Summary
CHAF1B is the p60 subunit of the chromatin assembly factor (CAF1) complex, which is responsible for assembly of histones H3.1/H4 heterodimers at the replication fork during S phase. Here we report that CHAF1B is required for normal hematopoiesis while its overexpression promotes leukemia. CHAF1B has a pro-leukemia effect by binding chromatin at discrete sites and interfering with occupancy of transcription factors that promote myeloid differentiation, such as CEBPA. Reducing Chaf1b activity by either heterozygous deletion or overexpression of a CAF1 dominant negative allele is sufficient to suppress leukemogenesis in vivo without impairing normal hematopoiesis.
Introduction
Chromatin Assembly Factor 1B (CHAF1B) is the p60 subunit of the heterotrimeric Chromatin Assembly Factor 1 (CAF1) complex that also includes a large subunit CHAF1A (p150) and a small subunit RBBP4 (RbAp48, p48) (Smith and Stillman, 1989; Stillman, 1986). This complex is localized to the nucleoplasm and frequently contains a histone H3/H4 heterodimer, each component in a 1:1:1:1 stoichiometry (Hu et al., 2006; Verreault et al., 1996). The canonical function of CAF1 is to facilitate the assembly of H3-H4 tetramers at the replication forks during S-phase (Krude, 1995; Marheineke and Krude, 1998).
CHAF1A is a multi-domain protein that has a replication linked nucleosome assembly activity as well as a replication independent function in the stabilization of heterochromatic regions. The C-terminal region of CHAF1A contains the primary PCNA-interacting motif responsible for tracking the CAF1 complex to the replication fork, an internal acidic region, and a large region at the carboxyl end responsible for direct interaction with CHAF1B (Dong et al., 2001; Shibahara and Stillman, 1999). Previous studies demonstrated that shRNA-mediated knockdown of CHAF1A results in loss of expression of CHAF1B due to degradation of the proteins (Ye et al., 2003). RBBP4 is a 7× WD-repeat protein with two α-helical domains at both ends of the peptide that facilitate its direct interaction with histone H4 (Qian and Lee, 1995; Qian et al., 1993; Zhang et al., 2013). RBBP4 also tightly interacts with HDAC1. Although RBBP4 has no enzymatic activity on its own, it is widely considered to act as a critical scaffold component of the larger HDAC1 complex (Song et al., 2008; Taunton et al., 1996).
CHAF1B is a 7× WD-repeat protein that is responsible for mediating the interaction between ASF1A/H3/H4 and CHAF1A within the CAF1 complex (Mattiroli et al., 2017a; Mattiroli et al., 2017b; Smith and Stillman, 1989; Tyler et al., 2001). In this way, CHAF1B is a central facilitator of multiple S-phase-linked CAF1 functions: (1) CHAF1A-directed localization to the replication fork via interaction with PCNA, (2) H3/H4 chaperone function by direct interaction with ASF1A, and (3) potential HDAC1 complex-mediated functions through RBBP4. CHAF1B also has several reported functions outside of canonical S-phase nucleosome assembly related to DNA-damage repair following UV irradiation damage through the nucleotide excision repair system (Gaillard et al., 1996; Martini et al., 1998; Polo et al., 2006).
Previous reports have also implicated a role for CAF1-mediated nucleosome assembly in determining cell fate by regulating transcription. For example, CHAF1A was implicated as an epigenetic silencing factor that maintains gene repression in an S-phase-dependent manner (Poleshko et al., 2010). The CAF1 complex was also reported to be critical in silencing of proviruses (Yang et al., 2015). Most notably, a study showed that knockdown of CHAF1A or CHAF1B potently enhanced the efficiency of somatic cell reprogramming through the opening of chromatin at specific sites, allowing transcription factor binding to enhancer regions of embryonic stem cell genes (Cheloufi et al., 2015).
CHAF1B is located within the Down syndrome (DS) critical region of chromosome 21, and thus its trisomy is potentially associated with DS-related pathologies (Blouin et al., 1996; Katsanis and Fisher, 1996). Our previous studies revealed that CHAF1B is more highly expressed in acute megakaryocytic leukemia (AMKL) cells from individuals with DS than in AMKL cells from those without trisomy 21 (Malinge et al., 2012). Furthermore, several solid tumor types show increased expression of CHAF1B, and in these cases CHAF1B expression is directly linked to metastasis and disease severity. Cancers with elevated CHAF1B expression include high-grade gliomas, melanomas, endometrial tumors, and prostate cancer (de Tayrac et al., 2011; Mascolo et al., 2010; Polo et al., 2010; Staibano et al., 2009; Staibano et al., 2011), though the mechanisms underlying this overexpression are unexplored.
Given that dysregulation of genes that regulate chromatin is frequently observed in hematologic malignancies, we investigated the role of CHAF1B in normal and malignant hematopoiesis.
Results
Chaf1b is required for hematopoiesis
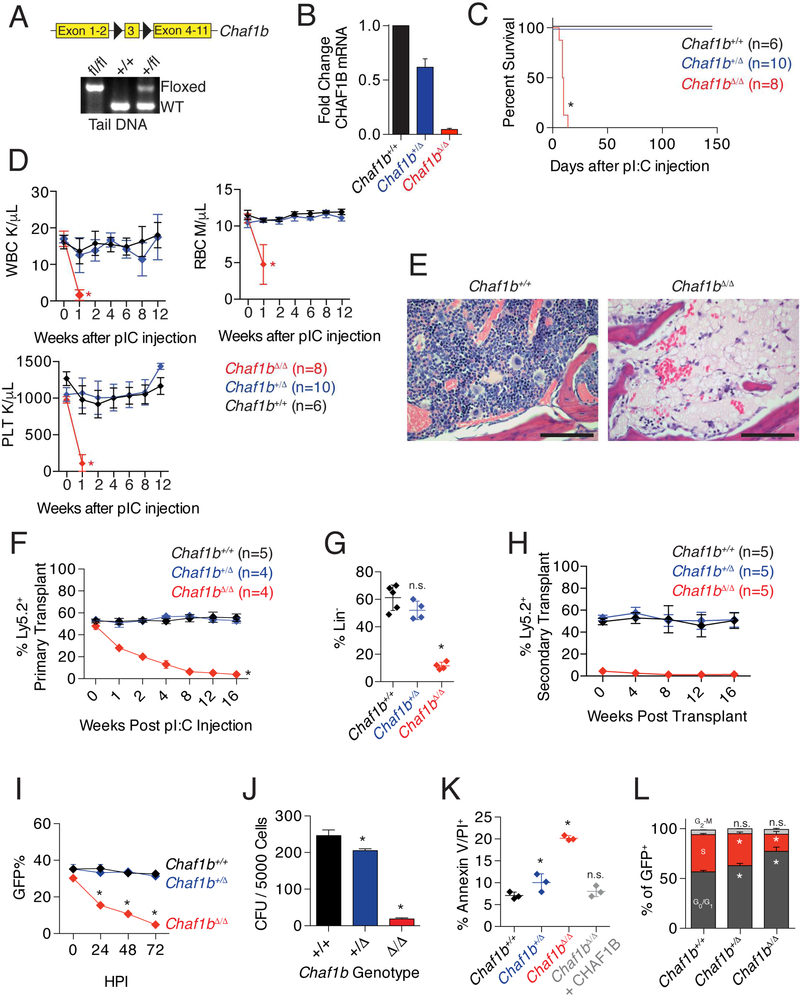
To determine the requirement for Chaf1b in normal hematopoiesis, we utilized a mouse strain generated by injecting embryonic stem cells containing a Chaf1b allele with floxed exon 3 into wild-type C57Bl/6 blastocysts (Figure 1A). We crossed the strain with Mx1-Cre transgenic mice and then induced gene deletion by treating Chaf1b floxed/Mx1-Cre animals with pIpC (Figure S1A). This process reduced CHAF1B expression in a dose-dependent manner by allele as measured by qRT-PCR (Figure 1B). Since CHAF1B is widely expressed throughout the hematopoietic system (Figure S1B), we predicted that homozygous loss of Chaf1b would be lethal. Indeed, Mx1-Cre/Chaf1bfl/fl mice (referred to as Chaf1b null or Chaf1bΔ/Δ) died within two weeks of pIpC injection due to pancytopenia and loss of hematopoietic cells in the bone marrow (Figure 1C-E). In addition to the complete loss of bone marrow cellularity in Chaf1bΔ/Δ mice, we observed a modest but statistically significant reduction in the cellularity of the bone marrow of pIpC-treated Mx1-Cre/Chaf1b+/fl (Chaf1b heterozygous deleted or Chaf1b+/Δ hereafter) mice (Figure S1C). Whereas bone marrow from Chaf1bΔ/Δ mice was unable to form colonies, Chaf1b+/Δ bone marrow gave rise to colonies, albeit fewer than mice with wild-type bone marrow (Figure S1D). Annexin V staining of bone marrow following pIpC treatment revealed a substantial increase in apoptosis in the Chaf1b null mice and a modest increase in the heterozygous deleted animals 10 days after injection (Figure S1E).
Figure 1: Chaf1b is required for hematopoiesis.
(A) Schematic of the floxed allele of Chaf1b and genotype confirmation in tail DNA. (B) qPCR of CHAF1B transcription in HSPCs after infection with MIGR1-Cre. (C) Survival curve of Mx1-Cre wild-type, Chaf1b heterozygous or Chaf1b homozygous floxed mice following pIpC treatment. (D) Peripheral blood white cells (WBC), red blood cells (RBC), and platelet counts (PLT). (E) Sternum bone marrow H&E stain 10 days following pIpC injection. Pictures are representative of three independent trials, scale bars represent 250 μm. (F) Contribution of Chaf1b wild-type, heterozygous and homozygous deficient cells to the peripheral blood following 1:1 competitive transplant. (G) Ly5.2 donor cell contribution to the bone marrow 4 months after transplant. (H) Contribution of Chaf1b wildtype, heterozygous and homozygous deficient cells to the peripheral blood of secondary transplant recipients. (I) In vitro competitive growth assay of HSPCs following introduction of Cre. (J) HSPC colony formation assay following in vitro deletion of Chaf1b. (K) Cell death following Chaf1b deletion by MIGR1-Cre. L) Cell cycle analysis of Chaf1b targeted cells by EdU/DAPI staining of HSPCs 72 hours post transduction. * indicates statistical significance (p<0.05) as determined by log-rank test (C), one-way ANOVA with Bonferroni correction (D, F-L). Results shown are representative of three independent biological replicates (E), are depicted as mean +/− SD from three independent biological replicates (B, I, J, L), are mean +/− SD from indicated numbers of mice (D, F, H), or mean +/− SD from 3–5 mice with points representing individual mice (G, K). See also Figures S1 and S2.
Flow cytometric analysis of bone marrow following pIpC injection (after 7 days for Chaf1bΔ/Δ and 60 days for Chaf1b+/Δ and Chaf1b+/+ mice) revealed that homozygous deletion of Chaf1b resulted in a depletion of hematopoietic stem and progenitor cells (Figure S1F-I). By contrast, Chaf1b+/Δ bone marrows displayed an increase in the percentage of LK cells without any significant skewing in myeloid progenitors, a decrease in LSK cells and an increased proportion of short-term SLAM -positive cells (Figure S1F-I), compared to Chaf1b+/+ controls. We confirmed the Chaf1b heterozygous deletion phenotype by analyzing Chaf1b-floxed mice that were crossed to the Vav-Cre strain (Figure S2A-F). Vav-Cre+Chaf1bfl/fl pups were never observed, supporting our hypothesis that Chaf1b is required for viability.
Loss of Chaf1b impairs hematopoietic reconstitution
To confirm a cell autonomous role for Chaf1b in hematopoiesis, we mixed equal numbers of wild-type Ly5.1 bone marrow cells with those from Ly5.2 Mx1-Cre/Chaf1b+/+, Mx1-Cre/Chaf1b+/fl, or Mx1-Cre/Chaf1bfl/fl mice, and then transplanted the cells to irradiated Ly5.1 recipients. As expected, there was a near complete loss of hematopoietic cells derived from homozygous Chaf1b null cells by 8 weeks after pIpC injection (Figure 1F). After four months, we observed an almost complete elimination of the Ly5.2+ Chaf1bΔ/Δ cells in the bone marrow (Figure 1G). Secondary transplants revealed that the heterozygous deletion, but not the homozygous deletion, was capable of serial reconstitution (Figure 1H).
