Abstract
The goal was to develop and optimize a simple, affordable, and effective bioassay to detect disease suppressive ability of a specific compost against soilborne fungus Rhizoctonia solani. R. solani is a pathogen of a wide range of plant hosts worldwide. The fungus survives in soils as a saprophyte and grows rapidly on simple water agar media. The plate assay is a rapid method to compare composts for their ability to slow the growth of R. solani. The assay also correlates well with suppression of other soilborne fungal pathogens that survive as saprophytes in soils such as Alternaria early blights, Fusarium wilt, Phytophthora root rot, and Pythium root rot.
Keywords: Biology, Issue 140, Biological control, soil microbial communities, Rhizoctonia solani, root diseases, soilborne fungal pathogens, bioassay
Introduction
Rhizoctonia represents a broad complex of fungi, of which Thanatephorus cucumeris (Frank) Donk (anamorph = Rhizoctonia solani Kühn) is the pathogen causing root rot and damping-off1. Rhizoctonia solani is an aggressive pathogen and a saprophyte that can survive as sclerotia under adverse environmental conditions1. As a result, it has a global distribution and can cause disease on wide range of plant hosts including Solanaceae, Fabaceae, Asteraceae, and Brassicaceae resulting in serious economic losses.
Compost has the capacity to harbor biocontrol agents for certain plant pathogens2. However, not all composts are alike nor do they affect all pathogens similarly3. Wood-based carbon has higher lignin to cellulose ratios than hay or straw-carbon based composts. R. solani prefers readily available carbon found in straw. In contrast, biological control fungi, such as Trichoderma spp., are more effective when carbon is less readily available. Beneficial fungi and bacteria into compost can suppress plant disease through competition, antagonism or regulating plant growth3. The proposed assay primarily detects antagonism created by production of antibiotics, ecoenzymes or chelators that are detrimental to the pathogen.
Plant bioassays are a gold standard to determine whether composts favor or deter plant growth4. However, plant bioassays are time-consuming (weeks to months) to complete which may be longer than desired and requires more labor to extract plants with roots to quantify severity of root systems. Comparably robust, but quicker (days) assays would be ideal for quality control programs. The goal of this paper is to demonstrate a relatively quick and accurate test to predict suppressive potential of compost. The method was patterned after Alfano et al.5 with two exceptions, compost extracts were diluted and water agar was used in place of potato dextrose agar (PDA). R. solani grows rapidly on simple water agar media whereas PDA promoted growth of bacteria and other fungi that contaminated the culture6.
This plate assay serves as an indicator of suppression that applies to a range of plant pathogens that survive in soil as saprophytes including R. solani7, Alternaria early blights, Fusarium wilt, Phytophthora root rot, and Pythium root rot. The plate competition assay is useful to screen a range of compost products for containing communities of microbes that serve as biological control agents of soil pathogens. The assay was one of the most consistent indicators of disease suppression in commercial compost products6,8. Products were chosen for their variation in recipe, maturity, and production process.
Protocol
1. Prepare in Advance
- Master fungal culture (test organism)
- Order from the American Type Culture Collection, Microbiology Collection9, by its teleomorph Thanatephorus cucumeris (Frank) (ATCC 10154) or MYA 4031.
- Alternatively: Collect local isolates as done by the authors. Seedlings of red radish (Raphanus sativus) work well as a bait plant; collect soil from a location known to have a history of damping-off or root rot caused by R. solani. Sow seeds in the soil and isolate from root lesions when seedlings are 3 to 4 weeks old.
- Remove seedlings from soil and rinse with tap water. Look for brown tissue in the hypocotyl region between the leaves and the root (Figure 1).
- Use a single edged razor blade to cut 1 cm segments of the hypocotyl or root that have a brown color. Use flame-sterilized forceps to dip the segments in 10% solution of household bleach for 1 min, followed by a 10 s rinse in sterile water. NOTE: The top and bottom of a sterile Petri dish are useful containers for this step.
- Use flame-sterilized forceps to transfer the plant pieces to the inside of a paper towel to pat dry and then place on the surface of a Petri dish with water agar (15 g in 1 L).
- Place the Petri dish inside a plastic container with a lid and incubate at room temperature (approximately 20 °C). Wipe the interior of the container with 10% bleach or 75% ethanol and let it air dry before inserting the dish.
- Keep the isolates in long-term storage on a minimal media of corn meal agar (17 g in 1 L) slants (at 5 °C).
