Abstract
Background
Usher syndrome is a genetically heterogeneous disorder featured by combined visual impairment and hearing loss. Despite a dozen of genes involved in Usher syndrome having been identified, the genetic basis remains unknown in 20–30% of patients. In this study, we aimed to identify the novel disease-causing gene of a distinct subtype of Usher syndrome.
Methods
Ophthalmic examinations and hearing tests were performed on patients with Usher syndrome in two consanguineous families. Target capture sequencing was initially performed to screen causative mutations in known retinal disease-causing loci. Whole exome sequencing (WES) and whole genome sequencing (WGS) were applied for identifying novel disease-causing genes. RT-PCR and Sanger sequencing were performed to evaluate the splicing-altering effect of identified CEP78 variants.
Results
Patients from the two independent families show a mild Usher syndrome phenotype featured by juvenile or adult-onset cone–rod dystrophy and sensorineural hearing loss. WES and WGS identified two homozygous rare variants that affect mRNA splicing of a ciliary gene CEP78. RT-PCR confirmed that the two variants indeed lead to abnormal splicing, resulting in premature stop of protein translation due to frameshift.
Conclusions
Our results provide evidence that CEP78 is a novel disease-causing gene for Usher syndrome, demonstrating an additional link between ciliopathy and Usher protein network in photoreceptor cells and inner ear hair cells.
INTRODUCTION
Primary cilium is a microtubule-based sensory organelle originating from centrioles. It widely exists in multiple cell types of human body. Primary cilium plays important roles in various cell signalling processes, thus essential for normal development and maintenance of tissue functioning.1,2 The cilia proteome is highly heterogeneous and dynamic, containing dozens of proteins involved in ciliary transport, structural maintenance, as well as a number of tissue-specific cargo proteins. Correspondingly, mutations in these ciliary proteins cause a wide range of genetic disorders collectively referred to as ciliopathy. Defects in primary cilia lead to a series of clinical phenotypes such as retinal dystrophy, hearing loss, neurodevelopmental defects and kidney disease.3
Usher syndrome is a genetic disorder characterised by combined retinitis pigmentosa and sensorineural hearing loss. It is caused by disruption of protein components in the supramolecular Usher protein network. There is growing evidence showing that this protein network is associated with cilia proteome.4 In retina photoreceptor cells, the Usher protein complex has been found to loca-lise at the periciliary region, which is critical for structural maintenance and ciliary transport.5 In addition, some specific interactions between Usher protein components and ciliary proteins were also identified, including DFNB31-RPGR and USH1G-CEP290.4,6 Usher syndrome has a prevalence ranging from 3.2 to 6.2 in 100 000,7–9 and it can be classified into three major categories, namely USH I, II and III, depending on the age of onset and severity. Until now, 13 genes have been associated with Usher syndrome (RetNet, the Retinal Information Network).10 However, mutations in these genes can only account for 70–80% of cases,11–13 suggesting additional disease-causing loci are yet to be identified.
In this study, by whole exome sequencing (WES) and whole genome sequencing (WGS), we identified mutations in CEP78, a ciliary gene,14 in two independent consanguineous families presenting a distinct Usher syndrome phenotype featured by cone–rod dystrophy and progressive sensorineural hearing loss. One allele affects the canonical splicing acceptor site of exon 14 and the other allele locates adjacent to the exon 10 splicing donor site. RT-PCR and Sanger sequencing confirmed that the two mutations cause splicing defects and lead to premature stop codons. Therefore, these results linked a new gene to Usher syndrome and provided more hints for understanding Usher/ciliary protein network in photoreceptor cells and inner ear hair cells.
METHODS
Clinical diagnosis
The patients in Family 1 were ascertained at the Department of Ophthalmology, Peking Union Medical College Hospital, Beijing, China. They underwent detailed ophthalmic evaluation, including best correct visual acuity (BCVA), slit-lamp bio-microscopy, dilated indirect ophthalmoscopy, fundus photography, visual field tests (Octopus, Interzeag, Schlieren, Switzerland), optical coherence tomography (OCT) (Heidelberg HRT II, Heidelberg, Germany) and electroretinograms (ERGs) (RetiPort ERG system, Roland Consult, Wiesbaden, Germany) using corneal ‘ERGjet’ contact lens electrodes. Auditory examinations including pure tone and speech audiometry were conducted by otolaryngologists. The patients in Family 2 were recruited at the Department of Ophthalmology, Ningxia Eye Hospital, Ningxia People’s Hospital. They also underwent complete ophthalmic examinations, including assessments of BCVA, slit-lamp examination, funduscopy, colour vision test, visual field tests (Humphrey Perimetry, ZEISS, Germany), ERG, OCT and fundus fluorescein angiography (FFA) examinations. Genomic DNA was isolated from peripheral leucocytes using QIAamp DNA Blood Midi Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. This study adhered to the Tenets of the Declaration of Helsinki.