We next examined the effect of Chaf1b deletion on c-Kit+ hematopoietic stem and progenitor cells (HSPCs) in vitro. First, we deleted one or both alleles of Chaf1b by transduction of the floxed HSPCs with MIGR1-Cre and found that these cells had a competitive disadvantage in culture over time (Figure 1I). Heterozygous Chaf1b loss resulted in a modest reduction in colony formation, while the homozygous deletion resulted in an almost complete block in colony formation (Figure 1J). Although Chaf1b heterozygous deleted HSPCs expanded as well as control cells, there was a significant increase in cell death as measured by annexin V/PI staining 72 hours post infection (Figure 1K). Apoptosis of MIGR1-Cre/Chaf1bfl/fl HSPCs was completely abrogated by expression of CHAF1B, confirming that the phenotype is specifically due to Chaf1b loss (Figure 1K). Cell cycle analysis further demonstrated that the proportion of cells in S-phase was reduced upon Chaf1b deletion, suggesting failure to enter S-phase from G0/G1 (Figure 1L).
CHAF1B overexpression enhances proliferation of HSPCs
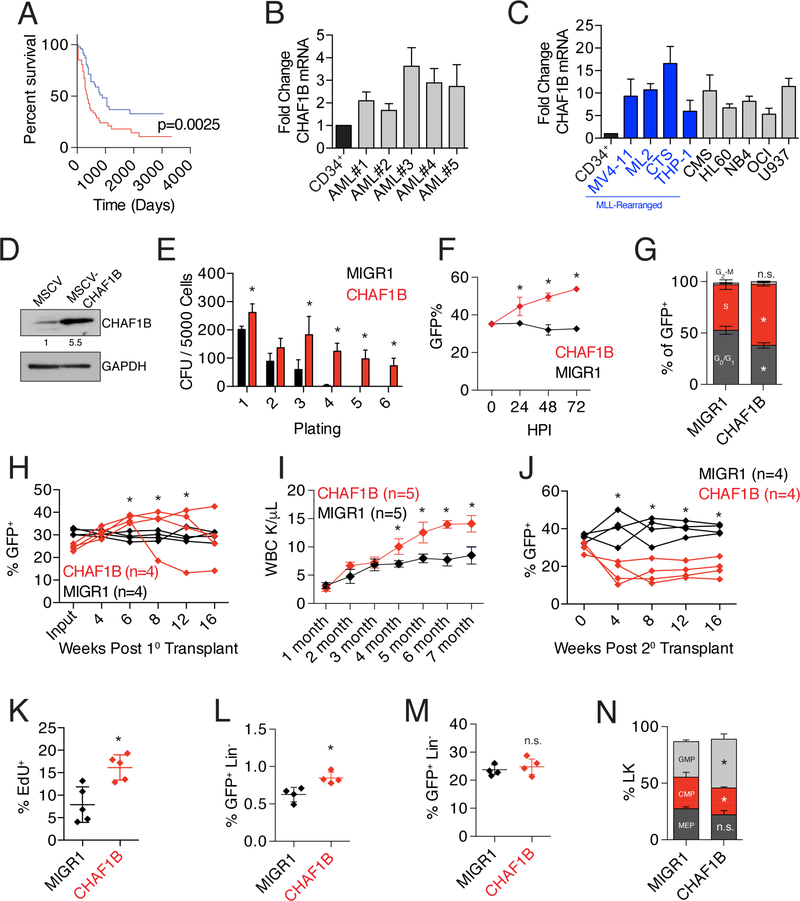
We previously reported that CHAF1B is overexpressed in AMKL patients with DS, consistent with its gene amplification via trisomy 21 (Malinge et al., 2012). In order to more generally determine the contributions of CHAF1B to AML, we analyzed the AML dataset from The Cancer Genome Atlas (TCGA) and found that higher expression of CHAF1B correlates with poor prognosis (Figure 2A). Further, our analysis of five primary AML patient samples showed these tumors consistently express CHAF1B at 2–4 times the level of healthy CD34+ cells (Figure 2B, Table S1). This increased expression was also observed in nine different human AML cell lines, with some of the highest expressers of CHAF1B also having MLL-rearrangements (Figure 2C). According to the Cancer Cell Line Encyclopedia (CCLE), CHAF1B is overexpressed in nearly every malignancy (Figure S3A). However, the tumors with the highest levels of CHAF1B overexpression were predominantly leukemias. Further analysis of CHAF1B expression in different FAB subtypes of AML in TCGA did not reveal any clear specificity for subtype (Figure S3B).
Figure 2: CHAF1B overexpression promotes the proliferation of hematopoietic cells.
(A) Survival analysis of TCGA LAML dataset comparing the top (red line) and bottom (blue line) 1/3rd of patients based on CHAF1B expression. (B) Measurement of CHAF1B expression in AML patient samples relative to proliferating CD34+ cells by qRT-PCR. (C) Measurement of CHAF1B expression in AML cell lines relative to proliferating CD34+ cells by qRT-PCR. Blue bars depict MLL rearranged cell lines. (D) Western blot of CHAF1B in MSCV or MSCVCHAF1B expressing HSPCs. Value shown is CHAF1B expression level relative to MSCV control. (E) Colony replating assay in methylcellulose with HSPCs following CHAF1B overexpression. GFP+ cells were sorted between each plating. (F) In vitro competition assay measuring the percentage of GFP+ cells in suspension culture over time. (G) Cell cycle analysis (EdU/DAPI) of HSPCs 72 hours after transduction with CHAF1B. (H) In vivo competitive reconstitution assay using HSPCs transduced with CHAF1B. Each line depicts percent of peripheral blood GFP+ cells and represents an individual mouse. (I) Peripheral white blood cell count of mice receiving CHAF1B-overexpressing HSPCs. (J) Percentage of GFP+ cells in the peripheral blood of secondary recipients of bone marrow from mice in (E). Each line represents an individual mouse (K) EdU incorporation in Lin- BM MNCs of mice 4 weeks following transplantation with CHAF1B-overexpressing HSPCs. (L,M) LSK (L) and LK (M) populations in the hind limb bone marrow of primary recipient mice 4 months after transplant. (N) Distribution of myeloid progenitors from the LK population of recipient mice 4 months after transplant. * indicates statistical significance (p<0.05) when compared to MIGR1 control as determined by log-rank test (A), one-way ANOVA with Bonferroni correction (B, E-G, H-J, N), Mann-Whitney test (C), or two-tailed t-test with Welch’s correction (D, K-M). Results shown are quantifications of at least three independent biological replicates or lines/points represent individual mice. Bar graphs depict mean +/− SD. See also Figure S3 and Table S1.
To determine the consequences of increased expression of CHAF1B in the hematopoietic system, we overexpressed CHAF1B in HSPCs and performed in vitro colony replating assays. We found that 5-fold overexpression of CHAF1B in HSPCs enhanced colony replating and imparted a significant growth advantage in culture, with overexpressing cells outcompeting control-transduced cells almost 2:1 by 72 hours (Figure 2D-F). The enhanced competitive capacity in vitro was associated with increased entry into S-phase of the cell cycle (Figure 2G).
We next transplanted Ly5.2 hematopoietic progenitor cells overexpressing CHAF1B or GFP alone (MIGR1) to irradiated Ly5.1 recipient mice and monitored engraftment and contribution to hematopoiesis over time. CHAF1B overexpression resulted in a progressive increase in contribution of the CHAF1B-overexpressing HSPCs to the peripheral blood (Figure 2H). This was accompanied by an increase in the peripheral white blood cell count (Figure 2I). By contrast, cells with CHAF1B overexpression were less competitive in secondary transplants (Figure 2J). This phenotype may be the result of the excessive proliferation of hematopoietic progenitors, which was observed in lineage-negative bone marrow MNCs two weeks posttransplantation (Figure 2K). We also observed modest but statistically significant increase in the percentage of lin−Sca1+c-kit+ (LSK) cells (Figure 2L). Analysis of primary recipients revealed no increase in the percentage of lin−c-kit+ (LK) cells in the bone marrow (Figure 2M), but precocious myeloid differentiation patterning, skewed towards production of granulocyte/macrophage progenitors (GMPs) (Figure 2N).
CHAF1B overexpression enhances leukemogenesis in an MLL-AF9 model of AML
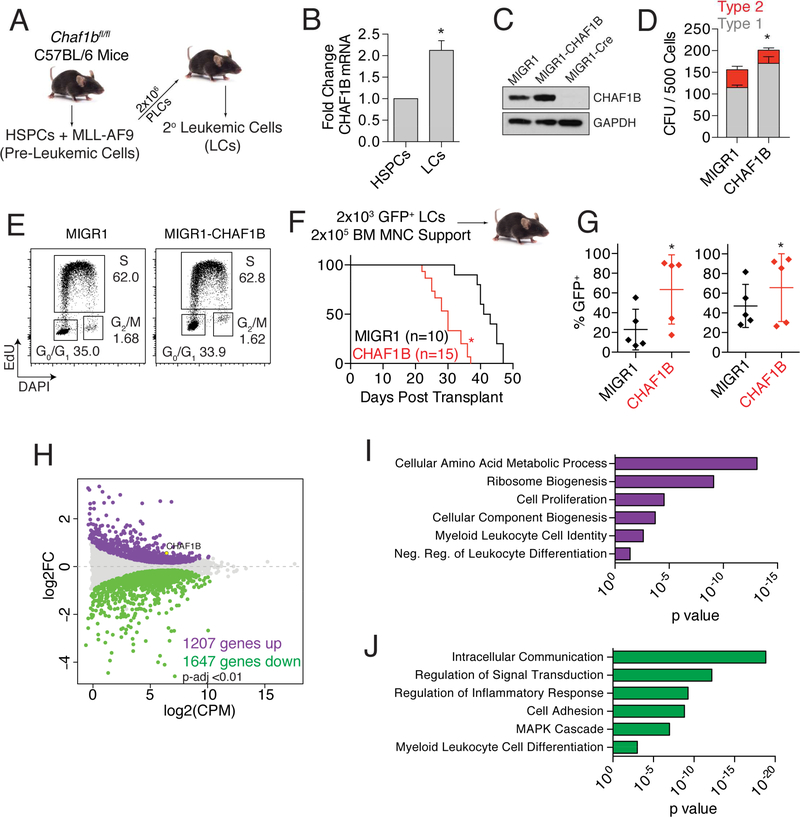
In order to determine the contribution of CHAF1B to promoting leukemogenesis, we turned to the MLL-AF9 model of murine AML (Figure 3A). MLL-AF9 leukemic cells expressed CHAF1B at twice the level of their non-transformed HSPC counterparts (Figure 3B), putting them within the range observed in primary AML patient samples (Figure 2B). Further overexpression of CHAF1B in MLL-AF9 leukemic cells (Figure 3C), increased colony formation, with a notable enrichment in less-differentiated Type 1 colonies (Figure 3D). We did not detect an increase in actively-cycling cells (Figure 3E), but this may be due to the highly proliferative state of these cells. We then transplanted MLL-AF9 leukemic cells overexpressing CHAF1B or eGFP alone into irradiated recipients and measured survival over time. Mice engrafted with MLL-AF9 cells that overexpress CHAF1B succumbed to disease significantly faster than those transplanted with control-transduced MLL-AF9 cells (Figure 3F). Analysis of the transplanted mice at 30 days revealed CHAF1B-overexpressing MLL-AF9 leukemic cells had overtaken the bone marrow and the spleen, with CHAF1B-overexpressing leukemic cells accounting for almost 90% of mononucleated cells in both organs (Figure 3G).
Figure 3: CHAF1B enhances leukemic development.
