Summary
Somatic reprogramming, which was first identified in rodents, remains poorly described in non-mammalian species. Here, we generated avian reprogrammed cells by reprogramming of chicken and duck primary embryonic fibroblasts. The efficient generation of long-term proliferating cells depends on the method of delivery of reprogramming factors and the addition of NANOG and LIN28 to the canonical OCT4, SOX2, KLF4, and c-MYC gene combination. The reprogrammed cells were positive for several key pluripotency-associated markers including alkaline phosphatase activity, telomerase activity, SSEA1 expression, and specific cell cycle and epigenetic markers. Upregulated endogenous pluripotency-associated genes included POU5F3 (POUV) and KLF4, whereas cells failed to upregulate NANOG and LIN28A. However, cells showed a tumorigenic propensity when injected into recipient embryos. In conclusion, although the somatic reprogramming process is active in avian primary cells, it needs to be optimized to obtain fully reprogrammed cells with similar properties to those of chicken embryonic stem cells.
Keywords: avian species, somatic reprogramming, embryonic stem cells, NANOG, pluripotency, chicken, duck, embryos, blastoderm, phylogenic comparison
Highlights
-
•
NANOG is required for avian somatic reprogramming
-
•
NANOG is necessary for long-term establishment of avian reprogrammed cells
-
•
Avian reprogrammed cells express pluripotency markers
-
•
Avian cells are only partially reprogrammed
In this article, the authors show that the NANOG gene plays a crucial role in the long-term establishment of avian reprogrammed cells that have some of the specific markers of stem cells but that do not present all the developmental properties of induced pluripotent stem cells such as described in mammals.
Introduction
In 2006, the somatic reprogramming process was established, and induced pluripotent stem cells (iPSCs) were generated from fibroblasts (Takahashi and Yamanaka, 2006, Takahashi and Yamanaka, 2016, Karagiannis and Eto, 2016). The reprogramming approach, which was first demonstrated in mice and then in humans, rats, and non-human primates, has been used with variable success in a large number of mammalian species (Koh and Piedrahita, 2014) including rabbits (Honda et al., 2013, Osteil et al., 2013), sheep and bovines (Liu et al., 2012, Sumer et al., 2011), pigs (Ezashi et al., 2009), horses (Nagy et al., 2011), and dogs (Shimada et al., 2010), and even in some endangered species (Verma et al., 2013). In most species, full characterization of the reprogrammed cells, including their developmental ability, was rarely documented. In non-mammalian species, the available information is scarce, with only a few reports for avian cells (Katayama et al., 2018, Lu et al., 2012, Lu et al., 2015, Yu et al., 2014, Susta et al., 2016, Rosselló et al., 2013) and other species (Rosselló et al., 2013) in the literature. Non-mammalian pluripotency-associated genes from zebrafish and chicken (Theunissen et al., 2011), or from axolotl, medaka, and xenopus (Tapia et al., 2012), have been used successfully to reprogram mammalian cells.
The efficacy of the reprogramming process is determined by the generation of reprogrammed cells with properties similar to those of embryonic stem cells (ESCs), including long-term self-renewal, differentiation into the three embryonic lineages, and the ability to contribute efficiently to embryogenesis when injected into a recipient embryo. Ultimately, iPSCs should demonstrate efficient germline contribution, a property presently reserved to naive pluripotent ESCs, which have been defined and characterized in rodents (Smith, 2017, Nichols and Smith, 2009). Epiblast stem cells derived from post-implantation embryos are the archetypes of the primed pluripotent state and do not contribute efficiently to the embryo (Wu et al., 2016, De Los Angeles et al., 2015, Tesar et al., 2007). In chicken, ESCs are derived and maintained for in vitro long-term culture (Pain et al., 1996, Aubel and Pain, 2013, Jean et al., 2013) and contribute to morphogenesis when injected back into a recipient embryo. Even if those cells show the plasticity to express the germ cell program in the presence of the CVH (Chicken Vasa homolog) master gene (Lavial et al., 2009), the chicken ESC (cESC) germline contribution is almost absent (Pain et al., 1996, Petitte et al., 2004); however, it can be expected with respect to the preformation origin of the avian germline (Extavour and Akam, 2003, Tsunekawa et al., 2000, Lee et al., 2016). cESCs have been characterized at the transcriptomic and epigenetic levels (Acloque et al., 2001, Acloque et al., 2012, Lavial et al., 2007, Jean et al., 2015, Kress et al., 2016), leading to the identification of pluripotency-associated genes. Different long-term proliferating cells with specific phenotypes and properties have been generated from ESCs, demonstrating the unique plasticity of these cells and their pluripotency-associated properties (Lavial et al., 2009, Boast and Stern, 2013, Couteaudier et al., 2015, Vautherot et al., 2017).
In mice, the combined action of the four transcription factors, Oct4, Sox2, Klf4, and c-Myc, also known as the OSKM cocktail, allows a fibroblast to become an iPSC. Various gene combinations have been identified, such as the OSNL (Oct4, Sox2, Nanog, and Lin28) combination (Yu et al., 2007). Additional actors including Nr5a2 (Heng et al., 2010), Esrrb (Feng et al., 2009), Zic3 (Declercq et al., 2013), Tbx3 (Han et al., 2010), microRNAs (miR-302 cluster) (Anokye-Danso et al., 2011), and Zpf296 (Fischedick et al., 2012) participate directly in the reprogramming process or increase reprogramming efficiency (Stadtfeld and Hochedlinger, 2010). The first validations were performed using integrated and stable retroviral constructs; alternative delivery systems were later developed, resulting in a large panel of reprogramming methods including the use of lentiviruses and transposons (Woltjen et al., 2009, Kaji et al., 2009). Non-integrative strategies were also established through the use of transfected RNAs, adenoviral vectors, episomal plasmids, and Sendai viruses.
In this study, we reprogrammed primary chicken (CEF) and duck (DEF) embryonic fibroblasts and compared different transgene delivery methods. The resulting cells were characterized by comparing them with spontaneously established cESCs at the molecular and developmental levels. The results obtained in two independent avian species showed that the reprogramming process was incomplete regardless of the delivery system and the gene combinations, that the reprogrammed cells are also transformed despite the expression of several unambiguously key ESC-specific markers, and that they presently lack the developmental in vivo properties to be fully considered as iPSCs.
Results
The Delivery of the Canonical OSKM Combination Affects the Generation of Long-Term Proliferating iPS-like Cells
The canonical OSKM gene combination, which was initially defined as an efficient gene cocktail to obtain mouse iPSCs, was first tested on CEFs.
First, the reprogramming gene combination was delivered using the pLent polycistronic lentiviral vector carrying GFP as an infection reporter, at various MOI. After a few days, colonies emerged (Figure 1A) from infected CEFs with both GFP- (Figure 1B) and alkaline phosphatase (AP)-positive cells; however, individual colonies were unable to expand, proliferate, and develop into cells with ES-like morphology and properties, in contrast with infected mouse embryonic fibroblasts (MEFs) used as positive control for reprogramming efficiency (data not shown).
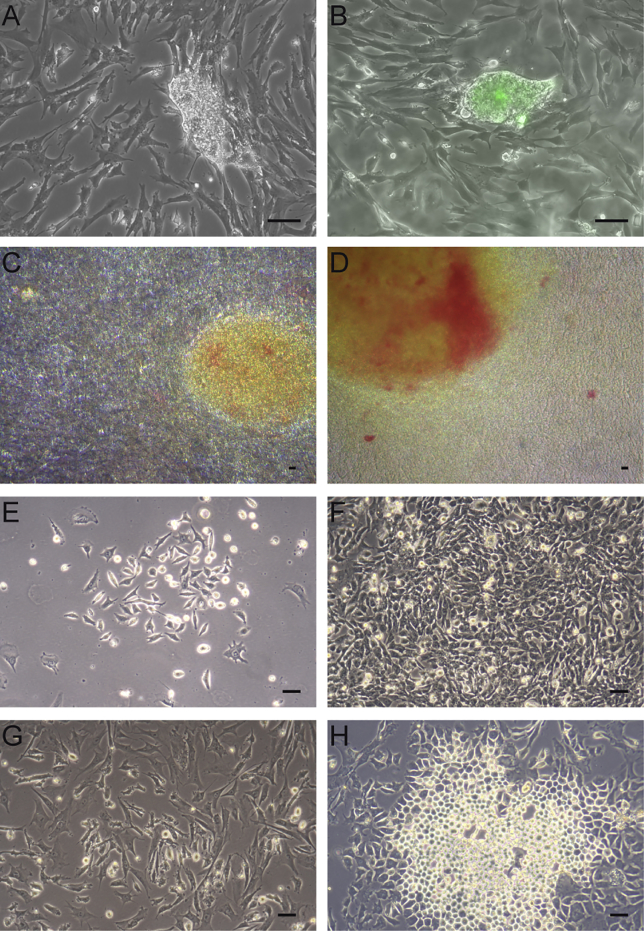
Figure 1.