Target capture sequencing, WES, WGS and bioinformatic analysis
Precapture Illumina libraries were generated as in previous literature.15–17 The targeted DNA was captured, washed and recovered using Agilent Hybridization and Wash Kits (Agilent Technologies, Santa Clara, California, USA). The genes included in target capture panel were listed in previous literature.18 WES was performed by capturing the DNA with NimbleGenSeqCap EZ Hybridization and Wash Kit. Captured DNA libraries were sequenced on Illumina HiSeq 2000 (Illumina, San Diego, California, USA). For WGS, the DNA was prepared using the TruSeq Nano DNA HT Sample Prep Kit (Illumina) and the sequencing was performed on Hiseq X Ten (X10) platform (Illumina). After the sequencing step, the reads were obtained and aligned to human hg19 genome using Burrows-Wheeler Aligner (BWA) V.0.6.1.19 Base quality recalibration and local realignment were done by the Genome Analysis Tool Kit V.3.6. Atlas-SNP2 and Atlas-Indel2 were used for variant calling.20 Variant frequency data were obtained from a series of public and internal control databases, including Exome Aggregation Consortium (ExAC) database, CHARGE Consortium,21 ESP-650022 and 1000 Genome Project.23 The consanguineous nature of these two families and the absence of disease pheno-types in other generations strongly suggest an autosomal recessive inheritance model; thus, variants with a minor allele frequency higher than 1/200 were filtered out. Synonymous and deep intronic (distance >10 bp from exon–intron junctions) variants were also excluded from the following analysis. Annotate Variation (ANNOVAR) (V.11/12/2014) and dbNSFP suite (V.2.9, contains SIFT, PolyPhen-2, LRT, MutationTaster, MutationAssessor, etc) were used to annotate protein-altering changes. Known retinal disease-causing alleles were detected based on the Human Gene Mutation Database (HGMD) Professional database (V.11/15/2014).
RT-PCR and Sanger sequencing
Total RNAs were extracted from the periphery blood samples of the patients and unaffected control subjects, using the QIAamp RNA Blood Mini Kit (QIAGEN). cDNA were synthesised from 8 mg total RNA using GoScript Reverse Transcription System (Promega, Madison, USA) according to manufacturer’s protocol. RT-PCR primers were designed on Primer 3 software (http://bioinfo.ut.ee/primer3-0.4.0/): RT-exon-9F (5′-GTGTCTGGTTTCTTGCCGTG-3′); RT-exon-11R (5′-GGCACTTCATGAACAC TCTCT-3′); RT-exon13F (5′-GCCCTTCATGCACAGTCATT-3′); RT-exon16R (5′-ACAGGAAAGGAGTCGAGAGG-3′). The 50 μL reaction system contained 40 ng cDNA, 10 pmol of each primer and 25 μL 2×Taq PCR Master Mix (TransGen Biotech, Beijing, China). DNA amplifications were performed with denaturing at 95°C for 5 min, followed by 33 cycles of a denaturing step at 95° C, an annealing step at 60°C and an extension step at 72°C, each for 30 s. A final extension step at 72°C was performed for 7 min. After purification, amplicons were sequenced using forward and reverse primers on an ABI 3730 Genetic Analyzer (ABI, Foster City, California, USA). Sequences were assembled and analysed using Lasergene SeqMan software (DNASTAR, Madison, Wisconsin, USA).