(A) Schematic representation of the method of generating MLL-AF9 leukemic cells. (B) CHAF1B mRNA levels in MLL-AF9 leukemic cells compared to HSPCs. (C) Western blot of CHAF1B in Chaf1bfl/fl MLL-AF9 LCs expressing MIGR1-CHAF1B or MIGR1-Cre. (D) Colony forming unit assay of leukemic cells following transduction with MIGR1-CHAF1B. Type 1 vs Type 2 colonies are represented. (E) Cell cycle analysis of leukemic cells following transduction with MIGR1-CHAF1B. One representative plot is shown of three independent experiments. (F) Survival curve of mice engrafted with MIGR1 or MIGR1-CHAF1B expressing leukemic cells. (G) Leukemic cell burden within the bone marrow (left) and spleen (right) 30 days after transplantation. Horizontal line indicates mean, verticle line indicates SD, and each dot represents an individual mouse. (H) MDS plot of genes significantly changed in CHAF1B overexpressing leukemic cells vs control (p<0.01). (I,J) GO pathway analysis of genes that are increased (I) or decreased (J) after CHAF1B overexpression in leukemic cells. * indicates statistical significance (p<0.05) as determined by log-rank test (F), two-tailed t-test with Welch’s correction (B, G), or one-way ANOVA with Bonferroni’s correction (D). Results shown are indicative of at least three biological replicates, or absolute numbers are notated. Bar graphs depict mean +/− SD.
Transcriptome analysis of CHAF1B-overexpressing MLL-AF9 leukemia cells revealed a substantial effect on gene expression (Figure 3H). Pathway analysis of the 1207 upregulated genes revealed numerous leukemic stem cell-associated pathways including those involved in metabolic processes, biogenesis, and negative regulation of leukocyte differentiation (Figure 3I). The most enriched pathways for the 1647 downregulated genes included those involved in cell signaling, responses to MAPK cascade, and promotion of leukocyte differentiation (Figure 3J).
Chaf1b is required to maintain the undifferentiated state of MLL-AF9 leukemic cells and for leukemia progression
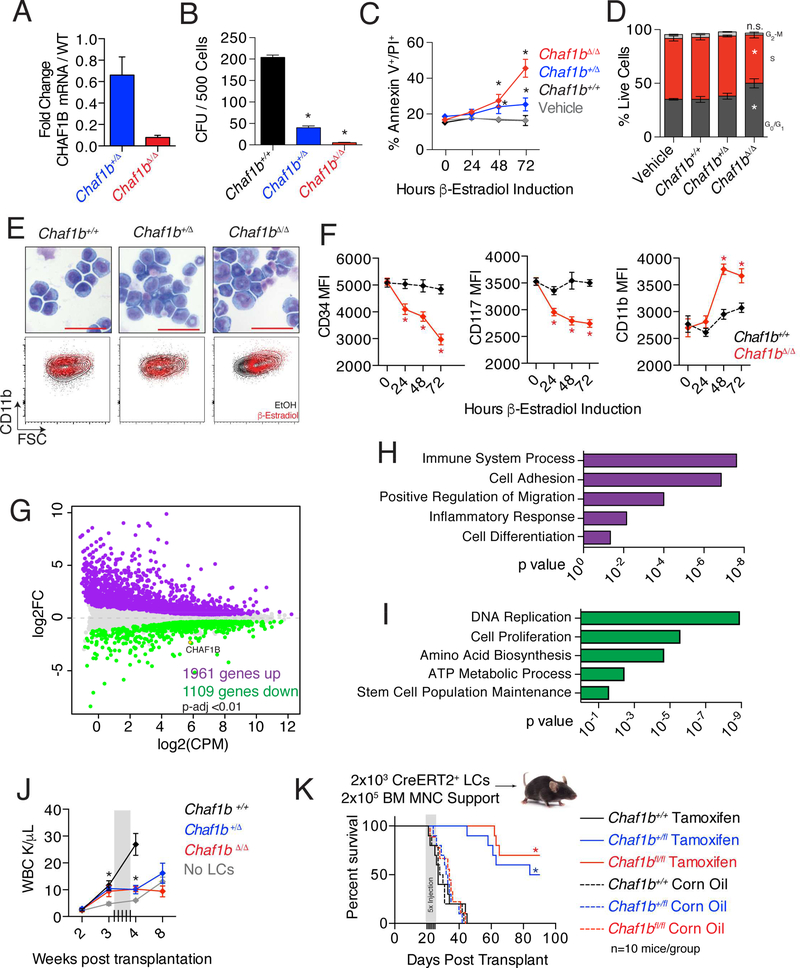
We next derived stable clones of MLL-AF9 Chaf1bfl/fl leukemic cells harboring MSCVCre-ERT2 or MSCV-puro. Treatment of the cells with β-estradiol led to 40% and >95% decreases in CHAF1B expression in heterozygous and homozygous targeted cells within 48 hours, respectively (Figure 4A). Homozygous deletion of Chaf1b completely abrogated the colony forming capacity of the leukemic cells, and notably, the heterozygous deletion also substantially reduced colony formation (Figure 4B). Deletion was associated with increased cell death, although the increase was modest in the heterozygous cells despite the profound loss of colonies (Figure 4C). Neither heterozygous nor homozygous deletion had a significant effect on cell proliferation at 72 hours (Figure 4D). Since the loss of colony formation could not be fully explained by cell death or cell cycle arrest, we assayed the differentiation status of these cells. We found that Chaf1b depletion was sufficient to induce differentiation of leukemia cells as determined by increased cell size, loss of blast-like nuclear/cytoplasmic morphology, and increased CD11b staining (Figure 4E). Additionally, we observed progressive reductions in surface staining for CD34 and CD117 (markers of immature hematopoietic cells), as well as an increase in CD11b (marker of myeloid differentiation) on MLL-AF9 leukemic cells following Chaf1b deletion (Figure 4F). This effect of Chaf1b downregulation is consistent with the observation that CHAF1B expression declines during normal hematopoietic cell differentiation (Figures S1B, S4A).
Figure 4: Loss of Chaf1b induces differentiation of MLL-AF9 leukemic cells.
(A) qRT-PCR analysis of MLL-AF9 leukemic cells (LCs) with CreERT2 for Chaf1b mRNA expression after Cre induction with estradiol. (B) Colony forming unit assay with CreERT2 MLL-AF9 LCs 24 hours after induction of Chaf1b deletion. (C) Annexin V/PI stain of LCs after Cre induction with estradiol. (D) Cell cycle analysis of LCs 72 hours after Cre induction with estradiol. (E) WrightGiemsa stain of leukemic cells 72 hours following Chaf1b deletion. Scale bar indicates 50 μm. Representative FACS plots (FSC vs CD11b) of each condition are shown beneath the images. (F) Mean fluorescence intensity of CD34, CD117 and CD11b in leukemic cells following Chaf1b deletion. (G) MDS plot showing genes with significantly (p<0.01) altered expression 48 hours after Chaf1b deletion. (H, I) GO Pathway analysis of genes that are increased (H) or decreased (I) 48 hours following Chaf1b deletion in leukemic cells. (J) Peripheral circulating leukocyte counts measured by complete blood count. Gray box indicates dated of tamoxifen injections. (K) Survival of recipient mice following injection with tamoxifen (solid lines) or vehicle (dotted lines). * indicates statistical significance (p<0.05) as determined by One-way ANOVA with Bonferroni’s correction (A-D, F, J), or log-rank test (K). Unless otherwise indicated, results shown are representative of three independent biological replicates. Bar graphs depict mean +/− SD. See also Figure S4.
RNA-seq analysis revealed that the expressions of 1961 genes were increased and 1109 decreased following Chaf1b deletion, suggesting CHAF1B may have an additional role in repressing transcription (Figure 4G). We confirmed our RNA-seq results by qRT-PCR for the top differentially regulated genes (Figure S4B). Of note, heterozygous deletion of Chaf1b was associated with an intermediate change in expression of the same genes (Figure S4B). The activated genes were enriched in pathways associated with immune system processes, regulation of migration, and cell differentiation (Figure 4H). By contrast, the pathways that were negatively enriched upon Chaf1b knockout are those involved in metabolic processes and DNA replication, the latter two consistent with the notion that loss of CHAF1B leads to defects in chromatin organization (Figure 4I). However, we expect that Chaf1b null leukemic cells were still able to assemble chromatin due to rescue of assembly by HIRA, because our RNA-seq analysis revealed that CHAF1B and HIRA are expressed at relatively similar levels, and the H3 variant H3.3 (the preferred H3 for HIRA) was substantially upregulated in Chaf1b null leukemic cells (Figure S4C-D).
Previous studies have shown that DNA damage can drive differentiation in the hematopoietic system (Santos et al., 2014). To determine if replication-linked DNA damage was a possible driver of differentiation in our MLL-AF9 leukemic cells, we induced Chaf1b deletion in Chaf1bfl/fl HSPCs and MLL-AF9 LCs and measured DNA damage over time. We found progressive incorporation of γH2A.X in Chaf1bΔ/Δ HSPCs. However, there was no change in γH2A.X incorporation in Chaf1bΔ/Δ MLL-AF9 leukemic cells (Figure S4E). Increased H3.3 transcription in MLL-AF9 leukemic cells following Chaf1b deletion led us to hypothesize that HIRA may be functionally compensating for CHAF1B loss in DNA replication as previously demonstrated in Candida albicans (Lopes da Rosa et al., 2011; Stevenson and Liu, 2013). We assayed this role for HIRA in Chaf1bfl/fl MLL-AF9 leukemia cells using shRNA to knockdown HIRA and were able to observe DNA damage after Chaf1b deletion (Figure S4F, G).
Finally, to determine if Chaf1b deletion-mediated differentiation could block leukemogenesis in vivo, we engrafted Chaf1b-floxed CreERT2+ MLL-AF9 leukemic cells into irradiated recipient mice and monitored the peripheral blood for evidence of disease. At 3 weeks post-transplant, a time when we observed elevated white blood cell counts in recipient mice (Figure 4J), we initiated Chaf1b deletion by injecting mice with tamoxifen. Both heterozygous and homozygous deletion of Chaf1b in LCs resulted in almost complete elimination of the disease as measured by survival (Figure 4K) and resolution of white blood cell levels in the peripheral blood (Figure 4J). Several mice succumbed to leukemia after 50–60 days as a result of tumors that escaped Chaf1b deletion.
CHAF1B occupies discrete regions of chromatin in leukemic cells
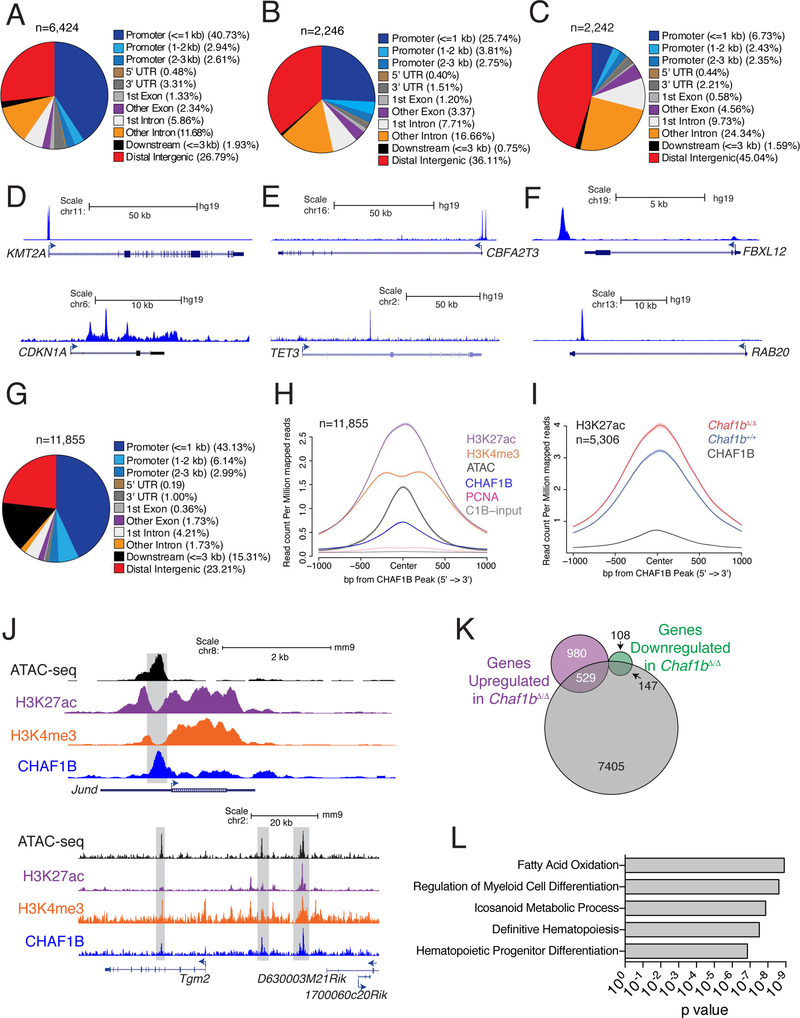
In order to determine the mechanism by which CHAF1B maintains the undifferentiated state of leukemic cells, we analyzed CHAF1B chromatin occupancy in three human leukemia cell lines: MOLM13 (AML, MLL-Rearranged), U937 (AML, non-MLL-Rearranged), and JURKAT (T-ALL) and found CHAF1B occupied discrete regions of the chromatin associated with promoters and distal intergenic elements in each cell type (Figure 5A-F). We confirmed the specificity of our CHAF1B antibody for ChIP-seq analysis in U937 cells expressing CHAF1B shRNA, which had reduced signal genome-wide following knockdown (Figure S5A-C). Genomic regions bound by CHAF1B in MOLM13 cells also tended to co-occupancy with CHAF1A. GO analysis of CHAF1B-bound regions showed enrichment for genes associated with myeloid differentiation (Figure S5D-F). Analysis of the Leucegene AML RNA-seq database (http://mistic.iric.ca/) further confirmed that expression of CHAF1B and CHAF1A are highly corelated (Figure S5G). We observed similar occupancy of CHAF1B near differentiation genes in U937 and JURKAT cells (Figure S5H, I).
