Abstract
Glioma is a highly aggressive form of brain cancer, with some subtypes having 5-year survival rates of less than 5%. Tumour cell invasion into the surrounding parenchyma seems to be the primary driver of these poor outcomes, as most gliomas recur within 2 cm of the original surgically-resected tumour. Many current approaches to the development of anticancer therapy attempt to target genetic weaknesses in a particular cancer, but may not take into account the microenvironment experienced by a tumour and the patient-specific genetic differences in susceptibility to treatment. Here we demonstrate the use of complementary approaches, 3D bioprinting and scaffold-free 3D tissue culture, to examine the invasion of glioma cells into neural-like tissue with 3D confocal microscopy. We found that, while both approaches were successful, the use of 3D tissue culture for organoid development offers the advantage of broad accessibility. As a proof-of-concept of our approach, we developed a system in which we could model the invasion of human glioma cells into mouse neural progenitor cell-derived spheroids. We show that we can follow invasion of human tumour cells using cell-tracking dyes and 3D laser scanning confocal microscopy, both in real time and in fixed samples. We validated these results using conventional cryosectioning. Our scaffold-free 3D approach has broad applicability, as we were easily able to examine invasion using different neural progenitor cell lines, thus mimicking differences that might be observed in patient brain tissue. These results, once applied to iPSC-derived cerebral organoids that incorporate the somatic genetic variability of patients, offer the promise of truly personalized treatments for brain cancer.
Keywords: 3D bioprinting, Cerebral organoid, Confocal microscopy, Connexin43, Gap junctions, Glioma
Introduction
The host environment surrounding cancer cells intimately affects the malignancy of human tumours. Most cancer treatments aim to destroy tumour cells by taking advantage of their accelerated growth, but much less effort is dedicated to addressing the contribution of host tissue in mediating the resistance of tumour cells to therapeutic treatment. Tumour cells interact dynamically with one another and with the surrounding environment so as to support tumour cell invasion (Nevo et al. 2014).
Glioma is a particularly aggressive cancer, with some subtypes having a 5-year survival rate of less than 5% despite surgical intervention (Goodenberger and Jenkins 2012). One prominent feature of glioma pathology is significant gliosis, an inflammatory response consisting in part of reactive astrocytes in and around the tumour (Lee et al. 2011; Colombo and Farina 2016). In addition to astrocytes, microglia and peripherally-derived cells such as macrophages and dendritic cells are also prominent in the brain tumour environment (Quail and Joyce 2017). We recently showed that expression of connexin43 (Cx43), a major gap junction protein in astrocytes, is significantly enhanced in glioma-associated astrocytes, especially at the region adjacent to the tumour core (Sin et al. 2016), further implicating the tumour microenviroment in cancer development.
The tissue environment that envelops the glioma cells not only affects the dissemination of cancer cells, but also affects the susceptibility of cancer cells to therapeutic intervention (McMillin et al. 2013). Increasing evidence suggest that somatic alterations of the genome in host tissue contributes to tumour invasion (Eng et al. 2009; Carter et al. 2017). Therefore, the need is not only to be able to culture patient-derived tumour cells, but also to simultaneously culture normal host tissue from the same patients to achieve the ultimate goal of personalized medicine. Indeed, others have found that sequencing to identify genomic mutations alone is insufficient to identify the best therapeutic options for patients; on the other hand, a tumour organoid culture will retain enough cellular complexities to accurately assess therapeutic response (Pauli et al. 2017). An early study indicates that 90% of recurrent gliomas occur within 2 cm of the resected tumour (Hochberg and Pruitt 1980), well within the corresponding region in mice that shows active gliosis with an increased number of reactive astrocytes (Sin et al. 2016).
In order to investigate the mechanisms of glioma invasion in a defined multicellular system with a tumour boundary that mimics the intact brain environment, we set out to develop a scaffold-free 3D culture-based platform that accounts for both the tumour and its interaction with the surrounding tissue, for potential translation to patient-specific drug screening. Such a platform would also allow us to identify and study other mechanisms regulating invasiveness of gliomas. Our ultimate objective is to optimize a scaffold-free 3D platform with the potential for translational application; we will accomplish this by co-culturing patient-derived glioma cells with normal tissues generated from patient iPSCs to mimic the invasive niche we observed at the tumour-stromal interface in vivo (Sin et al. 2016).
Materials and methods
iPSC Cell culture and spheroid formation
iPSC-derived human neural progenitor cells were purchased from Axol Bioscience and cultured according to the manufacturer’s instructions. GFP labeled U118 human glioma cells (Aftab et al. 2015) were cultured in DMEM-HG containing 10% fetal bovine serum (FBS). Spheroid formation was performed with 96-well U-bottom plates (PrimeSurface 96 U Plate, #MS-9096 U; Sumitomo Bakelite Co., Ltd.). iPSC-derived human neural progenitor cells (40,000 cells/well) were seeded in 96-well plates and cultured for 48 h. GFP- labeled U118 human glioma cells (10,000 cells/well) were seeded in another 96-well plate and cultured for 48 h. The diameter of both kinds of spheroids were around 500 μm.
