Abstract
Background and Aims
The aim of this study was to investigate the genome-wide DNA methylation status in treatment-naïve ulcerative colitis [UC], and to explore the relationship between DNA methylation patterns and gene expression levels in tissue biopsies from a well-stratified treatment-naïve UC patient group.
Methods
Mucosal biopsies from treatment-naïve patients [n = 10], and a healthy control group [n = 11] underwent genome-wide DNA bisulfite sequencing. Principal component analysis [PCA] and diverse statistical methods were applied to obtain a dataset of differentially methylated genes. DNA methylation annotation was investigated using the UCSC Genome Browser. Gene set enrichments were obtained using the Kyoto Encyclopaedia of Genes and Genomes [KEGG] and PANTHER.
Results
Of all significantly differentially expressed genes [DEGs], 25% correlated with DNA methylation patterns; 30% of these genes were methylated at CpG sites near their transcription start site [TSS]. Hyper-methylation was observed for genes involved in homeostasis and defence, whereas hypo-methylation was observed for genes playing a role in immune response [i.e. chemokines and interleukins]. Of the differentially DNA methylated genes, 25 were identified as inflammatory bowel disease [IBD] susceptibility genes. Four genes [DEFFA6, REG1B, BTNL3, OLFM4] showed DNA methylation in the absence of known CpG islands.
Conclusions
Genome-wide DNA methylation analysis revealed distinctive functional patterns for hyper-and hypo-methylation in treatment-naïve UC. These distinct patterns could be of importance in the development and pathogenesis of UC. Further investigation of DNA methylation patterns may be useful in the development of the targeting of epigenetic processes, and may allow new treatment and target strategies for UC patients.
Keywords: Genome-wide DNA methylation, ulcerative colitis [UC]
1. Introduction
Ulcerative colitis [UC] is one of the two most common conditions that constitute inflammatory bowel disease [IBD] in addition to Crohn’s disease [CD]. Whereas CD can affect any area in the gastrointestinal tract, UC affects the mucosa and submucosa of the colon and rectum.1,2 Due to chronic inflammation, patients affected by UC have a higher risk of developing colorectal cancer.3 Approximately 20% of IBD cases can be explained by known genetic variants, suggesting a more complex pathogenesis.4–6 The current knowledge of the underlying causes of UC is still incomplete. A complex interplay between genetic variation, host immune system, environmental factors and intestinal microbiota has been suggested.4 Therefore, it has been implied that epigenetic mechanisms may play an important role in disease development of UC.7–11 Epigenetic processes regulate gene expression via modifications of DNA, histone proteins and chromatin, and are known to play a role in complex disease phenotypes.12 Epigenetic modifications, such as DNA methylation, are believed to have a role in the immune dysfunction associated with IBD.13 They are influenced by several environmental factors such as diet14,15 and smoking16 which are known to be associated with inflammatory diseases.17,18 Gene-specific changes in DNA methylation in the pathogenesis of IBD have been recently reported.19–22 DNA methylation plays a regulatory role in gene transcription, either by activation of proteins that interfere with the suppression of gene transcription, or by inhibiting transcription factors from binding to DNA.23–25 That is why it isimportant to examine the interaction between gene expression and DNA methylation.
Therefore, in the present study we applied genome-wide methylation profiling by using bisulfite sequencing in order to obtain DNA methylation patterns at a single base-pair resolution.26,27 This method is a more quantitative approach in producing data with genome-wide coverage than other technologies.28,29 In addition, DNA methylation has been correlated to transcriptional levels of genes in order to demonstrate possible regulatory DNA methylation features of relevance for UC.
2. Materials and Methods
2.1. Patient material
A standardised sampling method was used to collect mucosal biopsies from the colon of newly diagnosed, treatment-naïve UC patients with mild to moderate disease activity [n = 10] and controls [n = 11]. For controls, biopsies from subjects undergoing cancer screening with normal colonoscopy and normal colonic histological examination were used. UC was diagnosed based upon established clinical, endoscopic and histological criteria as defined by the ECCO guidelines.30 The grade of inflammation was assessed during colonoscopy using the UC disease activity index [UCDAI] endoscopic sub-score, with 3 to 10 for mild to moderate disease.31 All biopsies were taken from the sigmoid part of the colon and the case biopsies from a site of active inflammation. Tumour necrosis factor alpha [TNF-α] mRNA expression levels were measured by real-time quantitative polymerase chain reaction PCR [qPCR], thereby indicating the grade of UC activity.32 This study is part of a larger, already published study, where the gene expression using transcriptome data was assessed.33 The samples were taken from an established biobank approved by the Norwegian Board of Health. The participants signed an informed and written consent form. The study was approved by the Regional Ethics Committee of North Norway and the Norwegian Social Data Services [REK Nord 2012/1349].