Figure 5: CHAF1B accumulates at discrete sites in the chromatin of leukemic cells.
(A-C) Distribution of CHAF1B peaks in MOLM13 (A), U937 (B), and JURKAT (C). (D-F) Track examples of CHAF1B occupancy in MOLM13 (D), U937 (E), and JURKAT (F). (G) Distribution of CHAF1B occupancy in MLL-AF9 leukemia cells. (H) Meta-analysis of ATAC-seq, H3K4me3, H3K27ac, PCNA, and input peaks localized to CHAF1B peaks in MLL-AF9 LCs. (I) Meta-analysis of H3K27ac peaks that increase at CHAF1B occupied sites before and after Chaf1b deletion. (J) Track examples of Jund and Tgm2. Gray boxes indicate areas of interest. (K) Venn diagram of unique genes with discrete CHAF1B peaks compared to genes upregulated or downregulated 48 hours after Chaf1b deletion. (L) GREAT analysis of genes and pathways localized near peaks determined in (K). See also Figure S5.
Next, we extended the human cell ChIP-seq data to primary mouse MLL-AF9 LCs overexpressing CHAF1B (Figure 5G, S5J). Analysis of the CHAF1B signature in these murine cells revealed occupancy at promoters and distal intergenic regions similar to MOLM13 cells (compare Figures 5A, G). Additionally, we found co-occupancy with other regions of protranscriptional chromatin including H3K4me3 regions, K3K27ac regions, and accessible regions by ATAC-seq (Figure 5H-J). Interestingly, we did not see strong co-occupancy with PCNA, suggesting the rest of the DNA replication complex is likely not present at these sites of accumulation (Figure 5H). A substantial proportion of genes whose expression changed with Chaf1b loss were occupied by CHAF1B (Figure 5K). Consistent with the observation of transcriptional upregulation by RNA-seq of differentiation genes, we also observed over 5000 unique sites in the chromatin where there was a statistically significant increase in H3K27ac signature at CHAF1B-bound chromatin following Chaf1b deletion (Figure 5I). These were predominantly associated with differentiation of hematopoietic cells (Figure 5L). The specificity of the CHAF1B murine antibody was validated by comparing the degree of chromatin occupancy in wild-type versus MLL-AF9 cells in which Chaf1b was deleted (Fig S5K).
Changes in chromatin accessibility do not predict expression of CHAF1B bound genes
Because CHAF1B is a chromatin assembly factor and a previous study reported that depletion of CHAF1B led to enhanced chromatin accessibility at stem cell enhancers (Cheloufi et al., 2015), we turned to ATAC-seq to determine if deletion of Chaf1b caused dysregulation of gene expression via changes in chromatin accessibility. We assayed the unique ATAC-seq peaks that co-occupied with a CHAF1B peak, and to our surprise, observed a general reduction in accessibility at CHAF1B binding sites after Chaf1b deletion (Figure S6A). Further analysis revealed that although there was a global reduction in ATAC-seq signal at CHAF1B-occupied regions, approximately 1100 and 1224 peaks were associated with the most decreased or the most increased ATAC signals respectively (Figure S6B-C). In fact, a very small percentage of the genes whose accessibility changed the most upon Chaf1b depletion overlapped with genes whose transcriptional levels changed (Figure S6D-E). For example, although Hira and Becn1 were among the genes with the greatest changes in chromatin accessibility following Chaf1b deletion (Figure S6F-G), there were no concomitant changes in expression of either gene (Figure S6H).
CHAF1B inhibits CEBPA-mediated differentiation of leukemic cells
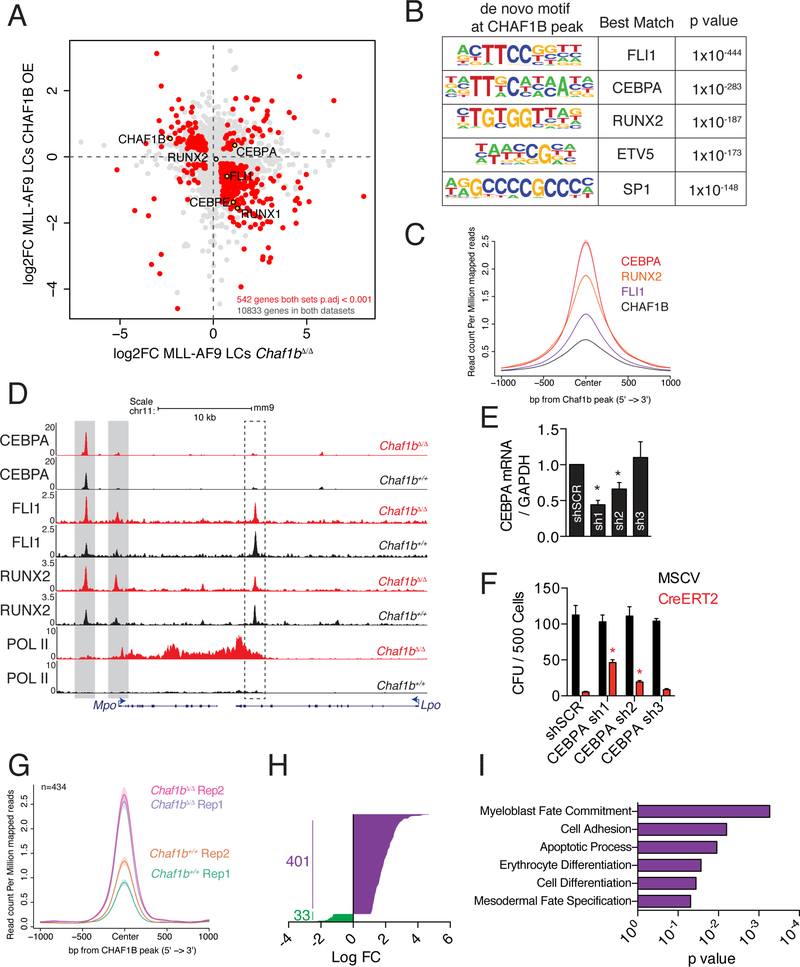
Analysis of RNA-seq data from Chaf1b deleted or CHAF1B-overexpressing leukemic cells revealed an abundance of genes that appeared to be repressed by CHAF1B. This class included notable myeloid differentiation transcription factors including CEBPE, FLI1, and RUNX1 (Figure 6A). The DNA-binding motifs of these factors, as well as CEBPA, RUNX2, ETV5, and SP1, were also strongly enriched at CHAF1B-binding sites genome-wide (Figure 6B). We pursued the top three motif hits further and found CEBPA, RUNX2, and FLI1 cooccupied chromatin sites bound by CHAF1B (Figure 6C). All three factors showed increased occupancy at the promoter and a proximal enhancer of Mpo, one of the top up-regulated genes in MLL-AF9 leukemic cells following Chaf1b deletion (Figure 6D). Lpo, a gene not changed in expression after Chaf1b deletion, did not show such changes in transcription factor binding (Figure 6D). These findings led us to hypothesize that CHAF1B may block differentiation by interfering with the occupancy of transcription factors at myeloid differentiation genes. Therefore, we expected that knockdown of the relevant transcription factor should restore leukemogenic capacity to Chaf1b deleted LCs. To test this hypothesis, we knocked down CEBPA and FLI1 in LCs and induced Chaf1b deletion before plating in methylcellulose (Figure 6E). While knockdown of FLI1 did not rescue colony formation (data not shown), we found that CEBPA knockdown partially restored colony formation induced by Chaf1b-deletion in a dosedependent manner (Figure 6F). We pursued possible interplay between CHAF1B and CEBPA by ChIP-seq by identifying 434 sites of significantly altered CEBPA chromatin occupancy following Chaf1b deletion in leukemic cells (Figure 6G). The vast majority (401 out of 434) of these peaks were increased following Chaf1b deletion (Figure 6H), with these peaks being proximal to genes associated with differentiation (Figure 6I). ChIP-seq analysis in human leukemia cell lines confirmed this effect, as MOLM13 and U937 cells (AML cell lines) showed inversed occupancy of CHAF1B with CEBPA (Figure S7A, B) while CHAF1B and GATA3 showed inversed occupancy in JURKAT cells (Figure S7C).
Figure 6: CHAF1B interferes with CEBPA to drive differentiation in leukemic cells.
(A) RNA-seq analysis in leukemic cells overexpressing CHAF1B or deleted for Chaf1b. (B) De novo motif analysis of DNA bound by CHAF1B peaks. (C) Metaplot of CEBPA, RUNX2, and FLI1 occupancy centered on CHAF1B peaks as determined by ChIP-seq in MLL-AF9 LCs. (D) Track example of Mpo and Lpo. Gray/dashed boxes indicate regions of interest in Mpo/Lpo respectively. (E) CEBPA mRNA levels after shRNA knockdown in LCs as measured by qRT-PCR. (F) Colony assay in leukemic cells with CEBPA shRNA following Chaf1b homozygous deletion by CreERT2. (G) Metaplot of significantly altered CEBPA peaks before and after Chaf1b deletion centered on CHAF1B peaks. (H) Log fold change in significantly altered CEBPA peaks. (I) Gene ontology analysis based on the nearest TSS to peaks identified in G and H. * indicates p<0.01 as determined by one-way ANOVA with Bonferroni’s correction. Results shown are mean +/− SD in E and F. C is a compilation of two biological replicates. Individual replicates are shown in G. See also Figures S6 and S7.
CHAF1B maintains MLL-AF9 leukemic cells through its replication-dependent nucleosome assembly function
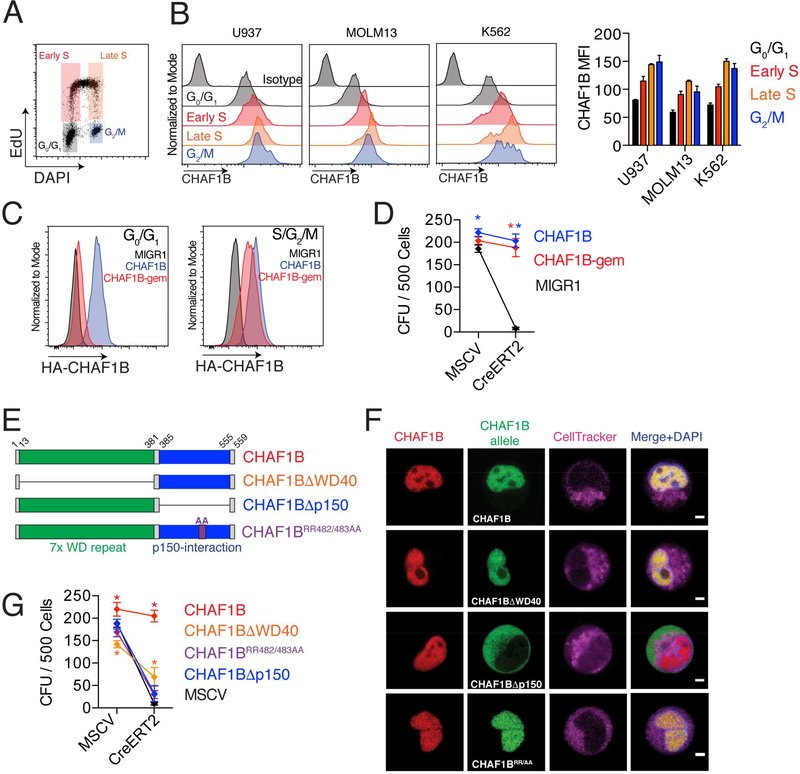
Since CHAF1B is a replication-dependent nucleosome assembly factor, we first measured CHAF1B expression during cell cycle (Figure 7A). We found that CHAF1B expression increased with cell cycle progression, with late S and M phase cells expressing the highest levels (Figure 7B). To determine the requirement for CHAF1B during the phases of the cell cycle, we expressed a geminin-degron tagged CHAF1B (CHAF1B-gem) that is stabilized in S/G2/M and is absent from G0/G1 cells (Figure 7C). We found that CHAF1B-gem was sufficient to rescue the anti-differentiation activity of Chaf1b (Figure 7D), suggesting that a G0/G1 function CHAF1B is not necessary to maintain MLL-AF9 leukemic cells.
