest models employ a set of weighted decision trees. The target used in this protocol was response under the qEASL criterion. Other targets, such as years of disease-free survival or quality of life years, may be chosen depending on the desired outcome under question.
The main limitation of the protocol is the need to manually define and obtain the features under consideration. This entails a significant amount of manual work such as parsing clinical notes, segmenting tumor volumes and counting the number of lesions. Deep machine learning attempts to automatically derive features from the raw source data, but requires significantly more training data. This growing technology has been shown to be superior to supervised learning in a variety of contexts, and will likely be the next evolution of predictive models for patients undergoing image-guided procedures.
Disclosures
A.A. works as a software consult for Health Fidelity, Inc. that employs similar machine learning techniques on clinical notes for optimizing medical reimbursement.
J.F.G. receives personal fees from Guerbet Healthcare, BTG, Threshold Pharmaceuticals (San Francisco, California), Boston Scientific, and Terumo (Elkton, Maryland); and has a paid consultancy for Prescience Labs (Westport, Connecticut).
None of the other authors have identified a conflict of interest.
Acknowledgments
A.A. received funding support from the Office of Student Research, Yale School of Medicine.
L.J.S. receives grants from the National Institutes of Health (NIH/NCI R01CA206180), Leopoldina Postdoctoral Fellowship, and the Rolf W. Guenther Foundation of Radiological Sciences (Aachen, Germany).
J.C. receives grants from the National Institutes of Health (NIH/NCI R01CA206180), Philips Healthcare, and the German-Israeli Foundation for Scientific Research and Development (Jerusalem, Israel and Neuherberg, Germany); and scholarships from the Rolf W. Guenther Foundation of Radiological Sciences and the Charite Berlin Institute of Health Clinical Scientist Program (Berlin, Germany).
J.S.D. and M.L. receive grants from the National Institutes of Health (NIH/NCI R01CA206180) and Philips Healthcare (Best, The Netherlands).
J.F.G. receives grants from the National Institutes of Health (NIH/NCI R01CA206180), Philips Healthcare, BTG (London, United Kingdom), Boston Scientific (Marlborough, Massachusetts), and Guerbet Healthcare (Villepinte, France)
References
- Benson A, et al. NCCN clinical practice guidelines in oncology: hepatobiliary cancers. J National Comprehensive Cancer Network. 2009;7(4):350–391. doi: 10.6004/jnccn.2009.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Miller K, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- Bruix J, et al. Clinical management of hepatocellular carcinoma. Conclusions of the Barcelona-2000 European Association for the Study of the Liver conference. Journal of Hepatology. 2001;35(3):421–430. doi: 10.1016/s0168-8278(01)00130-1. [DOI] [PubMed] [Google Scholar]
- Eisenhauer E, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) European Journal of Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Gillmore R, et al. EASL and mRECIST responses are independent prognostic factors for survival in hepatocellular cancer patients treated with transarterial embolization. Journal of Hepatology. 2011;55(6):1309–1316. doi: 10.1016/j.jhep.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Lin M, et al. Quantitative and volumetric European Association for the Study of the Liver and Response Evaluation Criteria in Solid Tumors measurements: feasibility of a semiautomated software method to assess tumor response after transcatheter arterial chemoembolization. Journal of Vascular and Interventional Radiology. 2012;23(12):1629–1637. doi: 10.1016/j.jvir.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacher V, et al. Comparison of Existing Response Criteria in Patients with Hepatocellular Carcinoma Treated with Transarterial Chemoembolization Using a 3D Quantitative Approach. Radiology. 2016;278(1):275–284. doi: 10.1148/radiol.2015142951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- Bishop C. Pattern recognition and machine learning. New York: Springer; 2006. p. 738. [Google Scholar]
- Alpaydin E. Introduction to machine learning. Third edition. Cambridge, Massachusetts: The MIT Press; 2014. p. 613. [Google Scholar]
- Kim S, Cho K, Oh S. Development of machine learning models for diagnosis of glaucoma. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son Y, Kim H, Kim E, Choi S, Lee S. Application of support vector machine for prediction of medication adherence in heart failure patients. Healthcare Informatics Research. 2010;16(4):253–259. doi: 10.4258/hir.2010.16.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Summers R. Machine learning and radiology. Medical Image Analysis. 2012;16(5):933–951. doi: 10.1016/j.media.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abajian A, et al. Predicting Treatment Response to Intra-arterial Therapies for Hepatocellular Carcinoma with the Use of Supervised Machine Learning-An Artificial Intelligence Concept. Journal of Vascular and Interventional Radiology. 2018. [DOI] [PMC free article] [PubMed]


