Abstract
Currently, there is an incomplete understanding of the molecular pathogenesis of meningiomas, the most common primary brain tumor. Several familial syndromes are characterized by increased meningioma risk, and the genetics of these syndromes provides mechanistic insight into sporadic disease. The best defined of these syndromes is neurofibromatosis type 2, which is caused by a mutation in the NF2 gene and has a meningioma incidence of approximately 50%. This finding led to the subsequent discovery that NF2 loss-of-function occurs in up to 60% of sporadic tumors. Other important familial diseases with increased meningioma risk include nevoid basal cell carcinoma syndrome, multiple endocrine neoplasia 1 (MEN1), Cowden syndrome, Werner syndrome, BAP1 tumor predisposition syndrome, Rubinstein-Taybi syndrome, and familial meningiomatosis caused by germline mutations in the SMARCB1 and SMARCE1 genes. For each of these syndromes, the diagnostic criteria, incidence in the population, and frequency of meningioma are presented to review the relevant clinical information for these conditions. The genetic mutations, molecular pathway derangements, and relationship to sporadic disease for each syndrome are described in detail to identify targets for further investigation. Familial syndromes characterized by meningiomas often affect genes and pathways that are also implicated in a subset of sporadic cases, suggesting key molecular targets for therapeutic intervention. Further studies are needed to resolve the functional relevance of specific genes whose significance in sporadic disease remains to be elucidated.
Keywords: Familial meningioma, Meningioma, Meningioma genetics, Meningioma pathogenesis
ABBREVIATIONS
- AKT1
v-Akt murine thymoma viral oncogene homolog 1
- BCC
basal cell carcinoma
- CS
Cowden syndrome
- IHC
immunohistochemistry
- KLF4
Kruppel-like factor 4
- MBAIT
melanocytic BAP1-mutated atypical intradermal tumors
- MEN1
multiple endocrine neoplasia 1
- NBCCS
nevoid basal cell carcinoma syndrome
- NF2
neurofibromatosis type 2
- PHTS
PTEN hamartoma tumor syndrome
- PI3K
phosphatidylinositol 3-kinase
- PTEN
phosphatase and tensin homolog
- SHH
sonic hedgehog
- SMO
smoothened
- TRAF7
TNF receptor-associated factor 7
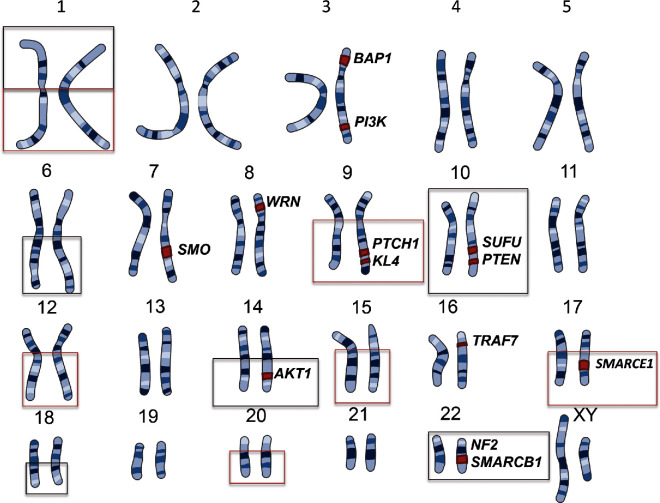
Meningiomas are the most common primary brain tumor reported in the United States, comprising approximately 36% of brain tumors.1,2 Estimates from recent data predict that more than 20 000 new meningiomas will be diagnosed each year, with an incidence of 7.5 per 100 000.2 While the majority of these tumors are sporadic, there are many familial syndromes that place a patient at increased risk for meningioma development. The most well characterized and documented of these genetic syndromes is neurofibromatosis type 2 (NF2). Identification of the NF2 gene led to the investigation of molecular genetic mutations in sporadic meningiomas, in which up to 60% of patients have also been found to have somatic inactivation of NF2.3,4 Recent whole genome sequencing and whole exome sequencing studies in non-NF2 mutated meningiomas have further delineated additional mutations in TRAF7, KLF4, AKT1, PI3K, and SMO; however, many sporadic tumors still have not been found to have a genetic basis (Figure 1).5,6 Greater understanding of the familial syndromes associated with meningiomatosis may elucidate further molecular mechanisms behind the development of sporadic meningiomas. Below, we present a review of familial syndromes associated with meningioma predisposition.
FIGURE 1.
Locations of known mutations in meningioma. Loss of chromosome regions shown in black, gain of chromosome regions shown in red.
METHODS
A review of the PubMed database was conducted to identify published studies on known familial meningioma syndromes up until June 2017. Combinations of search terms used included “meningioma,” “familial meningiomatosis,” “familial meningioma,” “nevoid basal cell carcinoma syndrome,” “Gorlin syndrome,” “multiple endocrine neoplasia 1,” “Cowden syndrome,” “Werner syndrome,” “BAP1 tumor predisposition syndrome,” “SMARCB1,” “SMARCE1,” “SUFU,” and “Rubinstein-Taybi syndrome.” Only studies available in English were included. All identified abstracts from these searches were analyzed, and those that reviewed or examined familial syndromes associated with meningiomas were included.
RESULTS
A review of PubMed abstracts from the described search criteria resulted in 46 studies that met inclusion (see Supplemental Digital Content). The results from these studies are presented below and include NF2, nevoid basal cell carcinoma syndrome (NBCCS, also known as Gorlin syndrome), multiple endocrine neoplasia 1 (MEN1), Cowden syndrome (CS), Werner syndrome, BAP1 tumor predisposition syndrome, Rubinstein-Taybi syndrome, and familial meningiomatosis caused by germline mutations in the SMARCB1 and SMARCE1 genes (Table 1).
TABLE 1.
List of Syndromes, Mutations, Pathways, and Sporadic Relationships
| Syndrome | Mutated gene | Involved pathway | Relationship to sporadic tumors |
|---|---|---|---|
| NF2 | NF2 tumor suppressor | EGFR, YAP/TAZ, MAPK, cytoskeletal architecture | Mutated in 40%-60% |
| Werner | WRN | DNA processing, maintenance and repair | Methylated with decrease in expression |
| BAP1-TPDS | BAP1 | Chromatin Remodeling, DNA repair | Found in high-grade rhabdoid meningiomas |
| SMARCE1 | SMARCE1 | Nucleosome remodeling, apoptosis | Found in Clear cell meningiomas |
| SMARCB1 | SMARCB1 | Nucleosome remodeling, apoptosis | Mutated in 3% |
| Gorlin | PTCH1 | SHH – PTCH1/SMO/SUFU/GLI-1,2,3 | SMO mutated in 5% |
| Cowden | PTEN | PI3K – RTK/AKT/PI3K/mTOR/PTEN | AKT mutated in 14%; PIK3CA mutated in 7% |
Neurofibromatosis Type 2
NF2 is an autosomal dominant, 100% penetrant condition characterized by a constellation of schwannomas and meningiomas.7 NF2 is clinically diagnosed using the Manchester criteria, the details of which are in Table 2. The overall prevalence of NF2 in the population is approximately 1 in 60 000.8
TABLE 2.
