Abstract
Interferon gamma (IFN-γ), is a cytokine, which is an important regulator of host defense system by mediating both innate and adaptive immune responses. IFN-γ signaling is primarily associated with inflammation and cell-mediated immune responses. IFN-γ is also represented as antitumor cytokine which facilitates immunosurveillance in tumor cells. In addition, IFN-γ mediated signaling also elicits pro-tumorigenic transformations and promotes tumor progression. Impact of IFN-γ signaling in mammalian cells has been widely studied which indicate that IFN-γ orchestrates distinct cellular functions including immunomodulation, leukocyte trafficking, apoptosis, anti-microbial, and both anti- and pro-tumorigenic role. However, a detailed network of IFN-γ signaling pathway is currently lacking. Therefore, we systematically curated the literature information pertaining to IFN-γ signaling and develop a comprehensive signaling network to facilitate better understanding of IFN-γ mediated signaling. A total of 124 proteins were catalogued that were experimentally proven to be involved in IFN-γ signaling cascade. These 124 proteins were found to participate in 81 protein-protein interactions, 94 post-translational modifications, 20 translocation events, 54 activation/inhibiton reactions. Further, 236 differential expressed genes were also documented in IFN-γ mediated signaling. IFN-γ signaling pathway is made freely available to scientific audience through NetPath at (http://www.netpath.org/pathways?path_id=NetPath_32). We believe that documentation of reactions pertaining to IFN-γ signaling and development of pathway map will facilitate further research in IFN-γ associated human diseases including cancer.
Electronic supplementary material
The online version of this article (10.1007/s12079-018-0486-y) contains supplementary material, which is available to authorized users.
Keywords: Interferon gamma receptor, Signal transducer and activator of transcription, Interferon response factors, Interferon-gamma activated sequence and PathVisio
Introduction
Interferon gamma (IFN-γ or IFN-gamma or IFNG), a soluble dimerized cytokine is one of the central regulator of host defense system which mediates both innate and adaptive immune response. IFN-γ is the sole “type II interferon” identified and described by E. F. Wheelock (Wheelock 1965). Biological active form of IFN-γ is a homodimer which is formed by anti-parallel inter-locking of IFN-γ monomer. Each IFN-γ monomer consists of a core of six alpha helices and an extended C-terminal region (Ealick et al. 1991). Gene encoding IFN-γ protein is located on chromosome 12q15 in humans (Naylor et al. 1983) and in mouse genome it is found on chromosome 10D2 (Naylor et al. 1984). The human gene for IFN-γ consists of four exons and three intervening regions, covering 5.4 kb (Liu et al. 2015). IFN-γ is primarily known to be produced by T helper cell type 1 (Th1) lymphocytes, CD8 lymphocytes, B cells, NKT cells, and antigen-presenting cells (monocytes, macrophages and dendritic cells). Cytokines IL-12 and IL-18 induces IFN-γ production, while IL-4, IL-10, transforming growth factor-beta and glucocorticoids are the negative regulators of IFN-γ production (Tominaga et al. 2000; Almawi et al. 1998).
IFN-γ binds to the IFN-γ receptor (IFNGR) complex which comprises of two distinct chains, high affinity IFNGR1 (alpha) and a low affinity IFNGR2 (beta) (Pestka et al. 2004). Binding of IFN-γ to its cognate receptors induce conformation change in the receptor allowing recruitment of JAK1 and JAK2 to the receptor complex (Lasfar et al. 2014). IFN-γ binding induce phosphorylation of JAK2 which further transphosphorylate JAK1. This in turn induces conformation change in receptor making docking site for STAT1. Further, JAK2 phosphorylates transcription factor STAT1. Phosphorylated STAT1 dimer translocates to nucleus and induces transcription of the interferon stimulated genes (ISGs) including several other transcription factors such as interferon response factor 1 (IRF1), IRF9 and others (Decker et al. 1997). These transcription factors further activate number of secondary IFN-γ regulated genes. IFN-γ signaling regulates several biological processes which are primarily known to regulate inflammation and cell-mediated immune responses, including regulation of innate and acquired immune response, apoptosis and cell cycle (de Weerd and Nguyen 2012). IFN-γ also activates macrophages, which in turn induces production of cytokines to facilitate accumulation of immune cell at the site of inflammation (Hu et al. 2008). IFN-γ mediates activation and differentiation of immune cells. Specifically, IFN-γ is involved in upregulation of the major histocompatibility (MHC) Class I molecules which aid in presentation of antigens to antigen presenting cells (APCs), promote activity of natural killer cells, regulate Th1 cells development and activation, expression of Class II major histocompatibility complex (MHC) molecules and regulation of B cells functions (Schroder et al. 2004).. Kaufmann and Booty et al., reported that decreased expression of IFN-γ renders an individual highly susceptible to mycobacterial and fungal infections, which underscores the indispensable role of IFN-γ in the maintenance of host defense system (Fenimore and Young 2016). IFN-γ is key modulator of hematopoiesis, its overexpression can mediate bone marrow suppression and number of bone marrow related disorders such as myelodysplasic syndromes and aplastic anemia (Yang et al. 2005). Studies have shown IFN-γ mediates the process of neurogenesis and neuronal differentiation via activation of ERK1/2 Pathway (Song et al. 2005). IFN-γ plays a vital role in macrophage metabolism via mTORC1 and MNK kinases, both converge on translation initiation factor 4E (eIF4E) (Su et al. 2015). Owing to the activation of a diverse range of downstream effector molecules, IFN-γ signaling regulates varied biological functions related to anti-viral and anti-bacterial defense system.