3D bioprinting and maturation of neural organoid
A 3D bioprinter (Regenova; Cyfuse Biomedical K.K., Tokyo, Japan) was used to assemble spheroids. The procedure has been described elsewhere (Kizawa et al. 2017). Briefly, the 3D bioprinter is fitted with a 9 × 9 array of needles. The needles have a diameter of 0.17 mm, and the needle bed has a pitch (the distance between needles) of 0.40 mm, for a total usable size of 3.20 mm square × 10.0 mm height. The spheroids were individually picked up from 96-well plates by a robotically controlled fine suction 27-gauge nozzle (inner diameter 0.19 mm), and stuck into a needle. The 3D schematic (top view) is shown in Fig. 1a. At first, 8 neurospheres were skewered on the needle array (1st printing) and cultured in neural maintenance medium for 3 weeks in a sterilized container, when the neurospheres had fused and matured to a neural organoid. Thereafter, a GFP-labeled U118 glioma spheroid was printed on the top of the neural organoid (2nd printing) and co-cultured for several weeks to allow invasion. Finally, the neural organoid with glioma was removed from the needle array and evaluated.
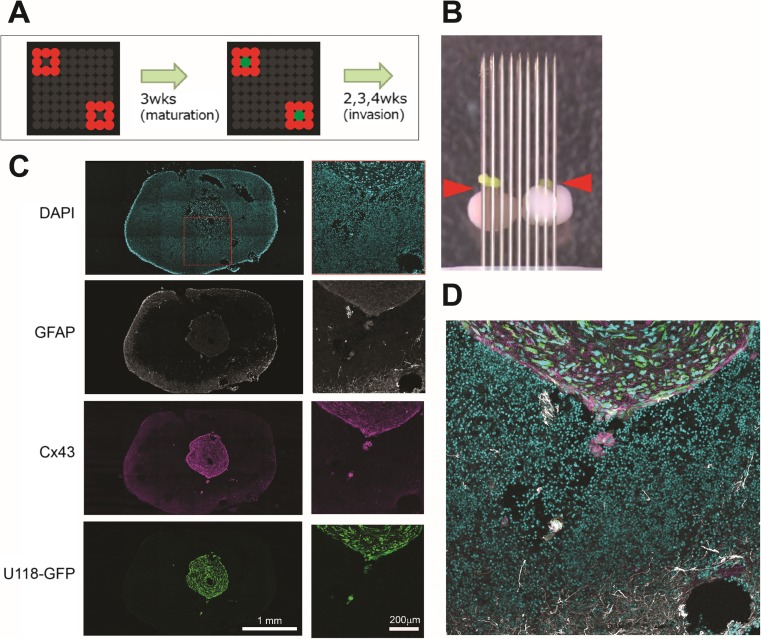
Fig. 1.
Using the “Kenzan” method to develop a 3D model of glioma invasion. A Experiment schematic. iPSC-derived human neurospheres were arrayed on fine needles and allowed to mature for 3 weeks. Then, a spheroid of U118 human glioma cells expressing a GIPZ-GFP construct was placed on top and allowed to invade for a number of weeks. B Photograph of the needle array, taken immediately after U118 docking. U118 cells express GFP, and are indicated by the red arrowheads. C Confocal image of one section of a cell mass similar to that shown in B, with staining for DNA (DAPI), glia (GFAP), Cx43, and GFP-labelled U118 cells. D Higher magnification on the red box shown in c, indicating invasion of U118 cells into the neural spheroids
Histological Preparation and immunofluorescence in needle array-prepared spheroids
Neural/glioma organoids were fixed with 4% paraformaldehyde at 4 °C for 3 h and then rinsed twice with 15% sucrose in PBS. The organoids were frozen in Tissue-Tek OCT compound (Sakura Finetechnical) prior to sectioning with a cryostat to generate 10 μm slices. Cryosections were blocked for 1 h at room temperature in 2% BSA and 0.3% Triton X-100. Sections were then incubated overnight at 4 °C in 1% BSA and 0.1% Triton X-100 with either rabbit anti-Cx43 (Sigma, 1:800) or mouse anti-GFAP (Sigma, 1:800). Sections were then probed with Alexa Fluor 543-conjugated anti-mouse and Alexa Fluor 647-conjugated anti-rabbit secondary antibodies (Invitrogen, 1:500), mounted in Prolong antifade mounting media (Invitrogen) with 4′-6-diamidino-2-phenylindole (DAPI) (Invitrogen) and visualized with a Leica TCS SP5 II Basic VIS confocal system.
Scaffold-free 3D cultures
Brain tumour stem cells GBM4 (Wakimoto et al. 2009; Wakimoto et al. 2012) were cultured in NeuroCult media (STEMCELL Technologies) according to the manufacturer’s instructions. Mouse neural stem cells were isolated from embryonic day 16 brain following the same procedure as the isolation of cortical neurons, as previously described (Sin et al. 2009) and modified (Ahlenius and Kokaia 2010). Briefly, isolated mouse cells were triturated in 200 μl DMEM/F12 without serum and plated 2 X 106 cells in a T25 flask in complete DMEM/F12 media containing 2 mM glutamine (Invitrogen), B27 supplement (Invitrogen), 20 ng/ml EGF (Sigma) and 10 ng/ml bFGF (Sigma). Neurospheres were trypsinized and expanded when they reached 100 μm in diameter. Neurospheres were cryopreserved as 50 μm spheroids in complete media with 10% DMSO. All breeding and animal procedures were approved by The University of British Columbia Animal Care Committee and performed in accordance with the guidelines established by the Canadian Council on Animal Care.
