Abstract
Chicken Mx1 gene, as a positive antiviral gene, has been reported to provide resistance to several viruses especially avian influenza virus. In present research, the genotype frequency contributions of chicken Mx1 polymorphisms were characterized in five lowly selected as well as one moderately selected Sichuan native chicken populations and two highly selected commercial chicken breeds. Together with two newly-identified mutation sites (r.8A > G and r.1257T > C), a total of 13 single nucleotide polymorphisms (SNPs), including seven nonsynonymous mutation and six synonymous mutation, were found in the coding region of chicken Mx1 gene. Local Chinese chicken populations exhibited higher nucleotide diversity than commercial populations. Moreover, amino acid substitution sites as well as positive selection sites were located only in the domain not determined and GTPase domain, implying that amino acids mutations were likely needed in the modulatory and structural regions to better adapt the environment. Collectively, our results suggest that different selection pressures greatly influenced the genotype frequency contributions of chicken Mx1 gene. Understanding the interaction between genetic diversity and artificial selection may help us to better select and breed superior domestic chickens.
Keywords: Genotype, Mx1 gene, Chicken, Association
Introduction
Host immunity, particularly innate immunity, plays a functional role in the prevention of infection (Akira et al. 2006). During viral infections, a subset of the most prominent cytokines produced are the interferons (IFNs), which served as inducers, regulators, and effectors in innate antiviral mechanisms (Malmgaard 2004). Myxovirus resistance (Mx) proteins are dynamin-like, large GTPases in response to IFNs stimuli (Haller et al. 2007). They are highly potent antiviral factors of the innate immune system that are able to inhibit the multiplication of a diverse range of viruses, including Orthomyxoviridae, Rhabdoviridae, Paramyxoviridae, Bunyaviridae, Togaviridae, and Hepadnaviridae (Haller et al. 2015; Ewald et al. 2011). In most vertebrates, at least two distinct Mx genes have been described (Haller and Kochs 2002; Thomas et al. 2006). However, chicken only have one Mx gene, which is known as chicken Mx1 gene. Studies demonstrated that a polymorphism at amino acid position 631(Ser/Asn) owing to a G-to-A nucleotide substitution (r.2032G > A) led to the positively antiviral function of chicken Mx1 gene. Chicken Mx1 protein with the 631Asn conferred positive antiviral activities, whereas the Mx1 protein carrying the 631Ser was not capable of inhibiting viral growth (Ko et al. 2004; Sasaki et al. 2013).
The increasing demands for eggs and poultry meat to meet the nutritional requirement are urging domestic chicken breeds more artificially selected (Muchadeyi and Dzomba 2017). In the meantime, disease resistance is neglected under excessive selection for growth traits and reproduction traits in domestic chicken populations, ultimately leading to serious threats to poultry industry (Niraj et al. 2016). Environment, selection, and negative correlations between commercial traits and disease resistance could contribute to highly skewed frequencies of the mutation among different populations (Li et al. 2006).
Chicken Mx1 gene, considered as a marker of chicken resistance to avian influenza virus, is composed of 20,766 bp linear DNA, 13 coding exons and 2118 bp coding sequence (Sartika et al. 2011). At present, more than 200 single-nucleotide polymorphisms (SNPs) have been identified in the full region of chicken Mx1 gene. The polymorphisms of chicken Mx1 gene at the Ser631Asn mutation have been reported in many chicken breeds, in which the results shows that the frequency of the allele with antiviral activity in native breeds is higher than that in commercial lines (Li et al. 2006; Sartika et al. 2011; Ko et al. 2002; Balkissoon et al. 2007; Seyama et al. 2006). However, whether the proportion of chicken Mx1 genotype frequency in the full coding region is associated with different selection pressures remains to be studied. In this study, we detected the genotype frequency contributions in the coding region of chicken Mx1 gene in eight populations with different selection pressures, including five lowly selected as well as one moderately selected Sichuan native chicken populations and two highly selected commercial chicken breeds. The present findings may lead to a better understanding of the effect of artificial selection on the allelic variation in domestic chicken populations and may be helpful for the selection of superior native chickens.
Materials and methods
Sampling and DNA Isolation
A total of 240 samples from eight chicken breeds were used to screen the polymorphisms in the coding sequence of chicken Mx1 gene. The chicken breeds are Caoke Chicken (CK), Guanyuan Grey Chicken (GYG), Jiuyuan Black Chicken (JYB), Green Shell Hen (GSH), Tibetan Chicken (TC), Daheng Boiler (DHB), Cobb Boiler (CB) and Lohman Laying Hen (LLH). The specific information of each chicken breed is shown in Table 1. Each breed used in the present study was composed of 30 unrelated individuals obtained from local poultry breeding company. The samples of each breed line were selected from one site only. Venous blood samples were collected from under the wing of the chickens, prepared for genomic DNA extraction. Total genomic DNA was extracted from chicken blood using the TIAN amp Genomic DNA Kit in accordance with the manufacturer’s instructions. DNA samples were stored at − 20 °C until use. The quality and purity were assessed by a Nucleic Acid Protein Analyzer. These experiments were approved by the Committee on the Care and Use of Laboratory Animals of the State-level Animal Experimental Teaching Demonstration Center of Sichuan Agricultural University (No. DKY-S20163656).
Table 1.