These studies indicate that IFN-γ signaling has significant impact on several biological processes including tumor progression and regression and hence it is necessary to develop IFN-γ signaling network to understand its divergent role. In this study, we have curated literature information pertaining to IFN-γ signaling and developed a pathway map to facilitate better understanding of IFN-γ induced signaling.
Methods
Literature survey and curation of signaling events mediated by IFN-γ
An extensive literature search was carried out in PubMed with key search terms including ‘Interferon gamma signaling’. We have screened over 10,565 articles related to IFN-γ signaling. In order to relate information for protein-protein interactions (PPIs), post-translational modifications (PTMs), activation/inhibition reactions, protein transport and gene expression events were curated from experimental papers. These reactions were incorporated under IFN-γ stimulation by exogenous and/or endogenous ligands. The entries were documented into ‘PathBuilder’, an annotation tool developed in-house for the manual curation of signaling events (Kandasamy et al. 2009). NetPath annotation pipeline was followed to develop IFN-γ signaling pathway (Kandasamy et al. 2010), which has been described previously by other groups at our institute, to develop several signaling pathways including oxytocin pathway (Chatterjee et al. 2016) and aryl hydrocarbon receptor (Yelamanchi et al. 2016) signaling pathways. Information regarding cell lines used in the experiment, PTM with site and residue information were also curated. Data was curated for gene expression from human cells in both normal and diseased conditions. We also included suggestions provided by an expert in the field (pathway authority) in order to improve confidence of the documented signaling events.
Generation of IFN-γ map
Pathway map tool ‘PathVisio’ were used to pictorially represent the reactions mediated by IFN-γ (http://www.PathVisio.org) (van Iersel et al. 2008). Moreover, a subset of highly confident IFN-γ mediated signaling events were documented in NetSlim map according to the criteria provided in the NetSlim database (http://www.netpath.org/netslim/criteria.html). The reactions stimulated by IFN-γ have been topologically organised from ligand binding onto IFN-γ receptor to target genes regulated by its induction. Pathway modules other than STAT1 mediated such as mTOR signaling, MAPK signaling and PI3K/AKT signaling which are regulated by IFN-γ are also been represented in the pathway map. The NetSlim version of the IFN-γ pathway map can be downloaded in various formats such as .gpml, and .pdf formats.
Results and discussion
Data integration and IFN-γ signaling map
We have screened a total of 10,565 articles from PubMed for generating the IFN-γ signaling map. Out of 10,565 articles which were screened, 699 had the relevant information pertaining to IFN-γ pathway and were used to develop a map (Fig. 1). A total of 124 proteins were catalogued that are experimentally proven to be involved in IFN-γ signaling cascade. These 124 proteins that correspond to 81 protein-protein interactions (PPIs), 94 post translational modifications (PTMs), 20 translocational events, 54 activation/inhibiton reactions were documented in NetPath. Further, 236 differential gene expressions were documented in response to IFN-γ stimulation (Supplementary Table 1).
Fig. 1.