Neurospheres (GBM4, CD1, and C57BL) were cultured and maintained in DMEM/F12 supplemented with 1X B27 supplement (Life Technologies), 10 ng/L bFGF (Sigma), and 20 ng/L rhEGF (STEMCELL Technologies). GBM4 cell cultures were supplemented with heparin (0.0002% w/v); CD1 and C57BL cell cultures were supplemented with 1X penicillin-streptomycin (Life Technologies). GBM4 cells were subcultured by trituration of spheroids. CD1 and C57BL cells were subcultured using Accutase (STEMCELL Technologies) according to the manufacturer’s instructions. To generate spheroids for invasion experiments, 105 isolated cells were cultured in 6-well plates with gentle rotational shaking at approximately 34 rpm and allowed to form spheroids for 1–3 days, typically with staining (see below), prior to combining. Combination was achieved by transferring spheroids to a 2.0 mL Eppendorf tube using a sterile plastic eyedropper, washing once in PBS, then combining the spheroids of interest at the bottom of a 1.5 mL Eppendorf tube for 1 h. Aggregated spheroids were then returned to shaking for the desired amount of time, or prepared for live imaging (see below).
Fixation and staining of self-assembled spheroids
Resuspended cells were stained with CellTracker® Red CMTPX or CellTracker® Blue CMAC (Molecular Probes) and allowed to self-assemble into spheroids prior to aggregation. Cell aggregates were fixed in 4% paraformaldehyde/PBS for 30 min with gentle inversion, then washed 3X in PBS and stained with 100 ng/mL Hoechst dye overnight prior to imaging.
Imaging
3D confocal imaging was conducted using a Leica TCS SP5 II Basic VIS system (Leica Microsystems, Concord, ON, Canada). For live cell imaging, aggregates were mounted in 0.6% noble agar/PBS that had been cooled to 39 °C prior to imaging in a 35 mm glass bottom dish (MatTek Corporation). The mounted aggregates were incubated in complete medium supplemented with HEPES pH 7.4 to 10 mM. A 37 °C heater was used to heat the imaging chamber beginning at least 3 h prior to the start of live cell imaging. Images were analyzed, including color merging, maximal projections, and 3D reconstructions, using ImageJ software (National Institutes of Health, USA).
Cryosectioning of self-assembled spheroids
Cell aggregates were incubated overnight in 30% sucrose/PBS + 0.05% NaN3. The aggregate was then embedded into OCT compound (Tissue-Tek) and subjected to cryosectioning to generate 20 μm thick slices.
Results
We used a scaffold-free human glioma model that involves encompassing glioma cells with normal tissue because of the possibility that matrix proteins used for scaffolding are bioactive and affect the invasiveness of tumour cells (Pickup et al. 2014). As a proof-of-concept, we collaborated with Cyfuse Biomedical K.K., (Tokyo, Japan) to use their Bio 3D printer Regenova® based on their proprietary ‘Kenzan’ method (https://www.cyfusebio.com/en/product/device/) to produce a scaffold-free human neural microenvironment. Human neurospheres consisting of iPSC-derived human neural progenitor cells (Axol Bioscience) were placed in fine needle arrays and merged to form a neural organoid (Fig. 1a, b). Using cryosectioning and confocal imaging, we were able to observe the implanted U118 human glioma cells expressing a GIPZ-GFP construct (Aftab et al. 2015) within the human neural tissue invading into the organoid (Fig. 1c, d). However, immunostaining for glial fibrillary acidic protein (GFAP) and Cx43, markers of astrogliosis (Ridet et al. 1997; Fawcett and Asher 1999; Sofroniew 2009; Theodoric et al. 2012), shows that, contrary to what we found in mouse brains (Sin et al. 2016), no gliosis is observed surrounding the tumour or invading cells.