RESULTS
Clinical findings
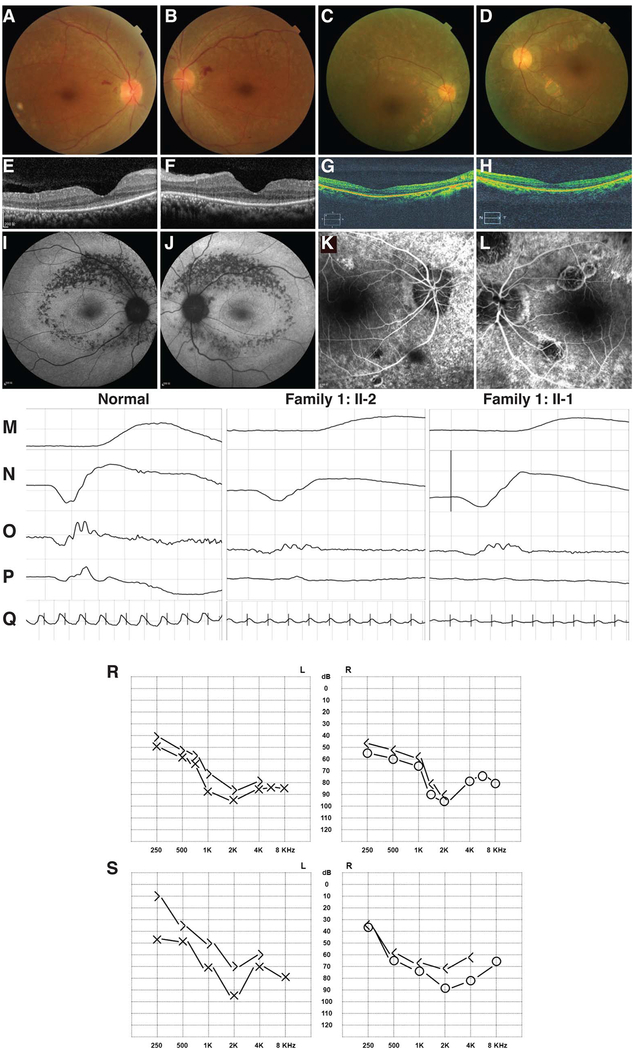
Family 1 is a consanguineous family from Fujian Province of South China with Han Chinese ancestry. The proband is a 33-year-old woman suffering from progressive vision loss for over 20 years. She currently had a visual acuity of counting finger oculus dexter (OD) and hand motion oculus sinister (OS). She also failed colour vision test and presents photophobia. Progressive sensorineural hearing loss was noticed in recent years (figure 1R). The proband has an affected elder sister at the age of 41, with over 30 years of progressive vision loss. She has a visual acuity of 20/1000 OD and 20/400 OS. Photophobia, impaired colour vision and recent hearing loss were also presented. The clinical findings in ERG tests, colour fundus photos, fundus autofluorescence and OCT are similar in two patients: both of them show moderately reduced rod responses and remarkably reduced cone responses in ERG tests (figure 1M–Q). Funduscopy results showed greyish retinal pigments mottling along the retinal vessels with attenuated retinal arteries (figure 1A, B and online supplementary figure S2A, B). The fundus autofluorescence demonstrated annular hypofluorescence patches around the posterior pole with an inner hyperfluores-cent ring around the macular region (figure 1I, J and online supplementary figure S2I, J). OCT results revealed reduced thickness of macular region and disappeared ellipsoid zone (figure 1E, F and online supplementary figure S2E, F). In addition, the proband also presented dilated retinal veins with retinal haemorrhage, probably confounded by her hypertension (170/110 mm Hg). The two patients were diagnosed as a distinct subtype of Usher syndrome, with an autosomal recessive inheritance due to the consanguinity of this family.
Figure 1.
Clinical manifestations of the patients with atypical Usher syndrome. Fundus images of Family 1: II-2, OD (A) and OS (B), Family 2: V-1, OD (C) and OS (D). Optical coherence tomography images of Family 1: II-2, OD (E) and OS (F), Family 2: V-1, OD (G) and OS (H). Fundus autofluorescence results of Family 1: II-2, OD (I) and OS (J). Fundus fluorescein angiography of Family 2: V-1, OD (K) and OS (L). The results of scotopic electroretinograms (ERGs) 0.01 (M), scotopic ERG 0.3 (N), oscillatory potential ERG (O), photopic ERG 3.0 (P) and flicker ERG 30 Hz (Q) of normal control (left panel), Family 1: II-2 (middle panel) and Family 1: II-1 (right panel). Audiogram images of Family 1: II-2 (R) and Family 2: V-1 (S). In the audiogram images, cross or circle labels indicate air-conduction hearing, and right angle labels indicate bone-conduction hearing.