Attempts to Obtain Chicken Reprogrammed Cells with Various Delivery Systems
Primary chicken embryonic fibroblasts (CEFs) were infected with the pLent polycistronic lentivirus. Colonies emerged (A) and expressed GFP (B), indicating that they were infected, although they failed to be amplified. More than 50 colonies were isolated in several independent experiments. Similarly, CEFs were infected with a cocktail of individual lentiviruses carrying mouse Oct4, Sox2, Klf4, and c-Myc cDNAs. Compact colonies with AP-positive cells were obtained (C and D) but could not be established long term. In another attempt, CEFs were infected with pMX retroviruses carrying mouse Oct4, Sox2, Klf4, and c-Myc cDNAs. The titer was determined using pMX-GFP as shown in Figure S1. Morphological changes were rapidly observed (E), and colonies expanded once individually picked (F). The phenotypic changes were not stable, and cells reverted to a fibroblast phenotype (G) after four to six passages. Similarly, CEFs infected with Sendai viruses carrying human OCT4, SOX2, KLF4, and c-MYC cDNAs exhibited morphological changes (H) but failed to be established and reverted to a fibroblast phenotype after a few passages.
Scale bars, 50 μm.
Second, we used individual lentiviruses, each carrying a reprogramming factor. With an MOI of ≥5, the CEFs were efficiently infected as determined by the GFP reporter lentivirus (data not shown). Morphological changes were observed during the selection process, and colonies emerged with AP-positive cells (Figures 1C and 1D). However, individually picked resistant colonies did not proliferate for more than four to five passages, and rapidly reverted to a fibroblast phenotype, suggesting a transient and unstable phenotypic change. No clones were established for long-term culture.
Third, we tested the infection of CEFs by pMX pantropic retroviruses expressing the OSKM genes in the presence of a GFP reporter retrovirus as a control for efficient infection of the cells. CEFs were efficiently infected, as determined by the 80%–85% GFP-positive cells obtained with the control vector (Figure S1). Morphological changes were rapidly observed starting at 3–4 days after the infection (Figure 1E). Colonies emerged 5–8 days after infection (Figure 1F), and were picked and allowed to proliferate in ESA medium. Despite the presence of promising morphology and a rapid initial growth of the clones, the cells presenting an ES-like morphology did not maintain this phenotype after 4–5 passages and progressively reverted to the fibroblast phenotype, entering into senescence at 50–60 days after the reprogramming infection (Figure 1G). None of the 50 picked clones and the initially infected pooled cells could be established long term, in sharp contrast to the MEFs that gave rise to iPSCs with high efficiency using the same constructs (data not shown).
Fourth, we evaluated the effect of non-integrative Sendai viruses carrying human cDNAs. Infection of CEFs, as validated by the detection of the viruses as recommended (data not shown), led to morphological changes characterized by the appearance of round yellowish cells clustered in small colonies at 7–10 days after infection (Figure 1H). These phenotypic changes did not evolve into an ES-like morphology or long-term establishment, and cells progressively reverted to a fibroblast morphology. Under similar conditions, human embryonic fibroblasts were fully and efficiently reprogrammed (data not shown).
Additional Genes Are Required for the Long-Term Establishment of Reprogrammed Chicken Cells
Next, we investigated the effect of transposons as expression vectors on CEFs using the canonical OSKM gene combination and additional genes in the reprogramming cocktail. Some of the tested gene combinations produced morphological changes, with the emergence of AP- and SSEA-1-positive colonies 10 days after the selection process (Figures 2A–2D). As shown by side scatter (SSC)/forward scatter (FSC) density plot analyzed by flow cytometry, global morphological changes were observed during the reprogramming process (Figure 2C) leading to various colony morphologies, including fibroblast-, scale-, and primary ES-like cells (Figures 2B and 2E). The proportions of different morphologies varied according to the gene combinations (Figure 2F). The SOX2 gene can be substituted by SOX3 in different combinations. In cases in which the OS2NL and OS3NL gene combinations alone did not produce morphological changes, the OS2KM or OS3KM gene combinations yielded proliferating cells, some of which showed ES-like morphology. However, the resulting cells reverted progressively to a fibroblast morphology and stopped proliferating after 30–40 generations depending on the clones (Figures 3A and 3B). The LIN28 addition increased the initial proportion of colonies with ES-like morphology, although it did not modify the relative yield or long-term establishment (Figure 3C). Only the addition of NANOG to the cocktail generated an important number of colonies with the potential for long-term establishment (Figure 3D). This led to the establishment of growing clones for more than 150 days and 40 passages, with at least 100 generations and an average doubling time of 28–29 hr compared with 16–18 hr for the cESCs and more than 40 hr for the CEFs, which finally entered into senescence after 20–25 generations. The emergence of long-term proliferating clones varied from one isolate to another, ranging from 15 to 25 days after the transduction process, when the more rapidly emerging colonies among the other combinations failed to be established.
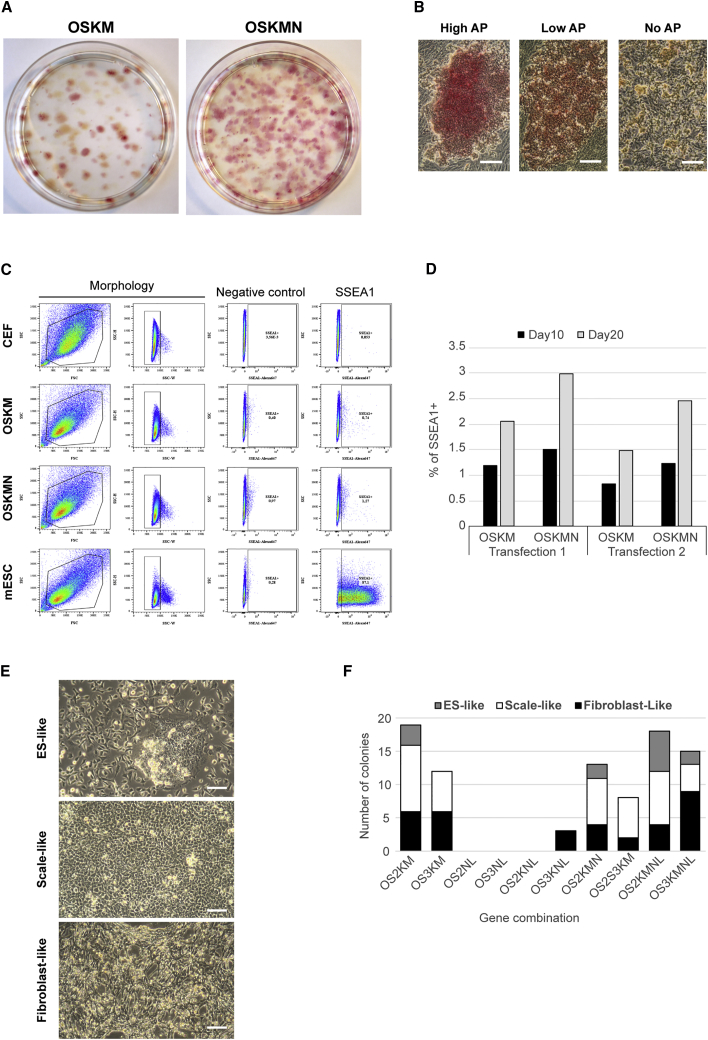
Figure 2.