Summary of eight chicken populations
| Breed | Abbreviation | Location | Population size | Type |
|---|---|---|---|---|
| Shimian caoke chicken | CK | Sichuan China | 30 | Native (lowly selected) |
| Guanyuan grey chicken | GYG | Sichuan China | 30 | Native (lowly selected) |
| Jiuyuan black chicken | JYB | Sichuan China | 30 | Native (lowly selected) |
| Green shell hens | GSH | Sichuan China | 30 | Native (lowly selected) |
| Tibetan chicken | TC | Sichuan China | 30 | Native (lowly selected) |
| Daheng boilers | DHB | Sichuan China | 30 | Boiler (moderately selected) |
| Cobb boilers | CB | American | 30 | Boiler (highly selected) |
| Lohman laying hens | LLH | German | 30 | Layer (highly selected) |
Design of PCR primers and sequencing
Primer pairs and conditions for the amplification of the coding sequence of chicken Mx1 gene (containing 13 exons) are given in Table 2. The primers were designed by Primer Premier v6.0 and Oligo v7.37 softwares according to NCBI reference sequence (NC_006088.5). Polymerase chain reaction (PCR) was carried out using a Gene Amp PCR System 9700 (Bio-Rad, USA) thermal cycler in a final volume of 25 µL containing 1.0 µl (100 ng/µl) of template, 0.8 µL of each primer (10 pmol/µl), 13 µL 2 × Taq PCR Master Mix and 9.4 µL ddH2O. The PCR programme were as, initial denaturation at 96 °C for 4 min, followed by 35 cycles of 95 °C for 30 s, annealing at prescribed annealing temperature for 1 min, and primer extension 72 °C for 90 s, and a final extension at 72 °C for 10 min. The PCR products were analyzed using electrophoresis in 1% agarose gel and sequenced by Chain Termination Method in Beijing TSINGKE Biological Technology Corporation.
Table 2.
Primer information for detecting SNPs in the coding sequences of Mx1 gene
| Primer sequences (5′-3′) | Annealing Temperature (℃) | Product(bp) | |
|---|---|---|---|
| Exon1 | F: CAGGAGGTAGAAGCATTGT | 61.2 | 431 |
| R: GGTGAGCATGGAGAGTATC | |||
| Exon2 | F: AGCATGAAGGTGCAAGGAT | 55.4 | 398 |
| R: TGAGTTTGTACCTTTGTCCCT | |||
| Exon3 | F: CAACTGTCACCTCTGTCTT | 61.2 | 274 |
| R: GTGGCTATAATTCCTGTGTTAG | |||
| Exon4 | F: ATCGTGTTCCCACTTCAC | 59.6 | 400 |
| R: CAGCACTGCCTTCCCAAA | |||
| Exon5 | F: TGGTGGCTTGTACTGCTA | 56.5 | 288 |
| R: ACACTACTTCAGACCTTCAC | |||
| Exon6 | F: CTTCCACCTTCAACTTCTG | 53.0 | 286 |
| R: TACTTTGGGTTGGGTTCT | |||
| Exon7 | F: CCCCAGAAGGTGACTTAG | 53.0 | 295 |
| R: GGAAGAAGGGTTGGATTT | |||
| Exon8 | F: GTTCAGCCTACCTAAGAGTCCT | 57.8 | 258 |
| R: CCCTCAATGACAGATTCCAG | |||
| Exon9 | F: CAGAGCAAAGCAACCAAG | 60 | 371 |
| R: AAAGCCAGGGGATAGAAT | |||
| Exon10 | F: AAGGCACTGAGCAATGAG | 53.2 | 418 |
| R: TTGGTCCATCAATGAGAACA | |||
| Exon11 | F: GTCCTAAAGGCGTATTCT | 57.8 | 405 |
| R: TAGTGCCATCACAAACAT | |||
| Exon12 | F: TCCGCTTCACCTTCCTGAG | 60 | 482 |
| R: GGCTTGCCATCATCCAGATAT | |||
| Exon13 | F: CTCTCCTTGTAGGGAGCAAG | 55 | 331 |
| R: GTTGCTGCTAATGGAGGATT |
Statistical analysis
The unripe sequences were processed by Bioedit v7.0.9 (Alzohairy et al. 2011). All the 240 sequences and reference sequence (NCBI Reference Sequence: NC_006088.5) were aligned using Clustal X program (Thompson et al. 1997). The nucleotide substitutions were identified using MEGA 7 (Kumar et al. 2016). The genotype frequencies were calculated by direct counting method. Genetic diversity parameters including number of variable sites (S), haplotypes (h), haplotype diversity (Hd) and nucleotide diversity (π) were estimated by DnaSP V5.0 (Librado and Rozas 2009). Pairwise FST values among populations were computed in Arlequin v3.5 (Excoffier and Lischer 2010). Haplotypes were constructed based on all the SNPs in each experimental animal by Haploview program (Barrett et al. 2005). The frequencies of each haplotype in populations were calculated by PHASE program v2.0. Chi-square test, a statistical significance test, was used in analysis of genotype frequencies among populations. The selection pressure analysis of chicken Mx1 gene was examined using HyPhy package as implemented in Datamonkey web-server (http://www.datamonkey.org/) (Pond et al. 2005). The positive selection codons were detected using five codon-specific selection methods (FEL, IFEL, SLAC, FUBAR, and MEME methods). FEL, IFEL, SLAC, and FUBAR models use a maximum-likelihood (ML), ML, ML, and counting and Bayesian approach to infer nonsynoymous (dN) and synonymous (dS) substitution rates on a per-site basis, respectively (Pond and Frost 2005; Murrell et al. 2013). MEME employs a mixed-effects ML approach to detect sites evolving under positive selection under a proportion of branches (Murrell et al. 2012). Sites identified by FEL, IFEL, SLAC, and MEME with p < 0.05 and FUBAR with posterior probability > 0.90 were considered to be under positive selection with high confidence levels.
