Abstract
The whitefly Bemisia tabaci (Gennadius) is an invasive pest of considerable importance, affecting the production of vegetable and ornamental crops in many countries around the world. Severe yield losses are caused by direct feeding, and even more importantly, also by the transmission of more than 100 harmful plant pathogenic viruses. As for other invasive pests, increased international trade facilitates the dispersal of B. tabaci to areas beyond its native range. Inspections of plant import products at points of entry such as seaports and airports are, therefore, seen as an important prevention measure. However, this last line of defense against pest invasions is only effective if rapid identification methods for suspicious insect specimens are readily available. Because the morphological differentiation between the regulated B. tabaci and close relatives without quarantine status is difficult for non-taxonomists, a rapid molecular identification assay based on the loop-mediated isothermal amplification (LAMP) technology has been developed. This publication reports the detailed protocol of the novel assay describing rapid DNA extraction, set-up of the LAMP reaction, as well as interpretation of its read-out, which allows identifying B. tabaci specimens within one hour. Compared to existing protocols for the detection of specific B. tabaci biotypes, the developed method targets the whole B. tabaci species complex in one assay. Moreover the assay is designed to be applied on-site by plant health inspectors with minimal laboratory training directly at points of entry. Thorough validation performed under laboratory and on-site conditions demonstrates that the reported LAMP assay is a rapid and reliable identification tool, improving the management of B. tabaci.
Keywords: Environmental Sciences, Issue 140, Bemisia tabaci, LAMP, loop-mediated isothermal amplification, point of entry diagnostics, plant health, rapid diagnostics, quarantine organisms
Introduction
The whitefly Bemisia tabaci (Gennadius) is an invasive insect pest affecting the yield of many economically important crops including ornamental plants, vegetables, grain legumes, and cotton1,2. Beside damage caused through direct phloem-feeding, the homopteran species harms plants indirectly by the excretion of large amounts of honeydew onto the surfaces of leaves and fruits, as well as by the transmission of numerous plant pathogenic viruses1,3,4. Recent genetic studies comparing DNA sequences of the mitochondrial gene cytochrome c oxidase 1 (COI) revealed that B. tabaci is a species complex of at least 34 morphocryptic species3,4. Two highly invasive and damaging members within this complex, biotype B originating from the Middle East and the Asian Minor region, as well as biotype Q originating from the Mediterranean region, have been dispersed globally through international trading activities with plant products, particularly by the transportation of ornamentals1,5,6. Due to its worldwide pest status, the International Union for the Conservation of Nature and Natural Resources (IUCN) listed B. tabaci as one of the "world's 100 worst invasive alien species" and members of the species complex are regulated organisms by many countries1,3,4.
In the European Union (EU), B. tabaci is listed in the Plant Health Directive 2000/29/EC Annex 1AI as a quarantine organism whose introduction from non-EU countries and its dissemination within the EU are banned4. An essential prevention measure against the spread of quarantine organisms is the inspection of plant shipments at points of entry (POEs) such as airports and seaports7,8. In the case a quarantine organism is found, the National Plant Protection Organization (NPPO) in charge takes action by either rejecting or treatment (including destruction) of the infested shipment9. However, officers inspecting the imports often do not have the taxonomic expertise to accurately identify the vast range of pest species associated with global trade9. Especially the identification of immature life stages (e.g., eggs and larvae) without distinct morphological keys is virtually impossible for non-taxonomists8,9,10. Consequently, to enable implementation of quarantine measures with minimal delay, there is a need for alternative, rapid on-site identification assays9.
A candidate method is the loop-mediated isothermal DNA amplification (LAMP) technology that has recently been shown to be a suitable technology for the identification of plant pathogens11,12,13. LAMP is highly specific because the method uses at least two primer pairs recognizing six distinct DNA target sequences14. Due to the DNA strand displacement activity of the Bst DNA polymerase, LAMP reactions are performed under isothermal conditions14. Hence, in contrast to conventional polymerase chain reaction (PCR)-based assays there is no need for a thermal cycler13,14. Another advantage over PCR-based assays is its resilience against potential inhibitors in the DNA extract, circumventing the need for a DNA purification step13. Due to the protocol's speed and simplicity, LAMP may even be performed under on-site conditions using a portable, battery driven real-time detection device8,15.