Enhanced Production of Chicken Reprogrammed Stem Cells with OSKMN Combination
(A) CEFs were transfected with four or five transposons expressing the chicken OCT4, SOX2, KLF4, c-MYC genes (OSKM) plus NANOG gene (OSKMN). Clones emerged 10 days after transfection and more alkaline-positive colonies were obtained with the OSKMN combination than with OSKM combination.
(B) Colonies with high, low, and no AP staining display different morphologies. Scale bars, 50 μm.
(C) Transfected CEFs undergo morphological changes 10 days after transfection as revealed by the SSC/FSC profile and start to express SSEA-1 as a typical mESC marker as analyzed by flow cytometry.
(D) The percentages of SSEA-1-positive cells were quantified in two independent transfections of CEF with OSKM and OSKMN combination after 10 and 20 days of transfection revealing the emergence of reprogrammed cells.
(E) Picked colonies from different gene combinations display three different morphotypes: ES-, scale-, and fibroblast-like morphologies. Scale bars, 100 μm.
(F) Among picked colonies, ES-, scale-, and fibroblast-like colony ratio have been determined for each gene combination.
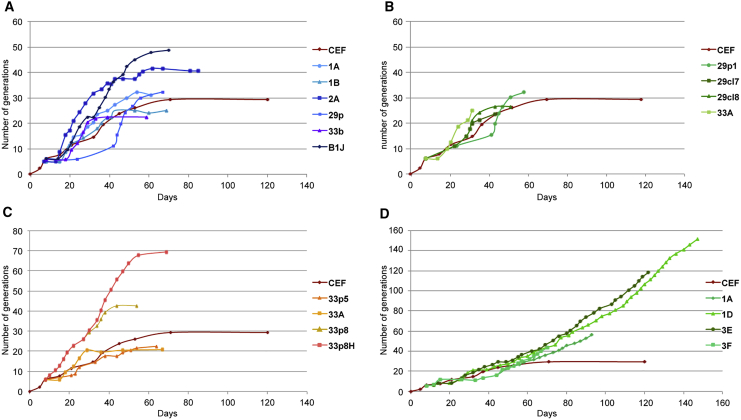
Figure 3.
Long-Term Cultured Established Reprogrammed Clones Proliferated at a Stable and Constant Rate
The proliferation rate and generation number of the different reprogrammed independent clones were determined after cell counting at each passage. CEFs entered into senescence after a few passages and <20–25 generations, which is a common feature of somatic cells. The reprogramming process resulted in the escape from programmed senescence.
(A–D) OS2KM (A) and OS3KM (B) clones started to proliferate, although long-term establishment failed beyond 30–40 generations. The presence of LIN28 (C) did not improve the rate of establishment. In contrast, even if the emergence of the OSKMN clones was variable, clones showed a more stable proliferation rate after a few passages and a long-term establishment (D).
The Established Reprogrammed Clones Shared Most of the Properties of cESCs
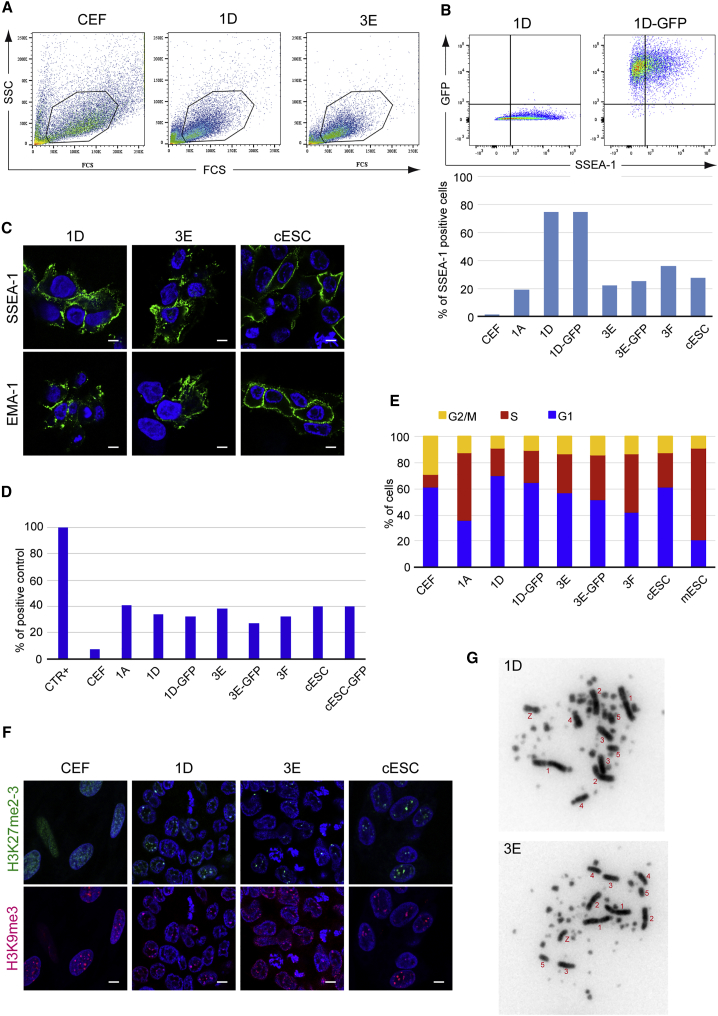
Different criteria are presently used to characterize ESCs and iPSCs, and numerous ones are present on the reprogrammed cells. First, those cells exhibit a morphology typical of that of a stem cell with a round shape, a prominent nucleolus (Figure 4A), and with a smaller size than initial CEF as shown by SSC/FSC density plot (Figure 5A). Electronic microscopy analysis confirmed the similarities between reprogrammed and cESCs in sharp contrast with the CEF (Figure 4B). Second, the different morphotypes and established reprogrammed cells were also able to form embryoid bodies (EBs) (Figure 4C) that expressed some markers of differentiation such as Brachury and Otx2 after 4 and 10 days of differentiation (Figure 4D) and that became Vimentin-positive 5 days after plating (Figure S2). Specific antibodies such as SSEA-1 and EMA-1 are also among the specific tools used to indicate the ES-like nature of cells. cESCs are positive for SSEA-1, SSEA-2, SSEA-3, and EMA-1, but negative for SSEA-4. The long-term cultured reprogrammed clones showed positive SSEA-1 and EMA-1 staining, as detected by flow cytometry and immunochemistry at various levels (Figures 5B and 5C). Endogenous telomerase activity (ETA) is another good criterion to characterize ES and stem cells, as these cells do not enter into senescence, unlike primary cells. Both cESCs and reprogrammed clones showed robust ETA (Figure 5D) and a specific stem cell-like cell-cycle profile, with a high percentage of cells in the S phase. This is in contrast to the CEF cell cycle, which is characterized by a high percentage of cells in G1 phase (Figure 5E). Regarding the epigenetic markers that distinguish pluripotent from differentiated cells (Kress et al., 2016), the reprogrammed clones were more similar to cESCs than to CEFs, as shown by the high levels of trimethylated histone H3 on lysine 27 (H3K27me3) colocalized with the foci of the heterochromatic mark H3K9me3 (Figure 5F). Finally, the karyotypes of the reprogrammed clones were similar to those of cESCs, as indicated by the presence of a regular number of macrochromosomes and the absence of supernumerous minichromosomes, a usual feature of transformed avian cells (Figure 5G).