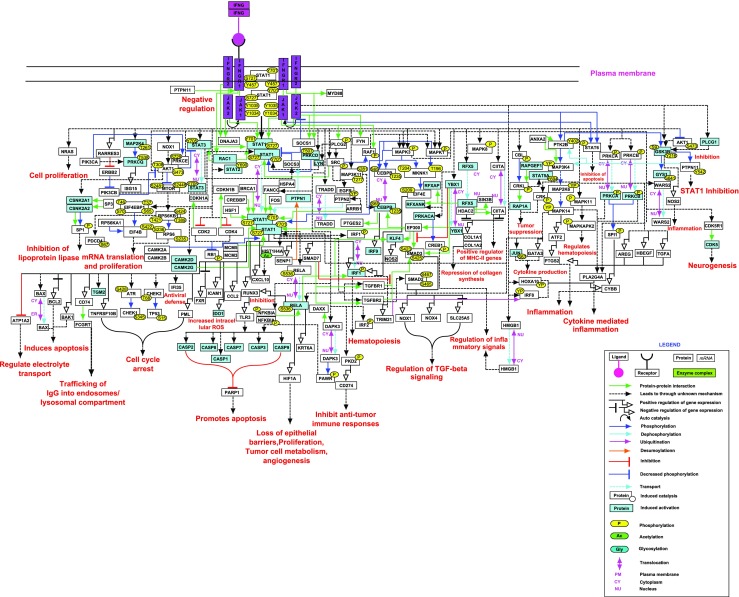
A graphical representation of IFN-γ signaling pathway. The pathway map represents reactions of IFN-γ signaling pathway that are documented in NetPath. Different types of reactions and molecules are distinguished with different colors and arrows as provided in the legend. Solid arrows represent direct reactions and a dashed arrow represents reactions whose mechanisms are currently unknown. Post-translational modifications with site and residues are also provided in the map
Development of IFN-γ mediated signaling network
We have submitted the IFN-γ pathway data to NetPath (http://www.netpath.org/pathways?path_id=NetPath_32). The web page for IFN-γ pathway in NetPath provides a concise description of IFN-γ signaling pathway, molecules and reaction statistics of the pathway map along with different downloadable formats. All molecules documented in NetPath are linked to a molecular page. For additional information on the pathway curated data, molecular pages have been linked to external protein resources including NCBI RefSeq, Human Protein Reference Database (HPRD) (Prasad et al. 2009), Entrez gene (Maglott et al. 2011), Swiss-Prot (Boeckmann et al. 2003) and OMIM databases (Hamosh et al. 2005). The molecular events are graphically represented for IFN-γ signaling pathway using PathVisio tool and is shown in Fig. 1. The curated data presented in NetPath related to this pathway will be periodically updated.
Data formats and availability
The pathway information and IFN-γ signaling pathway map is available in NetPath. The signaling pathway information is attuned with various international data exchange formats which includes Proteomics Standards Initiative for Molecular Interaction (PSI-MI) (Hermjakob et al. 2004), Biological PAthway eXchange (BioPAX level 3) (Demir et al. 2010) and Systems Biology Markup Language (SBML 3) (Hucka et al. 2003). The information present in these formats can be further used in pathway analysis software tools such as Cytoscape and Ingenuity pathway analysis. The data for IFN-γ signaling pathway can be downloaded in the above mentioned formats from NetPath (http://www.netpath.org).
Summary of IFN-γ mediated signaling pathway and regulation
IFN-γ signaling employs activation of JAK-STAT signaling cascade to drive expression of IFN-γ regulated genes. Majority of cytokines, hormones and other growth factors use JAK-STAT pathway to regulate the expression of effector genes (Nicolas et al. 2013). IFN-γ signaling cascade begins with binding of IFN-γ to its cognate receptors, which induces conformation change in receptors, wherein box 1 domains of receptors (IFNGR1 and IFNGR2) come in close proximity of each other allowing recruitment of JAK1 and JAK2, to IFNGR1 and IFNGR2 receptor chains, respectively (Blouin et al. 2016). IFN-γ binding to receptor induces autophosphorylation and activation of JAK2, which further phosphorylates JAK1. Activated JAK1 in turn phosphorylates IFNGR1 at specific tyrosine(Y) residue (Y440) which allows formation of docking site specific for SH2 domain of signal transducer and activator of transcription protein 1 (STAT1) (Chapgier et al. 2006). Further, STAT1 dimer recruited at the receptor site is phosphorylated by JAK2 at site Y701. Phosphorylated STAT1 pair dissociates from receptor site and translocates to nucleus (Schroder et al. 2004). STAT1 also interacts with other transcription factors such as STAT2 and IRF-9 to form heterodimer or heterotrimers and induce transcription of effector genes (Fink and Grandvaux 2013). IFN-γ stimulation is also known to induce nuclear localization of receptor IFNGR1 along with STAT1 and IFN-γ (Ahmed and Johnson 2006). In nucleus, pSTAT1 binds at promoter region to initiate or suppress transcription of interferon stimulated genes (ISGs). Phosphorylated STAT1 dimer specifically binds to DNA at gamma interferon activated site (GAS) bearing consensus sequence TTCN (2–4) GAA (Decker et al. 1997). T cell protein tyrosine phosphatase (TCP45) dephosphorylates nuclear STAT1, which recycles STAT1 back to cytoplasm, thus maintaining a reversible negative feedback loop (Kramer et al. 2009). Histone acetylasetranferase such as CREB-binding protein (CBP) negatively regulates STAT1 activation, while histone deacetylase such as histone deacetylase (HDAC3) enhances STAT1 activation (Kramer and Heinzel 2010). Many IFN-γ stimulated genes are transcription factors which includes interferon response factor 1 (IRF1), IRF2, IRF8, IRF9, RELA, JUN and others. These transcription factors further drive activation of secondary set of interferon stimulated genes (ISGs) which regulates expression of genes involved inflammatory response, apoptosis, cell proliferation and differentiation, hematopoiesis, neurogenesis, cellular metabolism and homeostasis amongst others.