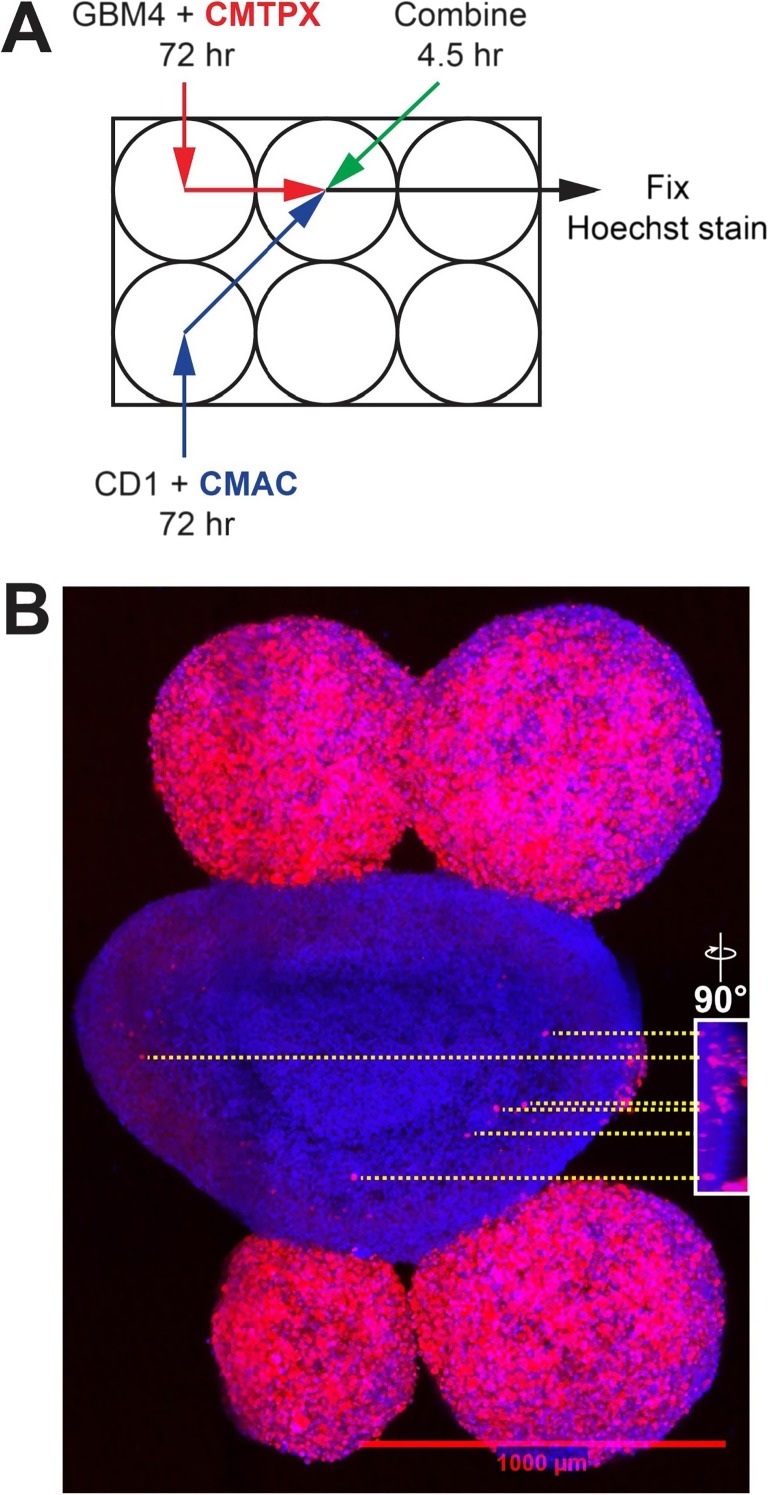
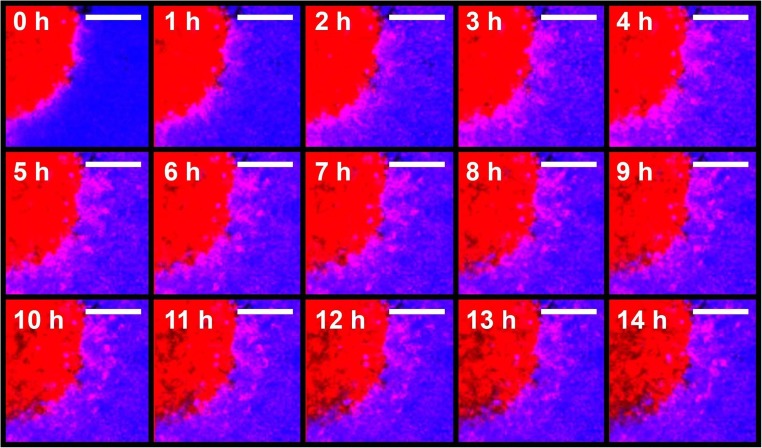
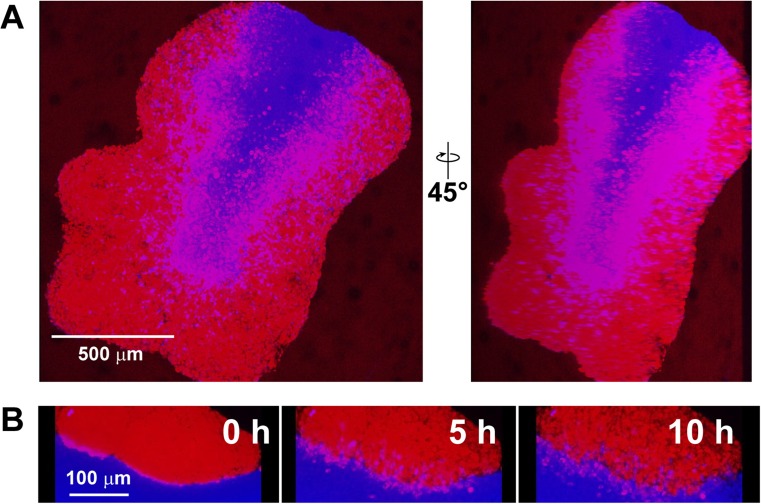
As our main objective is to develop a brain cancer model that addresses tumour-stromal interactions, it is not necessary to have the precise spatial control made possible by the needle array. Thus, we sought to develop an alternative method adapted from a protocol optimized for culture of large cerebral organoids (Lancaster and Knoblich 2014) without the need for a bioprinter. We sought to develop a simple system that could follow glioma invasiveness quickly and with minimal post-processing of acquired images. As a proof-of-principle, we first asked whether human glioma cells, when allowed to self-assemble into tumour-like spheroids, would invade similarly self-assembled neural progenitor cell spheroids. We selected mouse embryos as a source of neural progenitor cells due to their availability and ease of culture. The use of mouse cells also opens up the possibility of using neural stem cells derived from transgenic mice to confirm findings concluded from in vivo intracranial mouse models. The human glioma cell line GBM4 (Wakimoto et al. 2009; Wakimoto et al. 2012) and mouse neural progenitor cells from the CD1 background were incubated with gentle rotational shaking and allowed to self-assemble into spheroids (Fig. 2A). After 72 h of incubation, spheroids were combined for 4.5 h, then fixed and imaged using 3D confocal microscopy (Fig. 2B). We observed that GBM4 cells invaded into the CD1 neural progenitor cell-derived spheroid. We measured the invading distance of the 6 cells indicated in Fig. 2B to be between approximately 100 and 200 μm in the xy direction, and between approximately 5 and 30 μm in the z direction. In order to follow glioma cell invasion in real time, we carried out time-lapse imaging with living spheroids. Snapshots from this time lapse series (Fig. 3) show the invasion of GBM4 cells into a CD1 spheroid in real time. These images have been compiled into Supplementary Video 1 (https://drive.google.com/file/d/1vR9lG18FDh78JviosrHbCVe2H5HK8CbF/view?usp=drivesdk). To ensure that invasion of human glioma cells into mouse neural progenitor cell-derived spheroids was a general phenomenon, we carried out similar experiments using neural progenitor cells derived from the C57BL strain. Similar to CD1, we observed invasion of GBM4 cells into C57BL neural progenitor cell-derived spheroids (Fig. 4A), which we verified again using live cell-based, time-lapse imaging (Fig. 4B). Taken together, we are able to observe and capture 3D invasion of glioma cells into mouse neural spheroids using 3D confocal microscopy.
Fig. 2.