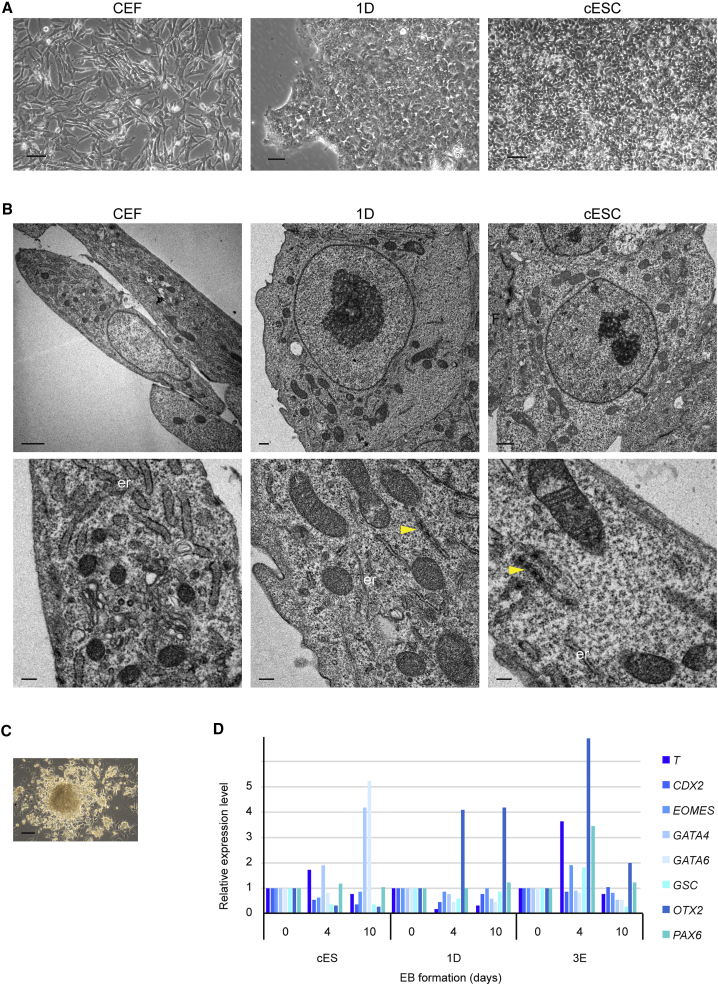
Figure 4.
Characterization of the Reprogrammed Cells
(A and B) Reprogrammed cells exhibit a typical stem cell morphology with a small round compacted shape similar to the cESC, grow in colonies in sharp contrast with the CEFs (A). Scale bars, 50 μm. (B) Ultrastructural analysis of reprogrammed cells by electron microscopy showed that their morphology is no longer the morphology of CEF. Their cytoplasms and nuclei are typically round, like that of cESC. The ER is not dilated as in CEF, and some annulate lamellae could be seen in their cytoplasm, a stem cell specificity (Underwood et al., 2017). Scale bars, 1 μm and 0.2 μm (lower panel).
(C) Reprogrammed cells are able to form embryoid bodies (EBs) Scale bar, 50 μm.
(D) The EBs express various markers of differentiation as detected by qRT-PCR, 4 and 10 days of induction. Results are of a representative experiment.
Figure 5.
Reprogrammed Cells Exhibit Stem Cells Features
(A–C) Long-term established reprogrammed clones present a more compact morphology as shown by lower SSC and FSC density plot for 1D and 3E clones compared with CEF by flow cytometry analysis. The reprogrammed clones (1A, 1D, 3E, and 3F), the GFP-labeled homologs (1D, GFP; 3E, GFP), and the established cESCs as controls for avian stem cells were positive for SSEA-1 and EMA-1, as detected by both fluorescence-activated cell sorting analysis (B) and confocal microscopy after immunofluorescence (C). Scale bars, 5 μm.
(D) Endogenous telomerase activity was measured in the same cells and in CEFs as negative control cells used as the substrates for reprogramming. CTR+ is the positive control provided by the manufacturer.
(E) Analysis of cell-cycle phases reveals a stem cell profile with a short G2/M phase and a long S phase for the chicken reprogrammed clones (1A, 1D, 3E, and 3F), the GFP-labeled homologs (1D, GFP; 3E, GFP), and for the established cESCs and mESCs as controls for avian and murine ESCs, and in CEFs and DEFs as somatic negative control cells.
(F) Reprogrammed cells (1D and 3E) were probed for the presence of large nuclear foci of trimethylated histone H3 on lysine 27, which is typical for cESCs and not observed in CEFs. These foci which colocalize with heterochromatin foci containing trimethylated histone H3 on lysine 9 were present in 66%–98% of the nuclei, depending on the cell clone. Scale bars, 5 μm.
(G) The karyotype analysis of the reprogrammed clones (1D and 3E) reveals a normal chicken karyotype with macrochromosomes and minichromosomes.
Taken together, these criteria suggest that the reprogrammed clones shared most of the key properties of ESCs, and cESCs in particular.
Molecular Characterization of the Established Reprogrammed Clones
To define the full transcriptomic landscape of the reprogrammed cells, deep RNA sequencing (RNA-seq) was performed on four independent reprogrammed clones in parallel with other types of chicken cells, including the CEFs used as starting material for the reprogramming, cESCs from two independent isolates, blastoderm cells (BCs) derived from non-incubated stage X-XII embryos, from which the cESCs were established in vitro, and long-term cultured primordial germ cells (PGCs), as cells able to colonize the germline.
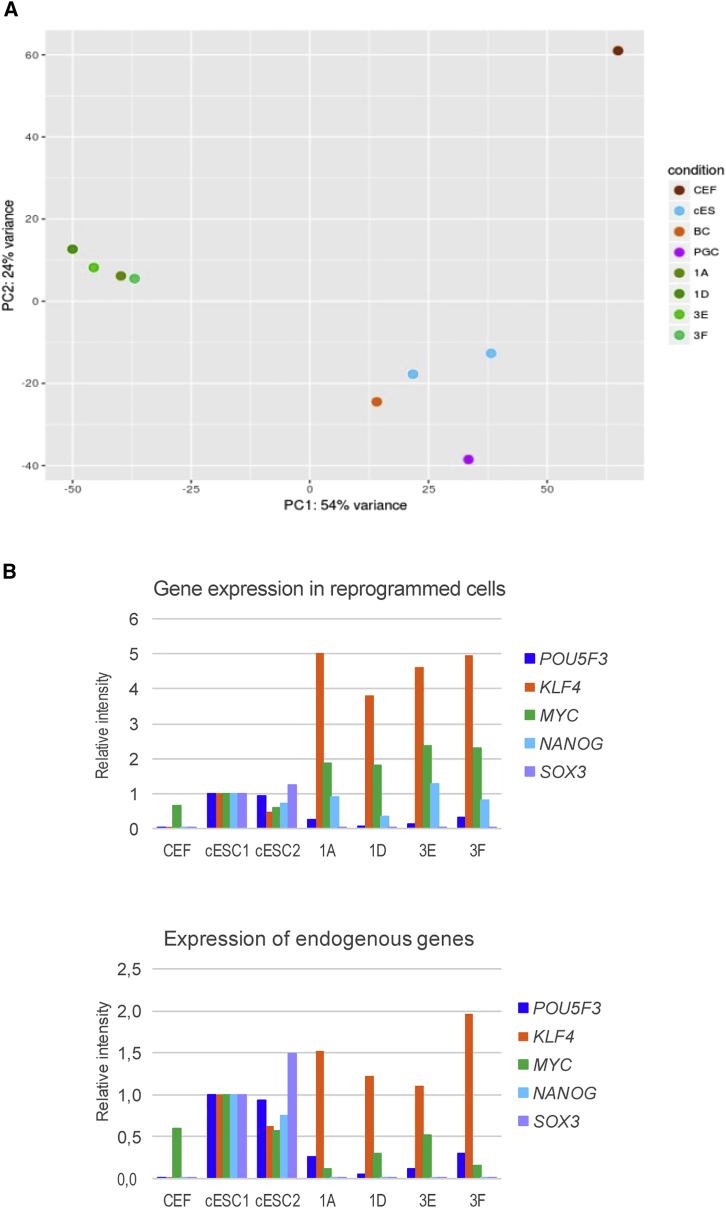
First, principal-component analysis performed with all the expressed genes indicated that the reprogrammed clones showed distinct expression profiles from those of the different stem cell types and the starting fibroblasts (Figure 6A). This suggests that the reprogramming process gave rise to a new cell type with stem cell properties that differed from those of the cESCs.
Figure 6.
Molecular Features of the Reprogrammed Clones
(A) Principal-component analysis revealed that the reprogrammed clones (1) were different at the molecular level from the starting CEFs, (2) clustered together leading to a common cell type generated by the reprogramming process, and (3) differed from spontaneously established cESCs (two independent isolates, cESC-1 and cESC-2, derived from BCs). Primordial germ cells clustered with the previous cell types (cESCs and BCs) and helped define the chicken pluripotent stem cell molecular signature.