Figure 7: CHAF1B maintains leukemic cells through its replication-dependent nucleosome assembly function.
(A) Schematic of U937 cell cycle analysis. (B) CHAF1B expression at different stages of the cell cycle. Representative plots are shown with mean +/SD of the mean fluorescence intensity quantification on the right. (C) Expression of CHAF1B or CHAF1B-gem at different stages of the cell cycle as measured by flow cytometry. (D) CFU assay of LCs expressing CHAF1B-gem after excision of endogenous Chaf1b. (E) Schematic of CHAF1B deletion mutants. (F) Live cell imaging of the localization of the endogenous CHAF1B (red) and the ectopically expressed CHAF1B alleles (green) in MLL-AF9 leukemia cells, cell tracker for plasma membrane (magenta) and DAPI to visualize the nucleus (blue). Scale bars indicate 2.5 μm. G) CFU assay in LCs expressing various CHAF1B deletion mutants after excision of endogenous Chaf1b. * indicates p<0.05 as determined by two-way ANOVA with Bonferroni’s correction (D, G). Results shown are mean +/− SD (D, G) or representative of three independent biological replicates (A-D, F).
We next assayed CHAF1B nucleosome-assembly function by overexpressing mutant alleles of CHAF1B in CreERT2+ Chaf1bfl/fl leukemic cells (Figure 7E) and compared their ability to rescue the knockout phenotype of differentiation. CHAF1B has two major protein domains: a 7× WD40 repeat domain, and a p150-interacting region that is required for replication-dependent nucleosome assembly by directly interacting with CHAF1A and ASF1A. Residues RR482/483 (contained within a β-sheet in the p150-interacting region) are critical for CHAF1B interaction with ASF1A (Tang et al., 2006). CHAF1BΔWD40 and CHAF1BRR482AA localized to the nucleus similar to wild-type CHAF1B, whereas CHAF1BΔp150 localized to the cytoplasm (Figure 7F). Interestingly, CHAF1BΔwd40 was able to partially rescue the Chaf1b deletion, suggesting that the WD40 domain is partially needed to prevent differentiation, though its exact contributions remain unclear. By contrast, the CHAF1BΔp150 and CHAF1BRR482AA alleles were unable to rescue Chaf1b deletion, indicating that replication-dependent nucleosome assembly is critical to CHAF1B function in leukemic cells (Figure 7G).
CHAF1B is a potential target for leukemic therapy
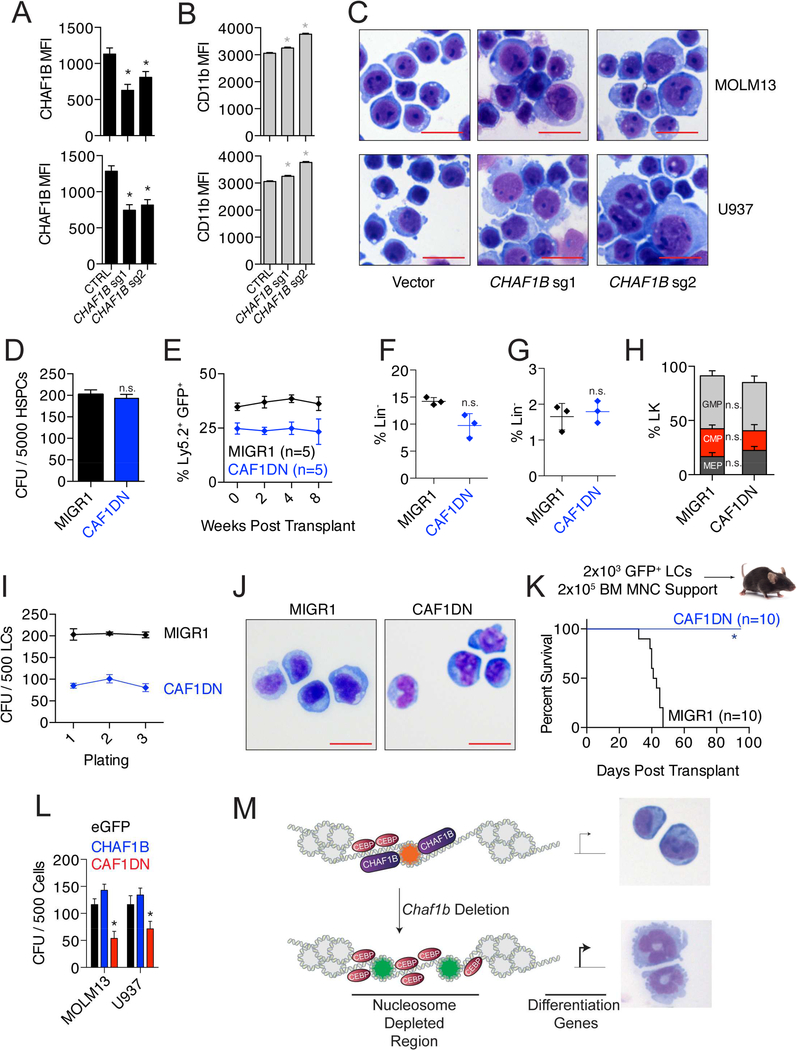
Our studies with MLL-AF9 cells has demonstrated that depletion of CHAF1B leads to myeloid differentiation. To confirm this function of CHAF1B in other cell types, we induced CHAF1B deletion in human AML cell lines MOLM13 and U937 using CRISPR with two separate sgRNAs (Figure 8A) We observed substantial differentiation in these cells 96 hours after CHAF1B deletion induction as measured by surface expression of CD11b and changes in morphology (Figure 8B, C). We also confirmed the anti-tumor effect of Chaf1b deletion in a conditional KrasG12D model of myeloproliferative disease (MPD). Upon induction with pIpC, Mx1-Cre+KrasG12Dlsl mice succumbed to MPD within 100 days. This survival was substantially improved when one allele of Chaf1b was simultaneously deleted due to restoration of normal bone marrow function and multi-lineage differentiation (Figure S8A,B).
Figure 8: Disruption of CHAF1B is a potential anti-leukemic strategy.
(A) Confirmation of CRISPR-mediated CHAF1B deletion by intracellular flow cytometry. (B) Surface expression of CD11b by flow cytometry as determined by the MFI. (C) Morphology of leukemia cells 96 hours after induction of CHAF1B deletion. Scale bars indicate 25 μm. (D) CFU assay of HSPCs overexpressing CAF1DN. (E) Contribution of CAF1DN-expressing HSPCs to peripheral blood. (F-H) Analysis of GFP+ LK (F), LSK (G), and myeloid progenitor (H) population in bone marrow of mice reconstituted with MIGR1 or CAF1DN-expressing HSPCs. Horizontal lines indicate the mean, verticle lines indicate SD. (I) Colony replating assay with leukemic cells overexpressing MIGR1 or CAF1DN. (J) Morphology of leukemic cells after third plating from G. Scale bars represent 25 μm. (K) Survival curve of recipient mice receiving 2000 LCs overexpressing CAF1DN. (L) CFU assay in sorted MOLM13 and U937 cells expressing empty vector, CHAF1B, or CAF1DN. (M) Proposed mechanism of CHAF1B-mediated gene expression. * indicates statistical significance (p<0.05) as determined by one-way ANOVA with Bonferroni’s correction (A, B, D, E, I, L), or Student’s t-test with Welch’s correction (F, G), or log-rank test (K). D, E, H, I, L are mean ± SD from three separate biological replicates, individual points symbolize individual mice in F and G. See also Figure S8.
We found heterozygous deletion of Chaf1b in leukemic cells was sufficient to block leukemogenesis in vivo (Figure 4K), while heterozygous deletion of Chaf1b in healthy hematopoietic tissues did not impair reconstitution in vivo (Figure 1C, F, H). This suggests there may be a therapeutic window for inhibition of CHAF1B as an anti-leukemic strategy. As a proof of concept, we introduced a CAF1 dominant negative allele of CHAF1A (CAF1DN) that binds to CHAF1B and prevents interactions with PCNA and the replication fork (Ye et al., 2003) in HSPCs and leukemic cells. The CAF1DN did not affect HSPC colony formation in vitro (Figure 8D), hematopoietic reconstitution in vivo (Figure 8E), or the proportions of healthy stem and progenitor cells in vivo (Figure 8F-H). While this construct had no discernable effect on leukemic cells in suspension culture, we observed a reduction in colony formation in vitro (Figure 8I). These cells were still able to replate, though the reduction in colony potential could have been due to partial differentiation of leukemic blasts driven by the CAF1DN in methylcellulose (Figure 8J). Of note, introduction of the CAF1DN into MLL-AF9 leukemia cells was sufficient to completely block leukemia formation in vivo (Figure 8K). We also confirmed the activity of the CAF1DN in human AML cell lines MOLM13 and U937, and observed a similar repression in colony number (Figure 8L). Together these data led us to our proposed model that CHAF1B is recruited to the chromatin through its canonical replication-linked nucleosome assembly function, but the maintenance of the leukemic stem cell transcription program occurs through an extra-canonical function characterized by competition with transcription factors for chromatin occupancy (Figure 8M).
Discussion
The chromatin assembly complex has an essential role in facilitating nucleosome assembly on newly replicating DNA and also participates in DNA damage repair (Martini et al., 1998; Nabatiyan and Krude, 2004; Zhu et al., 2009). Though loss of any component of the CAF1 complex in yeast leads to dysregulation of Okazaki fragment length (Smith and Whitehouse, 2012) and reduction in CAF1 activity can lead to DNA damage and S-phase arrest in U2OS cells (Ye et al., 2003). For example, animals deficient for the large CHAF1A subunit exhibit lethality at the 16 cell stage due to defects in pericentric heterochromatin organization (Houlard et al., 2006). More recent studies using haripins directed to the CAF1 complex have reported additional functions of the complex in preserving somatic cell identity (Cheloufi et al., 2015) and chromatin compaction during spermatogenesis (Doyen et al., 2013). While CHAF1A and CHAF1B are overexpressed in multiple cancer types (Gevaert and Plevritis, 2013; Shah et al., 2014), the mechanisms leading to this increase in expression are not known. Based on our work in AML, we hypothesize that the CAF1 complex may play a role in maintaining cancer stem cell fate. In fact, a recent study showed knockdown of CHAF1A was sufficient to induce differentiation of neuroblastoma cells through dysregulation of neuroblastoma genes and a loss of metabolic gene expression due to global reduction in H3K9me3 modifications (Barbieri et al., 2014). However, the mechanism by which the CAF1 complex regulates gene expression and contributes to tumorigenesis during replication-dependent nucleosome assembly, especially in the hematopoietic system, has not been demonstrated.