Neurofibromatosis Type 2 Diagnostic Criteria
| A diagnosis can be made if an individual meets one of the following: | |
|---|---|
| Bilateral vestibular schwannomas | |
| A first degree relative with NF2 + unilateral vestibular schwannoma or 2 NF2-associated lesions* | |
| Unilateral vestibular schwannoma + 2 NF2-associated lesions* | |
| Multiple meningiomas + unilateral vestibular schwannoma or 2 other NF-2 associated lesions* |
*Meningioma, schwannoma, glioma, neurofibroma, posterior subcapsular lenticular opacities.
NF2 is caused by germline mutations in the NF2 tumor suppressor gene, located at 22q12.2. The protein product is called merlin or schwannomin.9 Within the cell, merlin has been found to interact with several proteins that affect PI3K, YAP/TAZ, and mitogen-activated protein kinase (MAPK) signaling. All 3 pathways are important in cell growth and cellular proliferation.8,10,11 Given its location at the cell membrane-cytoskeletal interface, several other potential functions have been proposed, including contact-dependent inhibition of EGFR, effects on cell-to-cell adhesion, and regulation of cytoskeletal architecture.12-14
The role of NF2 inactivation in sporadic meningiomas was investigated due to the high frequency of meningiomas in NF2 patients. The frequency of NF2 inactivation is estimated to be present between in 40% and 60% of cases of sporadic meningiomas based on several studies, and this mutation is thought to be the causative mutation in these cases.4 Interestingly, NF2-mutated meningiomas have certain location predilections.5 Those with NF2 mutations and/or chromosome 22 loss were found preferentially in the hemispheres. For meningiomas of the skull base, those located laterally and posteriorly were significantly more likely to have NF2 mutations and/or chromosome 22 loss, whereas medially located skull base tumors were more commonly non-NF2 mutated. The work on NF2 has provided an excellent example of how investigating a familial syndrome with meningiomas can lead to establishing the genetic basis for sporadic disease.
Nevoid Basal Cell Carcinoma Syndrome
Nevoid basal cell carcinoma syndrome, also known as basal cell nevus syndrome or Gorlin syndrome, is an autosomal dominant syndrome that is classically characterized by multiple basal cell carcinomas (BCC), jaw keratocysts, and bifid ribs.15 The complete list of major clinical criteria required for diagnosis and cited by several studies is detailed in Table 3. 16,20 The estimated prevalence is between 1 in 30 000 and 1 in 256 000 and equally affects men and women.16,17 Additional tumors that are associated with the syndrome include meningiomas and rhabdomyosarcomas.16,18
TABLE 3.
Nevoid Basal Cell Carcinoma Syndrome Diagnostic Criteria
| A diagnosis can be made if an individual meets 2 major criteria or 1 major + 2 minor criteria | |
|---|---|
| Major criteria | Minor criteria |
| Basal cell carcinomas: ≥5 in a lifetime or 1 diagnosed before age 30 | Childhood medulloblastoma |
| Lamellar calcification of the falx before the age of 20 | Lympho-mesenteric or pleural cysts |
| Jaw odontogenic keratocyst | Macrocephaly (occipitofrontal circumference (OFC) > 97th percentile) |
| Palmar or plantar pits: ≥2 | Cleft lip/palate |
| First-degree relative with NBCCS | Vertebral or rib anomalies (bifid/splayed/extra ribs, bifid vertebrae) |
| Preaxial or postaxial polydactyly | |
| Ovarian or cardiac fibromas | |
| Ocular anomalies (cataract, developmental defects, pigmentary changes of the retinal epithelium) | |
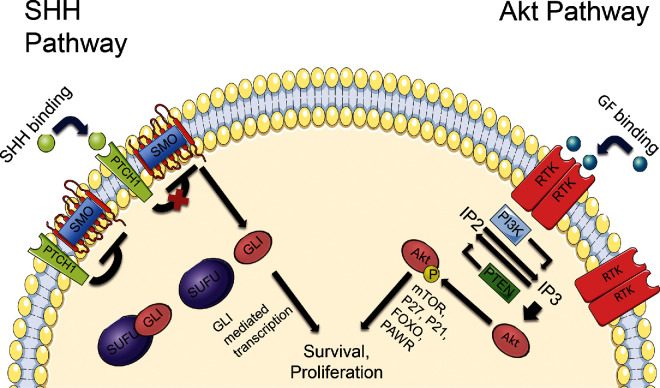
Specifically, meningiomas were reported in 5% of NBCCS patients in 2 studies.19,20 Given that meningiomas commonly arise from the falx cerebri, it is interesting to note that NBCCS is characterized by prominent calcifications of this area.16,19 The genetic causes behind NBCCS are pathogenic mutations in genes of the sonic hedgehog (SHH) signaling pathway including PTCH1, PTCH2, and SUFU, located at 9q22.32, 1p34.1, and 10q24.32, respectively, with variations in PTCH1 being the most common.21,22 The activation of this pathway drives cell proliferation in both normal neural development and in tumor development.23 In BCC and a subset of medulloblastoma, the loss of PTCH1 or SUFU and the activation of SMO have been implicated in tumor initiation and maintenance.24,25 In fact, mutations in PTCH1 or SMO are sufficient to drive tumorigenesis in BCC and medulloblastoma mouse models.23,26,27 The PTCH1 gene codes for a protein that constitutes the ligand-binding component of the SHH receptor complex. The details of the SHH pathway are depicted in Figure 2.28,29
FIGURE 2.
Details of the SHH and Akt pathways. Without SHH binding, PTCH1 renders smoothened (SMO) inactive and inhibits it from signaling downstream targets. When SHH binds to PTCH1, SMO is released from this inhibition, allowing it to interact with SUFU. This results in the activation and nuclear translocation of glioma-associated oncogene homologue 1 (GLI1) and (GLI2), and the degradation of GLI3. In the Akt pathway, PTEN normally targets IP3 to inhibit the phosphorylation of Akt. When growth factor (GF) binding to receptor tyrosine kinases (RTK) occurs, PI3K signaling is increased, leading to IP3 accumulation, phosphorylation of Akt and activation of downstream targets.