Results
Nucleotide and amino acid variations of chicken Mx1 gene
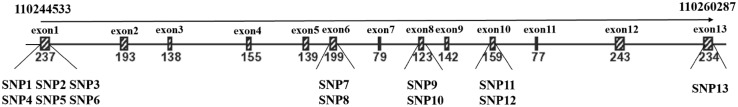
The genomic structure of chicken Mx1 gene in the coding region is illustrated in Fig. 1. A total of 13 SNPs in the coding region were detected, including seven nonsynonymous mutations and 6 synonymous mutations. The specific information of these SNPs is listed in Table 3, along with their locations, coding region positions, nucleotide changes and amino acid changes. At the nucleotide level, six SNPs, two SNPs, two SNPs, two SNPs, and one SNP were harbored in the exons 1, 6, 8, 10, and 13 of chicken Mx1 gene, respectively (Fig. 1). In the meantime, patterns of diversity at the amino acid level were primarily located in the domain not determined and GTPase domain, while all of the SNPs located in the middle domain and GTPase effector domain were synonymous mutations. Coupled with the observations at the nucleotide level, we implied that tolerance for new genetic variants was different at different coding region of the chicken Mx1 gene. Importantly, two novel SNPs at r.8A > G and r.1257T > C were identified in chickens for the first time. However, we failed to detect the aforementioned Ser631Asn mutation caused by the SNP at r.2032G > A, which indicated that all the populations used in the present study were likely sensitive to viral infection.
Fig. 1.
Genomic structure of chicken Mx1 gene in the coding region. The number in each exon is the length of each exon and the number on the top is the location from the first coding exon to the last coding exon (NCBI reference sequence: NC_006088.5)
Table 3.
Detail information for the SNPs of chicken Mx1 gene
| Markers | Location | Coding region position | Amino acid position | Variation | AA change | Domain1 |
|---|---|---|---|---|---|---|
| SNP1 | exon1 | 8 | 3 | A/G | N3S | ND |
| SNP2 | exon1 | 13 | 5 | C/T | R5W | ND |
| SNP3 | exon1 | 62 | 21 | G/A | R21Q | ND |
| SNP4 | exon1 | 122 | 41 | C/G | P41R | ND |
| SNP5 | exon1 | 125 | 42 | T/C | L42S | ND |
| SNP6 | exon1 | 156 | 52 | T/A | – | ND |
| SNP7 | exon6 | 922 | 308 | G/A | V308I | GD |
| SNP8 | exon6 | 1015 | 339 | A/G | T339A | GD |
| SNP9 | exon8 | 1248 | 416 | A/G | – | MD |
| SNP10 | exon8 | 1257 | 419 | T/C | – | MD |
| SNP11 | exon10 | 1455 | 485 | C/T | – | MD |
| SNP12 | exon10 | 1545 | 515 | G/A | – | MD |
| SNP13 | exon13 | 2019 | 673 | G/A | – | GED |
ND domain not determined, GD GTPase domain, MD Middle domain, GED GTPase effector domain
aMx protein domain
The genotype frequencies of these 13 SNPs in each chicken population, which could intuitively display the effect of intensive selections on chicken Mx1 gene, are shown in Table 4 (the frequency distribution of SNP1, 2, 10, 13 did not satisfy the conditions of Chi-square test and, therefore, were excluded from the statistical significance test). The results showed that there are significant differences between lowly selected native chicken populations and highly selected commercial birds. For example, at the SNP4 and SNP5 sites, there are no significant differences in the frequency distribution between CB and LLH (P > 0.05), but it is significantly different between these commercial chickens and the majority of the Chinese local chicken populations (P < 0.05). Simultaneously, in the LLH population, the frequency of genotype at the SNP3, SNP9, and SNP11 showed extreme difference from other populations (P < 0.01), while the CB chickens had significantly distinct frequencies of alleles at the SNP7 and SNP12 sites (P < 0.05). As for the two newly-identified SNP sites which were not included in the Chi-square test, the mutation type of SNP 1 (r.8A > G) was only detected in four Chinese native populations (CK, GYG, TC, DHB) and all of the individuals used in this study possessed at least one copy of the wild type. These observations implied that the r.8A > G mutation at this site may have occurred recently and be only specific to certain Chinese native chicken populations. In contrast, the r.1257T > C of chicken Mx1 gene was presented in Chinese native populations as well as highly selected LLH. However, the homozygote of mutation type was only observed in two Chinese native populations. Collectively, these results suggest that artificial selection potentially influenced the genotype frequency of chicken Mx1 gene.
Table 4.
Genotype frequencies of SNPs of Mx1 SNPs in eight chicken populations
| CK | GYG | JYB | GSH | TC | DHB | LLH | CB | |
|---|---|---|---|---|---|---|---|---|
| SNP1 | ||||||||
| AA | 80% | 93.33% | 100% | 100% | 96.67% | 93.33% | 100% | 100% |
| AG | 20% | 6.67% | 0% | 0% | 3.33% | 6.67% | 0% | 0% |
| GG | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% |
| SNP2 | ||||||||
| CC | 46.67% | 86.67% | 76.67% | 80% | 90% | 33.33% | 100% | 63.33% |
| CT | 43.33% | 13.33% | 23.33% | 20% | 10% | 46.67% | 0% | 36.67% |
| TT | 10% | 0% | 0% | 10% | 0% | 20% | 0% | 0% |
| SNP3 | ||||||||
| AA | B 56.67% | A 26.67% | B 66.67% | A 36.67% | A 40% | B 56.67% | C 3.33% | B 60% |