In Family 2, consanguineous marriage was reported, while no family history of visual problem was recorded. The proband developed night blindness since early childhood. She also noticed hypochromatopsia and mild hearing loss since age 8 (figure 1S). She suffered from visual field decrease and central vision loss since age 10. At her latest visit at age 35, her BCVAs dropped to 20/125 OD and 20/200 OS. Hypochromatopsia was confirmed by colour vision test. Funduscopy features include attenuated vessels, waxy optic disc and pigment deposits in the mid-peripheral retina (figure 1C, D). Macular region was also involved. Consistent with the fundus photography, FFA also revealed aberrant vascular arcades and speckled changes of increased fluorescence in the mid-peripheral retina (figure 1K, L). Attenuated outer nucleus layer, retinal pigment epithelium (RPE) and loss of ellipsoid zone were suggested by OCT presentations (figure 1G, H). Both scotopic and photopic ERG responses were abolished (see online supplementary figure S1A–C). She has an affected brother with similar ophthalmo-logical findings (see online supplementary figure S2, C, D, G, H, K, L) as well as hearing loss. According to these abnormalities, the patients in Family 2 were also clinically diagnosed as an atypical Usher syndrome phenotype.
Genetic findings
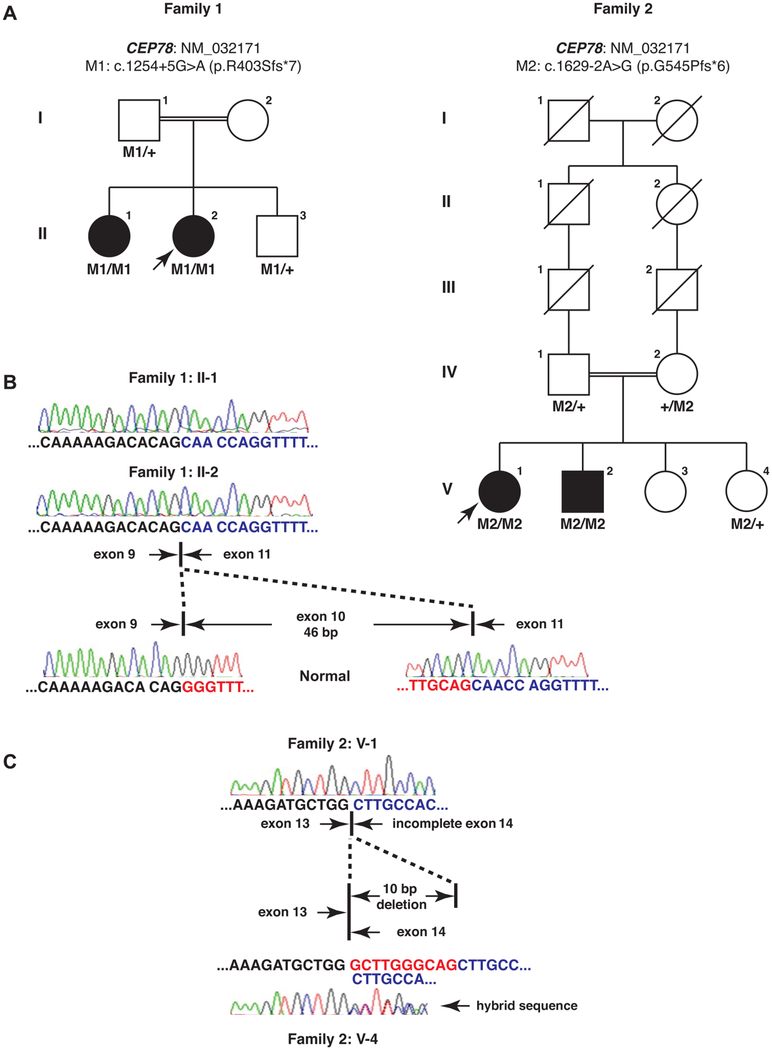
To identify the causative mutation underlying this phenotype, we initially performed target capture sequencing of known retinal disease genes on the proband of Family 1 with high quality (mean coverage: 160.65×; regions with >20× coverage: 95.74%). However, no candidate causative variants were identi-fied, suggesting the possibility that the patient carries mutations in a new disease-causing locus. Therefore, WES was performed on both patients. A mean WES coverage of 86.21× and 99.86× for the proband and her affected sister was obtained, respectively. Upon filtering, 397 and 411 variants were remained and 9 biallelic variants were shared by both patients (see online supplementary table S1). After variant exclusion and prioritisation, one CEP78 homozygous variant was identified as the top candidate due to its possible loss-of-function nature and the functional link between CEP78 and cilia14,24,25 (see online supplementary table S1). This variant (NM_032171, c.1254 +5G>A, p.?) is near the splice donor site of exon 10/intron 10 junction. It is extremely rare in control population with a frequency of 1/30 990 in the ExAC population database. Although this variant is not at the canonical splicing site, two computational methods that predict the splicing-altering effect (AdaBoost and random forests)26 give scores of 0.603 and 0.528, respectively, suggesting that the variant is likely to affect the pre-mRNA splicing of CEP78 transcript. Sanger sequencing was performed to validate the variant and confirm the cosegregation of this variant with disease phenotype in this family (figure 2A and online supplementary figure S3). For Family 2, WGS was performed on the proband as described and a mean coverage of 30× was achieved. A total of 352 rare protein-altering variants were remained after filtering and we also focused on homozygous variants due to the consanguinity (see online supplementary table S2). Interestingly, among them, a homozygous variant that affects the canonical splicing acceptor site of exon 14 (c.1629–2A>G, p.?) of CEP78 was identified. It was not observed in any public control databases. Sanger sequencing also confirmed the genotype–phenotype cosegregation in this family (figure 2A and online supplementary figure S3). Genetic screening in additional 71 unsolved patients with Usher syndrome did not identify additional cases with biallelic CEP78 mutations.