(B) The expression profile of exogenous and endogenous reprogramming genes reveals the expression of endogenous POU5F3, KLF4, and c-MYC, but an absence of NANOG endogenous expression as also detected by the deep sequencing analysis.
Second, analysis of the genes expressed in the reprogrammed clones in comparison with those of the cESC established lines, and the BCs and PGCs as two other chicken stem cells, identified a set of 76 commonly expressed genes forming a pluripotency gene network centered on both NANOG and POU5F1 (POU5F3 in chicken) (Table S1A). DNMT3B, SALL1/SALL4, EOMES, SOX17, and FOXA2 are other key transcription factors participating in this network. A second node centered on CDH1 cluster molecules involved in cell adhesion and cell-cell interaction, such as CLDN1, OCLN, GJB1, F11R, VTN, and EPCAM, was identified (Figure S3A). The main gene ontology (GO) terms to characterize those common genes are stem cell differentiation, apical plasma membrane, and protein binding (Table S1B). A set of 148 common genes was identified by restricting the analysis to cESCs instead of cESCs, BCs, and PGCs as the basis for the comparison (Table S2A). The main GO terms to characterize those genes are WNT signaling pathway, bicellular tight-junction, and protein binding (Table S2B). The pluripotency and CDH1 nodes were reinforced with additional linked genes, and two additional nodes emerged, one centered on the cKIT proto-oncogene and one on WNT3. This could suggest the importance of this signaling pathway for avian ESCs. These genes were not expressed in CEFs, illustrating the reprogramming effect (Figure S3B). By contrast, another set of 150 genes was absent or expressed at low levels in the reprogrammed clones compared with their expression in chicken stem cells (Table S3A). The main GO terms to characterize those genes are cell adhesion, heterotrimeric G-protein complex, and G-protein β/γ subunit complex binding (Table S3B). In addition, five main nodes centered on SOX2, EFNB2, MYL3, GNAI1, and AGTR1 were identified, and each included candidate genes to improve the reprogramming process (Figure S3C).
Third, exogenous genes were often still expressed in the long-term established clones. Regarding the expression of pluripotency-associated genes, some of the endogenous genes such as POU5F3 and KLF4 were expressed, as determined by both qRT-PCR and by the presence of the corresponding 3′ UTRs in the aligned sequences when endogenous NANOG was not upregulated (Figures 6B and S4A–S4D). The c-MYC gene was also still strongly expressed from the exogenous transduced vector. SOX2 (exogenous and endogenous) was hardly detectable, as mentioned previously, and SOX3 was among the genes detected only in the chicken stem cells, with almost undetectable levels in the reprogrammed clones. Taken together, these expression profiles indicated that endogenous genes were not fully upregulated to assume the pluripotency of the cells independently from the introduced exogenous reprogramming factors. This suggested that the reprogramming process was incomplete when compared with the strict reprogramming process described in rodent cells. Endogenous LIN28A expression was not upregulated in the four analyzed clones, whereas LIN28B was expressed at higher levels than in CEFs but at lower levels than in cESCs. In clones derived from the OSKMNL combination, the LIN28A gene was not downregulated, indicating that this gene is required to maintain the ESC-like phenotype (data not shown).
Developmental Properties of the Reprogrammed Avian Cells
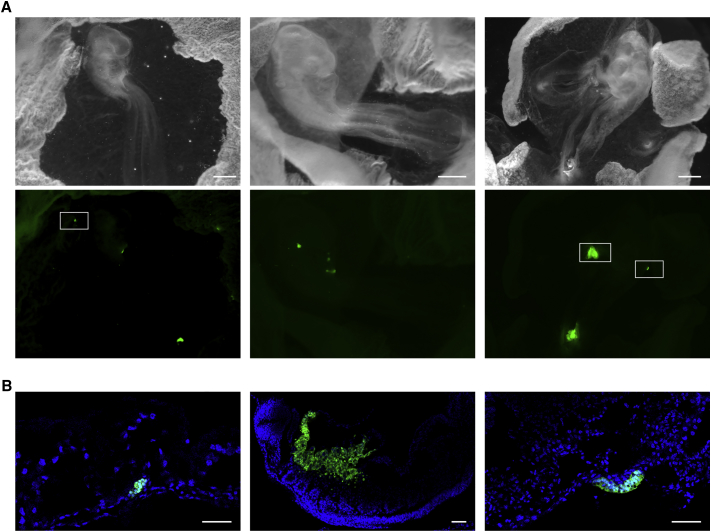
One of the key properties of an ESC is its ability to contribute to development and morphogenesis when injected back into recipient embryos. To test this, two independent clones were first genetically labeled using a GFP reporter expressed under control of a ubiquitous CAG promoter. The selected sub-clones showed homogeneous GFP expression and were positive for the SSEA1 and EMA-1 markers, as well as showing other specific features (cell cycle, epigenetic marks, and ETA) present in their parental clones (Figure 5). Once injected into a stage X-XII embryo using a previously described procedure, the cESC-GFP-positive cells were detected in various parts of the embryos a few days after the injection in a scattered pattern throughout recipient embryonic and extra-embryonic tissues (Figure 7A), but were almost always found in large aggregates similar to tumor-like compact structures (Figures 7A, 7B, S5, and S6). This phenomenon was not restricted to one clone or a unique embryo; instead, it was observed in most of the embryos injected with both 1D and 3E GFP-labeled clones (not shown). This suggested that the injected cells were able to survive in the host, to colonize the embryonic tissues and the vitelline membrane, but that they also are unable to contribute to the surrounding tissues and to differentiated properly in vivo. This observed transformed phenotype was confirmed by performing a chorioallantoic membrane (CAM) assay, which demonstrated that, with the noticeable exception of clone 3F, the three other clones formed tumor-like structures in more than one-third of the embryos (Table S4). Cells dissociated from those structures were cultured a second time and showed growth and morphology patterns identical to those of the grafted cells (data not shown), reflecting their transformed nature.
Figure 7.
Developmental Properties of the Reprogrammed Clones
The 1D reprogrammed clone was GFP labeled and injected into stage X-XII recipient embryos as described previously (Aubel and Pain, 2013) to evaluate its developmental potential.
(A) Three injected embryos showing a contribution of the injected cells to various parts of the embryos, including the vitelline membrane, but mass-like structures were almost always observed in the injected embryos, reflecting a lack of in vivo differentiation of the reprogrammed cells. Bright-light embryo observations (upper panel) and corresponding GFP detection (lower panel). Scale bars, 1 mm.
(B) GFP and Hoechst detection by confocal microscopy on embryo cryosections. Scale bars, 50 μm.
Reprogramming Ability of Duck Cells
Duck is an avian species that diverged genetically from the chicken more than 60 million years ago. Long-term cultured clones were obtained with a procedure similar to that used for primary DEFs using transposon transfection, which resulted in a slower emergence of the reprogrammed clones up to 30–40 days after transfection. Some of the clones could be established long term once picked, but only with the OSKMN gene combination and not with OSKM. These clones were positive for the same markers as those detected in chicken clones, including ETA, SSEA-1, and EMA-1, and specific cell cycle and epigenetic marks similar to those of ESCs (Figure S7); however, they were less tumorigenic than chicken cells (Table S4). Molecular analysis of the duck reprogrammed cells also reveals the importance of the WNT3 pathway as for the chicken cells, but the poor duck genome annotation presently impairs a complete comparison with the chicken reprogrammed cells.
Discussion
Somatic reprogramming was first described in mice and then successfully performed in other mammalian species including non-human primates and humans. Because different pluripotency states have been described in mammalian species, the use and choice of various criteria to define and characterize the reprogrammed cells obtained in these species remain a challenge.