IFN-γ signaling in diseases
Effects of IFN-γ are cell and tissue specific so it becomes imperative to study IFN-γ signaling in the context of different diseases (Lin and Young 2013). IFN-γ has been shown to promote autoimmune diseases due to its proinflammatory functions. Notable inflammatory disorders associated with IFN-γ are Rheumatoid Arthritis (RA), systemic lupus erythematous (SLE) and multiple sclerosis (MS) (Pollard et al. 2013; Lees and Cross 2007). IFN-γ has shown anti-angiogenic and anti-tumorigenic potential in cancer cells (Beatty and Paterson 2001). In addition, IFN-γ signaling has shown tumor surveillance functions by enhancing immunogenicity in tumor cells. Low expression of major histocompatibility complex (MHC) antigen help tumor cells to evade host immune response against tumor cells (Garrido et al. 1997). Restoring IFN-γ-activated MHC antigen by silencing transforming growth factor beta1 (TGF-β1) signaling and/or by exogenous addition of IL6 that has potent anti-TGF-β1 activity promote tumor regression (Hsiao et al. 2008). IFN-γ signaling facilitates tumor cell recognition and elimination by recruiting cytotoxic T lymphocytes to tumor cells. IFN-γ sharpens immune response by inducing expression of tumor suppressive factors such as MIG-1 and GBP-1 and was shown to suppress growth of malignant mammary carcinoma (Lipnik et al. 2010; Walser et al. 2007). IFN-γ treatment is reported to induce the expression of PDL1 and PDL2 in many immune cells and patient derived cells in various cancers (Maleki Vareki et al. 2017; Lyford-Pike et al. 2013).
It has been shown that IFN-γ induces expression of nitric oxide synthase 2 (NOS2) and consequently production of nitric oxide (NO) in renal cell carcinoma which results in growth inhibition of tumor cells and angiogenesis (Tate et al. 2012). IFN-γ treatment significantly increased expression of pro-apoptotic factors such as caspase 1 (CASP1) and induces cleavage of poly-(ADP ribose) polymerase (PARP1) which facilitates suppression of tumor growth in pancreatic cancer cells (Detjen et al. 2001). IFN-γ induced AKT/mTOR/ p70 S6 kinase axis is required for translation of IFN-stimulated genes (ISGs) and generation of antiviral effects mediated by IFN-γ (Kaur et al. 2008). IFN-γ mediates activation of TGFβ/SMAD signaling module, which induces transcription of NADPH oxidases such as NOX1 and NOX4. These oxidases contribute to generation of reactive oxygen species (ROS) which triggers DNA damage and tumor cell senescence (Hubackova et al. 2016). Despite the role of IFN-γ in tumor immune surveillance, pro-tumoregenic effect of IFN-γ also has been reported (Zaidi and Merlino 2011).
IFN-γ facilitates papilloma development by up-regulating proinflammatory cytokines and a local T helper 17 (Th17) response (Xiao et al. 2009). Mice lacking negative regulator of IFN-γ, suppressor of cytokine signaling-1 (SOCS1) has been associated with development of colorectal cancer in an IFN-γ dependent manner (Hanada et al. 2006). Exogenous expression of interferon gamma in mouse model triggers wave of inflammatory responses accompanied with increased rate of proliferation and gastric neoplasia (Syu et al. 2012). It has been reported that IFN-γ induces expression of PD-L1 and PKD2 in oral squamous carcinoma cells in both time and dose dependent manner (Chen et al. 2012). Increased expression of PD-L1 in tumor cells is associated with tumor evasion from host T cells and favours growth and progression of cancer. Reports indicate significant upregulation of CXC chemokines such as CXCL3, CXCL5, CXCL9, CXCL10 and CXC11 in response to IFN-γ treatment (Yuzawa et al. 2008; Zhou et al. 2006; Bukowski et al. 1999). It has been shown CXCL10, CXCL9 and CXCL11 is significantly upregulated in oral lichen planus (OLP) compared to normal oral mucosa and was shown to be involved in epithelial to mesenchymal transition (EMT) in pre-malignant OLP (Marshall et al. 2017; Liu et al. 2017). Thus, IFN-γ plays a vital role in cellular processes such as cell cycle, differentiation, apoptosis, and pro- and anti-tumorigenesis along with immunomodulatory functions.