2.2. DNA and RNA isolation
Genomic DNA was isolated using the Allprep DNA/RNA Mini Kit from Qiagen and the QIAcube instrument [Qiagen, Hilden, Germany], according to the manufacturer’s protocol. DNA quantity and purity were assessed by using the NanoDrop ND-1000 spectrophotometer [Thermo Fisher Scientific, Wilmington, DE, USA]. The DNA samples were kept at -80°C until further handling.
2.3. Quantitative polymerase chain reaction [qPCR]
The TNF-α levels in biopsies were measured using qPCR. RNA quantity was assessed with NanoVue Plus [GE Healthcare, UK]. Synthesis of cDNA was performed using the QuantiTect Reverse Transcription Kit [Qiagen], and the QuantiNova Probe PCR Kit [Qiagen, Hilden, Germany]. Beta-actin [β-actin] was used as housekeeping gene. For the detection, a CFX Connect Real Time PCR Detection System [Bio-Rad, USA] was used. The results were measured in copies/µg protein. Values <7000 copies/µg protein are considered as non-inflamed tissues, and values >7000 copies/µg protein are considered as inflamed tissues.32
2.4. Library preparation and next-generation sequencing
DNA libraries were prepared with the SeqCap Epi CpGiant Enrichment kit [Roche, Switzerland]. The DNA was bisulfite-converted using the EZ DNA Methylation-lightning Kit [Zymo Research, USA] before the hybridisation step and according to the manufacturer’s instructions. Transcriptome libraries were prepared as described previously.33 The amount of input material was 1060 ng of genomic DNA per sample. The Bioanalyzer 2100 and the Agilent DNA 1000 kit [Agilent Technologies, Santa Clara, USA] were used to assess the quality of DNA libraries. DNA libraries with an average fragment size of 329 bp were generated, then diluted to 2 nM, and subsequently sequenced with the NextSeq 550 instrument [Illumina, USA] according to the manufacturer’s instructions.
2.5. Data analysis
The algorithm package STAR-2.5.2b [https://github.com/alexdobin/STAR] was used for down-stream analysis of the transcriptome.34 Transcripts were aligned to UCSC GRCh38/hg38 [http://hgdownload.cse.ucsc.edu/goldenPath/hg38/]. The count matrix was generated by HTSeq-count [https://htseq.readthedocs.io/en/release_0.9.1/], normalised by DESeq2 [https://bioconductor.org/packages/release/bioc/html/DESeq2.html]. Principal component analysis [PCA],35 Limma,36 and p-value adjustment methods37 in Bioconductor R [https://www.bioconductor.org/] were used to obtain and characterise a dataset of significant DEGs and for analysis of relative methylation in patient samples.
For DNA methylation analyses, the Bismark Bisulfite Mapper v0.16.0 [www.bioinformatics.bbsrc.ac.uk/projects/bismark/] was used and the same genome build as the transcriptome was used to generate methylation counts. The globalTest function from the BiSeq package [https://bioconductor.org/packages/release/bioc/html/BiSeq.html] was used to find significant differentially methylated regions between UC and normal samples. As whole-genome bisulfite sequencing [WGBS] data are extremely computationally expensive and rather large, areas of interest were reduced to the promoter regions of expressed transcripts [DESeq2] and transcripts whose normalised log2 [counts] were greater than 5. To find differentially methylated regions, Goeman’s Global test was used.38,39 A modified algorithm of the Goemans’s Global test in the BiSeq package [https://bioconductor.org/packages/release/bioc/vignettes/BiSeq/inst/doc/BiSeq.pdf] works with relative methylation data and score tests, with methylation levels as independent variables. Promoters with Goeman’s test p-values < 0.05 were kept for further use. Significant regions with low coverage [few methylation sites] or a poor Goeman’s statistic were removed. Differentially methylated regions [DMR] of interest were restricted to 200 bp downstream and 2000 bp upstream of a transcription start site [TSS]. DMR regions were further restricted to those containing a minimum of four methylation events. DMRs were investigated with the UCSC Genome Browser [https://genome.ucsc.edu/].