A meningioma has been reported in a patient with NBCCS with a confirmed germline PTCH1 mutation in which the genetic sequencing of the patient's meningioma revealed an additional somatic mutation in PTCH1. Using the Knudson's 2-hit theory, they considered the development of this meningioma to be caused by loss of function of PTCH1.30 Additionally, driver mutations in SMO have been identified as one of the most common non-NF2 mutations in sporadic meningiomas, and have a predilection for the olfactory groove. Meningiomas with an SMO mutation in the olfactory groove have been found to have a higher recurrence rate, especially late recurrences (>5 yr). Given this information and the significance with regard to prognostication, surgeons and neuro-oncologists should consider testing olfactory groove meningiomas specifically for SMO mutations and adjusting their follow-up schedule accordingly for these lesions, which is currently commercially available at multiple CLIA (Clinical Laboratory Improvement Amendments)-certified laboratories.5,31
Patients with NBCCS caused by SUFU mutations have been found to be significantly more likely to have a meningioma in comparison to NBCCS patients caused by PTCH1 or PTCH2 mutations.22SUFU mutations have also been investigated in a family with multiple meningiomas and the absence of other features of NBCCS.32 Genome-wide linkage analysis and exome sequencing in this family found that germline SUFU mutations segregated with disease in the family. Functional studies showed that the identified mutation led to significantly increased SHH pathway activation due to loss of the normal suppressor function of SUFU. This group also sequenced the coding exons and exon-intron boundaries of SUFU in blood/germline samples of 162 meningiomas (11 of these patients had a familial history of meningiomas and 40 had multiple meningiomas) from a national meningioma database; however, no pathogenic variants of SUFU were detected.
The SHH pathway is critical to the formation of meningiomas in some instances. Studies on patients with NBCCS and familial meningiomatosis due to germline SUFU mutations have provided important information into the mechanisms by which derangement of the SHH pathway can lead to these tumors. Further research into the other components of this pathway is important.
Cowden Syndrome
Cowden syndrome, part of the PTEN hamartoma tumor syndrome (PHTS), is an autosomal dominant disorder caused by germline mutations in phosphatase and tensin homolog (PTEN) on chromosome 10q23.31.33 Its diagnostic criteria include a host of major and minor criteria as well as pathognomonic findings listed in Table 4.34 CS is estimated to affect between 1 in 200 000 and 1 in 250 000 people.35,36 Although not part of the major or minor criteria, meningiomas are found in approximately 8% of patients.37 Other clinical syndromes that are classified as PHTS include Bannayan-Riley-Ruvalcaba and Proteus syndrome.33 Multiple case reports have also identified meningiomas in patients with Proteus syndrome.38,39
TABLE 4.
Cowden Syndrome Diagnostic Criteria
| A diagnosis can be made if an individual meets 3 major criteria (1 must be macrocephaly, LDD, or GI hamartomas) or 2 major + 3 minor criteria* | |
|---|---|
| Major criteria | Minor criteria |
| Breast cancer | Autism spectrum disorder |
| Endometrial cancer (epithelial) | Colon cancer |
| Thyroid cancer (follicular) | Esophageal glycogenic acanthosis: ≥ 3 |
| Gastrointestinal hamartomas (including ganglioneuromas, but excluding hyperplastic polyps): ≥ 3 | Lipomas: ≥ 3 |
| Adult Lhermitte-Duclos disease (LDD) | Intellectual disability (IQ ≤ 75) |
| Macrocephaly (occipitofrontal circumference (OFC) ≥ 97th percentile) | Renal cell carcinoma |
| Macular pigmentation of the glans penis | Testicular lipomatosis |
| Multiple mucocutaneous lesions (any of the following): | Thyroid cancer (papillary or follicular variant of papillary) |
| Trichilemmomas: ≥ 3, at least one biopsy proven | Thyroid lesions (adenoma, multinodular goiter) |
| Acral keratoses: ≥ 3 palmoplantar keratotic pits and/or acral hyperkeratotic papules) | Vascular anomalies (including multiple intracranial developmental venous anomalies) |
| Mucocutaneous neuromas: ≥ 3 | |
| Oral papillomas (particularly on tongue and gingiva): ≥ 3 or 1 biopsy proven/dermatologist diagnosed | |
*In an individual who has a relative who meets the above criteria or has a PTEN mutation, a diagnosis can be made if he or she meets 2 major criteria, 1 major + 2 minor criteria, or 3 minor criteria.
The PTEN protein acts as a phosphatase to dephosphorylate various cellular lipids and proteins. Dysfunctional PTEN leads to upregulation of the PI3K-AKT-mTOR (mammalian target of rapamycin) pathway, resulting in increases in cell proliferation, energy metabolism, and survival.40 It has been found to be important in breast and prostate cancer as well as glioblastoma multiforme.41 In mouse models of glioma, PTEN deletion results in earlier tumor onset and increases tumor grade.42,43 PTEN targets include phosphatidylinositol trisphosphate and phosphatidylinositol bisphosphate, both products of PI3K. This pathway is illustrated in Figure 2. As part of its protein phosphatase activity, PTEN has been shown to dephosphorylate focal adhesion kinase, which has negative effects on cell migration and cell spreading, as well as MAPK that regulates cell survival.44,45
In sporadic meningiomas, higher-grade tumors are associated with progressive changes at the chromosomal level including losses of chromosomes 1p, 9q, 10q, 14q, and 22q.46 Given this information and PTEN’s location at 10q23.3, it was investigated as a candidate gene in sporadic meningiomas.47 Using a group of 55 WHO grade I, 10 grade II, and 10 grade III meningiomas, Peters et al47 sequenced the entire coding sequence of the PTEN gene in tumor samples. No PTEN mutations were seen in the grade I tumors, but 1 grade III tumor harbored a somatic mutation in PTEN. These findings led them to hypothesize that mutations in PTEN are unlikely to be involved in the initiation and formation of low-grade meningiomas, but may contribute to malignant progression to higher-grade lesions. Such a conclusion supports other work in the chromosomal analysis comparing high-grade and low-grade sporadic meningiomas.46 This is similar to gliomas in that low-grade gliomas do not have as high of a frequency of PTEN loss as high-grade gliomas.48
Given PTEN’s role in regulation of the PI3K-AKT-mTOR pathway, and evidence of dysregulation of this pathway in many cancers, downstream targets of PTEN have also been investigated as potential oncogenic drivers in meningioma.49 Both AKT and PI3KA mutations have been found in sporadic meningiomas, comprising approximately 9% and 7% of non-NF2-mutant meningiomas, respectively.5,50 The complexity of this pathway suggests ample room for more investigation into the role of both PTEN and its downstream targets in meningioma pathogenesis. Previous research in patients with CS has provided some of the framework for our understanding of this pathway's role in cancer.
Werner Syndrome
Werner syndrome, also known as adult progeria, was first described in a set of siblings with short stature, “senile” appearance, premature graying of the hair, early-onset cataracts, dermatologic changes, and muscle atrophy.51 Biallelic mutations in the WRN gene (also known as RECQL2) were later established as the cause of the syndrome, which is inherited in an autosomal recessive manner.52 A set of cardinal and additional signs and symptoms has been designated and was updated most recently in 2013 (Table 5).53 A definite diagnosis is achieved when all 6 cardinal signs are present or a gene mutation and at least 3 cardinal signs are present. Interestingly, Japan has the highest frequency of carriers, at rates of 6 per 1000, while the disease prevalence in the overall population is estimated to be 1 in 1000 000 individuals based on publically available allele frequency databases.54,55 A patient with Werner syndrome is approximately 36.2 times Standardized Incidence Ratio (SIR); 95% confidence interval 17.3, 66.5) more likely to develop a meningioma than the general population and are more likely to develop them at a younger age.56 In addition, the proportion of meningiomas to other brain tumors in the Werner Syndrome is higher than expected.57
TABLE 5.