Conclusions
IFN-γ signaling map is important for understanding and deciphering various biological processes and diseases mediated by IFN-γ regulated transcription of several genes. This comprehensive and well curated map of IFN-γ signaling reactions will aid in understanding the role of various molecules in the context of normal physiological or pathological conditions such as cancers and other diseases in association with IFN-γ. IFN-γ signaling pathway map will lead to further research on IFN-γ associated biomarkers for various human diseases.
Electronic supplementary material
(XLSX 58 kb)
Acknowledgments
We thank the Department of Biotechnology (DBT), Government of India for research support to the Institute of Bioinformatics, Bangalore. MYB is a recipient of Senior Research Fellowship from Department of Biotechnology (DBT), Government of India. JA is recipient of Senior Research Fellowship from Council of Scientific and Industrial Research (CSIR), Government of India. AAK is recipient of Senior Research Fellowship from Indian Council of Medical Research (ICMR), New Delhi.
Abbreviations
- IFN-γ /IFNG
Interferon gamma
- IFNGR1
IFN-γ receptor subunit 1
- IFNGR2
IFN-γ receptor subunit 2
- STAT
Signal transducer and activator of transcription protein
- ISGs
Interferon stimulated genes
- PPI
Protein-protein interaction
- SBML
Systems Biology Markup Language
- BioPAX
Biological Pathway Exchange
- PTM
Post-translational modification
- HPRD
Human Protein Reference Database
- PSI-MI
Proteomics Standards Initiative for Molecular Interaction
References
- Ahmed CM, Johnson HM. IFN-gamma and its receptor subunit IFNGR1 are recruited to the IFN-gamma-activated sequence element at the promoter site of IFN-gamma-activated genes: evidence of transactivational activity in IFNGR1. J Immunol. 2006;177:315–321. doi: 10.4049/jimmunol.177.1.315. [DOI] [PubMed] [Google Scholar]
- Almawi WY, Hess DA, Rieder MJ. Multiplicity of glucocorticoid action in inhibiting allograft rejection. Cell Transplant. 1998;7:511–523. doi: 10.1177/096368979800700602. [DOI] [PubMed] [Google Scholar]
- Beatty GL, Paterson Y. Regulation of tumor growth by IFN-gamma in cancer immunotherapy. Immunol Res. 2001;24:201–210. doi: 10.1385/IR:24:2:201. [DOI] [PubMed] [Google Scholar]
- Blouin CM, Hamon Y, Gonnord P, Boularan C, Kagan J, Viaris de Lesegno C, Ruez R, Mailfert S, Bertaux N, Loew D, Wunder C, Johannes L, Vogt G, Contreras FX, Marguet D, Casanova JL, Gales C, He HT, Lamaze C. Glycosylation-dependent IFN-gammaR partitioning in lipid and actin Nanodomains is critical for JAK activation. Cell. 2016;166:920–934. doi: 10.1016/j.cell.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowski RM, Rayman P, Molto L, Tannenbaum CS, Olencki T, Peereboom D, Tubbs R, McLain D, Budd GT, Griffin T, Novick A, Hamilton TA, Finke J. Interferon-gamma and CXC chemokine induction by interleukin 12 in renal cell carcinoma. Clin Cancer Res. 1999;5:2780–2789. [PubMed] [Google Scholar]
- Chapgier A, Boisson-Dupuis S, Jouanguy E, Vogt G, Feinberg J, Prochnicka-Chalufour A, Casrouge A, Yang K, Soudais C, Fieschi C, Santos OF, Bustamante J, Picard C, de Beaucoudrey L, Emile JF, Arkwright PD, Schreiber RD, Rolinck-Werninghaus C, Rosen-Wolff A, Magdorf K, Roesler J, Casanova JL. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet. 2006;2:e131. doi: 10.1371/journal.pgen.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee O, Patil K, Sahu A, Gopalakrishnan L, Mol P, Advani J, Mukherjee S, Christopher R, Prasad TS. An overview of the oxytocin-oxytocin receptor signaling network. J Cell Commun Signal. 2016;10:355–360. doi: 10.1007/s12079-016-0353-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Feng Y, Lu L, Wang H, Dai L, Li Y, Zhang P. Interferon-gamma-induced PD-L1 surface expression on human oral squamous carcinoma via PKD2 signal pathway. Immunobiology. 2012;217:385–393. doi: 10.1016/j.imbio.2011.10.016. [DOI] [PubMed] [Google Scholar]