In avian species, previously identified and established cESCs exhibit various traits of mouse ESCs (mESCs), including the ability to contribute efficiently to embryonic morphogenesis; however, unlike mESCs, they do not give rise to functional germ cells in the recipient embryos (Pain et al., 1996, Petitte et al., 2004). This main difference could be attributed to the germ cell lineage determination in the early embryo, as avian germplasm appears to be predetermined according to different recent observations (Tsunekawa et al., 2000, Lee et al., 2016). Nevertheless, these cESCs are AP- and telomerase-positive, and SSEA-1- and EMA-1-positive, and express pluripotency-associated genes including POUV/POU5F3, NANOG, DNMT3B, CLDN1, ASTL, SOX3, ESRP2, EOMES, KRT19, LIN28 (A and B), TRIM71, SALL4, and CDH1, as previously reported and confirmed by RNA-seq analysis (Jean et al., 2015, Vautherot et al., 2017). Chicken and quail iPSCs can be derived from primary fibroblasts using the canonical mouse or human OSKM gene combination as a reprogramming cocktail (Lu et al., 2012, Lu et al., 2015, Yu et al., 2014, Rosselló et al., 2013). More recently, the OSKMNL gene combination was also reported to produce chicken iPSCs (Katayama et al., 2018). These reports do not clearly establish the nature of the reprogrammed cells, nor do they compare it with that of cESCs lines established from long-term cultured BCs. Regarding some of the morphological features described, the transformed status of the cells is never mentioned or reported. In this report, the OSKM gene combination alone, regardless of its delivery system, was unable to produce long-term proliferating cells with the set of markers present in cESC lines. Despite showing a transient change in their phenotype, the cells could not be established. Previous reports did not distinguish this initial and transient change from stable and long-term changes.
Our findings demonstrate that the reprogramming process is effective in avian species, as indicated by the changes in CEFs and DEFs at both phenotype and gene expression levels following the transduction of the OSKMN and OSKMNL gene combinations. The OSKM combination was inefficient for generating cells with the capacity for long-term culture, which is in agreement with previous reports (Katayama et al., 2018). The origin of the genes (chicken, mouse, or human) did not play a key role, although chicken genes were more efficient for the generation of reprogrammed cells. However, the gene delivery system was important to obtain stable reprogrammed cells, favoring transposon vectors for yielding numerous clones of reprogrammed avian cells. The reprogrammed cells expressed several key markers and showed features of ESCs including a stable phenotype, long-term and constant proliferating doubling time, AP and telomerase activities, cell cycle and epigenetic marks, and the ability to give rise to EBs, which displayed a higher expression level of some differentiation markers than undifferentiated counterpart. The molecular signature of the long-term established reprogrammed clones included some of the pluripotency-associated genes that govern the pluripotency molecular core and that are also found in chicken stem cells (Jean et al., 2015). However, according to the criteria for defining fully reprogrammed cells, which include the upregulation of endogenous pluripotency genes to sustain an operating molecular pluripotency core network, the avian reprogrammed clones were not fully reprogrammed cells. In addition, the developmental contribution was restricted to the formation of tumor-like structures in both embryonic and extra-embryonic tissues (vitelline membrane) but was not to the tissue itself. All together this lack of developmental potential also revealed the strong and spontaneous tumorigenic properties of the reprogrammed cells. The high level of exogenous c-MYC gene could indeed contribute to this property, as avian cells have been shown to be highly sensitive to the c-MYC level and its oncogenic potential.
Taken together, these observations and findings indicate that somatic reprogramming is indeed feasible in avian species; however, a factor is still missing for the generation of fully reprogrammed cells. This factor could be a specific gene and/or specific culture conditions to allow the emergence of stable, non-tumorigenic, fully reprogrammed avian iPSCs that would contribute with high efficiency to the recipient embryo. Identifying the culture conditions and the molecular network necessary for the establishment of avian ESCs capable of contributing to both somatic and germ lineages would be important to define, if it exists, the concept of naive cells in avian species.
Experimental Procedures
Primary and ESCs
CEFs and DEFs were prepared as described previously (Jean et al., 2015). In short, 11- to 12-day-old chick embryos, and 15- to 16-day-old Peking duck embryos, were mechanically collected, washed in PBS, beheaded, eviscerated, minced, and trypsinized. Dissociated cells (2 × 106) were plated at 38°C in primary medium (PM) consisting of DMEM/HamF12 medium with 10% tryptose phosphate broth, 8% fetal bovine serum, and 2% chicken serum. After 4–6 days, primary cells were dissociated and amplified in PM at 1 × 106 cells per 100-mm dish for 3–5 passages. Somatic reprogramming attempts were performed on early passages (from 3 to 6) to avoid the replicative senescence that occurs rapidly in avian primary cells. mESCs E14tg2a used as controls were maintained as described previously (Coronado et al., 2013), and cESCs were isolated and maintained in ESA medium as described previously (Aubel and Pain, 2013). Long-term cultured PGCs were established, grown, and characterized as described previously (Jean et al., 2015).
Reprogramming Vectors
The cDNAs from chicken POUV (POU5F3), SOX2, SOX3, KLF4, c-MYC, NANOG, and LIN28 were cloned, sequenced, inserted into the pGAE lentivirus or pPB transposon backbones, and deposited to Addgene. All constructs were generated using the Gibson Assembly Mix (NEB) and sequenced to validate the cDNA insertion. Viral ready-to-use stocks were purchased for the polycistronic pLentG-mKOSM lentiviral vector expressing mouse genes (Cell Biolabs) and for the Sendai viral vectors expressing the human genes (Cytotune Kit, Life Technologies). pMX reprogramming vectors were purchased from Addgene (nos. 13366, 13367, 13375, and 13370).
Viral Production and Infection
Lentiviral particles were obtained following transfection of the pGAE-CAG/WPRE backbone, a simian immunodeficiency virus-based vector, with pEMX2 and pMD2G expressing plasmids on 293T cells, as described previously (Wianny et al., 2008). Titration was performed with the pGAE-GFP-expressing lentivirus. pMX retroviral vectors were generated as vesicular stomatitis virus G (VSV-G) pseudotyped murine leukemia virus (MLV)-derived vectors. The MLV packaging (pTG5349) and the VSV-G envelope encoding (phCMV-G) plasmids were transiently co-transfected into 293T cells with each of the pMX reprogramming vectors encoding for mouse Oct4, Klf4, Sox2, c-Myc, and GFP using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. The supernatants containing viruses were collected 48 hr after transduction and filtered through a 0.45-μm filter, and 0.5 mL of each virus was used directly on 105 CEFs in the presence of 8 μg/mL polybrene. The pMX-GFP vector was used to estimate the viral infectious titer. Fluorescence-activated cell sorting analysis was performed on a Becton DIVA and analyzed using FlowJo (Milteny Biotec). The chosen figures are representative of at least two independent experiments.
Vector Transduction into CEFs and DEFs
Cultured CEFs and DEFs at early passages were dissociated, plated at a density of 2 × 105 cells per well in six-well plates, and transfected using liposomes (Fugene, Promega) or Lipofectamine (Life Technologies), or infected in the presence of 8 μg/mL Polybrene. For electroporation (Neon, Life Technologies), 1 × 106 cells were plated in two wells of a six-well plate. After transduction, fibroblasts were maintained in CEF medium for 2 days and then dissociated and plated in two 100-mm dishes in ESA complete medium, which is routinely used for the maintenance of cESCs (Aubel and Pain, 2013). For some vectors, transduced cells were selected in the presence of hygromycin at 75 μg/mL or puromycin at 1 μg/mL, according to the selection cassette in complete ESA medium for an average of 7 days. When present, colonies were picked at 8–12 days after transduction and maintained for long-term establishment and phenotype stabilization.
