Abstract
Quasi-metagenomics sequencing refers to the sequencing-based analysis of modified microbiomes of food and environmental samples. In this protocol, microbiome modification is designed to concentrate genomic DNA of a target foodborne pathogen contaminant to facilitate the detection and subtyping of the pathogen in a single workflow. Here, we explain and demonstrate the sample preparation steps for the quasi-metagenomics analysis of Salmonella enterica from representative food and environmental samples including alfalfa sprouts, ground black pepper, ground beef, chicken breast and environmental swabs. Samples are first subjected to the culture enrichment of Salmonella for a shortened and adjustable duration (4–24 h). Salmonella cells are then selectively captured from the enrichment culture by immunomagnetic separation (IMS). Finally, multiple displacement amplification (MDA) is performed to amplify DNA from IMS-captured cells. The DNA output of this protocol can be sequenced by high throughput sequencing platforms. An optional quantitative PCR analysis can be performed to replace sequencing for Salmonella detection or assess the concentration of Salmonella DNA before sequencing.
Keywords: Biology, Issue 140, quasimetagenomics, immunomagnetic separation (IMS), whole genome amplification, sequencing, detection, subtyping, Salmonella
Introduction
Metagenomics sequencing theoretically allows concerted detection and subtyping of foodborne pathogens. However, food samples present challenges to the pathogen analysis by direct sequencing of the food microbiome. First, foodborne pathogens are often present at low levels in food samples. Most of the commercially available rapid detection methods still require 8–48 h culturing to enrich pathogen cells to a detectable level1. Second, many foods contain abundant microflora cells and/or food DNA, making foodborne pathogen DNA a small fraction of food metagenome and an elusive target for detection and subtyping by direct metagenomic sequencing.
Modification of food microbiomes has been reported to allow substantial concentration of foodborne pathogen DNA to facilitate sequencing-based detection of Shiga toxin-producing Escherichia coli2,3 and Salmonella enterica4. Because modified food microbiomes are still mixtures of different microbial species, their sequencing is termed as quasi-metagenomic analysis4. Microbiome modification can be performed by culture enrichment alone2,3 or in combination with immunomagnetic separation (IMS) and multiple displacement amplification (MDA)4,5. IMS can selectively capture pathogen cells from the enrichment culture via antibody-coated magnetic beads. MDA can generate sufficient amounts of genomic DNA for sequencing through the highly efficient ɸ29 DNA polymerase6. IMS-MDA has allowed culture-independent pathogen detection from clinical samples7, and shortening of the culture enrichment for quasi-metagenomic detection and subtyping of Salmonella in food samples4.
The overall goal of this method is to prepare quasi-metagenomic DNA from food samples to allow targeted concentration of Salmonella genomic DNA and subsequent detection and subtyping of the Salmonella contaminant by sequencing. Compared to standard methods for Salmonella detection8,9 and subtyping10, the quasimetagenomic approach can substantially shorten the turnaround time from contaminated food and environmental samples to molecular subtypes of the pathogen by unifying the two typically separated analyses into a single workflow. This method is particularly useful for applications such as foodborne outbreak response and other trace back investigations where robust pathogen subtyping is required in addition to pathogen detection and rapid analytical turnaround is important.
Protocol
1. Sample Preparation
NOTE: Food samples are prepared for pre-enrichment according to Microbiology Laboratory Guidebook (MLG) of U.S. Department of Agriculture Food Safety and Inspection Service (USDA-FSIS)11 and Bacteriological analytical manual (BAM) of U.S. Food and Drug Administration (FDA)12.