Werner Syndrome Diagnostic Criteria
| A confirmed diagnosis can be made if an individual has all 6 cardinal signs or a WRN mutation + 3 cardinal signs. A diagnosis is suspected if an individual has 2 cardinal signs or 1 cardinal sign + additional signs | |
|---|---|
| Cardinal signs* | Additional signs |
| Premature greying or thinning of scalp hair | Abnormal glucose and/or lipid metabolism |
| Bilateral cataracts | Skeletal abnormalities (osteoporosis) |
| Characteristic dermatologic changes (atrophic skin, tight skin, clavus, callus, intractable skin ulcers) | Malignant tumors (nonepithelial tumors, thyroid cancer) |
| Soft-tissue calcification (Achilles tendon) | Parental consanguinity |
| ‘Bird-like’ facies | Premature atherosclerosis (angina pectoris, myocardial infarction) |
| Abnormal voice (high pitched, squeaky, hoarse) | Hypogonadism |
| Short stature and low bodyweight | |
*Age of onset between 10-40 years.
The WRN gene is located on chromosome 8p12 and encodes for a protein with DNA helicase activity that is a member of the RecQ family.52,58 The WRN protein product uses its helicase function to help resolve intermediate DNA structures generated during DNA metabolism and also functions in DNA repair (specifically base excision repair and double-stranded DNA break repair) replication, transcription, and telomere maintenance.59 Its loss of function in Werner syndrome patients leads to genomic instability and limited replicative capacity in somatic cells.60,61 The majority of pathogenic germline mutations leading to Werner syndrome are stop codons, small indels, or splicing mutations that cause loss of the nuclear localization of the protein. Other mutations lead to protein instability and loss of function, but essentially all WRN mutations are functional null alleles in Werner syndrome.62
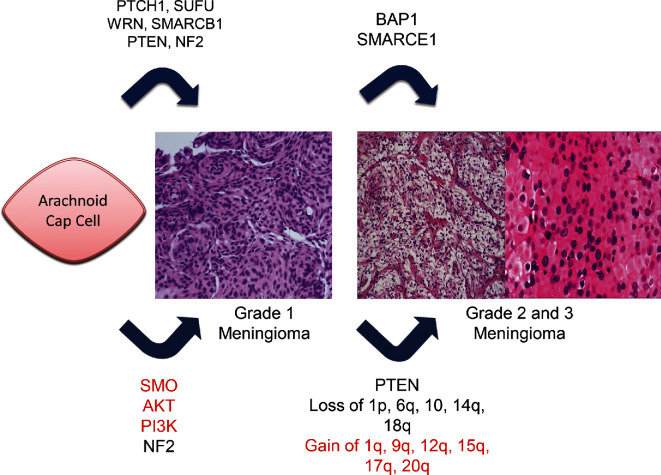
Sporadic meningiomas are characterized by a number of chromosomal copy number alterations, and this appears to increase with meningioma grade and tumor progression (Figure 3).63,64 The most common alteration is monosomy 22, followed by loss of chromosome 1p and 14q. Other chromosomal changes include loss of 6q, 10, and 18q as well as gain of chromosomes 1q, 9q, 12q, 15q, 17q, and 20q.65 In particular, combined 1p/14q deletions are significantly more likely in higher-grade meningiomas and 14q deletions are more likely in benign recurrent meningiomas.64 It has been postulated that mutations in WRN and other DNA helicases may result in such chromosomal changes, given their role in proper DNA replication and repair. WRN silencing through promoter methylation has been investigated.66 Li et al66 found that meningiomas had a significantly higher WRN methylation rate than did healthy arachnoid control tissue. Subsequently, WRN was expressed significantly less in meningioma tissue than in normal arachnoid tissue. WRN promoter hypermethylation has been investigated in other cancers, and was found to be most prevalent in colorectal, non-small cell lung, gastric, prostate, and thyroid cancers.67 In addition, loss of WRN function led to chromosomal instability in these cancers. Given that many sporadic meningiomas have DNA changes that could be related to dysfunction of WRN or WRN-like proteins, the role of DNA repair and replication enzymes in sporadic meningiomas may be a relevant topic for further investigation.
FIGURE 3.
Mutations involved in meningioma progression. Gain of function mutations shown in red, loss of function mutations in black. Familial mutations are shown on the top, sporadic mutations and chromosomal changes are shown on the bottom.
BAP1 Tumor Predisposition Syndrome
The oncogenicity of the BAP1 gene was originally discovered in the genomic assessment of families with uveal melanoma and malignant mesothelioma.68 Further studies on BAP1 indicated that several other neoplasms including meningioma are included in the syndrome (Table 6). A hallmark feature of these patients is the presence of melanocytic BAP1-mutated atypical intradermal tumors (MBAITs).69 MBAITs are skin-colored, dome-shaped papules that appear in up to two-thirds of carriers, typically before other associated malignancies develop. Given that this is a more recently discovered syndrome, prevalence and penetrance data are scarce, and the phenotypic spectrum of associated tumors continues to expand.68
TABLE 6.