- de Weerd NA, Nguyen T. The interferons and their receptors--distribution and regulation. Immunol Cell Biol. 2012;90:483–491. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T, Kovarik P, Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J Interf Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D'Eustachio P, Schaefer C, Luciano J, Schacherer F, Martinez-Flores I, Hu Z, Jimenez-Jacinto V, Joshi-Tope G, Kandasamy K, Lopez-Fuentes AC, Mi H, Pichler E, Rodchenkov I, Splendiani A, Tkachev S, Zucker J, Gopinath G, Rajasimha H, Ramakrishnan R, Shah I, Syed M, Anwar N, Babur O, Blinov M, Brauner E, Corwin D, Donaldson S, Gibbons F, Goldberg R, Hornbeck P, Luna A, Murray-Rust P, Neumann E, Ruebenacker O, Samwald M, van Iersel M, Wimalaratne S, Allen K, Braun B, Whirl-Carrillo M, Cheung KH, Dahlquist K, Finney A, Gillespie M, Glass E, Gong L, Haw R, Honig M, Hubaut O, Kane D, Krupa S, Kutmon M, Leonard J, Marks D, Merberg D, Petri V, Pico A, Ravenscroft D, Ren L, Shah N, Sunshine M, Tang R, Whaley R, Letovksy S, Buetow KH, Rzhetsky A, Schachter V, Sobral BS, Dogrusoz U, McWeeney S, Aladjem M, Birney E, Collado-Vides J, Goto S, Hucka M, Le Novere N, Maltsev N, Pandey A, Thomas P, Wingender E, Karp PD, Sander C, Bader GD. The BioPAX community standard for pathway data sharing. Nat Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detjen KM, Farwig K, Welzel M, Wiedenmann B, Rosewicz S. Interferon gamma inhibits growth of human pancreatic carcinoma cells via caspase-1 dependent induction of apoptosis. Gut. 2001;49:251–262. doi: 10.1136/gut.49.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ealick SE, Cook WJ, Vijay-Kumar S, Carson M, Nagabhushan TL, Trotta PP, Bugg CE. Three-dimensional structure of recombinant human interferon-gamma. Science. 1991;252:698–702. doi: 10.1126/science.1902591. [DOI] [PubMed] [Google Scholar]
- Fenimore J, Young AH. Regulation of IFN-γ expression. Adv Exp Med Biol. 2016;941:1–19. doi: 10.1007/978-94-024-0921-5_1. [DOI] [PubMed] [Google Scholar]
- Fink K, Grandvaux N. STAT2 and IRF9: beyond ISGF3. JAKSTAT. 2013;2:e27521. doi: 10.4161/jkst.27521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido F, Ruiz-Cabello F, Cabrera T, Perez-Villar JJ, Lopez-Botet M, Duggan-Keen M, Stern PL. Implications for immunosurveillance of altered HLA class I phenotypes in human tumours. Immunol Today. 1997;18:89–95. doi: 10.1016/S0167-5699(96)10075-X. [DOI] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Kobayashi T, Chinen T, Saeki K, Takaki H, Koga K, Minoda Y, Sanada T, Yoshioka T, Mimata H, Kato S, Yoshimura A. IFNgamma-dependent, spontaneous development of colorectal carcinomas in SOCS1-deficient mice. J Exp Med. 2006;203:1391–1397. doi: 10.1084/jem.20060436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, Moore S, Orchard S, Sarkans U, von Mering C, Roechert B, Poux S, Jung E, Mersch H, Kersey P, Lappe M, Li Y, Zeng R, Rana D, Nikolski M, Husi H, Brun C, Shanker K, Grant SG, Sander C, Bork P, Zhu W, Pandey A, Brazma A, Jacq B, Vidal M, Sherman D, Legrain P, Cesareni G, Xenarios I, Eisenberg D, Steipe B, Hogue C, Apweiler R. The HUPO PSI's molecular interaction format--a community standard for the representation of protein interaction data. Nat Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- Hsiao YW, Liao KW, Chung TF, Liu CH, Hsu CD, Chu RM. Interactions of host IL-6 and IFN-gamma and cancer-derived TGF-beta1 on MHC molecule expression during tumor spontaneous regression. Cancer Immunol Immunother. 2008;57:1091–1104. doi: 10.1007/s00262-007-0446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Chakravarty SD, Ivashkiv LB. Regulation of interferon and toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol Rev. 2008;226:41–56. doi: 10.1111/j.1600-065X.2008.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubackova S, Kucerova A, Michlits G, Kyjacova L, Reinis M, Korolov O, Bartek J, Hodny Z. IFNgamma induces oxidative stress, DNA damage and tumor cell senescence via TGFbeta/SMAD signaling-dependent induction of Nox4 and suppression of ANT2. Oncogene. 2016;35:1236–1249. doi: 10.1038/onc.2015.162. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novere N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Kandasamy K, Keerthikumar S, Raju R, Keshava Prasad TS, Ramachandra YL, Mohan S, Pandey A. PathBuilder--open source software for annotating and developing pathway resources. Bioinformatics. 