A recent study reported that CAF1 preserves somatic cell identity by suppressing expression of stem cell genes through control of chromatin accessibility (Cheloufi et al., 2015). Their findings suggest that CAF1 deficiency improves the efficiency of conversion to iPSCs through increased chromatin accessibility and SOX2 occupancy of distal enhancers of embryonic stem cell genes in fibroblasts. Our model differs in that the mechanism we propose by which CHAF1B controls transcription in leukemia cells is through direct accumulation at discrete sites in the chromatin rather than regulation of chromatin accessibility. Although we found that there were some ATAC-seq peaks that were elevated upon Chaf1b loss, the vast majority of changes at CHAF1B-bound regions resulted in decreased accessibility. It should be noted, however, that the genome wide profile of accessible regions is substantially different between leukemia cells (our study) and iPSCs (Cheloufi et al., 2015). Our findings are more consistent with a recent study that demonstrated that accessible regions of chromatin are a better indicator of cell identity than mRNA expression (Corces et al., 2016). Our data point to a model in which CHAF1B restricts differentiation and preserves the stem cell program by preventing recruitment of lineage specific transcription factors (e.g. CEBPA in AML cells, and GATA3 in T-ALL cells, possibly SOX2 in iPSCs) to DNA through what appears to be a mechanism of competition at accessible sites during replication. CHAF1B therefore has a cell type specific effect on gene regulation: in differentiated cells, CHAF1B preserves cell identity while in malignant cells, CHAF1B preserves the undifferentiated state. This differential activity of CHAF1B is likely mediated by the composition of lineage specific transcription factors and accessible regions of chromatin within the nucleus.
The CHAF1B deletion mutant study confirms that MLL-AF9 leukemic cells require the nucleosome assembly function of CHAF1B to maintain their undifferentiated state, because both the Δp150 (CHAF1A binding deficient) and the RR482/483AA (ASF1A binding deficient) mutants are unable to rescue colony formation following Chaf1b deletion. The function of the WD40 repeat region of CHAF1B has yet to be defined, though it is likely that this region serves as a scaffold for other protein complexes. Based on these gene complementation experiments, we propose that CHAF1B is recruited to the chromatin through its replication-dependent nucleosome assembly function and then remains on specific loci to preserve a cellular state. A recent study by Gao et al. demonstrated the ability of ASF1A to occupy promoters of lineagespecific differentiation genes in embryonic stem cells (Gao et al., 2018). Other components of the DNA replication machinery have similar discrete binding patterns within regions of DNAse/ATAC-accessible chromatin, including ORC2 and the MCM family of proteins, though the exact mechanism detailing how these proteins are retained at those loci is not fully understood (Miotto et al., 2016; Tsai et al., 2015). There is precedence for a dual role of transcription factors in DNA replication. For example, Roeder and colleagues demonstrated that the transcription factor OTF-1 and the DNA replication factor NF-III, are identical, indicating that one factor can have dual replication and transcription functions (O’Neill et al., 1988). Our findings demonstrate the dual effects of a DNA replication factor on transcription in cancer cells, and specifically is a demonstration of a member of the CAF1 complex having this dual function.
Given that expression of the CAF1DN allele led to differentiation of leukemia cells and complete block of leukemogenesis in vivo, but did not impair normal colony formation or hematopoietic reconstitution of normal hematopoietic cells, we conclude that targeted disruption of the CAF1 complex by targeting the PCNA:CHAF1A:CHAF1B interaction might provide a differentiation-driving strategy for MLL rearranged leukemias and other tumors with high CHAF1A/B levels. Recent studies provide important insights into the structure of the CAF1 complex in yeast (Mattiroli et al., 2017a; Mattiroli et al., 2017b). These data may facilitate development of differentiating inducing agents for MLL-rearranged leukemia.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, John Crispino (j-crispino@northwestern.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human subjects
Samples from individuals with acute myeloid leukemia were obtained from both male and female adult subjects (details provided in Table S1) with informed consent. The study was approved by the Wayne State University and the Northwestern University Institutional Review Boards.
Animals
Chaf1b-targeted C56Bl/6 embryonic stem cells were obtained from EUCOMM (European Conditional Mouse Mutagenesis). These cells were introduced into C57Bl/6 blastocysts to derive chimeras. Two male chimeras were bred to wild-type albino C57Bl/6 females, and pigmented offspring were checked for full insertion of critical portions of the targeting construct by PCR and sanger sequencing. Complete Chaf1b targeted mice were backcrossed to wild-type C57Bl/6J mice for six additional rounds of breeding. Targeted mice were then bred to C57Bl/6 Flp+ mice to create Chaf1b+/fl animals. Wild-type Ly5.2, Ly5.1, Mx1-Cre, and Vav-Cre mice were obtained from Jackson Laboratory. Both male and female mice were included in the study. For genotyping, tail-tips were collected at weaning, digested, then recovered. DNA was amplified using primers listed in Table S2. Mice were housed in barrier facility. All animal studies were performed with approval from the Northwestern University Institutional Animal Care and Use Committee and conducted in accordance with institutional and national regulatory standards.
Cell Lines and primary cultures
Human leukemia cell lines U937 (male, CALM10-AFF10) and MOLM13 (male, MLL-AF9, FLT3-ITD) were gifts from Dr. Ali Shilatifard at Northwestern University. Jurkat (male) cells were a gift from Dr. Panos Ntziachristos at Northwestern University. These cell lines were authenticated by short tandem repeat testing (IDEXX) prior to use, and cultured in RPMI-1640 (Gibco) supplemented with 10% FBS (Gibco), Penicillin/Streptomycin (Gibco), and LGlutamine (Gibco). CD34+ cells were obtained from Fred Hutchinson Cancer Center and cultured in StemSpan (StemCell Technologies) supplemented with 50 ng/ml SCF (StemCell), 10 ng/mL each of IL3, IL6, and Flt3L (StemCell Technologies). Primary murine hematopoietic stem and progenitor cells (HSPCs) were selected from flushed bone marrow of the hindlimbs using a CD117+ selection kit according to manufacturer’s protocol (StemCell Technologies). All primary HSPCs were maintained in StemSpan (StemCell Technologies) supplemented with 50 ng/mL recombinant murine SCF, 10 ng/mL recombinant murine IL3, 10 ng/mL recombinant murine IL6, and 1:200 human LDL. MLL-AF9 cells were maintained in RPMI-1640 (Gibco) supplemented with 10% FBS, L-glutamine, 100 ng/mL recombinant murine SCF, 50 ng/mL recombinant murine IL6, and 20 ng/mL recombinant murine IL3. All cells were cultured in 6-well flat-bottom plates at 37C with 5% CO2.
METHOD DETAILS
Bone marrow transplantation and treatment
Ly5.2 or Ly5.1 recipient mice were exposed to 9 Gy of ionizing radiation in a Gammacell 40 irradiator. Immediately prior to and following irradiation, mice were fed bactrim-supplemented water and injected with the indicated numbers of hematopoietic cells by the tail vein. pIpC (Invivogen) was administered at 12.5 mg/kg in a 200 μL bolus by IP injection at indicated time points following transplantation or birth. Tamoxifen (Sigma) was suspended in corn oil (Sigma) and administered 10 days following transplantation at 75 mg/kg in a 100 μL bolus by IP injection daily for five days.
Derivation of MLL-AF9 leukemia cells
HSPCs from indicated genotypes of mice were isolated as described above and spinoculated with MIGR1-MLLAF9-IRES-Neo retrovirus at 2500 RPM for 90 minutes at 32C. Two days following spinoculation, HSPCs were treated with G418 at a 1:125 concentration (Sigma). Once all non-transduced cells were eliminated, the remaining cells were transferred to leukemic cell (LC) media consisting of RPMI-1640 supplemented with 10% FBS, penicillin/streptomycin, l-glutamine, 100 ng/mL recombinant mSCF, 50 ng/mL recombinant mIL6, and 20 ng/mL recombinant mIL3 to expand. 2×106 MLL-AF9 pre-leukemic cells were transplanted into irradiated recipient mice via the tail vein along with 2×105 bone marrow support cells. Mice developed disease after 2–3 months, and spleens were harvested. Mononuclear cells from spleens of diseased mice were dissociated and cultured in LC media with G418 for an additional five days to eliminate non leukemia cells from the culture. These cells were used for all MLL-AF9 leukemic cell studies.
Retroviral and lentiviral experiments
10 μg of retroviral DNA backbone was transfected into Platinum-Eco cells using FuGene ExtremeGene-9 (Roche) transfection reagent. pLKO.1 shRNA lentiviral particles were generated with 293T cells according to the RNAi Consortium protocol. pRRL-eGFP lentiviral particles were generated with 10 μg of lentiviral backbone cotransfected with second generation packaging plasmids. Eighteen hours after transfection, cells were washed with fresh DMEM collection media and viral supernatant was collected at 48 and 72 hours after transfection. Viral particles were concentrated using Amicon Ultra-100 filter tubes by centrifugation. Lentivirus production and relative titer was confirmed with Lenti Go-Stix (Roche). Virus was then either used fresh or aliquoted and frozen at −80C in low-protein binding tubes (Eppendorf). For transduction, cells were placed in 12-well flat-bottom plates in 1 mL of growth medium. Polybrene (10 μg/mL, Millipore) was then added to 1 mL of concentrated virus before being mixed with the target cells. Cells were then spinoculated at 2250 RPM for 90 minutes at 32C. Following centrifugation, cells were rested in the incubator for 6 hours and then given an additional 1 mL of growth medium. Experiments were conducted the following days and cells were grown as described above. Cells were selected for retroviral expression by FACS sorting for GFP 24 hours following transduction with MIGR1, MIGR1-CHAF1B, MIGR1CAF1DN; or cells were selected for MSCV-puro, MSCV-CreERT2 expression by culturing for one week in 10 μg/mL puromycin (Gibco). Complete selection was confirmed by co-selecting non-transduced cells.
Cell sorting and flow cytometry
Cells were suspended in sterile FACS buffer (PBS, 0.5% BSA, 1 mM EDTA) and sorted for indicated markers using a FACS ARIA IIu (BD). All flow cytometry analysis was performed on a BD LSRII flow cytometer, acquired using BD FACSDiva, and analyzed using FlowJo (Treestar). The analysis was performed on single cells as determined by forward scatter or DNA stain (where appropriate), and all cell mixtures were stained with an appropriate color of Fixable Viability Dye (Life Technologies) to exclude dead cells from analysis. Following sorting, cells were maintained in complete media supplemented with 2.5 μg/mL gentamicin.
Measurement of cell cycle and apoptosis
For cell cycle analysis, 2.5×105 MLL-AF9 cells or normal HSPCs were cultured in growth conditions and pulsed with EdU at 10 nM final concentration (ThermoFisher) for 60 minutes at 37C. In vivo EdU incorporation was performed by injecting 50 mg/mL EdU (Abcam, resuspended in PBS) intraperitoneally into mice. Two hours after injection, mice were sacrificed and bone marrow cells were collected for further analysis. Cells were washed and incubated with FVD780 fixable viability dye (eBioscience) for 30 min at 4C, then washed twice with ice-cold PBS with 1 mM EDTA. Cells were fixed in 4% PFA for 15 minutes, and permeabilized with 0.05% Saponin wash buffer supplemented with BSA and EDTA. AF647 azide (Invitrogen) was affixed to EdU using Click-It chemistry via a kit from Life Technologies. DNA was stained using DAPI. Live single cells were analyzed for cell cycle using an LSRII flow cytometer. In vivo cell cycle was determined by treating mice with 50 mg/mL EdU (Abcam) in 200 μL of PBS and injected intraperitoneally. Two hours following injection, bone marrow was isolated, and lineagedepleted bone marrow cells were stained for EdU incorporation as described above. For the analysis of cell death, 2.5×105 MLL-AF9 cells or normal HSPCs were washed twice with PBS and incubated with Annexin V-APC/PI (BD Biosciences) according to manufacturer’s protocol and analyzed for cell death using an LSRII flow cytometer.
Immunofluorescence staining
MLL-AF9 cells were allowed to attached to the glass bottom dishes (MatTek) coated with 25 μg/ml of fibronectin for 1 hour at 37C. Cells were stained with Hoechst (Sigma-Aldrich) for 1 hour, washed with phosphate buffer saline and incubated with cell tracker dye (Molecular Probes) for 30 min to label the membrane. Fluorescent images were obtained using the Nikon A1R+ confocal microscope under a 60× Plan-Apochromat oil immersion lens. Image pseudocoloring was performed using Nikon Elements and ImageJ.