Tumors Associated With BAP1 Tumor Predisposition Syndrome
| Uveal melanoma | |
| Malignant mesothelioma | |
| Renal cell carcinoma | |
| Cutaneous melanoma | |
| Meningioma | |
| Melanocytic bap1-mutated atypical intradermal tumors |
BAP1 is located on chromosome 3p21.1 and encodes the BRCA1-associated protein 1, which functions in transcription, chromatin modification, and DNA damage response. Part of its tumor suppressive function is related to its interaction with BRCA1, which plays an important role in the repair of double-stranded DNA breaks and has been well described in breast cancer predisposition and development.68,70
Meningiomas were identified as part of the BAP1 tumor predisposition syndrome when examining a group of patients with uveal melanoma.71 In this study, a germline truncating mutation of BAP1 was identified in 5 individuals of the same family. One family member had a diagnosis of meningioma, and subsequent tumor tissue analysis revealed biallelic inactivation of BAP1. The BAP1 gene has also been found to be inactivated in a subset of high-grade rhabdoid meningiomas.72 Initially thought to be sporadic tumors, germline BAP1 mutations were identified in some of these patients, further supporting meningiomas as part of the BAP1 tumor predisposition syndrome spectrum. In this series, somatic BAP1 mutations were a predictor of clinically aggressive tumors, highlighting how genetic characterization of meningiomas can guide clinical management. It is also recommended that patients with somatic BAP1-mutated meningiomas be evaluated for BAP1 tumor predisposition syndrome, illustrating an instance where genetic testing in meningiomas should be completed. Germline genetic testing for BAP1 mutations is commercially available. A BAP1 antibody is available for immunohistochemistry (IHC) testing of patient samples, and in the study listed above all BAP1 mutants showed loss of BAP1 expression on IHC.72
SMARCE1
Germline mutations of SMARCE1 have been implicated in several families with familial meningiomatosis.73,74SMARCE1 is also known as BAF57, and is thought to be involved in loss of apoptosis in other types of cancer, including breast, ovarian, and prostate.75 Located on chromosome 17q21, SMARCE1 encodes for a subunit of the SWI/SNF complex, which regulates chromatin structure by nucleosome remodeling.76 Specifically, SMARCE1 is responsible for inducing apoptosis by stimulating expression of CYLD.77
SMARCE1 mutations have been investigated in the germline of families with a history of meningiomas as well as sporadic tumors.73 Smith et al73 examined SMARCE1 mutations in a group of patients with spinal meningiomas and a positive family history of meningiomas that tested negative for germline mutations in NF2. Whole exome sequencing with Sanger confirmation revealed heterozygous loss-of-function mutations in SMARCE1 in these patients. The histology on all of these tumors was of the clear-cell subtype, and the loss of function phenotype was thought to be consistent with a tumor suppressor mechanism. The authors concluded that the SWI/SNF complex plays a key role in the pathogenesis of meningiomas, and that they had identified a new discrete entity with a genetic basis for multiple meningiomas. This work has been further corroborated by the identification of a family with a germline SMARCE1 mutation and cranial meningiomas.74 Additionally, in 1 cohort of patients less than 25 years of age with a solitary meningioma, germline SMARCE1 mutations were identified in 14% (9/63) of patients.78 All affected individuals also had meningiomas of the clear cell type. These studies have important clinical management implications when a clear cell meningioma is encountered in clinical practice, especially in young patients or those with a family history of meningiomas. Germline genetic testing for SMARCE1 mutations should be considered in this subset of patients as part of their work-up. Individuals with an identified mutation should be followed more closely given their predilection for multiple lesions, and their family members should be offered predictive genetic testing for risk stratification. IHC investigating the SMARCE1 status of patient samples has been previously performed, and loss of SMARCE1 protein staining appears specific to clear cell histology.79,80 However, IHC and genomic profiling of tumor specimens do not routinely include evaluation of SMARCE1 at this time.
SMARCB1
Similar to SMARCE1, SMARCB1 (also known as INI1) encodes a protein that serves as a subunit of the SWI/SNF complex.81 It is located on chromosome 22q11.23, in close proximity to the NF2 gene. Germline inactivating mutations of SMARCB1 have been found to predispose patients to several cancers including malignant rhabdoid tumors and schwannomatosis, both independently and as part of a 4-hit mechanism with NF2.82-84
In a study examining a family with schwannomatosis and a germline SMARCB1 mutation, multiple family members were also noted to have 1 or more meningiomas, independent of an NF2 mutation.85 In a similar fashion to schwannomatosis, SMARCB1 mutations have been described as part of a 4-hit mechanism in a family with multiple meningiomas with a germline SMARCB1 mutation and somatic NF2 mutations.86 In this particular group of patients, this complement of mutations appeared to preferentially produce meningiomas of the falx cerebri. This study also screened newly found carriers of the SMARCB1 mutation for intracranial lesions by magnetic resonance imaging of the brain and found that 7 of 11 these patients had asymptomatic lesions. All of these lesions appeared to be meningiomas, and all of them were attached to falx. In a patient with a falcine meningioma and a family history of meningioma, germline genetic testing for SMARCB1 mutations should be considered, which would also allow for the subsequent risk stratification of family members. IHC is also readily available to test tissue samples for loss of SMARCB1(INI1) expression. However, the identified mutations in the studies above were missense mutations and it is unclear if they will affect overall protein abundance, potentially limiting the value of IHC.
The role of SMARCB1 has been investigated in sporadic tumors as well. In 1 series of sporadic meningiomas, somatic mutations in exon 9 of SMARCB1 were noted in 3% of tumors.87 An insertional mutation in exon 9 of SMARCB1 was also noted in 1 patient in an 80 patient series of sporadic meningiomas by a separate group.88
Other Familial Syndromes
Other syndromes including MEN1 and Rubinstein-Taybi syndrome have been reported to be associated with meningiomas, but have only been demonstrated in case reports and case series.89,90
DISCUSSION
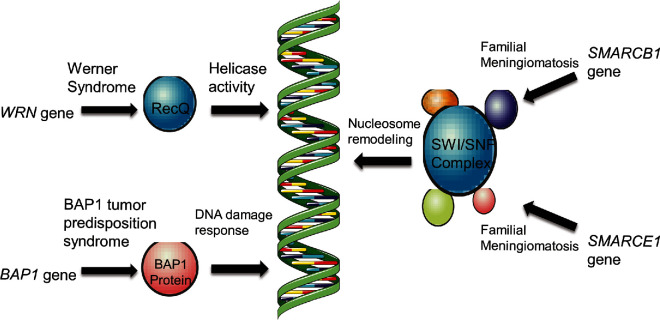
A set of common themes emerges when looking at the work done on familial disease and the extension of that work to examine sporadic cases. With the notable exception of NF2, most of these genes are not commonly mutated in sporadic disease. However, they are all involved in pathways or processes that appear essential to meningioma formation. These pathways include or involve SHH, AKT-PI3K-mTOR, and YAP/TAZ. SHH pathway mutations lead to uncontrolled cell growth and proliferation in many cancers, while AKT-PI3K-mTOR pathway malfunction leads mainly to increased survival.23,40 YAP/TAZ signaling dysfunction through merlin mutations produces increased proliferation through the loss of contact inhibition.12 Merlin is also involved with regulation of the mTOR pathway, a potential point of convergence in these important pathways in meningioma pathogenesis.91 Another common theme between the familial syndromes presented is that the protein products of the mutated genes commonly function in DNA maintenance and repair (Figure 4). This is particularly interesting given the chromosomal copy number and other large-scale genetic changes that occur in meningioma. These changes are even more prevalent in higher-grade tumors, suggesting an essential role in the pathogenesis of meningioma progression. While it is known that these changes occur, the exact mechanism by which they occur and the ultimate effects of these changes remains to be elucidated.
FIGURE 4.
Meningioma-associated mutations in genes involving chromatin structure and the DNA damage response. WRN codes for a protein that is a member of the RecQ family. Its helicase function serves broad functional roles in normal DNA metabolism as well as DNA repair. The BAP1 protein is involved in the DNA damage response, specifically functioning in double-stranded break repair. The SMARCB1 and SMARCE1 genes code for components of the SWI/SNF complex, which participates in chromatin remodeling to regulate transcription.