Figure 2.
Genetic findings in the patients with Usher syndrome. (A) CEP78 mutations were identified in two unrelated consanguineous families. The variants cosegregate with the disease phenotype. Arrows indicate probands. (B) RT-PCR and Sanger sequencing confirmed the splicing-altering effect of the variant c.1254+5G>A in Family 1. In two patients’ mRNA, the 46 bp exon 10 is completely skipped compared with normal control. (C) RT-PCR and Sanger sequencing confirmed the splicing-altering effect of the variant c.1629–2A>G in Family 2. In the probands’ mRNA, a 10 bp-long region in CEP78 exon 14 was deleted, while in the unaffected heterozygous carrier, wild-type/mutant hybrid sequence was observed.
To further demonstrate that the two identified CEP78 variants are bona fide splicing-disrupting, we collected the patient blood and performed RT-PCR with Sanger sequencing. In Family 1, we found that in both patients the 46 bp-long exon 10 was completely lost in CEP78 mRNA (figure 2B). For Family 2, the proband has a 10 bp deletion of CEP78 exon 14 (figure 2C), while a heterozygous carrier in this family shows wild-type/mutant hybrid sequence. Therefore, we confirmed that the two variants indeed affect the pre-mRNA splicing in vivo and they both lead to frameshifts and premature stop codons. Based on the Sanger sequencing results, the two variants are reannotated as c.1254+5G>A (p.R403Sfs*7) and c.1629–2A>G (p.G545Pfs*6).
DISCUSSION
CEP78 was originally identified as a centrosomal protein in a proteomic study.27 In planarians, RNAi knockdown of Smed-cep78 led to centriole-anchoring defects and slow locomotion of animals.24 Furthermore, CEP78 knockdown in human RPE1 cells causes a significant reduction of ciliated cell number,24 suggesting the functionally conserved and essential role of CEP78 in ciliogenesis. Further characterisation showed that CEP78 localises at the distal end of centriole and the leucine-rich repeats are critical for its localisation.25 The CEP78 localisation data are consistent with the functional linkage between centriole distal end and primary ciliogenesis.28,29
Additionally, some evidence have shown the potential involvement of CEP78 in retinal and hearing abnormalities: CEP78 was identified to interact with cilia protein CEP250,30 and the latter is a known Usher syndrome protein.31 In another study, researchers tried identifying crucial genes which show significantly altered expression in cochlea after noise-induced hearing loss and CEP78 was among the 15 genes with more than five-fold upregulation. These results suggest CEP78 as a strong candidate disease-causing gene in the families we investigated.
Currently, the involvement of Usher protein network in cilia formation and function remains largely elusive.4 The phenotypic overlapping between CEP78 and other Usher syndrome genes would provide more clues for understanding the physical and genetic interactions within this protein network. Specifically, CEP78-associated phenotype has an age of onset similar to or even milder than Usher syndrome type III, suggesting the role of CEP78 in structural and functional maintenance instead of during early development. On the other hand, CEP78 disruption distinctively affects cone photoreceptors earlier, while in all other Usher syndrome types, rod photoreceptor functions are initially compromised.32 This indicates a specialised role of CEP78 in cone photoreceptors.