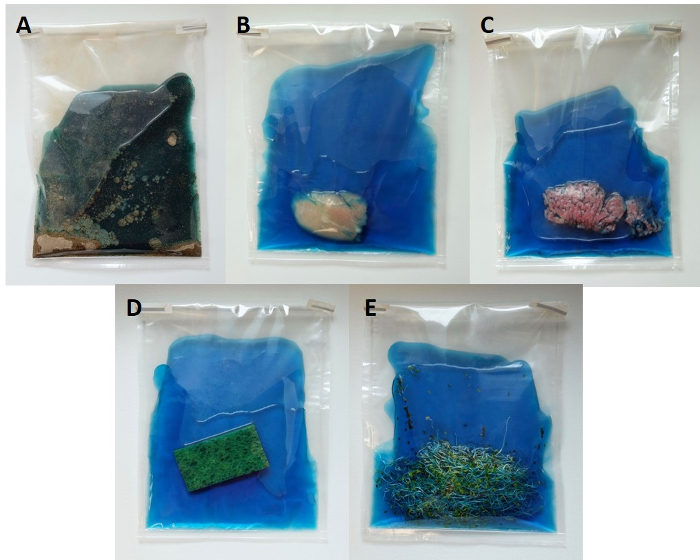
Aseptically place a 25 g portion of food sample such as black pepper, chicken breast, ground beef, and alfalfa sprouts or an environmental swab into a sterile laboratory blender bag with a built-in filter. Prepare an environmental swab by aseptically moistening a sponge with enrichment broth (Rappaport-Vassiliadis, RV). Then, drag the swab over entire surface or predetermined area and place it into a laboratory blender bag.
Figure 1: Representative food and environmental samples for quasi-metagenomics detection and subtyping of Salmonella. Samples are placed in sterile filter stomacher bags along with RV broth. Please click here to view a larger version of this figure.
Thoroughly mix each 25 g sample with 225 mL of RV enrichment broth using a laboratory blender or hand massage for 30 s. NOTE: Variants of RV broth as recommended by MLG and BAM may be used depending on sample types.
Incubate sample-enrichment broth mixture in a laboratory incubator at 42 °C for 4–24 h.
After incubation, collect a 50 mL subsample of enrichment broth from the filtered side of the bag in a separate 50 mL centrifuge tube.
Centrifuge the tube at 100 x g for 10 min to remove solid debris in the homogenate.
Recover the supernatant carefully to a new 50 mL centrifuge tube and centrifuge the tubes at 3,000–6,000 x g for 10 min to recover cell pellet.
(Optional) Discard the supernatant and wash the pellet by re-suspension in 5 mL of Buffered Peptone Water (BPW) and centrifuge the tubes at 3,000–6,000 × g for 10 min. NOTE: BPW can prepared by dissolving 10 g of peptone, 5 g of sodium chloride, 3.5 g of disodium phosphate, and 1.5 g of monopotassium phosphate into 1 L of distilled water. The final pH should be adjusted to 7.2 ± 0.2 at 25 °C. Autoclave at 121 ºC for 15 min. Commercially available BPW can also be used.
Discard supernatant and re-suspend the pellet in 5 mL of BPW.
2. Immunomagnetic Separation (IMS) and Multiple Displacement Amplification (MDA)
NOTE: To prevent cross-contamination between samples, work in a biosafety cabinet, change pipette tips for each tube, avoid touching the tube with the pipette, and do not place tubes close together in magnetic stand during the washing step.
Thaw the sample buffer and the reaction buffer from the MDA kit on ice or at 4 °C in advance.
Mix 1 mL of re-suspended cell pellet in BPW with 20 μL of anti-Salmonella beads in 1.5 mL microcentrifuge tubes and place it on the rotating mixer for 30 min at room temperature. NOTE: Competitive flora, fat particulates, and proteins in the resuspended pellet can interfere with IMS. Dilution of re-suspended cell pellet in BPW is helpful to reduce the loss of the bead-Salmonella complexes from the magnetic stand during washing steps.
Insert the tubes into the magnetic stand and allow 3 min for the proper recovery of beads by inverting the rack several times to concentrate the beads into a pellet on the side of the tube.
Keep the tubes on the magnetic stand. Aspirate and discard the supernatant from each tube, as well as the remaining liquid in each tube’s cap. NOTE: Be careful not to disturb the pellet of IMS beads on the side wall of the tube against the magnet.