This work supports the strategy of examining familial syndromes to identify candidate genes and pathways for further study in sporadic cases. The relationship of SMARCE1 mutations to the clear cell subtype and BAP1 mutations to the rhabdoid subtype demonstrate how familial studies also provide insight into the specific histopathologic subtypes of meningioma. The histologic grading of meningiomas is challenging given the multitude of types, and the discordance between histology and clinical behavior lessens its utility. Studies in which specific genetic mutations are associated with specific histologies are the beginning of a potential molecular classification of these tumors that could be used to better predict patient outcomes. Epigenetic classification schemes are also a potential method for classification, with a recently proposed scheme based on DNA methylation for meningiomas being an example.92 The authors found that methylation patterns profiled meningiomas into 2 major groups with several subclasses and that this profiling better predicted recurrence and prognosis than the current WHO classification. The authors included the familial genes SMARCE1, SMARCB1, PTEN, SUFU, and NF2 in their analysis of the subtypes of their classification scheme, with NF2 and SUFU being the only genes that segregated significantly. Investigating the other familial genes discussed in this review could have provided the authors further information. We agree with the authors and WHO that given the complex nature of these tumors, an amalgamated approach combining histology, genetics, epigenetics, and clinical findings will provide the best system for classification. The results of studies on familial syndromes combined with large-scale genetic studies on sporadic meningioma leave up to 20% of meningiomas without a genetic basis. Given the comprehensive examination of the genetic landscape of meningioma that these studies provide, one hypothesis that could be derived is that the remaining causes of meningioma lay outside of genetic mutations. Studies examining gene expression profiles, epigenetic regulation, microRNA, and long noncoding RNA will be important to gain a better understanding of meningioma pathogenesis.
CONCLUSION
Here we present a comprehensive review of the familial syndromes associated with meningiomas. Details of the implicated genes and their associated pathways are given to facilitate further understanding of the molecular pathogenesis of meningiomas and to support future research. As many of the studies on these familial syndromes have shown, the molecular understanding of these syndromes often leads to investigations that broaden our understanding of sporadic disease.
Disclosure
The authors have no personal, financial, or institutional interest in any of the drugs, materials, or devices described in this article.
Supplementary Material
Acknowledgments
Meenakshi B. Bhattarcharjee, MD for her pathology slides for Figure 3 and Leomar Y. Ballester, MD, PhD for his assistance in discussion of the clinical pathology elements of the manuscript.
Supplemental Digital Content. Appendix. List of Included Studies. This list provides the studies that were included after the literature review with the provided search terms.
COMMENT
This manuscript presents a summary of known genomic drivers of familial meningiomas and how they help in dissecting the pathogenesis of sporadic cases.
Stephen Yip
Vancouver, Canada
REFERENCES
- 1. McNeill KA. Epidemiology of brain tumors. Neurol Clin. 2016;34(4):981-998. [DOI] [PubMed] [Google Scholar]
- 2. Ostrom QT, Gittleman H, Liao P et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro Oncol. 2014;16(suppl 4):iv1-iv63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choy W, Kim W, Nagasawa D et al. The molecular genetics and tumor pathogenesis of meningiomas and the future directions of meningioma treatments. Neurosurg Focus. 2011;30(5):E6. [DOI] [PubMed] [Google Scholar]
- 4. Ruttledge MH, Sarrazin J, Rangaratnam S et al. Evidence for the complete inactivation of the NF2 gene in the majority of sporadic meningiomas. Nat Genet. 1994;6(2):180-184. [DOI] [PubMed] [Google Scholar]
- 5. Clark VE, Erson-Omay EZ, Serin A et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abedalthagafi M, Bi WL, Aizer AA et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. 2016;18(5):649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans DG, Baser ME, O’Reilly B et al. Management of the patient and family with neurofibromatosis 2: A consensus conference statement. Br J Neurosurg. 2005;19(1):5-12. [DOI] [PubMed] [Google Scholar]
- 8. Asthagiri AR, Parry DM, Butman JA et al. Neurofibromatosis type 2. Lancet. 2009;373(9679):1974-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rouleau GA, Merel P, Lutchman M et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature. 1993;363(6429):515-521. [DOI] [PubMed] [Google Scholar]
- 10. Rong R, Tang X, Gutmann DH, Ye K. Neurofibromatosis 2 (NF2) tumor suppressor merlin inhibits phosphatidylinositol 3-kinase through binding to PIKE-L. Proc Natl Acad Sci USA. 2004;101(52):18200-18205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Plouffe SW, Hong AW, Guan KL. Disease implications of the Hippo/YAP pathway. Trends Mol Med. 2015;21(4):212-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Curto M, Cole BK, Lallemand D, Liu CH, McClatchey AI. Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. J Cell Biol. 2007;177(5):893-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scherer SS, Gutmann DH. Expression of the neurofibromatosis 2 tumor supp-ressor gene product, merlin, in schwann cells. J Neurosci Res. 1996;46(5):595-605. [DOI] [PubMed] [Google Scholar]
- 14. Wiederhold T, Lee MF, James M et al. Magicin, a novel cytoskeletal protein associates with the NF2 tumor suppressor merlin and Grb2. Oncogene. 2004;23(54):8815-8825. [DOI] [PubMed] [Google Scholar]
- 15. Gorlin RJ, Goltz RW. Multiple nevoid basal-cell epithelioma, jaw cysts and bifid rib. N Engl J Med. 1960;262(18):908-912. [DOI] [PubMed] [Google Scholar]
- 16. Lo Muzio L. Nevoid basal cell carcinoma syndrome (gorlin syndrome). Orphanet J Rare Dis. 2008;3(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Evans DG, Howard E, Giblin C et al. Birth incidence and prevalence of tumor-prone syndromes: estimates from a UK family genetic register service. Am J Med Genet. 2010;152A(2):327-332. [DOI] [PubMed] [Google Scholar]
- 18. Mortimer PS, Geaney DP, Liddell K, Dawber RP. Basal cell naevus syndrome and intracranial meningioma. J Neurol Neurosurg Psychiatry. 1984;47(2):210-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kimonis VE, Mehta SG, Digiovanna JJ, Bale SJ, Pastakia B. Radiological features in 82 patients with nevoid basal cell carcinoma (NBCC or gorlin) syndrome. Genet Med. 2004;6(6):495-502. [DOI] [PubMed] [Google Scholar]