| AG | 40% | 60% | 26.67% | 50% | 40% | 33.33% | 70% | 40% |
| GG | 3.33% | 13.33% | 6.67% | 13.33% | 20% | 10% | 26.67% | 0% |
| SNP4 | ||||||||
| CC | A 23.33% | AB 26.67% | A 13.33% | A 13.33% | A 23.33% | A 20% | B 36.67% | B 46.67% |
| CG | 40% | 56.67% | 46.67% | 53.33% | 43.33% | 46.67% | 60% | 46.67% |
| GG | 36.67% | 16.67% | 40% | 33.33% | 33.33% | 33.33% | 3.33% | 6.67% |
| SNP5 | ||||||||
| CC | A 36.67% | AB 16.67% | A 40% | A 33.33% | A 33.33% | A 33.33% | B 3.33% | B 6.67% |
| CT | 40% | 56.67% | 46.67% | 53.33% | 43.33% | 46.67% | 60% | 46.67% |
| TT | 23.33% | 26.67% | 13.33% | 13.33% | 23.33% | 20% | 36.67% | 46.67% |
| SNP6 | ||||||||
| AA | A 60% | DE 16.67% | AB 56.67% | CD 33.33% | BC 36.67% | ABC 46.67% | E 3.33% | DE 16.67% |
| AT | 33.33% | 66.67% | 33.33% | 50% | 43.33% | 43.33% | 66.67% | 60% |
| TT | 6.67% | 16.67% | 10% | 16.67% | 20% | 10% | 30% | 23.33% |
| SNP7 | ||||||||
| AA | A 23.33% | B 6.67% | B 3.33% | D 0% | D 3.33% | A 40% | D 3.33% | C 36.67% |
| AG | 53.33% | 46.67% | 50% | 13.33% | 23.33% | 40% | 6.67% | 60% |
| GG | 23.33% | 46.67% | 46.67% | 86.67% | 73.33% | 20% | 90% | 3.33% |
| SNP8 | ||||||||
| AA | B 23.33% | BC 36.67% | C 46.67% | D 86.67% | D 73.33% | AB 20% | D 90% | E 3.33% |
| AG | 53.33% | 50% | 50% | 13.33% | 23.33% | 40% | 8.33% | 60% |
| GG | 23.33% | 13.33% | 3.33% | 0% | 3.33% | 40% | 1.67% | 36.67% |
| SNP9 | ||||||||
| AA | A 23.33% | B 46.67% | BC 46.67% | A 23.33% | B 56.67% | A 23.33% | D 6.67% | C 60% |
| AG | 56.67% | 36.67% | 46.67% | 51.67% | 30% | 46.67% | 70% | 40% |
| GG | 20% | 16.67% | 6.67% | 25% | 13.33% | 30% | 23.33% | 0% |
| SNP10 | ||||||||
| TT | 100% | 80% | 83.33% | 80% | 76.67% | 80% | 70% | 100% |
| TC | 0% | 20% | 13.33% | 20% | 20% | 20% | 30% | 0% |
| CC | 0% | 0% | 3.33% | 0% | 3.33% | 0% | 0% | 0% |
| SNP11 | ||||||||
| CC | AB 43.33% | A 40% | A 26.67% | A 26.67% | C 10% | B 53.33% | D 3.33% | A 30% |
| CT | 50% | 56.67% | 66.67% | 56.67% | 33.33% | 33.33% | 93.33% | 66.67% |
| TT | 6.67% | 3.33% | 6.67% | 16.67% | 56.67% | 13.33% | 3.33% | 3.33% |
| SNP12 | ||||||||
| AA | ABC 10% | A 3.33% | AB 6.67% | B 16.67% | D 43.33% | C 13.33% | AB 3.33% | E 3.33% |
| AG | 53.33% | 56.67% | 63.33% | 56.67% | 46.67% | 33.33% | 66.67% | 96.67% |
| GG | 36.67% | 40% | 30% | 26.67% | 10% | 53.33% | 30% | 0% |
| SNP13 | ||||||||
| AA | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% |
| AG | 10% | 10% | 10% | 1.67% | 3.33% | 3.33% | 33% | 0% |
| GG | 90% | 90% | 90% | 98.33% | 96.67% | 96.67% | 67% | 100 |
Label in the top left corner containing the same letter represents the difference was not significant peer data (P > 0.05), without the same letter represents significant difference (P < 0.05) in one line. The frequency distribution of SNP1, 2, 10, and 13 did not satisfy the conditions of Chi-square test
Population differentiation
To evaluate the population differentiations among the eight populations, pairwise FST values between each population were computed using Arlequin v3.5. As shown in Table 5, most of the pairwise FST values among the six native chicken populations were lower than the values between native chicken breeds and commercial chickens. These results thus indicated that all native chicken breeds have very close relationships among themselves, and relatively remote relationships to both commercial chicken breeds (CB and LLH). At the same time, we noticed that pairwise FST value between CB and LLH was higher than the other populations, which suggested that the variation of chicken Mx1 gene may associate with different selected purposes.
Table 5.
Pairwise FST between eight chicken breeds
| CK | GYG | JYB | GSH | TC | DHB | LLH | CB | |
|---|---|---|---|---|---|---|---|---|
| CK | – | 0.09494 | 0.03175 | 0.1511 | 0.1559 | 0.02079 | 0.3395 | 0.1206 |
| GYG | – | 0.07178 | 0.08891 | 0.06242 | 0.09860 | 0.1549 | 0.1337 | |
| JYB | – | 0.06242 | 0.09215 | 0.06078 | 0.2682 | 0.1841 | ||
| GSH | – | 0.05649 | 0.1725 | 0.1272 | 0.3191 | |||
| TC | – | 0.1869 | 0.2216 | 0.2711 | ||||
| DHB | – | 0.3590 | 0.1092 | |||||
| LLH | – | 0.4535 | ||||||
| CB | – |
Genetic diversity
Genetic diversity parameters of the eight chicken populations were calculated and are shown in Table 6. Consistent with the fact that native chicken breeds have seldom been subjected to intensive artificial selections on commercial traits, our results showed that local chicken populations exhibited relatively higher nucleotide diversity than highly selected commercial populations, which is partly due to intensive artificial selections on commercial traits. On the other hand, a total of 91 haplotypes have been identified in the 240 complete coding sequences with an overall haplotype diversity of 0.966. Of these chickens, CK exerted the highest haplotypes and haplotype diversity, while the highly-selected layer LLH possessed the lowest haplotypes and haplotype diversity among the eight populations.
Table 6.