A LAMP assay was designed in response to the demand for a rapid on-site identification method for B. tabaci8. The overarching aim was to develop a protocol that can be performed by plant health inspectors with limited laboratory training. A strong focus was, therefore, set on optimizing speed and simplicity of the protocol. While existing diagnostic tests have generally been developed for the identification of one or several biotypes of B. tabaci, the novel LAMP assay covers the whole B. tabaci species complex8,16,17,18. The problem of the pronounced genetic within-taxon diversity of the complex was solved by using combinations of different primer sets and the application of degenerate primers8. The novel B. tabaci LAMP assay is designed in such a way that the primers target a fragment at the 3' end of the mitochondrial COI gene8. This gene presents a suitable target for animal diagnostic assays because it harbors regions conserved enough to ensure diagnostic sensitivity for a specific species, while discriminating enough between closely related organisms19,20. Furthermore, the COI gene is often used as a genetic marker in population genetic studies and as a signature sequence in DNA barcoding analyses, resulting in numerous DNA sequence entries in open source databases such as GenBank and BOLD21,22. Beside the publicly available COI sequences from B. tabaci, COI sequences from closely related species (Aleurocanthus spp. [N = 2], Aleurochiton aceris, Aleurodicus dugesii, Bemisia spp. [N = 3], Neomaskellia andropogonis, Tetraleurodes acaciae, and Trialeurodes spp. [N = 4]) were included in the primer design of this study and used to assess diagnostic sensitivity and specificity in silico8.
Due to the accuracy of the method, its speed (<1 h) and the simplicity of the protocol, the assay has been shown to be suitable for on-site application when implemented as part of the import control procedure at a Swiss POE8.
Protocol
1. Preparations
- Preparing aliquots of alkaline DNA extraction solution.
- Produce a stock of alkaline DNA extraction solution using molecular grade water supplemented with 600 µM potassium hydroxide (KOH) and 2 µM Cresol Red. CAUTION: KOH is a strong base dissolved in water. Avoid spills, and skin and eye contact.
- Dispense 30 µL of alkaline DNA extraction solution (prepared in step 1.1.1) into 0.5 mL microcentrifuge tubes and store the aliquots at 4 °C. NOTE: Use the aliquoted DNA extraction solution within 1 year.
- Preparing B. tabaci positive amplification control (PAC).
- Generate PCR amplicons of the LAMP target DNA fragment. NOTE: An introduction into general PCR principles and practices is given by Lorenz23.
- Set up the PCR reaction as described in Table 1. Use DNA extract (see step 2.1) of a reference B. tabaci specimen as DNA template. NOTE: Optionally, it is possible to extract the B. tabaci DNA for the PAC using a commercial kit according to the manufacturer’s instructions.
- Program a thermal cycler using the following conditions: 15 min at 95 °C; 45 cycles of 40 s at 95 °C, 15 s at 45 °C, ramping over 60 s to 60 °C, 2 min at 72 °C; 7 min at 72 °C; hold at 4 °C.
- Clean the PCR amplification product using a commercial PCR clean-up kit according to the manufacturer’s protocol and elute the final product in molecular grade water.
- Use a commercial kit with DNA-intercalating dye to measure the DNA concentration of the PCR amplification product according to the manufacturer’s instructions and dilute with molecular grade water to a concentration of 1 ng/µL. Store the diluted PCR amplification product as PAC stock solution at -20 °C. NOTE: Use the PAC stock solution within 1 year.
- Supplement the PAC stock solution (prepared in step 1.2.1.5) with 0.6 µM KOH and dilute with molecular grade water to a concentration of 5 x 10-3 ng/µL. Store the product at 4 °C. NOTE: Use the PAC within 5 h for the preparation of the ready-to-use B. tabaci LAMP kits described in the next step.