- 20. Kimonis VE, Goldstein AM, Pastakia B et al. Clinical manifestations in 105 persons with nevoid basal cell carcinoma syndrome. Am J Med Genet. 1997;69(3):299-308. [PubMed] [Google Scholar]
- 21. Farndon PA, Del Mastro RG, Evans DG, Kilpatrick MW. Location of gene for gorlin syndrome. Lancet. 1992;339(8793):581-582. [DOI] [PubMed] [Google Scholar]
- 22. Evans DG, Oudit D, Smith MJ et al. First evidence of genotype-phenotype correlations in Gorlin syndrome. J Med Genet. 2017;54(8):530-536. [DOI] [PubMed] [Google Scholar]
- 23. Hatton BA, Villavicencio EH, Tsuchiya KD et al. The Smo/Smo model: Hedgehog-induced medulloblastoma with 90% incidence and leptomeningeal spread. Cancer Res. 2008;68(6):1768-1776. [DOI] [PubMed] [Google Scholar]
- 24. Epstein EH. Basal cell carcinomas: Attack of the hedgehog. Nat Rev Cancer. 2008;8(10):743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pak E, Segal RA. Hedgehog signal transduction: Key players, oncogenic drivers, and cancer therapy. Dev Cell. 2016;38(4):333-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nitzki F, Becker M, Frommhold A, Schulz-Schaeffer W, Hahn H. Patched knockout mouse models of Basal cell carcinoma. J Skin Cancer. 2012;2012:907543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277(5329):1109-1113. [DOI] [PubMed] [Google Scholar]
- 28. Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001;15(23):3059-3087. [DOI] [PubMed] [Google Scholar]
- 29. Teglund S, Toftgard R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 2010;1805(2):181-208. [DOI] [PubMed] [Google Scholar]
- 30. Kijima C, Miyashita T, Suzuki M, Oka H, Fujii K. Two cases of nevoid basal cell carcinoma syndrome associated with meningioma caused by a PTCH1 or SUFU germline mutation. Familial Cancer. 2012;11(4):565-570. [DOI] [PubMed] [Google Scholar]
- 31. Boetto J, Bielle F, Sanson M, Peyre M, Kalamarides M. SMO mutation status defines a distinct and frequent molecular subgroup in olfactory groove meningiomas. Neuro Oncol. 2017;19(3):345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aavikko M, Li SP, Saarinen S et al. Loss of SUFU function in familial multiple meningioma. Am J Hum Genet. 2012;91(3):520-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blumenthal GM, Dennis PA. PTEN hamartoma tumor syndromes. Eur J Hum Genet. 2008;16(11):1289-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eng C. Will the real cowden syndrome please stand up: revised diagnostic criteria. J Med Genet. 2000;37(11):828-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Starink TM, van der Veen JP, Arwert F et al. The cowden syndrome: A clinical and genetic study in 21 patients. Clin Genet. 1986;29(3):222-233. [DOI] [PubMed] [Google Scholar]
- 36. Nelen MR, Kremer H, Konings IB et al. Novel PTEN mutations in patients with Cowden disease: absence of clear genotype-phenotype correlations. Eur J Hum Genet. 1999;7(3):267-273. [DOI] [PubMed] [Google Scholar]
- 37. Yakubov E, Ghoochani A, Buslei R, Buchfelder M, Eyupoglu IY, Savaskan N. Hidden association of cowden syndrome, PTEN mutation and meningioma frequency. Oncoscience. 2016;3(5-6):149-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Asahina A, Fujita H, Omori T, Kai H, Yamamoto M, Mii K. Proteus syndrome complicated by multiple spinal meningiomas. Clin Exp Dermatol. 2008;33(6):729-732. [DOI] [PubMed] [Google Scholar]
- 39. Gilbert-Barness E, Cohen MM Jr, Opitz JM. Multiple meningiomas, craniofacial hyperostosis and retinal abnormalities in proteus syndrome. Am J Med Genet. 2000;93(3):234-240. [DOI] [PubMed] [Google Scholar]
- 40. Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13(5):283-296. [DOI] [PubMed] [Google Scholar]
- 41. Li J, Yen C, Liaw D et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943-1947. [DOI] [PubMed] [Google Scholar]
- 42. Wei Q, Clarke L, Scheidenhelm DK et al. High-grade glioma formation results from postnatal pten loss or mutant epidermal growth factor receptor expression in a transgenic mouse glioma model. Cancer Res. 2006;66(15):7429-7437. [DOI] [PubMed] [Google Scholar]
- 43. Kwon CH, Zhao D, Chen J et al. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 2008;68(9):3286-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weng LP, Smith WM, Brown JL, Eng C. PTEN inhibits insulin-stimulated MEK/MAPK activation and cell growth by blocking IRS-1 phosphorylation and IRS-1/Grb-2/Sos complex formation in a breast cancer model. Hum Mol Genet. 2001;10(6):605-616. [DOI] [PubMed] [Google Scholar]
- 45. Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280(5369):1614-1617. [DOI] [PubMed] [Google Scholar]
- 46. Lamszus K, Kluwe L, Matschke J, Meissner H, Laas R, Westphal M. Allelic losses at 1p, 9q, 10q, 14q, and 22q in the progression of aggressive meningiomas and undifferentiated meningeal sarcomas. Cancer Genet Cytogenet. 1999;110(2):103-110. [DOI] [PubMed] [Google Scholar]
- 47. Peters N, Wellenreuther R, Rollbrocker B et al. Analysis of the PTEN gene in human meningiomas. Neuropathol Appl Neurobiol. 1998;24(1):3-8. [DOI] [PubMed] [Google Scholar]
- 48. Louis DN, Ohgaki H, Wiestler OD et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bleeker FE, Felicioni L, Buttitta F et al. AKT1(E17K) in human solid tumours. Oncogene. 2008;27(42):5648-5650. [DOI] [PubMed] [Google Scholar]
- 50. Abedalthagafi M, Bi WL, Aizer AA et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. 2016;18(5):649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Epstein CJ, Martin GM, Schultz AL, Motulsky AG. A review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine. 1966;45(3):177-221. [DOI] [PubMed] [Google Scholar]
- 52. Yu CE, Oshima J, Fu YH et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272(5259):258-262. [DOI] [PubMed] [Google Scholar]
- 53. Takemoto M, Mori S, Kuzuya M et al. Diagnostic criteria for werner syndrome based on japanese nationwide epidemiological survey. Geriatr Gerontol Int. 2013;13(2):475-481. [DOI] [PubMed] [Google Scholar]
- 54. Yokote K, Chanprasert S, Lee L et al. WRN mutation update: mutation spectrum, patient registries, and translational prospects. Human Mutat. 2017;38(1):7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Satoh M, Imai M, Sugimoto M, Goto M, Furuichi Y. Prevalence of Werner's syndrome heterozygotes in Japan. Lancet. 1999;353(9166):1766. [DOI] [PubMed] [Google Scholar]
- 56. Lauper JM, Krause A, Vaughan TL, Monnat RJ Jr.. Spectrum and risk of neoplasia in werner syndrome: a systematic review. PLoS One. 2013;8(4):e59709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in werner syndrome (adult progeria). Cancer Epidemiol Biomarkers Prev. 1996;5(4):239-246. [PubMed] [Google Scholar]
- 58. Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 2010;18(2):177-187. [DOI] [PubMed] [Google Scholar]
- 59. Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83(1):519-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Oshima J, Campisi J, Tannock TC, Martin GM. Regulation of c-fos expression in senescing werner syndrome fibroblasts differs from that observed in senescing fibroblasts from normal donors. J Cell Physiol. 1995;162(2):277-283. [DOI] [PubMed] [Google Scholar]