Remove the tube holder from the magnetic stand and add 1 mL of wash buffer (PBS containing 0.05 % (v/v) polysorbate 20) and invert several times to remove non-specifically binding bacteria from the complex.
Repeat steps 2.3–2.5 twice.
After the 3rd wash, perform the final magnetic separation of beads by repeating steps 2.3–2.4. Remove tubes from magnetic rack and place them in a microfuge for 1 s to spin down beads. Then, place the tubes in the magnetic rack for 3 min before removing any residual wash buffer.
Re-suspend the bead-Salmonella complexes in 9 μL of Sample buffer and incubate at 95 °C for 3 min for denaturation.
After cooling to 4 °C on ice, combine with 9 μL of Reaction buffer plus 1 μL of enzyme mix and keep on ice.
In a thermal cycler, incubate tubes at 30 °C for 2 h for amplification, followed by heating at 65 °C for 10 min to inactivate the enzyme and cool to 4 °C on ice.
Assess the quantity and quality of the MDA products by measuring the DNA concentration and purity (260/280 ratio > 1.8) on a fluorospectrometer instrument. NOTE: Because of non-specific binding of IMS beads, genomic DNA from organisms other than Salmonella is likely to be present in the MDA products, but it does not affect downstream analysis.
Store the final products (20 μL) at -20 °C until use for real-time PCR and/or library preparation for quasimetagenomics sequencing.
3. Real-Time PCR
NOTE: This step is optional for 1) detecting Salmonella without subtyping, and 2) assessing sample quality prior to quasimetagenomics sequencing.
Prepare 18 μL of PCR mixture per sample, containing Universal PCR Master Mix (10 μL), forward primer (2 μL, 900 nM), reverse primer (2 μL, 900 nM), probe (2 μL, 250 nM), and molecular grade water (2 μL). Note: Salmonella-specific oligonucleotide primers (forward: CTCACCAGGAGATTACAACATGG, reverse: AGCTCAGACCAAAAGTGACCATC) and probe were designed to amplify a 94-bp sequence within the ttr gene (GenBank accession no. AF 282268)13.
Mix the MDA product from step 2.12 by gently pipetting up and down bead-Salmonella complexes along with liquid to create a suspension. Add 2 μL of the suspension into 18 μL of PCR mixture.
Run real-time PCR using an optimized real-time PCR protocol, specifying two holding periods, one at 50 °C for 2 min and another at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s.
Calculate the threshold cycle (Ct), which is the number of cycles required for the fluorescence from the amplified DNA to cross the threshold line. NOTE: The threshold line is set in the linear phase between base line and plateau of the amplification plots of samples. Negative results correspond to Ct values over 40 or samples with Ct values higher than that of negative control.
4. Library Preparation for Quasimetagenomic Sequencing
NOTE: The MDA products can be sequenced by both short read and long read (nanopore) sequencing platforms. Use the latest version of DNA library prep kits provided by the manufacturers of the sequencing platforms. Perform DNA library preparation according to manufacturers’ instruction. Use the 2D low-input genomic DNA protocol for library preparation for the long-read sequencing platform5. For library preparation for the short-read sequencing platform, minor modifications to the manufacturer’s protocol are provided below.
Follow standard library preparation methods by adding buffer solutions and reagents to MDA products. Transfer 40 μL of the sequencing prepped MDA products to a new plate and add 20 μL of PCR purification beads to each well.
Incubate the plate at room temperature for 10 min without shaking.
Wash the beads with 80% ethanol and dry the beads for 12 min.
Re-suspend dried beads in 53 μL of resuspension buffer and incubate for 2 min without shaking.
Dilute and pool libraries following manufacturer’s instruction. NOTE: A 10 pM pooled and denatured library is now ready to be sequenced.