2009;25:2860–2862. doi: 10.1093/bioinformatics/btp453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, Navarro JD, Mathivanan S, Pecquet C, Gollapudi SK, Tattikota SG, Mohan S, Padhukasahasram H, Subbannayya Y, Goel R, Jacob HK, Zhong J, Sekhar R, Nanjappa V, Balakrishnan L, Subbaiah R, Ramachandra YL, Rahiman BA, Prasad TS, Lin JX, Houtman JC, Desiderio S, Renauld JC, Constantinescu SN, Ohara O, Hirano T, Kubo M, Singh S, Khatri P, Draghici S, Bader GD, Sander C, Leonard WJ, Pandey A. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 2010;11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Sassano A, Dolniak B, Joshi S, Majchrzak-Kita B, Baker DP, Hay N, Fish EN, Platanias LC. Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc Natl Acad Sci U S A. 2008;105:4808–4813. doi: 10.1073/pnas.0710907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer OH, Heinzel T. Phosphorylation-acetylation switch in the regulation of STAT1 signaling. Mol Cell Endocrinol. 2010;315:40–48. doi: 10.1016/j.mce.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Kramer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Guhrs KH, Stauber RH, Bohmer FD, Heinzel T. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 2009;23:223–235. doi: 10.1101/gad.479209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasfar A, Cook JR, Cohen Solal KA, Reuhl K, Kotenko SV, Langer JA, Laskin DL. Critical role of the endogenous interferon ligand-receptors in type I and type II interferons response. Immunology. 2014;142:442–452. doi: 10.1111/imm.12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees JR, Cross AH. A little stress is good: IFN-gamma, demyelination, and multiple sclerosis. J Clin Invest. 2007;117:297–299. doi: 10.1172/JCI31254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F-C, Young HA. The talented interferon-gamma. Adv Biosci Biotechnol. 2013;04(07):8. doi: 10.4236/abb.2013.47A3002. [DOI] [Google Scholar]
- Lipnik K, Naschberger E, Gonin-Laurent N, Kodajova P, Petznek H, Rungaldier S, Astigiano S, Ferrini S, Sturzl M, Hohenadl C. Interferon gamma-induced human guanylate binding protein 1 inhibits mammary tumor growth in mice. Mol Med. 2010;16:177–187. doi: 10.2119/molmed.2009.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Song Y, Shi W. IFN-gamma +874 T/A polymorphisms contributes to cervical cancer susceptibility: a meta-analysis. Int J Clin Exp Med. 2015;8:4008–4015. [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu G, Liu Q, Tan J, Hu X, Wang J, Wang Q, Wang X. The cellular character of liquefaction degeneration in oral lichen planus and the role of interferon gamma. J Oral Pathol Med. 2017;46:1015–1022. doi: 10.1111/jop.12510. [DOI] [PubMed] [Google Scholar]
- Lyford-Pike S, Peng S, Young GD, Taube JM, Westra WH, Akpeng B, Bruno TC, Richmon JD, Wang H, Bishop JA, Chen L, Drake CG, Topalian SL, Pardoll DM, Pai SI. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013;73:1733–1741. doi: 10.1158/0008-5472.CAN-12-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki Vareki S, Garrigos C, Duran I. Biomarkers of response to PD-1/PD-L1 inhibition. Crit Rev Oncol Hematol. 2017;116:116–124. doi: 10.1016/j.critrevonc.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Marshall A, Celentano A, Cirillo N, McCullough M, Porter S. Tissue-specific regulation of CXCL9/10/11 chemokines in keratinocytes: implications for oral inflammatory disease. PLoS One. 2017;12:e0172821. doi: 10.1371/journal.pone.0172821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor SL, Sakaguchi AY, Shows TB, Law ML, Goeddel DV, Gray PW. Human immune interferon gene is located on chromosome 12. J Exp Med. 1983;157:1020–1027. doi: 10.1084/jem.157.3.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor SL, Gray PW, Lalley PA. Mouse immune interferon (IFN-gamma) gene is on chromosome 10. Somat Cell Mol Genet. 1984;10:531–534. doi: 10.1007/BF01534857. [DOI] [PubMed] [Google Scholar]
- Nicolas CS, Amici M, Bortolotto ZA, Doherty A, Csaba Z, Fafouri A, Dournaud P, Gressens P, Collingridge GL, Peineau S. The role of JAK-STAT signaling within the CNS. JAKSTAT. 2013;2:e22925. doi: 10.4161/jkst.22925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- Pollard KM, Cauvi DM, Toomey CB, Morris KV, Kono DH. Interferon-gamma and systemic autoimmunity. Discov Med. 2013;16:123–131. [PMC free article] [PubMed] [Google Scholar]