Genes associated with the risk of IBD were downloaded from the genome-wide association studies [GWAS] catalogue,40 using the search term IBD [www.ebi.ac.uk/gwas]. Gene set enrichments were performed by using the PANTHER classification system [https:// pantherdb.org/], the Kyoto Encyclopaedia of Genes and Genomes [KEGG; www.genome.jp/kegg/]. For principal component analysis [PCA] of the transcription data, the top 5000 most variable of the DESeq2-Rlog variance stabilised transcripts were used. For the estimation of specific cell populations in patient transcription samples, all DESeq2-Rlog normalised transcripts with a log2 average mean >5 were included. The analysis was performed using the R/Bioconductor CellMix manual [http://web.cbio.uct.ac.za/~renaud/CRAN/web/CellMix/] with the IRIS weighted marker list characteristic for the different cell types41 and as described previously.33 To investigate the correlation between the cell deconvolution PCA and the methylation PCA scores, partial least squares regression [PLSR] [https://cran.r-project.org/web/packages/pls/index.html] was used.42 In order to visualise co-variation, the procrustes algorithm [https://www.rdocumentation.org/packages/vegan/versions/2.4-2/topics/procrustes] was applied.43
Fisher’s exact test44 was used to compare if the gene list depicted in Supplementary Data 2 [available as Supplementary data at ECCO-JCC online] associated with significant methylation changes to gene lists from previous microarray-based methylation studies.22,45,46
3. Results
3.1. Patients
Mucosal biopsies from treatment-naïve UC patients [n = 10] and controls [n = 11] were collected according to a standardised sampling method, as described in Materials and Methods [section 2]. The disease activity within UC patients was classified30 as mild to moderate as described by the UCDAI; the biopsies showed clinical scores of 7 ± standard deviation [SD] 2.6 and endoscopy scores of 1.9 ± SD 0.3. The biopsies from the control group showed normal colonoscopy, colon histology, and immunohistochemistry, with clinical and endoscopy scores = 0. All biopsies were taken from the sigmoid part of the colon. Gender distribution within both groups was almost identical, with seven males in the UC group, eight males in the control group, and three females in each group. The age distribution differed between the groups, at 37 ± SD 12 years in the UC group, and 52 ± SD 14 years in the control group. TNF-α mRNA expression levels were measured by qPCR to estimate the inflammatory status of UC.32 TNF-α measurements in UC group were estimated as 13,240 ± SD 6056, and for control group as 4291 ± SD 1878. A summary of all patient characteristics is listed in Table 1.
Table 1.
Patient characteristics.
| Characteristics | Control group [n = 11] | Ulcerative colitis [n = 10] |
|---|---|---|
| Male/female | 8/3 | 7/3 |
| Age mean ± SD | 52 ± 14 | 37 ± 12 |
| Endo score mean ± SD | 0 | 1.9 ± 0.3 |
| Clinical score ± SD | 0 | 7 ± 2.6 |
| TNF-α level ± SD | 4291 ± 1878 | 13,240 ± 6056 |
SD, standard deviation; TNF, tumour necrosis factor.