Representative Results
Prior to quasimetagenomic sequencing, the overall quantity and purity of IMS-MDA products can be evaluated by fluorospectrometer (Table 1).
| Enrichment time (h) | Ct value | Concentration (ng/ul) | Purity (260/280) | |
| Brand A | 4 | 24.71 | 812.91 | 1.89 |
| 8 | 25.80 | 900.36 | 1.87 | |
| 12 | 15.41 | 753.88 | 1.84 | |
| Brand B | 4 | 20.63 | 872.61 | 1.87 |
| 8 | 22.61 | 993.41 | 1.87 | |
| 12 | 19.08 | 954.44 | 1.87 |
Table 1: Quality and quantity assessment of DNA samples for quasi-metagenomic sequencing.Salmonella cells were inoculated in 25 g of raw chicken breast of two brands at 1 CFU/g. Inoculated samples were enriched in RV for 4, 8, and 12 h before IMS-MDA.
Targeted quantity assessment of Salmonella DNA can be performed by real-time PCR (Table 1 and Figure 2), which also serves as an alternative for Salmonella detection if subtyping is not needed.
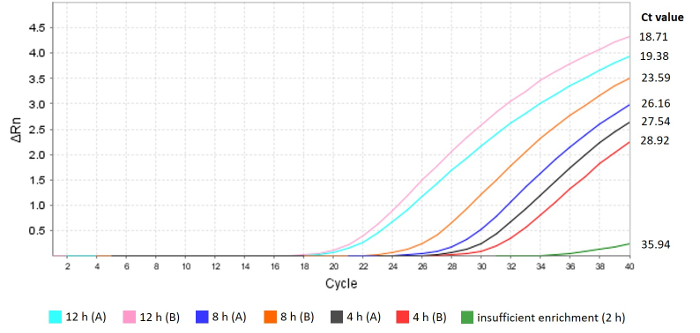
Figure 2: Real-time PCR for Salmonella detection or sample assessment for quasi-metagenomics sequencing. Amplification curves of real-time PCR of Salmonella DNA in IMS-MDA products after 4, 8 and 12 h of enrichment time are shown. A sample after only 2 h enrichment is also included. Salmonella cells (0.27 CFU/g) were inoculated 25 g of raw chicken breast from two brands A and B. Please click here to view a larger version of this figure.
Fluorospectrometer readings allow initial evaluation of sample preparation. Concentration of IMS-MDA products prepared from Salmonella contaminated chicken breast samples ranged from 701.54 to 945.86 ng/µL, suggesting that the sample DNA has been amplified. According to manufacturer's guidance, the minimum DNA sample concentration for short read and long read sequencing is 0.2 ng/µL and 1 µg, respectively. The 260/280 ratio ranged from 1.85 to 1.88 (Table 1), showing high purity of the DNA samples with low levels of protein contamination.
Lower Ct values indicate higher concentrations of Salmonella genomic DNA in the sample. As shown in Table 1 and Figure 2, Ct values of IMS-MDA products decrease as enrichment time increases. In our previous study4, when Ct values were lower than 25, the majority (>50%) of the SE genome was likely to be sequenced, and serotype prediction from raw sequencing reads was successful.
Unsuccessful culture enrichment or IMS can result in a relatively high Ct values indicating low concentration of Salmonella genomic DNA in the final sample as shown in Figure 2. This can happen despite a high DNA concentration and purity of the IMS-MDA products as suggested by fluorospectrometer readings.
Discussion
Because of the often-low abundance and in-homogenous presence of Salmonella in food and environmental samples, culture enrichment before IMS-MDA is still necessary for Salmonella detection and subtyping; it is therefore a critical step of the protocol. To identify optimal conditions to increase the abundance of Salmonella relative to sample background flora, different enrichment media may be evaluated for specific samples. According to MLG and BAM, both selective medium such as RV broth and non-selective medium such as buffered peptone water and lactose broth can be used for Salmonella enrichment from food samples. The use of RV at 42 °C has been evaluated in our previous study4 for Salmonella enrichment from raw chicken meat, iceberg lettuce and black peppercorns to support quasimetagenomic analysis of the pathogen. According to FDA BAM and AOAC precollaborative8,9and collaborative studies14,15, RV is recommended for the analysis of high microbial and low microbial load foods.