- Preparing ready-to-use B. tabaci LAMP kit (protocol for 20 units)
- Use scissors to cut 8-tube LAMP strips into two 4-tube LAMP strips.
- Label the tubes of the 4-tube LAMP strips according to the scheme shown in Figure 1.
- Prepare B. tabaci LAMP reaction mastermix (protocol for 80 reactions).
- Add 1195.1 µL of ready-to-use GspSSD isothermal master mix (containing GspSSD polymerase, pyrophosphatase, magnesium sulfate, deoxynucleotides, double strand binding DNA binding dye) and 717.4 µL of B. tabaci LAMP primer mix to a 2 mL microcentrifuge tube. Briefly vortex and pulse centrifuge.
- Dispense 22.5 µL of B. tabaci LAMP reaction mastermix (prepared in step 1.3.3.1) into each tube of the 4-tube LAMP strips (prepared in step 1.3.1) and pulse centrifuge.
- Vortex the B. tabaci LAMP PAC (prepared in step 1.2) quickly and pulse centrifuge. Then, add 2.5 µL into the tube labelled with “PAC” of each 4-tube LAMP strip (Figure 1).
- Close lids and store the ready-to-use B. tabaci LAMP kit units at -20 °C. NOTE: Use them within 1 year.

2. On-site LAMP Analysis
- DNA extraction
- Use sterile toothpicks to transfer the insect specimens into 0.5 mL microcentrifuge tubes containing 30 µL of DNA extraction solution (prepared in step 1.1.2). NOTE: Make sure that the insects are immersed in the extraction solution.
- Incubate the samples for 5 min at 95 °C in a thermomixer (300 rpm). Briefly vortex and pulse centrifuge.
- B. tabaci LAMP assay
- Thaw a ready-to-use B. tabaci LAMP kit prepared in step 1.3. Vortex quickly and pulse centrifuge. NOTE: With each kit, it is either possible to test two different specimens or to analyze the DNA extract of one specimen in duplicate.
- Add 2.5 µL of sample DNA extract (prepared in step 2.1) into the tubes labeled “S1” and “S2” of the ready-to-use B. tabaci LAMP kit (Figure 1).
- Add 2.5 µL of pure alkaline DNA extraction solution (prepared in section 1.1) into the tube labeled “NAC” for the negative amplification control (Figure 1).
- Vortex the ready-to-use B. tabaci LAMP kit quickly and pulse centrifuge.
- Insert the ready-to-use B. tabaci LAMP kits into the LAMP analysis device (with real-time fluorescence measurement) or a real-time PCR platform and perform an isothermal DNA amplification analysis at 65 °C for 60 min.
- Measure the melting temperatures of DNA amplification products by heating up to 98 °C with a subsequent cooling step (ramp rate of 0.05 °C/s) to 75 °C, while measuring fluorescence in real-time.
- LAMP assay read-out
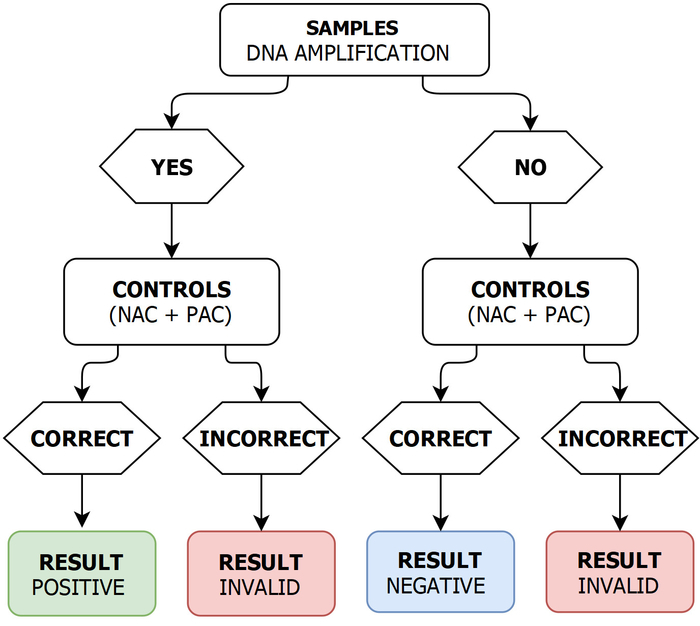
- Validate the LAMP read-out manually as follows.