Replication-dependent DNA damage assay
Chaf1bfl/fl HSPCs or Chaf1bfl/fl LCs were spinoculated with MSCV-Cre-eGFP to induce Chaf1b deletion. 24, 48 and 72 hours following spinoculation, cells were incubated with FVD510 (eBiosciences) for 30 minutes at 4C. After wash with FACS buffer, cells were fixed in 4% PFA in PBS for 15 minutes at room temperature. yH2A.X-APC antibody (BD) was diluted 1:75 in Saponin buffer (0.05% Saponin, 1% BSA, PBS) and 100 μL added to the fixed cells. Cells were incubated on ice for 1 hour before wash and were assayed with an LSRII (BD) flow cytometer using FlowJo software (TreeStar).
Intracellular staining for CHAF1B
MLL-AF9 LCs or human AML cell lines were incubated with appropriately colored viability dye for 30 minutes at 4C to exclude dead cells from analysis. Then cells were washed with FACS buffer and pellets were fixed in 4% PFA in PBS at room temperature for 15 minutes. Following fixation, cells were washed in FACS buffer and resuspended in 100 μL saponin buffer with intracellular antibodies. Cells were incubated for 1 hour on ice protected from light. After incubation, cells were washed in saponin buffer and pellets resuspended in FACS buffer for analysis on LSRII flow cytometer.
Western blots
2×106 cells were lysed in RIPA supplemented with PMSF (Cell Signaling Technologies) and HALT Protease Inhibitor Cocktail (Thermofisher). After brief sonication, debris was pelleted at 12000g for 10 minutes at 4C and supernatant transferred to a fresh tube. 4× LDS buffer and 10× sample reduction buffer (Invitrogen) was added to final concentration of 1×, and samples were heated at 70C for 30 minutes before running on NuPAGE™ 4–12% BisTris Protein Gels (Invitrogen). Dual-color protein ladder (BioRad) was run as size marker. Proteins were then transferred to 0.45μm pore Immobilon-P PVDF membrane (Millipore) for 2 hours in methanol. Protein transfer was confirmed by Ponceau stain (Sigma-Aldrich) and membranes were blocked in 5% milk for 1 hour before probing with primary antibody. All primary antibodies were resuspended 1:1000 in 4% BSA and probed overnight at 4C with gentle rocking. After 5 washes in TBS-T, membranes were probed with appropriate HRP-conjugated secondary antibody (Sigma Aldrich) at 1:5000 for one hour in 5% milk. HRP signal was catalyzed with Thermofisher Pico substrate and visualized with HyBlot CL radiography film (Denville). Band intensity was calculated relative to control with ImageJ.
qRT-PCR
2×106 MLL-AF9 or normal HSPCs were lysed in TRIzol (Invitrogen) and RNA was purified with the Direct-Zol RNA Miniprep kit with on-column DNAse digestion (Zymo Research). For qRT-PCR, cDNA was produced with the SuperScript IV kit (Invitrogen) from 250 ng of DNAse-digested RNA according to manufacturer specifications using random hexamers. Quantitative PCR was performed on an Applied Biosystems 7500 thermocycler using the following primer sequences listed in Table S1 and analyzed via ΔΔCT method.
Colony forming unit assays
2×104 BM mononuclear cells, 5×103 BM HSPCs, or 500 MLL-AF9 leukemia cells per dish were plated in M3434 (StemCell Technologies) and incubated for 5–7 days. On the last day colonies were counted by visualization through an inverted tissue culture microscope. Discrete clusters containing >100 cells were considered to be colonies. For replating, colonies were resuspended and washed in PBS, and 5×103 cells were plated into plates with fresh M3434 methylcellulose and the process was repeated up to 6 generations. Human AML cell lines were plated in complete M4100 (RPMI-1640 + 20% FBS) and counted in a similar manner to the mouse experiment.
Complete blood counts and blood smears
50 μL of blood from the tail vein was collected in EDTA-coated tubes (Fisher) and analyzed using a Hemavet950 (Drew Scientific). Remaining blood was smeared on a non-charged glass slide and stained with modified WrightGiemsa Three Step Stain Set (ThermoFisher). After the slides were dried, cover slips were fixed using Permount, and morphology observed by light microscopy (Leica DM4000B). Images were acquired using Leica Application Suite V4.4 software. This same methodology was applied to cytospun samples as well for visualization.
Tissue histology
Sternum, spleen, lung, and liver were fixed whole in normal buffered formalin for 48 hours before transfer to 70% ethanol (spleen, lung, liver) or normal buffered formalin (sternum) for H&E treatment and stain.
Deletion of Chaf1b in MLL-AF9 leukemia cells
MLL-AF9 cells were transduced with MSCV-CreERT2 and selected for three days with puromycin at 10 μg/mL or five days with blasticidin at 5 μg/mL (Invitrogen). After selection, LCs were treated with β-estradiol (SigmaAldrich) dissolved in ethanol at 10−7M final concentration. Cells were harvested at 24 hour intervals for four days and measured for morphology changes by cytopsin and Wright-Giemsa stain, cell death by Annexin V/PI stain, and surface markers (eBioscience: CD34, CD117, CD11b) by flow cytometry.
TCGA analysis
The TCGA LAML dataset was analyzed using UCSC cancer browser by first setting a signature for CHAF1B expression, and ranking patients based on CHAF1B expression level. Then the top and bottom third of CHAF1B expressing patients were plotted separately as a function of survival over time. Significance was calculated using a log-rank test.
ATAC-Seq
Pellets of 5×103 LCs or HSPCs were resuspended in 50 μL of ATAC buffer (25 μL 2× TD buffer, Nextera; 2.5 μL TDE1, Nextera; 0.5 μL Digitonin, Promega; 22 μL nuclease-free water) for 15 minutes and agitated at 300 RPM at 37C. After transposition, DNA fragments were collected using a Qiagen MinElute PCR purification kit. Sequencing libraries were prepared and controlled for quality as previously described (Buenrostro et al., 2013). Paired-end sequences were merged and aligned to the mouse (UCSC mm9). Alignments were processed with Bowtie version 1.1.2, first trimming the sequencing adaptors and then allowing only uniquely mapping reads with up to two mismatches within the 50 base pair read. The resulting reads were normalized to total reads aligned (reads per million, rpm).
RNA-Seq
48 hours after CHAF1B overexpression/knockout mouse hematopoietic stem and progenitor cells/leukemic cells were collected and lysed with Trizol reagent. Total RNA was extracted from Trizol using Direct-Zol RNA Miniprep kit with on-column DNAse digestion (Zymo Research). 500 ng RNA was used for library preparation with TruSeq Stranded Total RNA with Ribo-Zero Gold kit (Illumina, RS-123–2201) with ERCC spike in (Thermofisher). The sequenced reads were aligned to the mouse genome (UCSC mm9) with STAR aligner using gene annotations from Ensembl 72, and intronic reads were discarded. Sequencing result was normalized to ERCC spike in and differential gene expression performed with EdgeR (Empirical analysis of digital gene expression data in R) version 3.08 (Robinson et al., 2010). Adjusted p values were computed using the Benjamini-Hochberg method. Protein coding genes, long non-coding RNA and pseudogenes with adjusted p values less than 0.01 were used for the downstream analysis with DAVID.
ChIP-seq
MLL-AF9 leukemic cells overexpressing MIGR1-CHAF1B, murine MLL-AF9 LCs, or human AML/ALL cell lines were subjected to ChIP assays according to a published protocol (Liang et al., 2015). Briefly, cells were crosslinked with 1% paraformaldehyde for 15 minutes and were quenched with glycine for 5 minutes at room temperature. Fixed chromatin was sonicated with a Covaris Focused-ultrasonicator and immunoprecipitated with the indicated antibody. For samples utilized for ChIP-seq, libraries were prepared with the high throughput library preparation kit standard PCR amp module (KAPA Biosystems) for next-generation sequencing. ChIP-seq reads were aligned to the mouse (UCSC mm9) or human (UCSC hg19) reference genome. Alignments were processed with Bowtie version 1.1.2, allowing only uniquely mapping reads with up to two mismatches within the 50 bp read. The resulting reads were extended to 150 bases toward the interior of the sequenced fragment and normalized to total reads aligned (reads per million, rpm). For histone marks, ChIP-seq in MLL-AF9 cells peak detection was performed with MACS (model-based analysis of ChIP-Seq) version 1.4.2 using default parameters (Zhang et al., 2008). The average coverage (calculated using rpm tracks described above) across the entire region is shown in the boxplots where p values were calculated with the Wilcoxon signed-rank test. Heatmaps depict log2 fold change of coverage profiles in a 4 kb window around the merged peak center in 25 bp binned averages and sorted by total coverage in this window. Heatmaps and metagene plots of the genes with changed H3K27ac read coverage around the TSS (±3kb) after Chaf1b knockout were plotted with ngs.plot 2.47 and ranked by read intensity. The CHAF1B read coverage at these sites was plotted with the same order.
GREAT analysis
Annotated BED files from ChIP sequencing experiments were uploaded to GREAT (http://bejerano.stanford.edu/great/public/html/) for further pathway and peak localization analysis. Peaks were assumed to be affecting the nearest gene.
Plasmids, shRNA and sgRNA
MIGR1-CAF1DN was sub-cloned from pCDNA3-HA-p150c (gift from Peter D. Adams, UK Beatson, Glasgow). MIGR1-MLL-AF9-Neo was a gift from Dr. Jiwang Zhang at Loyola University Chicago. MIGR1-CHAF1B was subcloned from MSCV-CHAF1B (Addgene plasmid #34901). MSCV-CreERT2-puro (Addgene #22776) was a gift from Tyler Jacks, MSCV-CreERT2-blasticidin was generated by subcloning into MSCV-blast from MSCVCreERT2-puro. RNAi consortium shRNA in pLKO.1 were obtained from Sigma (shRNA to CHAF1B, CEBPA, HIRA). All plasmids were confirmed by restriction enzyme digest and sanger sequencing prior to use for this study. pLKO.1 and pLentiCrisprV2 constructs containing shRNA and sgRNA, respectively, were either obtained from Sigma (Mission shRNA) or sgRNA protospacers were generated in-house using the Broad Institute GPP Web Portal (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). sgRNA protospacers identified using the GPP web portal were cloned into pLentiCrisprV2-puro and packaged/infected as described above. Cells were selected in 1 μg/mL puromycin for 48 hours before validation of knockdown efficiency by western blot or QPCR. Sequences of the shRNAs are provided in Table S1.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analysis was performed with GraphPad Prism 6. Unless otherwise indicated in the figure legend, plots shown are representative of at least three independent biological replicates. In mouse experiments, each point represents one individual mouse. When comparing two normally-distributed populations, significance was determined by two-tailed t-test with Welch’s correction. When comparing two non-normally-distributed populations, significance was determined using the Mann-Whitney two-tailed U test. When comparing multiple populations, significance was determined using one-way ANOVA with Bonferroni post-hoc test for multiple comparison correction. Differences in survival in either patients or mice was determined by log-rank test. Power was calculated by β value of > 0.8 and an α value of <0.05 in order to determine the sufficient number of replicates for experiments. The nature of the statistical analysis performed for each dataset is included in the figure legends.
DATA AND SOFTWARE AVAILABILITY
All ChIP-seq, ATAC-seq and RNA-seq datasets were deposited with accession number GEO: GSE120063.
Supplementary Material
Highlights.
CHAF1B is elevated in multiple blood and solid tumor types
One allele of Chaf1b is sufficient for normal hematopoiesis
Chaf1b deletion differentiates AML cells and prevents leukemia development in vivo
CHAF1B impairs occupancy of CEBPA on elements along myeloid differentiation genes
Volk et al. show that overexpression of the chromatin assembly factor complex subunit CHAF1B promotes leukemogenesis by interfering with chromatin occupancy of transcription factors that promote myeloid differentiation. Reducing CHAF1B activity suppresses leukemogenesis without impairing normal hematopoiesis.
Significance.