In summary, our results identified a novel ciliary gene CEP78 involved in Usher syndrome. Further studies on additional cases and animal models would help to describe the CEP78-related phenotype more precisely and reveal the ciliary involvement in Usher protein network in photoreceptor cells and inner ear hair cells, thus pushing forward more efficient disease management and treatment for Usher syndrome.
Supplementary Material
Acknowledgements
We thank the patients and their family members for participating in our study. We thank the ExAC that provided exome variant data.
Funding NGS was conducted at the Functional Genomic Core (FGC) facility at Baylor College of Medicine supported by National Institutes of Health (NIH) shared instrument grant 1S10RR026550 to RC. This work was supported by grants from National Eye Institute (R01EY022356, R01EY018571, EY002520), Retinal Research Foundation, Foundation Fighting Blindness (BR-GE-0613–0618-BCM) to RC, grants from the Foundation Fighting Blindness USA (CD-CL-0808–0470-PUMCH and CDCL-0214–0631-PUMCH), the Ministry of Science and Technology of the People’s Republic of China (Grant No.: 2010DFB33430), National Natural Science Foundation of China (81470669) and Beijing Natural Science Foundation (7152116) to RS, National Key Basic Research Program of China (2013CB967500 to CZ), National Natural Science Foundation of China (81525006 to CZ, 81260154 to XS), Jiangsu Province’s Innovation Team (to CZ), the Fundamental Research Funds of the State Key Laboratory of Ophthalmology (to CZ) and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Footnotes
Competing interests None declared.
Patient consent Obtained.
Ethics approval Institutional Review Board of Baylor College of Medicine.
REFERENCES
- 1.Satir P, Pedersen LB, Christensen ST. The primary cilium at a glance. J Cell Sci 2010;123(Pt 4):499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singla V, Reiter JF. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science 2006;313:629–33. [DOI] [PubMed] [Google Scholar]
- 3.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med 2011;364: 1533–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorusch N, Wunderlich K, Bauss K, Nagel-Wolfrum K, Wolfrum U. Usher syndrome protein network functions in the retina and their relation to other retinal ciliopathies. Adv Exp Med Biol 2014;801:527–33. [DOI] [PubMed] [Google Scholar]
- 5.Maerker T, van Wijk E, Overlack N, Kersten FF, McGee J, Goldmann T, Sehn E, Roepman R, Walsh EJ, Kremer H, Wolfrum U. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum Mol Genet 2008;17:71–86. [DOI] [PubMed] [Google Scholar]
- 6.Wright RN, Hong DH, Perkins B. RpgrORF15 connects to the usher protein network through direct interactions with multiple whirlin isoforms. Invest Ophthalmol Vis Sci 2012;53:1519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hope CI, Bundey S, Proops D, Fielder AR. Usher syndrome in the city of Birmingham–prevalence and clinical classification. Br J Ophthalmol 1997;81:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenberg T, Haim M, Hauch AM, Parving A. The prevalence of Usher syndrome and other retinal dystrophy-hearing impairment associations. Clin Genet 1997;51:314–21. [DOI] [PubMed] [Google Scholar]
- 9.Espinós C, Millán JM, Beneyto M, Najera C. Epidemiology of Usher syndrome in Valencia and Spain. Community Genet 1998;1:223–8. [DOI] [PubMed] [Google Scholar]
- 10.BR SD, Greenberg J, Christoffels A, Hide W. RetNet. http://www.sph.uth.tmc.edu/RetNet (accessed 23 Jun 2016).