The optimal duration of culture enrichment depends on multiple factors, such as sample type, level of Salmonella contamination, and the physiological state of the Salmonella contaminant. For example, when low levels of Salmonella (e.g., < 1 CFU/g) cells are present in a meat sample and under refrigeration stress, longer enrichment time (e.g., > 12 h) may be needed to revive and enrich Salmonella cells. Sufficient abundance of Salmonella in the modified microbiome leads to adequate sequencing coverage of the Salmonella contaminant genome, which can allow strain-level single nucleotide polymorphism (SNP) typing4. Shorter culture enrichment (e.g., less than 12 h) is possible for rapid detection5 or serotyping of Salmonella4.
Higher levels of Salmonella contamination and/or longer culture enrichment of contaminated food samples can result in higher concentrations of Salmonella genomic DNA in modified food microbiomes, which lead to lower Ct values from the real-time PCR analysis. Ct values display a negative correlation with sequencing coverage of the Salmonella contaminant genome4, and therefore can be used for optimization and modification of the workflow. According to our experience, culture enrichment, especially the length of it, is often the focus of troubleshooting efforts. For example, the Ct value of the IMS-MDA product from a raw chicken breast sample was relatively high at 35.94 (Figure 1). This sample was inoculated with 2.5 CFU/g of SE cells and incubated in BPW at 37 °C for 2 h. Sequencing of this sample did not provide sufficient coverage of the SE genome to allow serotype prediction. In comparison, all the other samples with 4 h or longer enrichment were successfully serotyped from quasimetagenomic sequencing data (data not shown).
After sequencing, we recommend using Kraken16 to identify Salmonella reads for further bioinformatics analyses. CFSAN SNP pipeline17 and SeqSero18 can be used for SNP typing and serotyping prediction from raw sequencing reads, respectively.
This quasimetagenomic technique has several limitations. First, DNA concentration and purity of IMS-MDA products measured by a fluorospectrometer should be used only as a preliminary indicator of proper sample preparation. Due to the high efficiency of MDA, trace amounts of input DNA can be amplified to suffice whole genome sequencing19. Therefore, high quality and quantity DNA samples can still be generated even if culture enrichment or IMS fails to effectively concentrate Salmonella cells. The optional real-time PCR analysis provides a more targeted and informative assessment of the entire sample preparation workflow. Second, difficult-to-culture Salmonella cells, such as viable but nonculturable (VBNC) bacteria20, may not grow during culture enrichment. Therefore, these cells may not be detected by our method.
In summary, compared with traditional methods, our sample preparation method and quasi-metogenomic sequencing approach can allow substantial reduction of the analytical turnaround from Salmonella-contaminated food and environmental samples to robust subtyping of the Salmonella contaminants. In addition to Salmonella, this technique has the potential be applied to rapid and concerted detection and subtyping other foodborne pathogens.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
The authors would like to thank Mark Harrison and Gwen Hirsch of the University of Georgia for kindly providing the bacterial strain and other support to this study.