- If DNA amplifications were measured for the sample and the PAC, no DNA amplification was measured for the NAC, and the annealing temperature of the amplification products were between 80.0 and 85.5 °C, consider the LAMP results as POSITIVE (Figure 2).
- If there is no DNA amplification for the samples (i.e., tubes labeled S1 and S2) but for PAC and NAC then consider the LAMP result as NEGATIVE (Figure 2).
- If DNA amplification was measured for the samples, but the annealing temperatures of corresponding amplification products were outside the range 80.0 ‒ 85.5 °C, and/or PAC gave no DNA amplification, and/or NAC gave a DNA amplification, consider the LAMP result as INVALID (Figure 2).
- Optionally, validate the LAMP read-out using the LAMP validation application (Supplemental file 1).
- Define target species and define the number of tested samples. Click the "Generate Report" button.
- Transfer the read-out (DNA amplification yes/no, annealing temperature amplification product, results of PAC and NAC) from the on-site LAMP analysis device or real-time PCR platform to the corresponding input fields of the validation application. The result of the validation is immediately displayed after entering the data.
Representative Results
During the validation of the B. tabaci LAMP assay, insect specimens intercepted in the course of the regular Swiss import control process were analyzed8. The specimens originated from eight different countries (Canary Islands, Dominican Republic, Israel, Malaysia, Morocco, Singapore, Thailand, and Vietnam) and reflect the genetic diversity of B. tabaci found at European POEs8. All LAMP results were cross-validated by DNA barcoding8.
From a total of 80 specimens analyzed by LAMP, 75 specimens (93.8%) were correctly identified as B. tabaci (true-positives), two specimens (2.5%) were correctly identified as not being B. tabaci (true-negatives), and three specimens (3.8%) were wrongly identified as not being B. tabaci (false-negatives) (Table 2)8. The correct-negative results originated from two Trialeurodes vaporariorum specimens, a non-regulated species at high risk to be confused with B. tabaci at POEs for plant products8. Based on these results, the following measurements of diagnostic accuracy were calculated: test specificity (true-negative rate), 100%; test sensitivity (true-positive rate), 96.2%; test efficiency (percentage of correct test results), 96.3% (Table 2)8. When assessing the analytical sensitivity (detection limit), the B. tabaci LAMP assay successfully amplified sample DNA diluted to 100 fg/µL across three technical replicates (Table 3).
A subset of the assays (N = 13) was performed under on-site conditions at the Swiss POE Zurich Airport by plant health inspectors using the ready-to-use B. tabaci LAMP kits8. When cross-validated in the reference laboratory, all results from on-site testing were found to be correct (test efficiency = 100%)8. Assessing the on-site LAMP assay performance, the average time to positive (time until a positive results was available) was 38.4 ± 10.3 min (mean ± standard deviation)8. A representative DNA amplification plot and the corresponding annealing derivative from a B. tabaci LAMP analysis performed under on-site conditions are shown in Figure 3A and B. In this example, sample one and two were correctly identified as B. tabaci indicated by DNA amplification after approximately 30 min (Figure 3A) together with the expected annealing temperatures at approximately 82 °C (Figure 3B).