Overexpression of the chromosome 21 gene CHAF1B is observed in acute myeloid leukemia (AML), including cases with trisomy 21 or MLL rearrangements. Although AML in Down syndrome has a favorable prognosis, individuals with MLL rearrangements have a poor prognosis, and effective therapies are needed. We demonstrate that the chromatin assembly factor complex (CAF1) member CHAF1B blocks differentiation of leukemia cells by binding promoters of myeloid differentiation genes and inhibiting their expression. Reduction of CAF1 activity by overexpression of a dominant negative CAF1 mutant had no effect on healthy hematopoiesis but dramatically suppressed the growth of MLL-AF9 leukemia cells in vitro and in vivo. This work therefore suggests that targeting the CAF1 complex may be a therapeutic strategy for AML.
Acknowledgements
The authors thank Gina Kirsammer and Christian Marinaccio for advice and Peter Adams and Jiwang Zhang for providing critical reagents. The authors also would like to thank Guy Savageau for the Leucegene data and consultation regarding the expression of CHAF1A and CHAF1B. This study was supported by the Samuel Waxman Cancer Research Foundation (to JDC) and the NIH (R01 CA101774 to JDC; T32 CA080621 to AV). AV is supported by a Scholar Award from American Society of Hematology. Leukemia studies in the Shilatifard laboratory are supported by NCI R35 CA197569. EB is supported by R50 CA221848. This research was also supported by the Lurie Cancer Center. This work was supported by the Northwestern University Pathology Core and Flow Cytometry Facilities, funded by CCSG NCI CA060553, and cell sorting was performed on a BD FACSAria SORP system purchased with the support of NIH 1S10OD011996–01.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barbieri E, De Preter K, Capasso M, Chen Z, Hsu DM, Tonini GP, Lefever S, Hicks J, Versteeg R, Pession A, et al. (2014). Histone chaperone CHAF1A inhibits differentiation and promotes aggressive neuroblastoma. Cancer Res 74, 765–774. [DOI] [PubMed] [Google Scholar]
- Blouin JL, Duriaux-Sail G, Chen H, Gos A, Morris MA, Rossier C, and Antonarakis SE (1996). Mapping of the gene for the p60 subunit of the human chromatin assembly factor (CAF1A) to the Down syndrome region of chromosome 21. Genomics 33, 309–312. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Elling U, Hopfgartner B, Jung YL, Murn J, Ninova M, Hubmann M, Badeaux AI, Euong Ang C, Tenen D, et al. (2015). The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528, 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. (2016). Lineage-specific and singlecell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Tayrac M, Aubry M, Saikali S, Etcheverry A, Surbled C, Guenot F, Galibert MD, Hamlat A, Lesimple T, Quillien V, et al. (2011). A 4-gene signature associated with clinical outcome in high-grade gliomas. Clin. Cancer Res. 17, 317–327. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Lin W, Zhang CK, Xiong H, Fu G, Jin WR, Chen R, Chen Z, Qi ZT, and Huang GM (2001). Genomic sequence and expression analyses of human chromatin assembly factor 1 p150 gene. Gene 264, 187–196. [DOI] [PubMed] [Google Scholar]
- Doyen CM, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Rathke C, Renkawitz-Pohl R, and Verrijzer CP (2013). Subunits of the histone chaperone CAF1 also mediate assembly of protamine-based chromatin. Cell Rep. 4, 59–65. [DOI] [PubMed] [Google Scholar]
- Gaillard PH, Martini EM, Kaufman PD, Stillman B, Moustacchi E, and Almouzni G (1996). Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell 86, 887–896. [DOI] [PubMed] [Google Scholar]
- Gao Y, Gan H, Lou Z, and Zhang Z (2018). Asf1a resolves bivalent chromatin domains for the induction of lineage-specific genes during mouse embryonic stem cell differentiation. Proc. Natl. Acad. Sci. USA 115, E6162–E6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert O, and Plevritis S (2013). Identifying master regulators of cancer and their downstream targets by integrating genomic and epigenomic features. Pac Symp Biocomput, 123–134. [PMC free article] [PubMed] [Google Scholar]
- Houlard M, Berlivet S, Probst AV, Quivy JP, Hery P, Almouzni G, and Gerard M (2006). CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2, e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YH, Warnatz HJ, Vanhecke D, Wagner F, Fiebitz A, Thamm S, Kahlem P, Lehrach H, Yaspo ML, and Janitz M (2006). Cell array-based intracellular localization screening reveals novel functional features of human chromosome 21 proteins. BMC Genomics 7, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis N, and Fisher EM (1996). The gene encoding the p60 subunit of chromatin assembly factor I (CAF1P60) maps to human chromosome 21q22.2, a region associated with some of the major features of Down syndrome. Human Genet. 98, 497–499. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T (1995). Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res 220, 304–311. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Woodfin AR, Slaughter BD, Unruh JR, Box AC, Rickels RA, Gao X, Haug JS, Jaspersen SL, and Shilatifard A (2015). Mitotic Transcriptional Activation: Clearance of Actively Engaged Pol II via Transcriptional Elongation Control in Mitosis. Mol. Cell 60, 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes da Rosa J, Holik J, Green EM, Rando OJ, and Kaufman PD (2011). Overlapping regulation of CenH3 localization and histone H3 turnover by CAF-1 and HIR proteins in Saccharomyces cerevisiae. Genetics 187, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinge S, Bliss-Moreau M, Kirsammer G, Diebold L, Chlon T, Gurbuxani S, and Crispino JD (2012). Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of Down syndrome. J. Clin. Invest 122, 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marheineke K, and Krude T (1998). Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. J. Biol. Chem 273, 15279–15286. [DOI] [PubMed] [Google Scholar]
- Martini E, Roche DM, Marheineke K, Verreault A, and Almouzni G (1998). Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Bio 143, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascolo M, Vecchione ML, Ilardi G, Scalvenzi M, Molea G, Di Benedetto M, Nugnes L, Siano M, De Rosa G, and Staibano S (2010). Overexpression of Chromatin Assembly Factor-1/p60 helps to predict the prognosis of melanoma patients. BMC Cancer 10, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F, Gu Y, Balsbaugh JL, Ahn NG, and Luger K (2017a). The Cac2 subunit is essential for productive histone binding and nucleosome assembly in CAF-1. Sci Rep 7, 46274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F, Gu Y, Yadav T, Balsbaugh JL, Harris MR, Findlay ES, Liu Y, Radebaugh CA, Stargell LA, Ahn NG, et al. (2017b). DNA-mediated association of two histone-bound complexes of yeast Chromatin Assembly Factor-1 (CAF-1) drives tetrasome assembly in the wake of DNA replication. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Ji Z, and Struhl K (2016). Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc. Natl. Acad. Sci. USA 113, E4810–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabatiyan A, and Krude T (2004). Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Mol. Cell. Bio 24, 2853–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Zhao J, Ma J, Xie C, Edwards H, Wang G, Caldwell JT, Xiang S, Zhang X, Chu R, et al. (2016). Binding of Released Bim to Mcl-1 is a Mechanism of Intrinsic Resistance to ABT-199 which can be Overcome by Combination with Daunorubicin or Cytarabine in AML Cells. Clin. Cancer Res 22, 4440–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill EA, Fletcher C, Burrow CR, Heintz N, Roeder RG, and Kelly TJ (1988). Transcription factor OTF-1 is functionally identical to the DNA replication factor NF-III. Science 241, 1210–1213. [DOI] [PubMed] [Google Scholar]
- Poleshko A, Einarson MB, Shalginskikh N, Zhang R, Adams PD, Skalka AM, and Katz RA (2010). Identification of a functional network of human epigenetic silencing factors. J. Biol.Chem 285, 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Roche D, and Almouzni G (2006). New histone incorporation marks sites of UV repair in human cells. Cell 127, 481–493. [DOI] [PubMed] [Google Scholar]
- Polo SE, Theocharis SE, Grandin L, Gambotti L, Antoni G, Savignoni A, Asselain B, Patsouris E, and Almouzni G (2010). Clinical significance and prognostic value of chromatin assembly factor-1 overexpression in human solid tumours. Histopathology 57, 716–724. [DOI] [PubMed] [Google Scholar]
- Qian YW, and Lee EY (1995). Dual retinoblastoma-binding proteins with properties related to a negative regulator of ras in yeast. J. Biol.Chem 270, 25507–25513. [DOI] [PubMed] [Google Scholar]
- Qian YW, Wang YC, Hollingsworth RE Jr., Jones D, Ling N, and Lee EY (1993). A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature 364, 648–652. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MA, Faryabi RB, Ergen AV, Day AM, Malhowski A, Canela A, Onozawa M, Lee JE, Callen E, Gutierrez-Martinez P, et al. (2014). DNA-damage-induced differentiation of leukaemic cells as an anti-cancer barrier. Nature 514, 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seita J, Sahoo D, Rossi DJ, Bhattacharya D, Serwold T, Inlay MA, Ehrlich LIR, Fathman JW, Dill DL, and Weissman IL (2012). Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS One 7(7):e40321. doi: 10.1371/journal.pone.0040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah MA, Denton EL, Arrowsmith CH, Lupien M, and Schapira M (2014). A global assessment of cancer genomic alterations in epigenetic mechanisms. Epigenet. Chromatin 7, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, and Nestler E (2014). ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara K, and Stillman B (1999). Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585. [DOI] [PubMed] [Google Scholar]
- Smith DJ, and Whitehouse I (2012). Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, and Stillman B (1989). Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58, 15–25. [DOI] [PubMed] [Google Scholar]
- Song JJ, Garlick JD, and Kingston RE (2008). Structural basis of histone H4 recognition by p55. Genes Dev. 22, 1313–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staibano S, Mascolo M, Mancini FP, Kisslinger A, Salvatore G, Di Benedetto M, Chieffi P, Altieri V, Prezioso D, Ilardi G, et al. (2009). Overexpression of chromatin assembly factor-1 (CAF-1) p60 is predictive of adverse behaviour of prostatic cancer. Histopathology 54, 580–589. [DOI] [PubMed] [Google Scholar]
- Staibano S, Mascolo M, Rocco A, Lo Muzio L, Ilardi G, Siano M, Pannone G, Vecchione ML, Nugnes L, Califano L, et al. (2011). The proliferation marker Chromatin Assembly Factor-1 is of clinical value in predicting the biological behaviour of salivary gland tumours. Onc. Rep 25, 13–22. [PubMed] [Google Scholar]
- Stevenson JS, and Liu H (2013). Nucleosome assembly factors CAF-1 and HIR modulate epigenetic switching frequencies in an H3K56 acetylation-associated manner in Candida albicans. Eukaryot Cell 12, 591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B (1986). Chromatin assembly during SV40 DNA replication in vitro. Cell 45, 555–565. [DOI] [PubMed] [Google Scholar]
- Tang Y, Poustovoitov MV, Zhao K, Garfinkel M, Canutescu A, Dunbrack R, Adams PD, and Marmorstein R (2006). Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat. Struct. Mol. Biol. 13, 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, and Schreiber SL (1996). A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. (2015). Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 18, 723–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FL, Vijayraghavan S, Prinz J, MacAlpine HK, MacAlpine DM, and Schwacha A (2015). Mcm2–7 Is an Active Player in the DNA Replication Checkpoint Signaling Cascade via Proposed Modulation of Its DNA Gate. Mol. Cell. Biol 35, 2131–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Collins KA, Prasad-Sinha J, Amiott E, Bulger M, Harte PJ, Kobayashi R, and Kadonaga JT (2001). Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol 21, 6574–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, and Stillman B (1996). Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87, 95–104. [DOI] [PubMed] [Google Scholar]
- Yang BX, El Farran CA, Guo HC, Yu T, Fang HT, Wang HF, Schlesinger S, Seah YF, Goh GY, Neo SP, et al. (2015). Systematic identification of factors for provirus silencing in embryonic stem cells. Cell 163, 230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, and Adams PD (2003). Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell 11, 341–351. [DOI] [PubMed] [Google Scholar]
- Zhang W, Tyl M, Ward R, Sobott F, Maman J, Murthy AS, Watson AA, Fedorov O, Bowman A, Owen-Hughes T, et al. (2013). Structural plasticity of histones H3-H4 facilitates their allosteric exchange between RbAp48 and ASF1. Nat. Struct. Mol. Biol 20, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Wani G, Arab HH, El-Mahdy MA, Ray A, and Wani AA (2009). Chromatin restoration following nucleotide excision repair involves the incorporation of ubiquitinated H2A at damaged genomic sites. DNA Repair 8, 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.