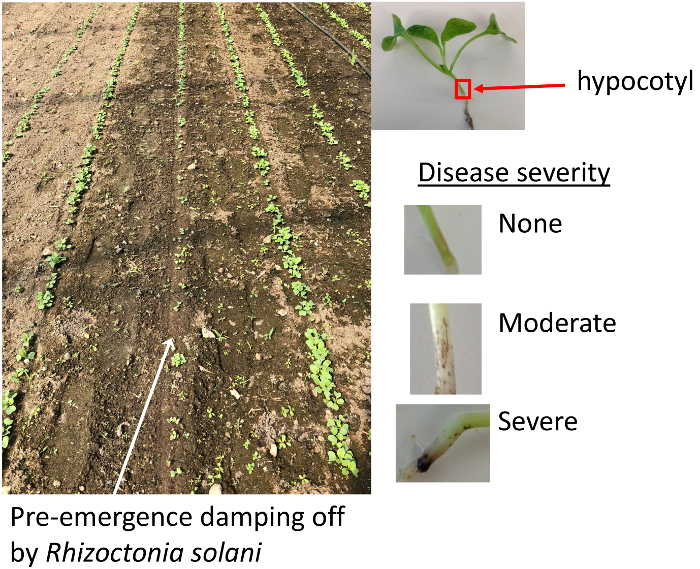
Figure 1: Disease symptoms. Soil containing R. solani results in patchy germination and establishment (left). Symptoms on radish seedlings occur as brown lesions at the hypocotyl (right). Please click here to view a larger version of this figure.
- Use potato dextrose agar plates (39 g in 1 L) for inoculum for this plate assay.
- Establish a daughter culture of Rhizoctonia culture on potato dextrose agar one or two days before the assay, to insure it is well established on the culture plate before the assay. Start the daughter culture by transferring a small piece (10 mm diameter) of R. solani from the master culture to the center of a fresh plate of potato dextrose agar (Figure 2). NOTE: Work in a laminar flow hood or wipe down a laboratory bench with 10% bleach or 75% ethanol to minimize risk of contamination.
Autoclave 25 mL test tubes with 10 mL aliquots of distilled water (2 x number of samples).
2. Preparation of Samples to Test
- Add two independent 0.5 g samples of each compost test sample to 10 mL of autoclaved water in 25 mL test tubes. Shake the test tubes overnight.
- Label the test tubes with unique sample numbers, and each member of the pair as A (reference) or B (sample).
After 24 h, add 1.5 g of plain agar to 90 mL of distilled water in two 125 mLconical flasks.
Autoclave both conical flasks with agar, and the A sample of each pair for 20 min with a slow exhaust setting.
After the autoclave, place the conical flasks with agar into a 45 °C water bath until they reach equilibrium (about 30 min). Do not put any sample slurries in the water bath until it has cooled to 45 °C.
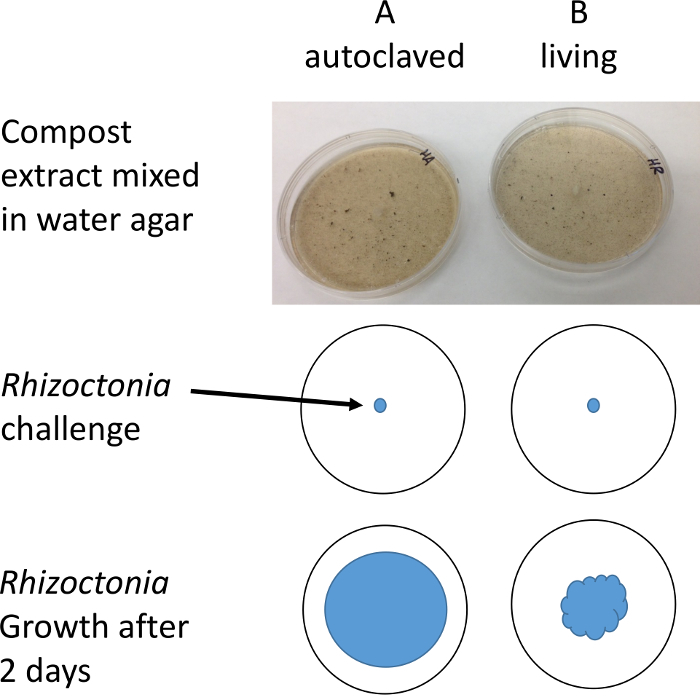
Figure 2: Illustration of protocol. Plugs of R. solani are placed on Petri dishes containing compost water extract. The diameter of mycelial growth is measured after 1-2 days with the aid of a stereo microscope to improve resolution and contrast. Please click here to view a larger version of this figure.
Pour the A sample (autoclaved reference) compost slurry into the molten agar of the A agar flask. Swirl to disperse compost into the agar.
Pour the B sample (living sample) compost slurry into the molten agar of the B agar flask. Swirl to disperse compost into the agar.
Pour the mixture of each flask into five plastic Petri dishes that are 100 mm in diameter.
Let the plates cool overnight so the agar hardens.