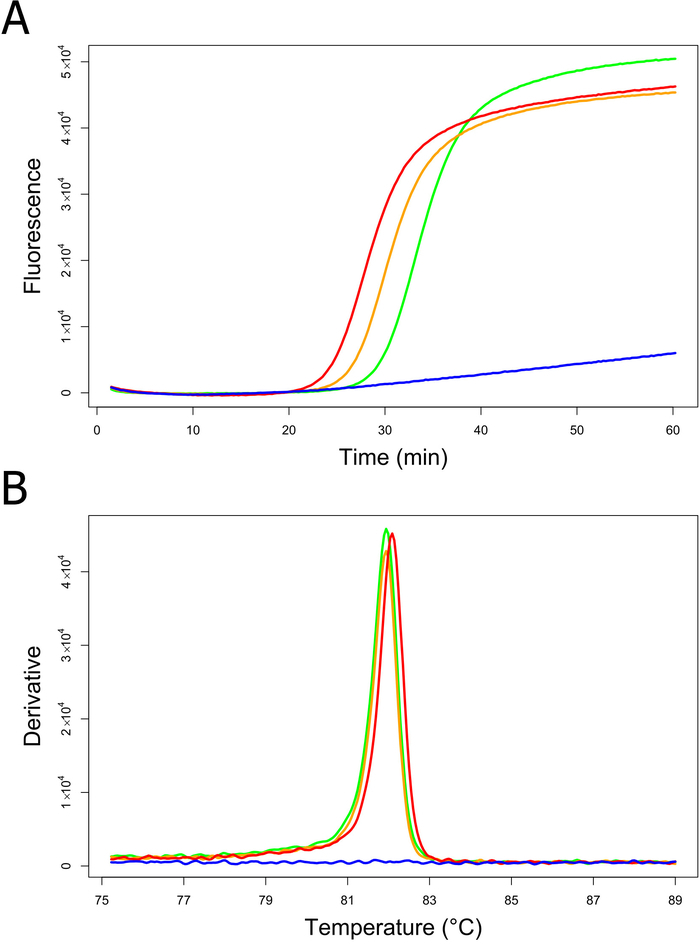
Figure 1: Visualization of the experimental set-up of a ready-to-use B. tabaci LAMP kit described in the protocol. S1, sample 1; S2, sample 2; PAC, positive amplification control; NAC, negative amplification control. Please click here to view a larger version of this figure.
Figure 2: LAMP read-out validation schema. PAC: positive amplification control; NAC: negative amplification control. Please click here to view a larger version of this figure.
Figure 3: DNA amplification plot (A) and annealing derivative (B) of a B. tabaci LAMP analysis performed under on-site conditions. Fluorescence was measured in relative intensity units. Green line, sample 1; orange line, sample 2; blue line, negative amplification control (NAC); red line, positive amplification control (PAC). Please click here to view a larger version of this figure.
| Component | Stock conc. | Final reaction conc. | Volume per reaction |
| Taq Polymerase Master Mix | 2x | 1x | 10 µL |
| Primer C1-J-2195 | 20 µM | 0.4 µM | 0.4 µL |
| Primer TL2-N-3014 | 20 µM | 0.4 µM | 0.4 µL |
| Molecular Grade Water | - | - | 8.2 µL |
| DNA Template | - | - | 1 µL |
Table 1: Preparation of PCR reaction mastermix for the B. tabaci positive amplification control. Components and concentrations needed to set up one PCR reaction. The final reaction volume is 20 µL. Primer sequences are shown in 1.2.1.1.
| N | NTP | NFP | NTN | NFN | SEN (%) | SPE (%) | EFF (%) |
| 80 | 75 | 0 | 2 | 3 | 96.2 | 100 | 96.3 |
Table 2: Results of the B.tabaci LAMP assay validation. N, number of analyses; NTP, number of true-positive results; NFP, number of false-positive results; NTN, number of true-negative results; NFN, number of false-negative results; SEN, diagnostic sensitivity; SPE, diagnostic specificity, EFF, test efficiency.