Genetic diversity parameters estimated for Mx1 gene of eight chicken populations
| Populations | Number of polymorphic sites | Nucleotide diversity | Number of Haplotypes | Haplotype diversity |
|---|---|---|---|---|
| CK | 12 | 0.00219 ± 0.00059 | 22 | 0.977 ± 0.014 |
| GYG | 13 | 0.00180 ± 0.00063 | 16 | 0.938 ± 0.026 |
| JYB | 12 | 0.00211 ± 0.00058 | 22 | 0.968 ± 0.019 |
| GSH | 11 | 0.00190 ± 0.00055 | 18 | 0.890 ± 0.052 |
| TC | 12 | 0.00218 ± 0.00059 | 21 | 0.972 ± 0.015 |
| DHB | 13 | 0.00217 ± 0.00063 | 20 | 0.940 ± 0.032 |
| LLH | 10 | 0.00103 ± 0.00051 | 13 | 0.883 ± 0.038 |
| CB | 10 | 0.00135 ± 0.00051 | 20 | 0.954 ± 0.023 |
| Total | 13 | 0.00216 ± 0.00035 | 91 | 0.966 ± 0.004 |
Haplotypes
A total of sixteen haplotypes (H1–H16) higher than 1% frequency were examined from the 240 chickens. The information of each haplotype and the corresponding frequencies of all samples are shown in Table 7, and the haplotype frequencies of each population are summarized in Table 8. The H1, H2, and H3 haplotypes were dominant (> 10%) in the 240 samples, and H1 is same to the reference sequence of NCBI. At the same time, haplotype H1, the largest shared haplotypes, was found in all chicken breeds. Furthermore, we observed that haplotype H1 and H2 were presented at an extremely high frequency in four native chicken populations (CK, GYG, JYB, GSH). The total proportion of H1 and H2 in GSH was even exceed 50%, whereas the frequencies of H1 and H2 were less than 10% in these two commercial lines (LLH and CB). Interestingly, sequences of both GYG and TC breeds harbored multiple variations and the majority of these sequences were not classified into the 16 common haplotypes (data not shown). Moreover, it was found that haplotype H4 was widely distributed in CB chickens (31.6%), but was not detected in LLH. In the same way, haplotype H10 made up 43.3% of the 30 LLH chicken but completely absent in CB. Taken together, it appeared that high artificial selection probably led to the decrease of haplotype diversity in the coding region of chicken Mx1 gene.
Table 7.
Haplotypes of Mx1 gene identified in 240 chickens and the corresponding frequencies
| Haplotypes | Position of variable sites in the coding sequence of chicken Mx1 gene | Frequency of 240 chickens | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | 13 | 62 | 122 | 125 | 156 | 922 | 1015 | 1248 | 1257 | 1455 | 1545 | 2019 | ||
| Reference sequence | A | C | G | C | T | T | G | A | A | T | C | G | G | |
| H1 | · | · | · | · | · | · | · | · | · | · | · | · | · | 0.135 |
| H2 | · | · | A | G | C | A | · | · | G | · | T | A | · | 0.132 |
| H3 | · | T | A | G | C | A | A | G | G | · | · | · | · | 0.124 |
| H4 | · | · | A | · | · | · | A | G | · | · | · | · | · | 0.082 |
| H5 | · | · | A | G | C | A | · | · | · | · | · | · | · | 0.070 |
| H6 | · | · | A | · | · | A | A | G | · | · | T | A | · | 0.064 |
| H7 | · | · | A | G | C | A | · | · | · | · | T | A | · | 0.054 |
| H8 | · | · | · | · | · | · | · | · | · | C | T | A | · | 0.051 |
| H9 | · | · | A | G | C | A | · | · | G | · | · | · | · | 0.037 |
| H10 | · | · | · | · | · | · | · | · | G | · | T | A | · | 0.037 |
| H11 | · | · | · | · | · | · | · | · | G | · | · | · | · | 0.021 |
| H12 | · | · | A | G | C | A | · | · | · | C | T | A | · | 0.019 |
| H13 | · | · | · | · | · | · | · | · | · | · | T | A | · | 0.017 |
| H14 | · | · | · | · | · | · | · | · | G | · | · | · | A | 0.016 |
| H15 | G | T | · | G | C | A | · | · | · | · | · | · | · | 0.014 |
| H16 | · | · | · | · | · | · | A | G | · | C | · | · | · | 0.012 |
Dot (.) denoted the same single nucleotide to the reference sequence; Position of variable sites in the coding sequence of chicken Mx1 gene was obtained using the sequence of Red Jungle Fowl genome as the reference sequence
Table 8.
Mx1 gene haplotype frequency of each population
| Haplotype | Chicken breed lines | |||||||
|---|---|---|---|---|---|---|---|---|
| CK | GYG | JYB | GSH | TC | DHB | LLH | CB | |
| H1 | 0.100 | 0.133 | 0.125 | 0.270 | 0.085 | 0.095 | 0.036 | 0.050 |
| H2 | 0.150 | 0.079 | 0.127 | 0.296 | – | 0.061 | 0.026 | 0.050 |
| H3 | 0.217 | – | 0.117 | 0.050 | – | 0.400 | – | 0.133 |
| H4 | 0.100 | 0.074 | 0.03 | – | – | 0.084 | – | 0.316 |
| H5 | – | 0.168 | 0.270 | 0.061 | 0.037 | – | 0.248 | 0.067 |
| H6 | 0.150 | – | 0.074 | – | – | 0.100 | – | 0.149 |
| H7 | – | 0.091 | 0.019 | 0.015 | 0.286 | – | 0.023 | – |
| H8 | – | – | 0.034 | – | – | 0.100 | – | 0.150 |
| H9 | 0.083 | 0.033 | – | 0.029 | 0.033 | 0.072 | – | – |
| H10 | – | – | 0.040 | 0.018 | 0.110 | – | 0.434 | – |
| H11 | 0.017 | 0.067 | – | 0.074 | – | – | – | – |
| H12 | – | – | 0.057 | 0.083 | – | – | – | – |
| H13 | – | – | – | 0.022 | 0.080 | 0.039 | – | – |
| H14 | – | – | – | – | – | – | 0.113 | – |
| H15 | 0.083 | – | – | – | – | 0.033 | – | – |
| H16 | – | 0.100 | – | – | – | – | – | – |
| Others | 0.1 | 0.255 | 0.107 | 0.082 | 0.369 | 0.016 | 0.12 | 0.085 |
Dashes (–) represented the absence of certain haplotype in the population
Positive selection sites
Positive selection is the main driving force of rapidly evolving genes. The best source of evidence for positive selection at the molecular level is the comparison of rates of synonymous and nonsynonymous nucleotide substitution (Hughes and Nei 1988). When nonsynonymous nucleotide substitutions per nonsynonymous site (dN) exceed the number of synonymous nucleotide substitutions per synonymous site (dS), positive diversifying selection is favored (Hughes 1999). Five codon-based methods in Datamonkey server have proved useful to infer positive selection and identify positive selection sites in protein-coding sequences of chicken Mx1 gene. Three sites (codon 5, 21, 339) were found under positive selection in Mx1 sequence samples by all methods. In addition, FEL also identified codon 42 as positive selection site and FUBAR identified seven positive selection sites (Table 9).