Characterization of Reprogrammed Cells
Reprogrammed cells were characterized for AP expression using an AP Detection Kit (Sigma), ETA using the TeloTAGGG telomerase PCR Elisa Kit (Roche), and reactivity toward SSEA-1, SSEA-3, and EMA-1 antibodies, as described previously (Aubel and Pain, 2013). Cell cycle was analyzed on exponentially growing cells as described previously (Coronado et al., 2013). In vitro differentiation potential was assessed by the ability to form EBs in hanging drops for 4 days as described previously (Aubel and Pain, 2013). The nuclear distribution of histone methylation marks was analyzed by immunofluorescence, as described previously (Kress et al., 2016), in four reprogrammed cell clones at different passage numbers (n > 200 nuclei). Real-time qPCR was performed as described (Jean et al., 2015), and all oligonucleotides (provided by Eurogentec) were designed with Primer 3 (Table S5) from sequences extracted from the Gallus_gallus-5.0 chicken genome. For each analysis, the chosen figure is representative of several independant experiments. The transcriptomic profile was analyzed by RNA-seq analysis (Helixio, http://helixio.com), as described previously (Vautherot et al., 2017), using CEFs and DEFs as starting material for the reprogramming and four independently established reprogrammed clones (1A, 1D, 3E, and 3F) for chicken and two (R71-2 and R71-5) for duck. Previously generated data (Vautherot et al., 2017) from cESCs, PGCs, and BCs as chicken stem cells were used for comparison. Data are available at the GEO under accession number GEO: GSE109970 as SuperSeries to group the chicken (GEO: GSE102353) and duck (GEO: GSE109969) data. The alignment was performed on the Gallus_gallus-5.0 chicken genome using Hisat, and differential expression was quantified using the R and DEseq2 packages (Love et al., 2014). String (https://string-db.org/) was used to identify the putative functional protein interaction networks and DAVID GO analysis tool (https://david.ncifcrf.gov/) to perform the GO analysis after the probe ID conversion and with a p value of 0.05 as a limit to define cluster of genes with predicted functions.
In Vivo Tests
The chick CAM assay was performed using 3 × 106 proliferating cells from the 1A, 1D, 3E, and 3F clones and cESCs as negative controls to evaluate the tumorigenic properties of the cells. Eggs were incubated for 6 days, and cells were placed on top of the membrane for 10 days; the membrane was then dissected, washed, and fixed for observation. The size of the nodules or cellular masses was measured. For injection into recipient embryos, different clones were genetically labeled by GFP using the lentiviral GAE vector and injected into the subgerminal cavity as described previously (Aubel and Pain, 2013). Injected embryos were analyzed on day 3 and observed using a fluorescent Leica (M165FC) binocular microscope.
Ethics Statements
Primary embryonic fibroblasts, CEFs, and DEFs were prepared from 11- to 12-day-old chick embryos and 15- to 16-day-old duck embryos. This procedure was performed in strict compliance with the French legislation for animal experiments, which states that the use of embryos from oviparous species before the last third of their development (i.e., before day 14 for chicken embryos) is not subject to regulation (Art. R.214-88). The CAM assay is performed on 6-day-old incubated chicken eggs by depositing the cells to be tested on the external embryonic membrane. The embryos were not touched, modified, or directly injected. At the end of the analysis, the eggs were cracked and the embryos were beheaded to collect the CAM membrane.
Author Contributions
A.F., G.M., P.A., C.J., S.R.-G., and C.K. performed the experiments and analyses. B.P., S.R.-G., and C.K. designed the experiments, analyzed data, and wrote the manuscript.
Acknowledgments
This work was supported by grants from ANR, the projects AviRepro - ANR 09-BLAN-0141, CRB-ANIM - ANR-11-INBS-0003, and ECLAIRE - ANR-14-CE16-0002-01 to B.P.
Published: October 11, 2018
Footnotes
Supplemental Information includes seven figures and five tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.09.005.
Supplemental Information
References
- Acloque H., Lavial F., Pain B. Astacin-like metallo-endopeptidase is dynamically expressed in embryonic stem cells and embryonic epithelium during morphogenesis. Dev. Dyn. 2012;241:574–582. doi: 10.1002/dvdy.23737. [DOI] [PubMed] [Google Scholar]
- Acloque H., Risson V., Birot A.M., Kunita R., Pain B., Samarut J. Identification of a new gene family specifically expressed in chicken embryonic stem cells and early embryo. Mech. Dev. 2001;103:79–91. doi: 10.1016/s0925-4773(01)00336-7. [DOI] [PubMed] [Google Scholar]
- Anokye-Danso F., Trivedi C.M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P.J., Epstein J.A., Morrisey E.E. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubel P., Pain B. Chicken embryonic stem cells: establishment and characterization. Methods Mol. Biol. 2013;1074:137–150. doi: 10.1007/978-1-62703-628-3_11. [DOI] [PubMed] [Google Scholar]
- Boast S., Stern C.D. Simple methods for generating neural, bone and endodermal cell types from chick embryonic stem cells. Stem Cell Res. 2013;10:20–28. doi: 10.1016/j.scr.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Coronado D., Godet M., Bourillot P.Y., Tapponnier Y., Bernat A., Petit M., Afanassieff M., Markossian S., Malashicheva A., Iacone R. A short G1 phase is an intrinsic determinant of naive embryonic stem cell pluripotency. Stem Cell Res. 2013;10:118–131. doi: 10.1016/j.scr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Couteaudier M., Trapp-Fragnet L., Auger N., Courvoisier K., Pain B., Denesvre C., Vautherot J.F. Derivation of keratinocytes from chicken embryonic stem cells: establishment and characterization of differentiated proliferative cell populations. Stem Cell Res. 2015;14:224–237. doi: 10.1016/j.scr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- De Los Angeles A., Ferrari F., Xi R., Fujiwara Y., Benvenisty N., Deng H., Hochedlinger K., Jaenisch R., Lee S., Leitch H.G. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- Declercq J., Sheshadri P., Verfaillie C.M., Kumar A. Zic3 enhances the generation of mouse induced pluripotent stem cells. Stem Cells Dev. 2013;22:2017–2025. doi: 10.1089/scd.2012.0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Extavour C.G., Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Ezashi T., Telugu B.P., Alexenko A.P., Sachdev S., Sinha S., Roberts R.M. Derivation of induced pluripotent stem cells from pig somatic cells. Proc. Natl. Acad. Sci. USA. 2009;106:10993–10998. doi: 10.1073/pnas.0905284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B., Jiang J., Kraus P., Ng J.H., Heng J.C., Chan Y.S., Yaw L.P., Zhang W., Loh Y.H., Han J. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat. Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- Fischedick G., Klein D.C., Wu G., Esch D., Höing S., Han D.W., Reinhardt P., Hergarten K., Tapia N., Schöler H.R., Sterneckert J.L. Zfp296 is a novel, pluripotent-specific reprogramming factor. PLoS One. 2012;7:e34645. doi: 10.1371/journal.pone.0034645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Yuan P., Yang H., Zhang J., Soh B.S., Li P., Lim S.L., Cao S., Tay J., Orlov Y.L. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463:1096–1100. doi: 10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng J.C., Feng B., Han J., Jiang J., Kraus P., Ng J.H., Orlov Y.L., Huss M., Yang L., Lufkin T. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Honda A., Hatori M., Hirose M., Honda C., Izu H., Inoue K., Hirasawa R., Matoba S., Togayachi S., Miyoshi H., Ogura A. Naive-like conversion overcomes the limited differentiation capacity of induced pluripotent stem cells. J. Biol. Chem. 2013;288:26157–26166. doi: 10.1074/jbc.M113.502492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean C., Aubel P., Soleihavoup C., Bouhallier F., Voisin S., Lavial F., Pain B. Pluripotent genes in avian stem cells. Dev. Growth Differ. 2013;55:41–51. doi: 10.1111/dgd.12021. [DOI] [PubMed] [Google Scholar]