- 11.Nakanishi H, Ohtsubo M, Iwasaki S, Hotta Y, Mizuta K, Mineta H, Minoshima S. Identification of 11 novel mutations in USH2A among Japanese patients with Usher syndrome type 2. Clin Genet 2009;76:383–91. [DOI] [PubMed] [Google Scholar]
- 12.Bonnet C, Grati M, Marlin S, Levilliers J, Hardelin JP, Parodi M, Niasme-Grare M, Zelenika D, Delepine M, Feldmann D, Jonard L, El-Amraoui A, Weil D, Delobel B, Vincent C, Dollfus H, Eliot MM, David A, Calais C, Vigneron J, Montaut-Verient B, Bonneau D, Dubin J, Thauvin C, Duvillard A, Francannet C, Mom T, Lacombe D, Duriez F, Drouin-Garraud V, Thuillier-Obstoy MF, Sigaudy S, Frances AM, Collignon P, Challe G, Couderc R, Lathrop M, Sahel JA, Weissenbach J, Petit C, Denoyelle F. Complete exon sequencing of all known Usher syndrome genes greatly improves molecular diagnosis. Orphanet J Rare Dis 2011;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang L, Liang X, Li Y, Wang J, Zaneveld JE, Wang H, Xu S, Wang K, Wang B, Chen R, Sui R. Comprehensive molecular diagnosis of 67 Chinese Usher syndrome probands: high rate of ethnicity specific mutations in Chinese USH patients. Orphanet J Rare Dis 2015;10:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunk K, Zhu M, Barenz F, Kratz AS, Haselmann-Weiss U, Antony C, Hoffmann I. CEP78 is a novel centriolar protein involved in Plk4-induced centriole overduplication. J Cell Sci 2016;129:2713–8. [DOI] [PubMed] [Google Scholar]
- 15.Salvo J, Lyubasyuk V, Xu M, Wang H, Wang F, Nguyen D, Wang K, Luo H, Wen C, Shi C, Lin D, Zhang K, Chen R. Next-generation sequencing and novel variant determination in a cohort of 92 familial exudative vitreoretinopathy patients. Invest Ophthalmol Vis Sci 2015;56:1937–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu M, Gelowani V, Eblimit A, Wang F, Young MP, Sawyer BL, Zhao L, Jenkins G, Creel DJ, Wang K, Ge Z, Wang H, Li Y, Hartnett ME, Chen R. ATF6 Is Mutated in Early Onset Photoreceptor Degeneration With Macular Involvement. Invest Ophthalmol Vis Sci 2015;56:3889–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tajiguli A, Xu M, Fu Q, Yiming R, Wang K, Li Y, Eblimit A, Sui R, Chen R, Aisa HA. Next-generation sequencing-based molecular diagnosis of 12 inherited retinal disease probands of Uyghur ethnicity. Sci Rep 2016;6:21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu M, Eblimit A, Wang J, Li J, Wang F, Zhao L, Wang X, Xiao N, Li Y, Wong LJ, Lewis RA, Chen R. ADIPOR1 Is Mutated in Syndromic Retinitis Pigmentosa. Hum Mutat 2016;37:246–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, Milosavljevic A, Gibbs RA, Yu F. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics 2012;13:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Psaty BM, O’Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, Uitterlinden AG, Harris TB, Witteman JC, Boerwinkle E. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2009;2:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 2012;337:64–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azimzadeh J, Wong ML, Downhour DM, Sanchez Alvarado A, Marshall WF. Centrosome loss in the evolution of planarians. Science 2012;335:461–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Javadi Esfehani Y CEP78, a novel centrosomal protein [Master Thesis]. University of Montreal, 2014. [Google Scholar]
- 26.Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res 2014;42:13534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 2003;426:570–4. [DOI] [PubMed] [Google Scholar]
- 28.Sillibourne JE, Hurbain I, Grand-Perret T, Goud B, Tran P, Bornens M. Primary ciliogenesis requires the distal appendage component Cep123. Biol Open 2013;2:535–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, Asara JM, Tsou MF. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev 2013;27:163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fogeron ML, Müller H, Schade S, Dreher F, Lehmann V, Kuhnel A, Scholz AK, Kashofer K, Zerck A, Fauler B, Lurz R, Herwig R, Zatloukal K, Lehrach H, Gobom J, Nordhoff E, Lange BM. LGALS3BP regulates centriole biogenesis and centrosome hypertrophy in cancer cells. Nat Commun 2013;4:1531. [DOI] [PubMed] [Google Scholar]
- 31.Khateb S, Zelinger L, Mizrahi-Meissonnier L, Ayuso C, Koenekoop RK, Laxer U, Gross M, Banin E, Sharon D. A homozygous nonsense CEP250 mutation combined with a heterozygous nonsense C2orf71 mutation is associated with atypical Usher syndrome. J Med Genet 2014;51:460–9. [DOI] [PubMed] [Google Scholar]
- 32.Petit C Usher syndrome: from genetics to pathogenesis. Annu Rev Genomics Hum Genet 2001;2:271–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.