GBM4 cells invade a CD1 neural progenitor cell-derived spheroid. A Schematic of experiment. GBM4 cells were stained with CellTracker® Red CMTPX dye. Cells were incubated with rotational shaking for 3 days to develop into spheroids. Spheroids were collected, combined and allowed to settle at the bottom of a 1.5 mL Eppendorf tube for 1 h, then were returned to rotational shaking for 3.5 h prior to fixation in 4% paraformaldehyde and staining with Hoechst dye. B Maximum projection of the spheroid aggregate. CD1 cells are stained blue, and GBM4 cells are stained red and blue. Invasion of GBM4 cells into CD1 spheroid is evident. Scale bar indicates 1000 μm. Inset: 3D reconstruction of the image stack used to generate b, rotated 90°. The relative locations of six selected cells are indicated with yellow dashed line
Fig. 3.
Time-lapse images of invasion of GBM4 cells into a CD1 neural progenitor cell-derived spheroid. GBM4 cells were stained with CellTracker® Red CMTPX dye, and CD1 cells were stained with CellTracker® Blue CMAC dye. Similar to 2A, cells were incubated with rotational shaking for 3 days to develop into spheroids. Spheroids were collected, combined and allowed to settle at the bottom of a 1.5 mL Eppendorf tube for 1 h and the aggregate was mounted in noble agar in a 35 mm imaging dish. Images were taken 1 h apart. Scale bar: 100 μm
Fig. 4.
Invasion of GBM4 cells into C57BL neural progenitor cell-derived spheroids. A Fixed imaging of invasion. GBM4 cells were stained with CellTracker® Red CMTPX dye, and C57BL cells were stained with CellTracker® Blue CMAC dye. Cells were grown into spheroids separately for 48 h, and then were allowed to adhere at the bottom of a 1.5 mL Eppendorf tube for 1 h. Aggregates were subsequently incubated with shaking for a further 24 h prior to fixation for 30 min with 4% PFA in PBS. B Time-lapse images of invasion of GBM4 cells into C57BL neural progenitor cell-derived spheroids. Cells were stained and incubated as in A, except that, after 1 h of incubation at the bottom of a 1.5 mL Eppendorf tube, the aggregate was mounted in noble agar in a 35 mm dish and subjected to live, time lapse imaging. Images were taken 1 h apart
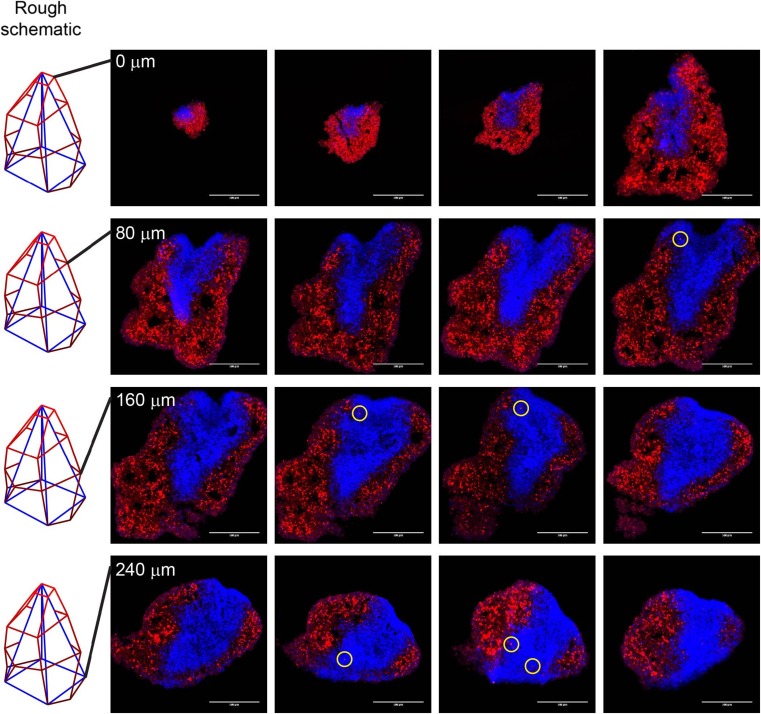
Finally, it is possible that what we see as “invasion” using 3D confocal microscopy is an artifact of glioma cells migrating across the surface of the mouse neural progenitor cell-derived spheroids. To rule this out and thereby validate our approach, we subjected the aggregate shown in Fig. 4A to conventional cryosectioning, with 20 μm thick sections (Fig. 5). The presence of GBM4 cells, surrounded by at least 20 μm of C57BL cells above and below, positively demonstrates that we are in fact seeing invasion deep into the mouse spheroids, as opposed to migration across the mouse spheroid surface.