3.2. Characterisation of DNA methylation in treatment-naïve UC
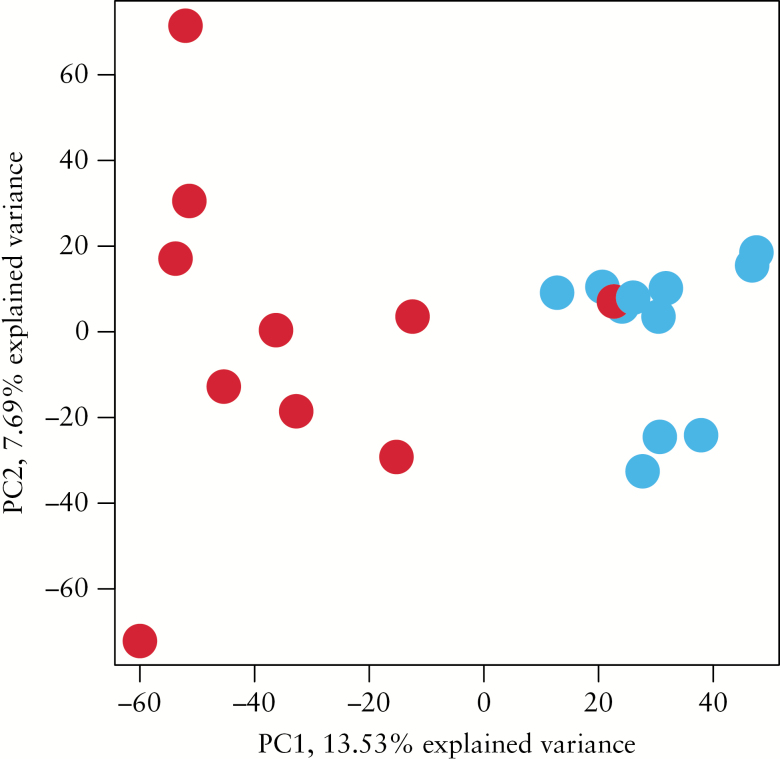
Pre-processing of the initial RNA and bisulfite sequencing data revealed expression of about 22000 transcripts which were used for initial principal component analysis [PCA] depicting relative methylation counts [0–100%] for over 9 million cytosine positions for the whole genome of all patient samples, both treatment-naïve UC and normal controls. PCA revealed a clear distinction between UC samples and normal control samples along the first component with a 13.5% explained variance [Figure 1]. Only one patient sample could not be distinguished from normal samples by this method [Figure 1]. A PCA plot indicating the age of participants showed no evidence for age clustering [Supplementary Data 1, available as Supplementary data at ECCO-JCC online].
Figure 1.
Unsupervised principal component analysis [PCA] depicting relative methylation counts [0–100%] for over nine M cytosine positions, including treatment-naïve ulcerative colitis [UC] [red; n = 10] and normal control [blue; n = 11] patient tissue samples with a variance of 13.5%.
The whole transcriptome of treatment-naïve UC has been recently established, and has been used as a basis for the interpretation of DNA methylation patterns in UC.33 Results show that 25% of the significantly DEGs [P-value < 0.05; log2 fold-change > 1.0; n = 357] correlated with the observed differential DNA methylation, which resulted in 30% hyper-methylated [n = 87] and 70% hypo-methylated [n = 270] genes [Supplementary Data 2]. Further analysis revealed that approximately 30% of the genes showed DNA methylation at CpG sites in the neighbourhood of their transcription start site [TSS], whereas the remaining 70% showed DNA methylation events at cis-acting elements like DNAse1 and enhancers. The relationship between the raw methylation data per sample, average difference between groups, and relationship to the TSS and transcript expression for all differentially methylated genes is depicted in Supplementary Data 3 [available as Supplementary data at ECCO-JCC online].
Of the differentially methylated genes, 25 have been related to the currently known 295 IBD susceptibility genes, of which 23 were hypo-methylated and two were hyper-methylated, and correlated with their direction of transcription [Table 2]. Gene annotation revealed their involvement in pathways for cell adhesion, intracellular signalling, metabolism, and transport. For six of the IBD susceptibility genes, neutrophil-activating protein 78 [CXCL5], fatty acid desaturase 1 [FADS1], intercellular adhesion molecule 1 [ICAM], solute carrier family 22 member 4 [SLC22A4], tumour necrosis factor receptor superfamily, member 5 [CD40; TNFRSF5], and TNF receptor superfamily member 4 [TNFRSF4] DNA methylation occurred at the transcription start site [TSS] and as indicated in Table 2.47,48
Table 2.
DNA methylated inflammatory bowel disease [IBD] susceptibility genes in treatment-naïve ulcerative colitis [UC].