| CDNA(fg/µL) | NPR | TP (min) (mean ± SD) | TA (°C) (mean ± SD) |
| 1 x 105 | 3 | 33.5 ± 2.9 | 81.3 ± 0.1 |
| 1 x 104 | 3 | 30.7 ± 1.1 | 81 ± 0.0 |
| 1 x 103 | 3 | 40.4 ± 3.9 | 81.1 ± 0.1 |
| 1 x 102 | 3 | 50.7 ± 1.6 | 81.1 ±0.1 |
| 1 x 101 | 0 | - | - |
| 1 x 100 | 0 | - | - |
Table 3: Analytical sensitivity (detection limit) of the B. tabaci LAMP assay. Each dilution was tested in triplicates. CDNA, DNA concentration per reaction; NPR, number of positive replicates; TP, time until a positive result was available; TA, annealing temperature; SD, standard deviation.
Discussion
The ability to accurately identify potentially harmful organisms without time delay represents a critical aspect for the management of pest species9,10,26. Besides being rapid, for plant import products, an ideal pest identification method should be simple to perform on-site at POEs8,26. This paper reports the protocol of a novel LAMP assay for the rapid identification of B. tabaci, a quarantine insect organism frequently intercepted at European borders (https://ec.europa.eu/food/sites/food/files/plant/docs/ph_biosec_europhyt_annual-report _2016.pdf).
The rationale behind the development of the diagnostic test was to design an easy-to-follow protocol which can be performed during the plant import control procedure by plant health inspectors with minimal laboratory training. In order to make on-site testing as rapid and simple as possible, the protocol is divided into two parts, the preparation of a ready-to-use kit and the actual performance of the LAMP assay. The first part may be done in an external laboratory so that the plant health inspector can perform the DNA extraction and LAMP assay on-site with only one pipetting step.
Though only one step, pipetting small amounts of liquid may be challenging for users with little or no laboratory experience. To address this issue, a dye (cresol red) is added to the extraction solution so that the operator can visually confirm the small amount (i.e., 2.5 µL) of DNA is correctly transferred to the respective tube. Another important simplification of the protocol is the validation application as it facilitates a reliable interpretation of the LAMP read-out (Supplemental file 1).
The novel B. tabaci LAMP assay has been validated under laboratory and on-site conditions by testing insect specimens intercepted during the regular import control process of Switzerland8. In total, 80 specimens from three continents, Africa, Eurasia, and North America, were analyzed by LAMP. Of the 80 specimens, only three (3.8%) were wrongly identified (false-negatives)8. When analyzing the primer target DNA sequences of the false-negative specimens, it was found that they were new B. tabaci haplotypes that have so far not been described8. Based on these results, the B. tabaci LAMP primer set has been modified and successfully re-validated8.
One major limitation of any DNA amplification-based method including LAMP is that they only identify pre-defined target DNA sequences8,27. A comprehensive knowledge of the genetic variation found in the primer target sequence is therefore crucial to ensure diagnostic accuracy8,27. However, such information is often very limited, especially in the case of newly emerging pest species8. Though rare, false-negative results caused by mutations in the target sequence are expected8. In the case of the present B. tabaci LAMP assay, a solution for this problem is the combination with a DNA barcoding-based technology, a strategy realized in the course of the implementation of this diagnostic test at the POE Zurich Airport8. Here, all LAMP-negative results were re-analyzed by DNA barcoding in an external laboratory8. In case a novel pest haplotype not yet described is encountered, the LAMP primers can be modified using the DNA sequence generated in the barcoding process8. Thereby, the resulting loss of speed in case of a negative LAMP result is compensated for the maximum diagnostic accuracy ensured in this two-stage process8.
The set-up costs for the current LAMP assay at a POE are approximately USD 25,000. With the increasing number of LAMP tests developed for plant pests (e.g., Erwinia amylovora, Flavescence dorée, and Guignardia citricarpa), such a one-time investment appears justified13,15,28. However, the protocol could potentially be modified to reduce these costs even further. For example, for the DNA extraction step at 95 °C the thermo mixer used here could be replaced by a less expensive water bath, or by performing this step directly in the real time LAMP device. Furthermore, the mixing steps on the vortex could probably be replaced by manually flicking the tubes, and in the DNA transfer step the pipettor might be replaced by sterile inoculation loops.