- Prasad TSK, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human protein reference database--2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- Song JH, Wang CX, Song DK, Wang P, Shuaib A, Hao C. Interferon gamma induces neurite outgrowth by up-regulation of p35 neuron-specific cyclin-dependent kinase 5 activator via activation of ERK1/2 pathway. J Biol Chem. 2005;280:12896–12901. doi: 10.1074/jbc.M412139200. [DOI] [PubMed] [Google Scholar]
- Su X, Yu Y, Zhong Y, Giannopoulou EG, Hu X, Liu H, Cross JR, Ratsch G, Rice CM, Ivashkiv LB. Interferon-gamma regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat Immunol. 2015;16:838–849. doi: 10.1038/ni.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syu LJ, El-Zaatari M, Eaton KA, Liu Z, Tetarbe M, Keeley TM, Pero J, Ferris J, Wilbert D, Kaatz A, Zheng X, Qiao X, Grachtchouk M, Gumucio DL, Merchant JL, Samuelson LC, Dlugosz AA. Transgenic expression of interferon-gamma in mouse stomach leads to inflammation, metaplasia, and dysplasia. Am J Pathol. 2012;181:2114–2125. doi: 10.1016/j.ajpath.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate DJ, Jr, Patterson JR, Velasco-Gonzalez C, Carroll EN, Trinh J, Edwards D, Aiyar A, Finkel-Jimenez B, Zea AH. Interferon-gamma-induced nitric oxide inhibits the proliferation of murine renal cell carcinoma cells. Int J Biol Sci. 2012;8:1109–1120. doi: 10.7150/ijbs.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga K, Yoshimoto T, Torigoe K, Kurimoto M, Matsui K, Hada T, Okamura H, Nakanishi K. IL-12 synergizes with IL-18 or IL-1beta for IFN-gamma production from human T cells. Int Immunol. 2000;12:151–160. doi: 10.1093/intimm/12.2.151. [DOI] [PubMed] [Google Scholar]
- van Iersel MP, Kelder T, Pico AR, Hanspers K, Coort S, Conklin BR, Evelo C. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399. doi: 10.1186/1471-2105-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser TC, Ma X, Kundu N, Dorsey R, Goloubeva O, Fulton AM. Immune-mediated modulation of breast cancer growth and metastasis by the chemokine Mig (CXCL9) in a murine model. J Immunother. 2007;30:490–498. doi: 10.1097/CJI.0b013e318031b551. [DOI] [PubMed] [Google Scholar]
- Wheelock EF. Interferon-like virus-inhibitor induced in human leukocytes by Phytohemagglutinin. Science. 1965;149:310–311. doi: 10.1126/science.149.3681.310. [DOI] [PubMed] [Google Scholar]
- Xiao M, Wang C, Zhang J, Li Z, Zhao X, Qin Z. IFNgamma promotes papilloma development by up-regulating Th17-associated inflammation. Cancer Res. 2009;69:2010–2017. doi: 10.1158/0008-5472.CAN-08-3479. [DOI] [PubMed] [Google Scholar]
- Yang L, Dybedal I, Bryder D, Nilsson L, Sitnicka E, Sasaki Y, Jacobsen SE. IFN-gamma negatively modulates self-renewal of repopulating human hemopoietic stem cells. J Immunol. 2005;174:752–757. doi: 10.4049/jimmunol.174.2.752. [DOI] [PubMed] [Google Scholar]
- Yelamanchi SD, Solanki HS, Radhakrishnan A, Balakrishnan L, Advani J, Raja R, Sahasrabuddhe NA, Mathur PP, Dutta P, Prasad TS, Korbonits M, Chatterjee A, Gowda H, Mukherjee KK. Signaling network map of the aryl hydrocarbon receptor. J Cell Commun Signal. 2016;10:341–346. doi: 10.1007/s12079-016-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzawa E, Imaizumi T, Matsumiya T, Yoshida H, Fukuhara R, Kimura H, Fukui A, Tanji K, Mori F, Wakabayashi K, Fujii S, Mizunuma H, Satoh K. Retinoic acid-inducible gene-I is induced by interferon-gamma and regulates CXCL11 expression in HeLa cells. Life Sci. 2008;82:670–675. doi: 10.1016/j.lfs.2007.12.025. [DOI] [PubMed] [Google Scholar]
- Zaidi MR, Merlino G. The two faces of interferon-gamma in cancer. Clin Cancer Res. 2011;17:6118–6124. doi: 10.1158/1078-0432.CCR-11-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Wang JX, Tang W, He PL, Yang YF, Li YC, Li XY, Zuo JP. (5R)-5-hydroxytriptolide inhibits IFN-gamma-related signaling. Acta Pharmacol Sin. 2006;27:1616–1621. doi: 10.1111/j.1745-7254.2006.00457.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 58 kb)