Table 9.
Sites found to be under positive selection using the FEL, IFEL, SLAC, FUBAR, and MEME methods implemented in Datamonkey server (http://www.datamonkey.org)
| Codon | FEL | IFEL | SLAC | FUBAR | MEME | ||||
|---|---|---|---|---|---|---|---|---|---|
| dN-dS | p value | dN-dS | p value | dN-dS | p value | dN-dS | Posterior probability | p value | |
| 3 | 445.025 | 0.105 | 462.228 | 0.125 | 27.567 | 0.277 | 8.409 | 0.965 | 0.131 |
| 5 | 1315.42 | 0.00738 | 1604.91 | 0.0061 | 83.664 | 0.0314 | 21.569 | 0.998 | 0.0124 |
| 21 | 1335.5 | 0.00191 | 1579.08 | 0.00246 | 81.175 | 0.0167 | 21.365 | 0.999 | 0.00356 |
| 41 | 394.713 | 0.106 | 270.06 | 0.219 | 25.803 | 0.257 | 10.399 | 0.984 | 0.132 |
| 42 | 509.563 | 0.0399 | 346.725 | 0.133 | 33.356 | 0.107 | 13.485 | 0.993 | 0.0568 |
| 308 | 665.947 | 0.061 | 795.337 | 0.0535 | 45.878 | 0.150 | 14.959 | 0.992 | 0.0821 |
| 339 | 896.401 | 0.0130 | 1045.32 | 0.013 | 67.468 | 0.026 | 19.902 | 0.999 | 0.0208 |
FEL, IFEL, SLAC, and MEME test significance levels are given as p values, while posterior probability are given for FUBAR
Sites found under positive selection were labeled in bold (p < 0.05, posterior probability > 0.95)
Discussion
To achieve maximum expression of the production traits, the commercial chicken breeds are mainly selected for higher growth or egg production. It seems that most production traits are being improved at the expense of immune competence (Zekarias et al. 2002). In contrast, lowly or moderately selected native chicken breeds always have greater ability than highly selected commercial populations to resist the virus controlled by antiviral genes (Sartika et al. 2011). However, whether the antiviral genes were also influenced after the target traits gained great genetic improvement has not been understood.
Chicken Mx1 gene is one of the best-characterized IFN-stimulated antiviral genes and has the ability to inhibit the replication of RNA virus (Ko et al. 2002), albeit the resistance property is restricted to certain genotypes. Here, we detected the functional contributions of chicken Mx1 polymorphisms of eight chicken breed lines with different selection pressures. In line with the earlier findings that nonsynonymous SNP of chicken Mx1 gene is primarily found in less well-defined regions (Balkissoon et al. 2007; Fulton et al. 2014), and five of the seven nonsynonymous sites in this research are located in domain not determined where the functional significance is not yet known. As for the other two nonsynonymous SNPs, they are harbored in the GTPase domain region, in which GTPase activity has been regarded as an essential role for Mx activity and may be responsible for pathogen recognition (Pitossi et al. 1993). Besides, five codon specific selection methods all suggested three codons (codon 5, 21 and 339) to be positively selected, indicating a strong signature of adaptive evolution. Of the three positively selected codons, codon 5 and codon 21 are located in the domain not determined. Lacking of the specific function of this less well-defined region, we could only speculate that this part may be particularly important in the long-term evolutionary history of antagonistic exposure to various viruses. Codon 339 appears to be located around the edge of the GTPase domain region. The signature of positive diversifying selection occurring on this site may potentially enhance the flexibility to recognize and adapt to a diverse array of evolving pathogens. Interestingly, all the variants identified in the middle domain and GTPase effector domain are synonymous mutations. It has been known that the amino acids in the middle domain and GTPase effector domain are essential for maintaining the quaternary structure of human MxA, and the GTPase effector domain acted as the effector domain in the antiviral activity (Gao et al. 2010, 2011). Thus, the domain with effector function may be structural conservation to maintain crucial function, whereas amino acids mutations were likely needed in the GTPase domain to obtain the regulation of new activity factors and even recognize a diverse array of pathogens. On the other hand, high frequency of amino acid change in domain not determined may give the opportunity to explore new function.