3. Add the Rhizoctonia Challenge
Using aseptic technique, transfer equal size pieces of R. solani to each pair of sample agar plates (3 to 5 mm corkborer works well to establish equal sized pieces). Take pieces of the colony from the outside margin of the colony to insure mycelium is actively growing. NOTE: Work in a laminar flow hood or wipe down a laboratory bench with 10% bleach or 70% ethanol.
Incubate the plates at room temperature for 1-2 days until the colony growth reaches about half way to the edge of the A plates.
4. Measure R. solani Growth
Measure the radius of the mycelium in each plate to the nearest 1 mm with a clear flat ruler using a stereo microscope with transmitted illumination. NOTE: Oblique or dark field illumination will make it easier to see and measure the transparent hyphae. At this point, zones of inhibition will be visible around compost fragments in the B plate if the compost is suppressive.
Record the radius at three places per plate and compute a mean of the three to serve as a representative measure.
Subtract the mean of the B plates from the A plates. If B < A then there are microbes in the B plates that are suppressive to the R. solani pathogen.
Divide the mean radius by the number of days of challenge to express units of relative suppression as a rate, mm mycelium per day.
Representative Results
Finished compost should be stable and mature, two terms that are often used interchangeably, so it can be safely packaged and transported, and not cause adverse effects during its end use4. Stability is a resistance to decomposition and is usually determined using indices of microbial activity. General measures of microbial respiration may measure compost stability but not necessarily disease suppression of an aggressive pathogen and saprophyte such as R. solani7. This study focused on maturity which infers the material is ready for a particular use, and, for horticultural purposes, is determined by plant germination and growth assays.
Extracts of the living composts suppressed R. solani growth significantly more than autoclaved samples (P ≤ 0.0001). Autoclaving kills off microbial activity, pointing to a microbially mediated suppression. This confirms that Plate A in the method serves as a negative control for microbially mediated biological control (Figure 3).
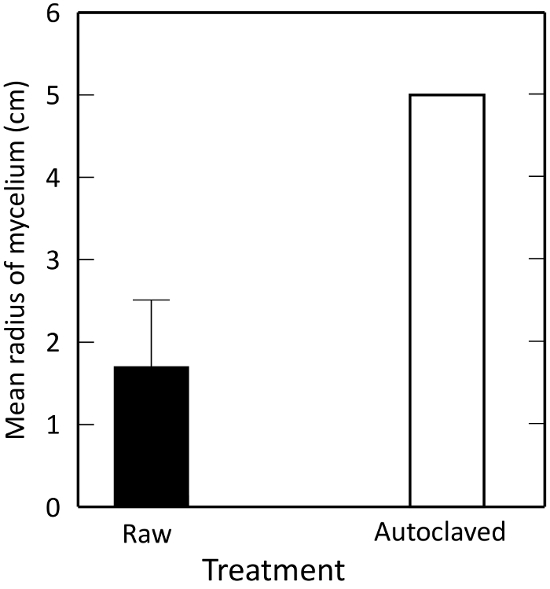
Figure 3: Effects of autoclaving on R. solani growth in vitro, as measured by percentage change in mycelial growth from control. A test material is 'suppressive' to the fungus if the radius of mycelial growth is less than an autoclaved control. Illustrated are means ± 1 standard error. The error bar for the autoclaved control is too small to see. Please click here to view a larger version of this figure.
Growth of R. solani mycelium on compost extract was affected by compost method and raw material in the recipe of compost but not duration of maturation or curing6,8. Growth was reduced by vermicompost and windrow processes and recipes containing hardwood bark (Figure 4). Suppressiveness represents a reduction in growth of R. solani.
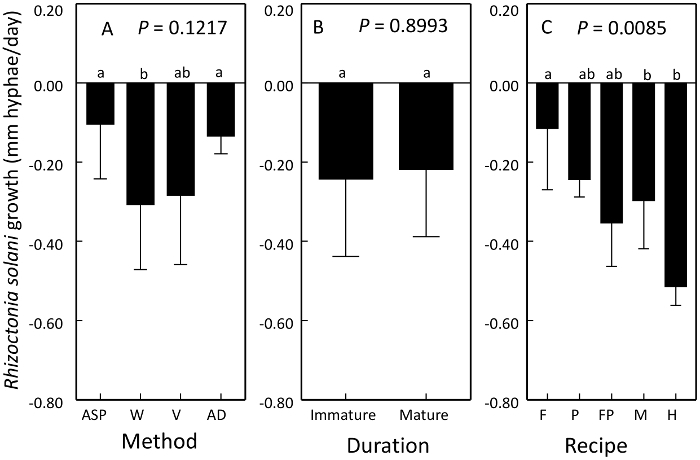
Figure 4: Use of plate competition assay to compare the suppressiveness of compost products. Means ± 1 standard error of the change in R. solani growth on an autoclaved reference plate. Both controls and treatment comparisons were inoculated with virulent R. solani. Values below zero represent suppressiveness. Suppressiveness of R. solani was affected by compost A) method, B) duration of curing or maturation, and C) recipe. Processes compared were aerated static pile (ASP), windrow (W), vermicompost (V) and anaerobic digestion (AD). Recipe ingredients compared were food waste (F), poultry manure (P), food waste and poultry manure (FP), dairy manure (M), and hardwood bark (H). Contrasting letters above bars denote treatments that are significantly different. This figure has been modified from Neher et al.8 Please click here to view a larger version of this figure.