Fig. 5.
Validation of confocal microscopy-based invasion assay using conventional cryosectioning. The GBM4 (red) and C57BL (blue) aggregate was subjected to cryosectioning with 20 μm thick slices. Sections are arranged in reading order (left to right, top to bottom). Red-stained cells bounded on all sides by blue cells of at least 20 μm depth are indicated by yellow circles
Discussion
In this work we have demonstrated two approaches to modeling glioma invasion into brain parenchyma: combining glioma cells and human neural spheroids using 3D bioprinting, and combining scaffold-free, self-assembled human glioma cell spheroids and mouse neural progenitor cell-derived spheroids. We have shown that both approaches can be used to follow the invasion of glioma cells into neural-like tissue, and have carried out proof-of-principle experiments to demonstrate that 3D tissue culture followed by 3D confocal microscopy can be used to follow invasion of glioma cells into neural-like tissue, both in fixed samples and in real time. This will form the basis of future experiments using human iPSC-derived neural progenitor cells in place of mouse embryonic neural progenitor cells, before ultimately moving towards modeling glioma invasion into cerebral organoids (Lancaster and Knoblich 2014).
Our work builds on similar work by others; for example, Dai and colleagues used 3D bioprinting of glioma cells in a gelatin-alginate-fibrinogen hydrogel to show that such techniques are conducive to the culture of glioma cells generally (Dai et al. 2016). However, the use of 3D culture and 3D bioprinting to generate brain-like tissues has inherent limitations: the main challenge, besides inclusion of a vascular element, is to generate normal neural tissue with a 3D architecture containing the proper cell types that will exhibit inflammatory responses similar to an intact brain. For example, our pilot experiment using neural-derived iPSCs (Fig. 1) did not exhibit reactive gliosis around the tumour core, as we observed in mouse brains (Sin et al. 2016). Similarly, gliosis was not observed in the mouse spheroid at the glioma – mouse cell interface (data not shown), suggesting other cell types, such as microglia, may be necessary for this process to occur. Progress has been made in demonstrating the successful generation of mature human astrocytes in 3D iPSC-derived spheroids showing the feasibility of generating tissue that resembles an intact brain (Sloan et al. 2017). Attempting to recapitulate in vivo tissue architecture with 3D bioprinting is an area of active research (Knowlton et al. 2018) and Krencik and colleagues recently developed a systematic means to co-culture human neurons and astrocytes that appears to recapitulate neuronal connections seen in vivo (Krencik et al. 2017). Thus, our future work will incorporate our experience obtained here and these latest advances to develop more representative human 3D neural cultures.
As discussed earlier, our current glioma/neural organoid body does not contain microglia, the immune cells of the brain and a prominent cell type in the glioma environment (Charles et al. 2011; Sin et al. 2016). As microglia have a very different embryonic origin (Ginhoux et al. 2010) and iPSC-derived microglia demand different culture conditions from neural cells (Abud et al. 2017), one option in our system would be to add well-characterized microglia-secreted cytokines, such as IL-1β and TNFα, into the culture medium of our glioma/neural tissue organoid to simulate the presence of reactive microglia (Cunningham 2013). Recent advances in the generation of iPSC-derived microglia (Muffat et al. 2016; Abud et al. 2017; Pandya et al. 2017) also suggest it may soon be possible to culture microglia and neural-derived cell types together. Finally, the proliferation of endothelial cells is critical for tumour-associated angiogenesis (Ezan et al. 2012; Boulay et al. 2016). Recent work has shown the feasibility of 3D bioprinting large tissues with complete blood vessels (Kolesky et al. 2016), and others have described the self-assembly of endothelial cell and smooth muscle progenitors into structures resembling blood vessels using the Kenzan method (Moldovan et al. 2017). Further work is necessary to determine whether the incorporation of commercially available human endothelial cells into a self-assembling platform such as the one described here will lead to biosimilar blood vessel formation.