Li et al. have reported a highly skewed Mx1 allele frequency at position 2032 among experimental lines and native Asian breeds of chickens. Results showed that frequency of allele A in native populations was much higher than those in commercial populations (Li et al. 2006). Balkissoon et al. confirmed a high frequency of the susceptibility allele presented in contemporary meat-type birds in 2007. The frequency of GG genotype in several broiler lines is 100% (Balkissoon et al. 2007). The Mx1 gene encoding Ser at amino acid position 631 is conserved in several birds. Seyama et al. investigated the amino acid sequences of Mx1 protein in other birds such as the Japanese quail, turkey, and goose, all of which carry the sensitive type of Ser at this position (Seyama et al. 2006). In the present study, the frequency distribution in SNP4 and SNP5 indicated that the degree of selection pressure influences the distribution of chicken Mx1 gene and it may be associated with higher adaption of local chicken populations. The frequency distribution in SNP3, SNP7, SNP8, SNP11 and SNP12 demonstrated that different purposes of artificial selection potentially causes great influence in the genotype distribution of chicken Mx1 gene. However, the specific variant (Ser631Asn) conferring antiviral activity was not detected. All of the 240 samples from eight chicken breeds all carried the sensitive GG genotype at the r.2032 position, this observation is probably due to the fact that all the indigenous chicken populations in Sichuan province have a very close relationship (Zhang et al. 2017) and limited sample were used in the two commercial breeds. To improve the disease resistance ability of these populations, chickens with the resistance alleles are needed to be introduced to Sichuan native chicken breeds. Alternatively, understanding the function of the newly-identified SNPs may also help us better breed these Chinese native chickens and commercial chickens.
Population genetic analyses of the present study indicated that all native chicken breeds had very close relationships among each other, while the genetic distance between indigenous and commercial chickens were much higher. The results support the hypothesis that chickens from distant geographic regions may have developed different adaptation mechanisms, which render them genetically distinct. The CB and LLH with different selected purpose had higher difference than among any other two compared breeds, which is consist with our previous results based on other antivirus genes (Li et al. 2017). The genetic diversity parameters also indicated that native chicken breeds exhibited relatively higher nucleotide diversity than CB and LLH, indicating that the improvement of commercial traits are associated with the reduction of nucleotide diversity of immune-related gene at least in the case of chicken Mx1 gene. On the other hand, higher diversity of Mx1 gene might help native chickens to better adapt variant local environments. Furthermore, the haplotype frequencies within the eight breed lines evaluated herein indicated that H1 and H2 might be beneficial to higher adaption to harsh environment, while H10 was potentially associated with egg production and H4 was potentially associated with meat production. These data highlight the need for a better understanding of the association between the haplotypes and both disease resistance and performance traits.
Native chicken populations are a source of adapted genotypes that are robust enough to survive in local environments with minimum human interventions (Khobondo 2015). It may be achieved by selectively increasing the frequencies of target alleles and haplotypes within specific marker-assisted schemes (Muchadeyi and Dzomba 2017). Recent study suggested that it is feasible to improve the productivity and enhance antibody responses towards viral infections simultaneously (Psifidi et al. 2016). Genomic tools such as linkage disequilibrium, comparative genomic analysis and transcriptome profiling to detect the selection footprints in the genomes of native chickens could point towards candidate genes for disease resistance (Lillie et al. 2017). The information on the selection hotspots in the native chicken genomes could potentially transfer the benefits to other commercial chicken breeds raised under similar production systems in further investigation.
Conclusion
Our results highlight that different selection pressures including degrees and purposes greatly influenced the genotype distribution of chicken Mx1 gene. The specific haplotypes showed potential association with performance traits.
Author contributions
JL designed and performed the experiments, analyzed the data, prepared tables, authored, or reviewed drafts of the paper. CY acquired materials and funding, authored or reviewed drafts of the paper, approved the final draft. JR conceived and designed the experiments, reviewed the manuscript, and approved the final draft. XJ acquired materials and funding, authored or reviewed drafts of the paper, and approved the final draft. HD authored or reviewed drafts of the paper, and approved the final draft. ZL performed the experiments and analyzed the data. YL authored or reviewed drafts of the paper. LZ conceived and designed the experiments, reviewed the manuscript, approved the final draft.
Funding
This research was supported by the Open Fund of Farm Animal Genetic Resources Exploration (Grant NO. 2016NYZ0043), Innovation Key Laboratory of Sichuan Province (Grant NO. 2017JZ0033) and Fundamental Research Funds of China West Normal University (Grant NO. 17E077).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
The authors declare that the experiments carried out in this work comply with current Chinese and International laws.
Data availability
The following information was supplied regarding data availability. The raw data are provided as a Supplemental File.
Contributor Information
Yiping Liu, Phone: +86-028-86290987, Email: liuyp578@sicau.edu.cn.