- 61. Melcher R, von Golitschek R, Steinlein C et al. Spectral karyotyping of werner syndrome fibroblast cultures. Cytogenet Genome Res. 2000;91(1-4):180-185. [DOI] [PubMed] [Google Scholar]
- 62. Oshima J, Sidorova JM, Monnat RJ. Werner Syndrome: Clinical Features, Pathogenesis and Potential Therapeutic Interventions. Ageing research reviews. 2017;33:105-114. doi:10.1016/j.arr.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bi WL, Zhang M, Wu WW, Mei Y, Dunn IF. Meningioma genomics: diagnostic, prognostic, and therapeutic applications. Front Surg. 2016;3:40 doi: 10.3389/fsurg.2016.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cai DX, Banerjee R, Scheithauer BW, Lohse CM, Kleinschmidt-Demasters BK, Perry A. Chromosome 1p and 14q FISH analysis in clinicopathologic subsets of meningioma: diagnostic and prognostic implications. J Neuropathol Exp Neurol. 2001;60(6):628-636. [DOI] [PubMed] [Google Scholar]
- 65. Weber RG, Bostrom J, Wolter M et al. Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci USA. 1997;94(26):14719-14724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li P, Hao S, Bi Z, Zhang J, Wu Z, Ren X. Methylation of werner syndrome protein is associated with the occurrence and development of invasive meningioma via the regulation of myc and p53 expression. Exp Ther Med. 2015;10(2):498-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Agrelo R, Cheng WH, Setien F et al. Epigenetic inactivation of the premature aging werner syndrome gene in human cancer. Proc Natl Acad Sci USA. 2006;103(23):8822-8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Murali R, Wiesner T, Scolyer RA. Tumours associated with BAP1 mutations. Pathology. 2013;45(2):116-126. [DOI] [PubMed] [Google Scholar]
- 69. Haugh AM, Njauw CN, Bubley JA et al. Genotypic and phenotypic features of BAP1 cancer syndrome. JAMA Dermatol 2017;(10). doi: 10.1001/jamadermatol.2017.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Johannsson OT, Idvall I, Anderson C et al. Tumour biological features of BRCA1-induced breast and ovarian cancer. Eur J Cancer. 1997;33(3):362-371. [DOI] [PubMed] [Google Scholar]
- 71. Abdel-Rahman MH, Pilarski R, Cebulla CM et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48(12):856-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shankar GM, Abedalthagafi M, Vaubel RA et al. Germline and somatic BAP1 mutations in high-grade rhabdoid meningiomas. Neuro Oncol. 2017;19(4):535-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Smith MJ, O'Sullivan J, Bhaskar SS et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet. 2013;45(3):295-298. [DOI] [PubMed] [Google Scholar]
- 74. Gerkes EH, Fock JM, den Dunnen WF et al. A heritable form of SMARCE1-related meningiomas with important implications for follow-up and family screening. Neurogenetics. 2016;17(2):83-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Garcia-Pedrero JM, Kiskinis E, Parker MG, Belandia B. The SWI/SNF chromatin remodeling subunit BAF57 is a critical regulator of estrogen receptor function in breast cancer cells. J Biol Chem. 2006;281(32):22656-22664. [DOI] [PubMed] [Google Scholar]
- 76. Kazantseva A, Sepp M, Kazantseva J et al. N-terminally truncated BAF57 isoforms contribute to the diversity of SWI/SNF complexes in neurons. J Neurochem. 2009;109(3):807-818. [DOI] [PubMed] [Google Scholar]
- 77. Wang L, Baiocchi RA, Pal S, Mosialos G, Caligiuri M, Sif S. The BRG1- and hBRM-associated factor BAF57 induces apoptosis by stimulating expression of the cylindromatosis tumor suppressor gene. Mol Cell Biol. 2005;25(18):7953-7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pathmanaban ON, Sadler KV, Kamaly-Asl ID et al. Association of genetic predisposition with solitary schwannoma or meningioma in children and young adults. JAMA Neurol. 2017;(9). doi: 10.1001/jamaneurol.2017.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Smith MJ, Ahn S, Lee JI, Bulman M, Plessis DD, Suh YL. SMARCE1 mutation screening in classification of clear cell meningiomas. Histopathology. 2017;70(5):814-820. [DOI] [PubMed] [Google Scholar]
- 80. Tauziede-Espariat A, Parfait B, Besnard A et al. Loss of SMARCE1 expression is a specific diagnostic marker of clear cell meningioma: A comprehensive immunophenotypical and molecular analysis. Brain Pathol. 2017. doi: 10.1111/bpa.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Smith MJ, Wallace AJ, Bowers NL et al. Frequency of SMARCB1 mutations in familial and sporadic schwannomatosis. Neurogenetics. 2012;13(2):141-145. [DOI] [PubMed] [Google Scholar]
- 82. Hadfield KD, Newman WG, Bowers NL et al. Molecular characterisation of SMARCB1 and NF2 in familial and sporadic schwannomatosis. J Med Genet. 2008;45(6):332-339. [DOI] [PubMed] [Google Scholar]
- 83. Hulsebos TJ, Plomp AS, Wolterman RA, Robanus-Maandag EC, Baas F, Wesseling P. Germline mutation of INI1/SMARCB1 in familial schwannomatosis. Am J Hum Genet. 2007;80(4):805-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sevenet N, Sheridan E, Amram D, Schneider P, Handgretinger R, Delattre O. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet. 1999;65(5):1342-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bacci C, Sestini R, Provenzano A et al. Schwannomatosis associated with multiple meningiomas due to a familial SMARCB1 mutation. Neurogenetics. 2010;11(1):73-80. [DOI] [PubMed] [Google Scholar]
- 86. van den Munckhof P, Christiaans I, Kenter SB, Baas F, Hulsebos TJ. Germline SMARCB1 mutation predisposes to multiple meningiomas and schwannomas with preferential location of cranial meningiomas at the falx cerebri. Neurogenetics. 2012;13(1):1-7. [DOI] [PubMed] [Google Scholar]
- 87. Schmitz U, Mueller W, Weber M, Sevenet N, Delattre O, von Deimling A. INI1 mutations in meningiomas at a potential hotspot in exon 9. Br J Cancer. 2001;84(2):199-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rieske P, Zakrzewska M, Piaskowski S et al. Molecular heterogeneity of meningioma with INI1 mutation. Mol Pathol. 2003;56(5):299-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Asgharian B, Chen YJ, Patronas NJ et al. Meningiomas may be a component tumor of multiple endocrine neoplasia type 1. Clin Cancer Res. 2004;10(3):869-880. [DOI] [PubMed] [Google Scholar]
- 90. Verstegen MJ, van den Munckhof P, Troost D, Bouma GJ. Multiple meningiomas in a patient with Rubinstein-Taybi syndrome. J Neurosurg. 2005;102(1):167-168. [DOI] [PubMed] [Google Scholar]
- 91. James MF, Han S, Polizzano C et al. NF2/merlin is a novel negative regulator of mTOR complex 1, and activation of mTORC1 is associated with meningioma and schwannoma growth. Mol Cell Biol. 2009;29(15):4250-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sahm F, Schrimpf D, Stichel D et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18(5):682-694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.