Discussion
We know from previous research, that certain composts are effective at suppressing R. solani and that the suppressive effects are due to the microbes living in the compost, not the abiotic properties of compost6,8. The use of autoclaving as a means to 'kill' microbiota has been criticized because it affects the carbon chemistry of the media10. We compared the use of autoclaving to vacuum filtration through Whatman No. 1 paper. Treatments that were neither filtered nor autoclaved showed the greatest suppression of R. solani growth6. Filtration removed the larger, solid particles that were apparently important to disease suppression by likely harboring antibiotic producing microbes.
Advantages of the method are the simplicity and affordability. Similar to the conclusions and recommendations of Alfano et al.2, the plate competition assay is a quick preliminary assessment of disease suppression but not reliable as a standalone assay because only the pathogen, and not the plant host, is present. Besides, disease would not establish if the environment was unfavorable; R. solani diseases are favored by warm and moist conditions1. Overall, there is more complexity in the soil and compost ecosystem than could be mimicked entirely by a single laboratory assay4,8. The plate assay is comparable to other indicators including a nematode index of compost maturity and three microbial ecoenzymes, phosphatase, β-1,4-glucosidase and β-1,4-N-acetylglucosaminidase8. The ecoenzymes reflects the state of decomposition and the carbon and nutrient needs of the microbial community. Confirmation of suppressiveness can be achieved with plant assays but they take longer to complete. Isolating and maintaining a pure culture of a fungus takes care to avoid contamination. ATCC9 offers tips and techniques for culturing yeasts and filamentous fungi.
The method is a potential candidate for commercial certification of disease suppressive properties. The result reflects suppression as a soil function. It does not specify mechanisms or specific microbial species responsible for disease suppression. A future application could be to cut out a portion of the inhibition zone for extraction of DNA for sequencing and identification of community members.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The Vermont Agricultural Experiment Station Competitive Hatch Program VT-HO1609 funded the research. Lynn Fang used the method as part of her M.S. thesis at University of Vermont6.
References
- Gonzalez Garcia G, Onco MAP, Susan VR. Review. Biology and systematics of the form genus Rhizoctonia. Spanish Journal of Agricultural Research. 2006;4(1):55–79. [Google Scholar]
- Bonanomi G, Antignani V, Pane C, Scala E. Suppression of soilborne fungal diseases with organic amendments. Journal of Plant Pathology. 2007;89:311–324. [Google Scholar]
- Noble R. Risks and benefits of soil amendment with composts in relation to plant pathogens. Australasian Plant Pathology. 2011;40:157–167. [Google Scholar]
- Wichuk KM, McCartney D. Compost stability and maturity evaluation - a literature review. Canadian Journal of Civil Engineering. 2010;37:1505–1523. [Google Scholar]
- Alfano G, Lustrato G, Lima G, Vitullo D, Ranalli G. Characterization of composted olive mill wastes to predict potential plant disease suppressiveness. Biological Control. 2011;3:199–207. [Google Scholar]
- Fang L. Biological indicators of compost-mediated disease suppression against the soilborne plant pathogen Rhizoctonia solani. University of Vermont; 2015. M.S. Thesis. [Google Scholar]
- Bonanomi G, Antignani V, Capodilupo M, Scala F. Identifying the characteristics of organic soil amendments that suppress soilborne plant diseases. Soil Biology and Biochemistry. 2010;42:136–144. [Google Scholar]
- Neher DA, Fang L, Weicht TR. Ecoenzymes as indicators of compost to suppress Rhizoctonia solani. Compost Science and Utilization. 2017;25(4):251–261. [Google Scholar]
- American Type Culture Collection. ATCC® Mycology Culture Guide. 2018. Available from: https://www.atcc.org/~/media/PDFs/Culture%20Guides/Mycology_Guide.ashx.
- Berns AE, Phillip H, Narres H-D, Burauel P, Vereecken H, Tappe W. Effect of gamma-sterilization and autoclaving on soil organic matter structure as studied by solid state NMR, UV and fluorescence spectroscopy. European Journal of Soil Science. 2008;59 [Google Scholar]