Future improvements for a rapid identification of B. tabaci and pest species in general could be an implementation of an on-site sequencing approach that would allow to perform DNA barcoding analyses at POEs. A promising candidate system for such an implementation is the nanopore sequencing technology. Indeed, the technology has recently been successfully implemented in an on-site DNA barcoding effort to assess the biodiversity of a rainforest8,29,30. An on-site DNA barcoding identification system can completely replace the need for the development of targeted diagnostic tests and their validation. Also it allows collecting additional information about pest characteristics such as pesticide resistance genes8. Nevertheless, until novel sequencing technologies will be implemented routinely, the B. tabaci LAMP assay represents a rapid (<1 h) and accurate identification method.
Disclosures
The author Michael Andreou is a shareholder of OptiGene Limited that produces reagents and instruments used in this article. The other authors have nothing to disclose.
Acknowledgments
The authors are grateful to Annette Grendelmeier, Aurelia Drenovac, Cornelia Studer, Daniel Frei, Elisabeth Razavi, Markus Oggenfuss, Seraina Vonzun, and Sven Moeller for participating in the validation of the B. tabaci LAMP assay.
References
- De Barro PJ, Liu SS, Boykin LM, Dinsdale AB. Bemisia tabaci: a statement of species status. Annual Review of Entomology. 2011;56:1–19. doi: 10.1146/annurev-ento-112408-085504. [DOI] [PubMed] [Google Scholar]
- Zhang GF, Lü ZC, Wan FH, Lövei GL. Real-time PCR quantification of Bemisia tabaci (Homoptera: Aleyrodidae) B-biotype remains in predator guts. Molecular Ecology Notes. 2007;7(6):947–954. [Google Scholar]
- Boykin LM, De Barro PJ. A practical guide to identifying members of the Bemisia tabaci species complex: and other morphologically identical species. Frontiers in Ecology and Evolution. 2014;2 [Google Scholar]
- Cuthbertson AGS, Vänninen I. The importance of maintaining protected zone status against Bemisia tabaci. Insects. 2015;6(2):432–441. doi: 10.3390/insects6020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmon A, Halkett F, Granier M, Delatte H, Peterschmitt M. Genetic structure of the invasive pest Bemisia tabaci: evidence of limited but persistent genetic differentiation in glasshouse populations. Heredity. 2008;100(3):316–325. doi: 10.1038/sj.hdy.6801080. [DOI] [PubMed] [Google Scholar]
- Dickey AM, Osborne LS, Shatters RG, Hall PAM, Mckenzie CL. Population genetics of invasive Bemisia tabaci (Hemiptera: Aleyrodidae) cryptic species in the United States based on microsatellite markers. Journal of Economic Entomology. 2013;106(3):1355–1364. doi: 10.1603/ec12512. [DOI] [PubMed] [Google Scholar]
- Bacon SJ, Bacher S, Aebi A. Gaps in border controls are related to quarantine alien insect invasions in Europe. PLOS ONE. 2012;7(10):e47689. doi: 10.1371/journal.pone.0047689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser S, et al. From laboratory to point of entry: development and implementation of a loop-mediated isothermal amplification (LAMP)-based genetic identification system to prevent introduction of quarantine insect species. Pest Management Science. 2018;74(6):1504–1512. doi: 10.1002/ps.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd R, Lima J, de Waard J, Humble L, Hanner R. Common goals: policy implications of DNA barcoding as a protocol for identification of arthropod pests. Biological Invasions. 2010;12(9):2947–2954. [Google Scholar]
- Armstrong KF, Ball SL. DNA barcodes for biosecurity: invasive species identification. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005;360(1462):1813–1823. doi: 10.1098/rstb.2005.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson JA, Boonham N, Dickinson M. Development and evaluation of a one-hour DNA extraction and loop-mediated isothermal amplification assay for rapid detection of phytoplasmas. Plant Pathology. 2010;59(3):465–471. [Google Scholar]