Long Zhang, Phone: +86-0817-2260685, Email: zlong4723@163.com, Email: long.zhang@cwnu.edu.cn.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Alzohairy AM. BioEdit: an important software for molecular biology. Gerf Bull Biosci. 2011;2:60–61. [Google Scholar]
- Balkissoon Devanand, Staines Karen, McCauley John, Wood James, Young John, Kaufman Jim, Butter Colin. Low frequency of the Mx allele for viral resistance predates recent intensive selection in domestic chickens. Immunogenetics. 2007;59(8):687–691. doi: 10.1007/s00251-007-0235-5. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Ewald SJ, Kapczynski DR, Livant EJ, Suarez DL, Ralph J, Mcleod S, Miller C. Association of Mx1 Asn631 variant alleles with reductions in morbidity, early mortality, viral shedding, and cytokine responses in chickens infected with a highly pathogenic avian influenza virus. Immunogenetics. 2011;63:363–375. doi: 10.1007/s00251-010-0509-1. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under linux and windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fulton JE, Arango J, Ali RA, Bohorquez EB, Lund AR, Ashwell CM, Settar P, O’Sullivan NP, Koci MD. Genetic variation within the Mx gene of commercially selected chicken lines reveals multiple haplotypes, recombination and a protein under selection pressure. PloS One. 2014;9:e108054. doi: 10.1371/journal.pone.0108054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature. 2010;465(7297):502–506. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- Gao S, von der Malsburg A, Dick A, Faelber K, Schröder GF, Haller O, Kochs G, Daumke O. Structure of myxovirus resistance protein a reveals intra-and intermolecular domain interactions required for the antiviral function. Immunity. 2011;35(4):514–525. doi: 10.1016/j.immuni.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Haller O, Kochs G. interferon-induced Mx proteins: dynamin-like GTPases with antiviral activity. Traffic. 2002;3:710–717. doi: 10.1034/j.1600-0854.2002.31003.x. [DOI] [PubMed] [Google Scholar]
- Haller O, Stertz S, Kochs G. The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect. 2007;9:1636–1643. doi: 10.1016/j.micinf.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Haller O, Staeheli P, Schwemmle M, Kochs G. Mx GTPases: dynamin-like antiviral machines of innate immunity ☆. Trends Microbiol. 2015;23:154–163. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Evolutionary diversification of the mammalian defensins. Cell Mol Life Sci CMLS. 1999;56:94–103. doi: 10.1007/s000180050010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335(6186):167. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Khobondo J. Genetic and nutrion development of indigenous chicken in Africa. Livest Res Rural Dev. 2015;27:3–6. [Google Scholar]
- Ko JH, Jin HK, Asano A, Takada A, Ninomiya A, Kida H, Hokiyama H, Ohara M, Tsuzuki M, Nishibori M. Polymorphisms and the differential antiviral activity of the chicken Mx gene. Genome Res. 2002;12:595–601. doi: 10.1101/gr.210702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Takada A, Mitsuhashi T, Agui T, Watanabe T. Native antiviral specificity of chicken Mx protein depends on amino acid variation at position 631. Anim Genet. 2004;35:119–122. doi: 10.1111/j.1365-2052.2004.01096.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Qu LJ, Yao JF, Yang N. Skewed allele frequencies of an Mx gene mutation with potential resistance to avian influenza virus in different chicken populations. Poultry Sci. 2006;85:1327–1329. doi: 10.1093/ps/85.7.1327. [DOI] [PubMed] [Google Scholar]
- Li JJ, Wang Y, Yang CW, Ran JS, Jiang XS, Du HR, Hu YD, Liu YP. Genotypes of IFIH1 and IFIT5 in seven chicken breeds indicated artificial selection for commercial traits influenced antiviral genes. Infect Genet Evol. 2017;56:54–61. doi: 10.1016/j.meegid.2017.10.019. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lillie M, Sheng Z, Honaker CF, Dorshorst BJ, Ashwell CM, Siegel PB, Carlborg Ö. Genome-wide standing variation facilitates long-term response to bidirectional selection for antibody response in chickens. BMC Genom. 2017;18:99. doi: 10.1186/s12864-016-3414-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmgaard L. Induction and regulation of IFNs during viral infections. J Interf Cytok Res. 2004;24:439–454. doi: 10.1089/1079990041689665. [DOI] [PubMed] [Google Scholar]
- Muchadeyi FC, Dzomba EF. Genomics tools for the characterization of genetic adaptation of low input extensively raised chickens. Poultry Sci. 2017;11:211–229. [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Pond SLK. Detecting individual sites subject to episodic diversifying selection. PLOS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Pond SLK, Scheffler K. FUBAR: A fast, unconstrained bayesian AppRoximation for inferring selection. Mol Biol Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niraj DK, Kumar P, Mishra C, Kashyap N, Narayan R, Bhattacharya TK, Bhushan B, Tiwari AK, Saxena VK, Sahoo NR. Nucleotide sequence analysis of Mx1 gene in Japanese quail. Indian J Anim Res. 2016;50:357–365. [Google Scholar]
- Pitossi F, Blank A, Schröder A, Schwarz A, Hüssi P, Schwemmle M, Pavlovic J, Staeheli P. A functional GTP-binding motif is necessary for antiviral activity of Mx proteins. J Virol. 1993;67(11):6726–6732. doi: 10.1128/jvi.67.11.6726-6732.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Psifidi A, Banos G, Matika O, Desta TT, Bettridge J, Hume DA, Dessie T, Christley R, Wigley P, Hanotte O. Genome-wide association studies of immune, disease and production traits in indigenous chicken ecotypes. Genet Sel Evol. 2016;48:74–90. doi: 10.1186/s12711-016-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartika T, Sulandari S, Zein MSA. Selection of Mx gene genotype as genetic marker for Avian Influenza resistance in Indonesian native chicken. BMC Proceedings. 2011;5:S37. doi: 10.1186/1753-6561-5-S4-S37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Yoneda A, Ninomiya A, Kawahara M, Watanabe T. Both antiviral activity and intracellular localization of chicken Mx protein depend on a polymorphism at amino acid position 631. Biochem Bioph Res Co. 2013;430:161–166. doi: 10.1016/j.bbrc.2012.11.053. [DOI] [PubMed] [Google Scholar]
- Seyama T, Ko JH, Ohe M, Sasaoka N, Okada A, Gomi H, Yoneda A, Ueda J, Nishibori M, Okamoto S. Population research of genetic polymorphism at amino acid position 631 in chicken Mx protein with differential antiviral activity. Biochem Genet. 2006;44:432–443. doi: 10.1007/s10528-006-9040-3. [DOI] [PubMed] [Google Scholar]
- Thomas Anne V., Palm Melanie, Broers Aurore D., Zezafoun Hussein, Desmecht Daniel J.-M. Genomic structure, promoter analysis, and expression of the porcine (Sus scrofa) Mx1 gene. Immunogenetics. 2006;58(5-6):383–389. doi: 10.1007/s00251-006-0109-2. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekarias B, Ter Huurne AA, Landman WJ, Rebel JM, Pol JM, Gruys E. Immunological basis of differences in disease resistance in the chicken. Veterinary Res. 2002;33:109–125. doi: 10.1051/vetres:2002001. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang P, Li Q, Gaur U, Liu Y, Zhu Q, Zhao X, Wang Y, Yin H, Hu Y. Genetic evidence from mitochondrial DNA corroborates the origin of tibetan chickens. PloS one. 2017;12:e0172945. doi: 10.1371/journal.pone.0172945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following information was supplied regarding data availability. The raw data are provided as a Supplemental File.