A personalized glioma platform utilizing iPSCs obtained from patients will account for both tumour microheterogeneity and genetic somatic variation. The development and translation of a 3D model that not only mimics the cellular architecture normally found in living tissues but that allows assessment of tumour response to anticancer therapy will complement the current initiative for personalized medicine. Although this work was initiated with brain cancer in mind, our 3D model can be easily adapted for personalized drug screening applicable to all kinds of cancer.
Acknowledgements
Part of this work was supported by a grant from Canadian Cancer Society (CCN, WCS). DvP was supported by the UBC Faculty of Medicine, the UBC Summer Student Research Program, and the Mach-Gaensslen Foundation. CCN holds a Canada Research Chair. We thank Dr. Hiroaki Wakimoto, Brain Tumor Research Center, Massachusetts General Hospital, for the kind gift of the GBM4 glioma stem cells. We thank John Bechberger for providing technical support for this project.
Abbreviations
- Cx43
connexin43
- GFAP
glial fibrillary acidic protein
- DAPI
4′-6-diamidino-2-phenylindole
- iPSC
induced pluripotent stem cell
References
- Abud EM et al. (2017) iPSC-derived human microglia-like cells to study neurological diseases. Neuron 94:278–293 e279 [DOI] [PMC free article] [PubMed]
- Aftab Q, Sin WC, Naus CC. Reduction in gap junction intercellular communication promotes glioma migration. Oncotarget. 2015;6:11447–11464. doi: 10.18632/oncotarget.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlenius Henrik, Kokaia Zaal. Methods in Molecular Biology. Totowa, NJ: Humana Press; 2010. Isolation and Generation of Neurosphere Cultures from Embryonic and Adult Mouse Brain; pp. 241–252. [DOI] [PubMed] [Google Scholar]
- Boulay AC, Cisternino S, Cohen-Salmon M. Immunoregulation at the gliovascular unit in the healthy brain: A focus on Connexin 43. Brain Behav Immun. 2016;56:1–9. doi: 10.1016/j.bbi.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Carter Hannah, Marty Rachel, Hofree Matan, Gross Andrew M., Jensen James, Fisch Kathleen M., Wu Xingyu, DeBoever Christopher, Van Nostrand Eric L., Song Yan, Wheeler Emily, Kreisberg Jason F., Lippman Scott M., Yeo Gene W., Gutkind J. Silvio, Ideker Trey. Interaction Landscape of Inherited Polymorphisms with Somatic Events in Cancer. Cancer Discovery. 2017;7(4):410–423. doi: 10.1158/2159-8290.CD-16-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia. 2011;59:1169–1180. doi: 10.1002/glia.21136. [DOI] [PubMed] [Google Scholar]
- Colombo Emanuela, Farina Cinthia. Astrocytes: Key Regulators of Neuroinflammation. Trends in Immunology. 2016;37(9):608–620. doi: 10.1016/j.it.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Cunningham C. Microglia and neurodegeneration: the role of systemic inflammation. Glia. 2013;61:71–90. doi: 10.1002/glia.22350. [DOI] [PubMed] [Google Scholar]
- Dai X, Ma C, Lan Q, Xu T. 3D bioprinted glioma stem cells for brain tumor model and applications of drug susceptibility. Biofabrication. 2016;8:045005. doi: 10.1088/1758-5090/8/4/045005. [DOI] [PubMed] [Google Scholar]
- Eng C, Leone G, Orloff MS, Ostrowski MC. Genomic alterations in tumor stroma. Cancer Res. 2009;69:6759–6764. doi: 10.1158/0008-5472.CAN-09-0985. [DOI] [PubMed] [Google Scholar]
- Ezan P, Andre P, Cisternino S, Saubamea B, Boulay AC, Doutremer S, Thomas MA, Quenech'du N, Giaume C, Cohen-Salmon M (2012) Deletion of astroglial connexins weakens the blood-brain barrier. J Cereb Blood Flow Metab [DOI] [PMC free article] [PubMed]
- Fawcett JW, Asher RA. The glial scar and central nervous system repair. Brain Res Bull. 1999;49:377–391. doi: 10.1016/S0361-9230(99)00072-6. [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205:613–621. doi: 10.1016/j.cancergen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Hochberg FH, Pruitt A. Assumptions in the radiotherapy of glioblastoma. Neurology. 1980;30:907–911. doi: 10.1212/WNL.30.9.907. [DOI] [PubMed] [Google Scholar]