- Kogovšek P, et al. LAMP assay and rapid sample preparation method for on-site detection of flavescence dorée phytoplasma in grapevine. Plant Pathology. 2015;64(2):286–296. doi: 10.1111/ppa.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Su TY, Lien YY, Sheu SC. The development of loop-mediated isothermal amplification (LAMP) assays for the rapid authentication of five forbidden vegetables in strict vegetarian diets. Scientific Reports. 2017;7:44238. doi: 10.1038/srep44238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühlmann A, et al. Erwinia amylovora loop-mediated isothermal amplification (LAMP) assay for rapid pathogen detection and on-site diagnosis of fire blight. Journal of Microbiological Methods. 2013;92(3):332–339. doi: 10.1016/j.mimet.2012.12.017. [DOI] [PubMed] [Google Scholar]
- Cavalieri V, Manglli A, Tiberini A, Tomassoli L, Rapisarda C. Rapid identification of Trialeurodes vaporariorum, Bemisia tabaci (MEAM1 and MED) and tomato-infecting criniviruses in whiteflies and in tomato leaves by real-time reverse transcription-PCR assay. Bulletin of Insectology. 2014;67(2):219–225. [Google Scholar]
- Baek JH, Lee HJ, Kim YH, Lim KJ, Lee SH, Kim BJ. Development of an antibody-based diagnostic method for the identification of Bemisia tabaci biotype B. Pesticide Biochemistry and Physiology. 2016;131:18–23. doi: 10.1016/j.pestbp.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Hsieh CH, Wang HY, Chen YF, Ko CC. Loop-mediated isothermal amplification for rapid identification of biotypes B and Q of the globally invasive pest Bemisia tabaci, and studying population dynamics. Pest Management Science. 2012;68(8):1206–1213. doi: 10.1002/ps.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejili M, et al. A PCR-based diagnostic assay for detecting DNA of the olive fruit fly, Bactrocera oleae, in the gut of soil-living arthropods. Bulletin of Entomological Research. 2016;106(5):695–699. doi: 10.1017/S000748531600050X. [DOI] [PubMed] [Google Scholar]
- Hebert PDN, Ratnasingham S, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings. Biological Sciences. 2003;270(Suppl 1):96–99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert PDN. bold: The Barcode of Life Data System (http://www.barcodinglife.org) Molecular Ecology Notes. 2007;7(3):355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Bank Wheeler DLGen. GenBank. Nucleic Acids Research. 2005;33:D34–D38. doi: 10.1093/nar/gki063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz TC. Polymerase chain reaction: basic protocol plus troubleshooting and optimization strategies. Journal of Visualized Experiments. 2012. p. e3998. [DOI] [PMC free article] [PubMed]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Annals of the Entomological Society of America. 1994;87(6):651–701. [Google Scholar]
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annual Review of Ecology, Evolution, and Systematics. 2006;37(1):545–579. [Google Scholar]
- Harper SJ, Ward LI, Clover GRG. Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications. Phytopathology. 2010;100(12):1282–1288. doi: 10.1094/PHYTO-06-10-0168. [DOI] [PubMed] [Google Scholar]
- Lauri A, Mariani PO. Potentials and limitations of molecular diagnostic methods in food safety. Genes & Nutrition. 2009;4(1):1–12. doi: 10.1007/s12263-008-0106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson JA, et al. A loop-mediated isothermal amplification-based method for confirmation of Guignardia citricarpa in citrus black spot lesions. European Journal of Plant Pathology. 2013;136(2):217–224. [Google Scholar]
- Branton D, et al. The potential and challenges of nanopore sequencing. Nature Biotechnology. 2008;26(10):1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz A, et al. Real-time DNA barcoding in a rainforest using nanopore sequencing: opportunities for rapid biodiversity assessments and local capacity building. GigaScience. 2018;7(4):giy033. doi: 10.1093/gigascience/giy033. [DOI] [PMC free article] [PubMed] [Google Scholar]