- Jean C., Oliveira N.M.M., Intarapat S., Fuet A., Mazoyer C., De Almeida I., Trevers K., Boast S., Aubel P., Bertocchini F. Transcriptome analysis of chicken ES, blastodermal and germ cells reveals that chick ES cells are equivalent to mouse ES cells rather than EpiSC. Stem Cell Res. 2015;14:54–67. doi: 10.1016/j.scr.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K., Norrby K., Paca A., Mileikovsky M., Mohseni P., Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagiannis P., Eto K. Ten years of induced pluripotency: from basic mechanisms to therapeutic applications. Development. 2016;143:2039–2043. doi: 10.1242/dev.138172. [DOI] [PubMed] [Google Scholar]
- Katayama M., Hirayama T., Tani T., Nishimori K., Onuma M., Fukuda T. Chick derived induced pluripotent stem cells by the poly-cistronic transposon with enhanced transcriptional activity. J. Cell Physiol. 2018;233:990–1004. doi: 10.1002/jcp.25947. [DOI] [PubMed] [Google Scholar]
- Koh S., Piedrahita J.A. From “ES-like” cells to induced pluripotent stem cells: a historical perspective in domestic animals. Theriogenology. 2014;81:103–111. doi: 10.1016/j.theriogenology.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress C., Montillet G., Jean C., Fuet A., Pain B. Chicken embryonic stem cells and primordial germ cells display different heterochromatic histone marks than their mammalian counterparts. Epigenetics Chromatin. 2016;9:5. doi: 10.1186/s13072-016-0056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavial F., Acloque H., Bachelard E., Nieto M.A., Samarut J., Pain B. Ectopic expression of Cvh (Chicken Vasa homologue) mediates the reprogramming of chicken embryonic stem cells to a germ cell fate. Dev. Biol. 2009;330:73–82. doi: 10.1016/j.ydbio.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Lavial F., Acloque H., Bertocchini F., Macleod D.J., Boast S., Bachelard E., Montillet G., Thenot S., Sang H.M., Stern C.D. The Oct4 homologue PouV and Nanog regulate pluripotency in chicken embryonic stem cells. Development. 2007;134:3549–3563. doi: 10.1242/dev.006569. [DOI] [PubMed] [Google Scholar]
- Lee H.C., Choi H.J., Lee H.G., Lim J.M., Ono T., Han J.Y. DAZL expression explains origin and central formation of primordial germ cells in chickens. Stem Cells Dev. 2016;25:68–79. doi: 10.1089/scd.2015.0208. [DOI] [PubMed] [Google Scholar]
- Liu J., Balehosur D., Murray B., Kelly J.M., Sumer H., Verma P.J. Generation and characterization of reprogrammed sheep induced pluripotent stem cells. Theriogenology. 2012;77:338–346.e1. doi: 10.1016/j.theriogenology.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., West F.D., Jordan B.J., Beckstead R.B., Jordan E.T., Stice S.L. Generation of avian induced pluripotent stem cells. Methods Mol. Biol. 2015;1330:89–99. doi: 10.1007/978-1-4939-2848-4_9. [DOI] [PubMed] [Google Scholar]
- Lu Y., West F.D., Jordan B.J., Mumaw J.L., Jordan E.T., Gallegos-Cardenas A., Beckstead R.B., Stice S.L. Avian-induced pluripotent stem cells derived using human reprogramming factors. Stem Cells Dev. 2012;21:394–403. doi: 10.1089/scd.2011.0499. [DOI] [PubMed] [Google Scholar]
- Nagy K., Sung H.K., Zhang P., Laflamme S., Vincent P., Agha-Mohammadi S., Woltjen K., Monetti C., Michael I.P., Smith L.C., Nagy A. Induced pluripotent stem cell lines derived from equine fibroblasts. Stem Cell Rev. 2011;7:693–702. doi: 10.1007/s12015-011-9239-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J., Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Osteil P., Tapponnier Y., Markossian S., Godet M., Schmaltz-Panneau B., Jouneau L., Cabau C., Joly T., Blachère T., Gócza E. Induced pluripotent stem cells derived from rabbits exhibit some characteristics of naive pluripotency. Biol. Open. 2013;2:613–628. doi: 10.1242/bio.20134242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain B., Clark M.E., Shen M., Nakazawa H., Sakurai M., Samarut J., Etches R.J. Long-term in vitro culture and characterisation of avian embryonic stem cells with multiple morphogenetic potentialities. Development. 1996;122:2339–2348. doi: 10.1242/dev.122.8.2339. [DOI] [PubMed] [Google Scholar]
- Petitte J.N., Liu G., Yang Z. Avian pluripotent stem cells. Mech. Dev. 2004;121:1159–1168. doi: 10.1016/j.mod.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Rosselló R.A., Chen C.C., Dai R., Howard J.T., Hochgeschwender U., Jarvis E.D. Mammalian genes induce partially reprogrammed pluripotent stem cells in non-mammalian vertebrate and invertebrate species. Elife. 2013;2:e00036. doi: 10.7554/eLife.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H., Nakada A., Hashimoto Y., Shigeno K., Shionoya Y., Nakamura T. Generation of canine induced pluripotent stem cells by retroviral transduction and chemical inhibitors. Mol. Reprod. Dev. 2010;77:2. doi: 10.1002/mrd.21117. [DOI] [PubMed] [Google Scholar]
- Smith A. Formative pluripotency: the executive phase in a developmental continuum. Development. 2017;144:365–373. doi: 10.1242/dev.142679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumer H., Liu J., Malaver-Ortega L.F., Lim M.L., Khodadadi K., Verma P.J. NANOG is a key factor for induction of pluripotency in bovine adult fibroblasts. J. Anim. Sci. 2011;89:2708–2716. doi: 10.2527/jas.2010-3666. [DOI] [PubMed] [Google Scholar]
- Susta L., He Y., Hutcheson J.M., Lu Y., West F.D., Stice S.L., Yu P., Abdo Z., Afonso C.L. Derivation of chicken induced pluripotent stem cells tolerant to Newcastle disease virus-induced lysis through multiple rounds of infection. Virol. J. 2016;13:205. doi: 10.1186/s12985-016-0659-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- Tapia N., Reinhardt P., Duemmler A., Wu G., Araúzo-Bravo M.J., Esch D., Greber B., Cojocaru V., Rascon C.A., Tazaki A. Reprogramming to pluripotency is an ancient trait of vertebrate Oct4 and Pou2 proteins. Nat. Commun. 2012;3:1279. doi: 10.1038/ncomms2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theunissen T.W., Costa Y., Radzisheuskaya A., van Oosten A.L., Lavial F., Pain B., Castro L.F., Silva J.C. Reprogramming capacity of Nanog is functionally conserved in vertebrates and resides in a unique homeodomain. Development. 2011;138:4853–4865. doi: 10.1242/dev.068775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunekawa N., Naito M., Sakai Y., Nishida T., Noce T. Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development. 2000;127:2741–2750. doi: 10.1242/dev.127.12.2741. [DOI] [PubMed] [Google Scholar]
- Underwood J.M., Becker K.A., Stein G.S., Nickerson J.A. The ultrastructural signature of human embryonic stem cells. J. Cell. Biochem. 2017;118:764–774. doi: 10.1002/jcb.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vautherot J.F., Jean C., Fragnet - Trapp L., Remy S., Chabanne– Vautherot D., Montillet G., Fuet A., Denesvre C., Pain B. ESCDL-1, a new cell line derived from chicken embryonic stem cells, supports efficient replication of mardiviruses. PLoS One. 2017;12:e0175259. doi: 10.1371/journal.pone.0175259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Liu J., Holland M.K., Temple-Smith P., Williamson M., Verma P.J. Nanog is an essential factor for induction of pluripotency in somatic cells from endangered felids. Biores. Open Access. 2013;2:72–76. doi: 10.1089/biores.2012.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wianny F., Bernat A., Huissoud C., Marcy G., Markossian S., Cortay V., Giroud P., Leviel V., Kennedy H., Savatier P., Dehay C. Derivation and cloning of a novel rhesus embryonic stem cell line stably expressing tau-green fluorescent protein. Stem Cells. 2008;26:1444–1453. doi: 10.1634/stemcells.2007-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K., Michael I.P., Mohseni P., Desai R., Mileikovsky M., Hämäläinen R., Cowling R., Wang W., Liu P., Gertsenstein M. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Yamauchi T., Izpisua Belmonte J.C. An overview of mammalian pluripotency. Development. 2016;143:1644–1648. doi: 10.1242/dev.132928. [DOI] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu P., Lu Y., Jordan B.J., Liu Y., Yang J.Y., Hutcheson J.M., Ethridge C.L., Mumaw J.L., Kinder H.A., Beckstead R.B. Nonviral minicircle generation of induced pluripotent stem cells compatible with production of chimeric chickens. Cell. Reprogram. 2014;16:366–378. doi: 10.1089/cell.2014.0028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.