- Kizawa H, Nagao E, Shimamura M, Zhang G, Torii H. Scaffold-free 3D bio-printed human liver tissue stably maintains metabolic functions useful for drug discovery. Biochem Biophys Rep. 2017;10:186–191. doi: 10.1016/j.bbrep.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton Stephanie, Anand Shivesh, Shah Twisha, Tasoglu Savas. Bioprinting for Neural Tissue Engineering. Trends in Neurosciences. 2018;41(1):31–46. doi: 10.1016/j.tins.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA. Three-dimensional bioprinting of thick vascularized tissues. Proc Natl Acad Sci U S A. 2016;113:3179–3184. doi: 10.1073/pnas.1521342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krencik Robert, Seo Kyounghee, van Asperen Jessy V., Basu Nupur, Cvetkovic Caroline, Barlas Saba, Chen Robert, Ludwig Connor, Wang Chao, Ward Michael E., Gan Li, Horner Philip J., Rowitch David H., Ullian Erik M. Systematic Three-Dimensional Coculture Rapidly Recapitulates Interactions between Human Neurons and Astrocytes. Stem Cell Reports. 2017;9(6):1745–1753. doi: 10.1016/j.stemcr.2017.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Borboa AK, Baird A, Eliceiri BP. Non-invasive quantification of brain tumor-induced astrogliosis. BMC Neurosci. 2011;12:9. doi: 10.1186/1471-2202-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillin DW, Negri JM, Mitsiades CS. The role of tumour-stromal interactions in modifying drug response: challenges and opportunities. Nat Rev Drug Discov. 2013;12:217–228. doi: 10.1038/nrd3870. [DOI] [PubMed] [Google Scholar]
- Moldovan Leni, Barnard April, Gil Chang-Hyun, Lin Yang, Grant Maria B., Yoder Mervin C., Prasain Nutan, Moldovan Nicanor I. iPSC-Derived Vascular Cell Spheroids as Building Blocks for Scaffold-Free Biofabrication. Biotechnology Journal. 2017;12(12):1700444. doi: 10.1002/biot.201700444. [DOI] [PubMed] [Google Scholar]
- Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, Bakiasi G, Tsai LH, Aubourg P, Ransohoff RM, Jaenisch R. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo I, Woolard K, Cam M, Li A, Webster JD, Kotliarov Y, Kim HS, Ahn S, Walling J, Kotliarova S, Belova G, Song H, Bailey R, Zhang W, Fine HA. Identification of molecular pathways facilitating glioma cell invasion in situ. PLoS One. 2014;9:e111783. doi: 10.1371/journal.pone.0111783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, Kim G, Brown MA, Elkahloun AG, Maric D, Sweeney CL, Gossa S, Malech HL, McGavern DB, Park JK. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci. 2017;20:753–759. doi: 10.1038/nn.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli Chantal, Hopkins Benjamin D., Prandi Davide, Shaw Reid, Fedrizzi Tarcisio, Sboner Andrea, Sailer Verena, Augello Michael, Puca Loredana, Rosati Rachele, McNary Terra J., Churakova Yelena, Cheung Cynthia, Triscott Joanna, Pisapia David, Rao Rema, Mosquera Juan Miguel, Robinson Brian, Faltas Bishoy M., Emerling Brooke E., Gadi Vijayakrishna K., Bernard Brady, Elemento Olivier, Beltran Himisha, Demichelis Francesca, Kemp Christopher J., Grandori Carla, Cantley Lewis C., Rubin Mark A. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine . Cancer Discovery. 2017;7(5):462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15:1243–1253. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail Daniela F., Joyce Johanna A. The Microenvironmental Landscape of Brain Tumors. Cancer Cell. 2017;31(3):326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridet JL, Malhotra SK, Privat A, Gage FH. Reactive astrocytes: cellular and molecular cues to biological function. Trends Neurosci. 1997;20:570–577. doi: 10.1016/S0166-2236(97)01139-9. [DOI] [PubMed] [Google Scholar]
- Sin WC, Moniz DM, Ozog MA, Tyler JE, Numata M, Church J. Regulation of early neurite morphogenesis by the Na+/H+ exchanger NHE1. J Neurosci. 2009;29:8946–8959. doi: 10.1523/JNEUROSCI.2030-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin WC, Aftab Q, Bechberger JF, Leung JH, Chen H, Naus CC. Astrocytes promote glioma invasion via the gap junction protein connexin43. Oncogene. 2016;35:1504–1516. doi: 10.1038/onc.2015.210. [DOI] [PubMed] [Google Scholar]
- Sloan Steven A., Darmanis Spyros, Huber Nina, Khan Themasap A., Birey Fikri, Caneda Christine, Reimer Richard, Quake Stephen R., Barres Ben A., Paşca Sergiu P. Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron. 2017;95(4):779-790.e6. doi: 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32:638–647. doi: 10.1016/j.tins.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoric N, Bechberger JF, Naus CC, Sin WC. Role of gap junction protein Connexin43 in astrogliosis induced by brain injury. PLoS One. 2012;7:e47311. doi: 10.1371/journal.pone.0047311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto H, Kesari S, Farrell CJ, Curry WT, Jr, Zaupa C, Aghi M, Kuroda T, Stemmer-Rachamimov A, Shah K, Liu TC, Jeyaretna DS, Debasitis J, Pruszak J, Martuza RL, Rabkin SD. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69:3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto H, Mohapatra G, Kanai R, Curry WT, Jr, Yip S, Nitta M, Patel AP, Barnard ZR, Stemmer-Rachamimov AO, Louis DN, Martuza RL, Rabkin SD. Maintenance of primary tumor phenotype and genotype in glioblastoma stem cells. Neuro-Oncology. 2012;14:132–144. doi: 10.1093/neuonc/nor195. [DOI] [PMC free article] [PubMed] [Google Scholar]