References
- Valderrama WB, Dudley EG, Doores S, Cutter CN. Commercially Available Rapid Methods for Detection of Selected Food-borne Pathogens. Critical Reviews in Food Science and Nutrition. 2016;56(9):1519–1531. doi: 10.1080/10408398.2013.775567. [DOI] [PubMed] [Google Scholar]
- Leonard SR, Mammel MK, Lacher DW, Elkins CA. Application of metagenomic sequencing to food safety: detection of Shiga Toxin-producing Escherichia coli on fresh bagged spinach. Applied and Environmental Microbiology. 2015;81(23):8183–8191. doi: 10.1128/AEM.02601-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard SR, Mammel MK, Lacher DW, Elkins CA. Strain-Level Discrimination of Shiga Toxin-Producing Escherichia coli in Spinach Using Metagenomic Sequencing. PLoS One. 2016;11(12):e0167870. doi: 10.1371/journal.pone.0167870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyeon JY, et al. Quasi-metagenomics and realtime sequencing aided detection and subtyping of Salmonella enterica from food samples. Applied and Environmental Microbiology. 2017. [DOI] [PMC free article] [PubMed]
- Hyeon JY, Deng X. Rapid detection of Salmonella in raw chicken breast using real-time PCR combined with immunomagnetic separation and whole genome amplification. Food Microbiology. 2017;63:111–116. doi: 10.1016/j.fm.2016.11.007. [DOI] [PubMed] [Google Scholar]
- Hosono S, et al. Unbiased whole-genome amplification directly from clinical samples. Genome Research. 2003;13(5):954–964. doi: 10.1101/gr.816903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith HM, et al. Whole-genome sequences of Chlamydia trachomatis directly from clinical samples without culture. Genome Research. 2013;23(5):855–866. doi: 10.1101/gr.150037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson AP, Gill VS, Irvin KA, Wang H, Hammack TS. Evaluation of methods to prepare samples of leafy green vegetables for preenrichment with the Bacteriological Analytical Manual Salmonella culture method. Journal of Food Protection. 2012;75(2):400–404. doi: 10.4315/0362-028X.JFP-11-196. [DOI] [PubMed] [Google Scholar]
- Jacobson AP, Hammack TS, Andrews WH. Evaluation of sample preparation methods for the isolation of Salmonella from alfalfa and mung bean seeds with the Bacteriological Analytical Manual's Salmonella culture method. Journal of AOAC International. 2008;91(5):1083–1089. [PubMed] [Google Scholar]
- Deng X, et al. Comparative analysis of subtyping methods against a whole-genome-sequencing standard for Salmonella enterica serotype Enteritidis. Journal of Clinical Microbiology. 2015;53(1):212–218. doi: 10.1128/JCM.02332-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous. Microbiology Laboratory Guidebook. 2018. Available from: https://www.fsis.usda.gov/wps/portal/fsis/topics/science/laboratories-and-procedures/guidebooks-and-methods/microbiology-laboratory-guidebook/microbiology-laboratory-guidebook.
- Andrews WH, Wang H, Jacobson A, Hammack T. Bacteriological Analytical Manual (BAM) Chapter 5: Salmonella. 2018. Available from: https://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070149.htm.
- Malorny B, et al. Diagnostic real-time PCR for detection of Salmonella in food. Applied and Environmental Microbiology. 2004;70(12):7046–7052. doi: 10.1128/AEM.70.12.7046-7052.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June GA, Sherrod PS, Hammack TS, Amaguana RM, Andrews WH. Relative effectiveness of selenite cystine broth, tetrathionate broth, and Rappaport-Vassiliadis medium for the recovery of Salmonella from raw flesh and other highly contaminated foods: precollaborative study. Journal of AOAC International. 1995;78(2):375–380. [PubMed] [Google Scholar]
- Hammack TS, Amaguana RM, June GA, Sherrod PS, Andrews WH. Relative effectiveness of selenite cystine broth, tetrathionate broth, and Rappaport-Vassiliadis medium for the recovery of Salmonella spp. from foods with a low microbial load. Journal of Food Protection. 1999;62(1):16–21. doi: 10.4315/0362-028x-62.1.16. [DOI] [PubMed] [Google Scholar]
- Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biology. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Pettengill JB, Luo Y, Payne J, Shpuntoff A, Rand H, Strain E. CFSAN SNP Pipeline: an automated method for constructing SNP matrices from next-generation sequence data. PeerJ Computer Science. 2015;1(e20) [Google Scholar]
- Zhang S, et al. Salmonella serotype determination utilizing high-throughput genome sequencing data. Journal of Clinical Microbiology. 2015;53(5):1685–1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue S, et al. Whole genome amplification and de novo assembly of single bacterial cells. PLoS One. 2009;4(9):e6864. doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy T, Ghosh A, Pazhani GP, Shinoda S. Current Perspectives on Viable but Non-Culturable (VBNC) Pathogenic Bacteria. Front Public Health. 2014;2:103. doi: 10.3389/fpubh.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]