| Gene symbol & annotation | % methyl | p < 0.05 methyl | #c | p < 0.05 transcript | Log2 fc > 1.0 transcript |
|---|---|---|---|---|---|
| Cell adhesion & intercellular signalling | |||||
| CD40a | 6,81 | 0,0426 | 94 | 1,12E-20 | 1,64 |
| CXCL5a | 19,21 | 1,38E-05 | 30 | 7,17E-20 | 4,62 |
| CXCL6 | 10,20 | 0,0041 | 18 | 5,46E-26 | 4,06 |
| CXCR5 | 16,18 | 0,0004 | 36 | 1,36E-07 | 2,06 |
| FCGR3A | 2,69 | 0,0036 | 30 | 2,02E-20 | 2,13 |
| ICAM1a | 2,65 | 0,0034 | 18 | 6,61E-28 | 1,96 |
| IFNG | 8,55 | 0,0057 | 10 | 1,54E-06 | 1,54 |
| IL12RB2 | 1,97 | 0,0035 | 39 | 7,15E-10 | 1,03 |
| IL2RA | 25,53 | 0,0001 | 24 | 2,51E-17 | 2,45 |
| ITGAL | 15,90 | 0,0002 | 10 | 2,64E-12 | 1,29 |
| OSM | 22,59 | 0,0002 | 36 | 2.00E-17 | 3,09 |
| SLAMF1 | 26,79 | 3,12E-06 | 11 | 1,72E-11 | 1,34 |
| SLAMF7 | 11,29 | 0,0062 | 7 | 1,11E-07 | 1,04 |
| TNFRSF9 | 0,40 | 0,0075 | 6 | 5,27E-24 | 2,91 |
| TNFRSF4a | 3,65 | 0,0348 | 76 | 3,46E-18 | 1,73 |
| TNFSF8 | 28,03 | 0,0002 | 23 | 1,71E-06 | 1,01 |
| Intracellular signalling | |||||
| APOBEC3G | 36,88 | 0,0241 | 55 | 1,68E-15 | 1,35 |
| CCDC88B | 17,29 | 5,89E-06 | 118 | 1,18E-23 | 1,95 |
| CD6 | 19,40 | 3,13E-05 | 48 | 2,34E-10 | 1,44 |
| DOK3 | 10,80 | 0,0082 | 30 | 5,03E-22 | 1,77 |
| UBASH3A | 5,39 | 0,0381 | 8 | 1,29E-06 | 1,05 |
| Metabolism | |||||
| ARHGAP30 | 36,89 | 2,22E-07 | 17 | 1,19E-12 | 1,13 |
| FADS1a | 0,18 | 0,0141 | 30 | 7,93E-12 | 1,30 |
| SULT1A2 | -8,85 | 0,0089 | 13 | 7,56E-09 | -1,67 |
| Transport | |||||
| SLC22A4a | -0,37 | 0,0092 | 172 | 1,24E-27 | -1,72 |
#c indicates number of methylated cytosines; % methyl indicates % difference of DNA methylation normal [N]-UC.
aCpG sites at their transcription start site [TSS].
The most down-regulated [P < 0.05 and log2FC > 1.5] and hyper-methylated genes in treatment-naïve UC are depicted in Table 3. Annotations revealed genes with the most possible relevance for UC: six members of the solute carrier family [SLC17A8, SLC22A4, SLC25A34, SLC30A10, SLC3A1, and SLC6A19], two guanylate cyclase activators [GUCA2A and GUCA2B], defensin B1 [DEFB1], intestinal alkaline phosphatase 1 [ALPI], two UDP-glucuronosyltransferases [UGTA8 and UGTA10], bone morphogenic protein/retinoic acid inducible neural-specific 3 [BRNP3], and proline rich acidic protein [PRAP1].
Table 3.
TOP down-regulated and hyper-methylated genes in treatment-naïve ulcerative colitis [UC]
| Gene symbol | % methyl | p < 0.05 | #c | p < 0.05 | Log2fc > 1.5 |
|---|---|---|---|---|---|
| methyl | transcript | transcript | |||
| ADIRF | -5,72 | 0,004 | 36 | 4,34E-18 | -1,88 |
| AGMO | -10,86 | 0,022 | 7 | 9,44E-09 | -1,57 |
| ALPI | -3,05 | 0,0102 | 34 | 1,10E-14 | -1,91 |
| ANKRD62 | -0,19 | 0,0011 | 91 | 2,47E-14 | -2,25 |
| BCHE | -4,17 | 0,0442 | 10 | 4,86E-12 | -1,69 |
| BRINP3 | -0,22 | 0,0407 | 29 | 6,76E-11 | -1,74 |
| CLDN8 | -0,45 | 0,0005 | 8 | 2,17E-05 | -1,9 |
| CYP3A4 | -15,45 | 0,0009 | 2 | 3,87E-21 | -3,58 |
| DEFB1 | -7,09 | 0,0334 | 16 | 2,16E-11 | -1,94 |
| FABP1 | -16,56 | 1,52E-05 | 18 | 1,36E-07 | -1,66 |
| FAM151A | -3,59 | 0,0091 | 16 | 4,28E-12 | -1,59 |
| FRMD1 | -2,58 | 0,0076 | 109 | 4,49E-14 | -2,26 |
| GBA3 | -14,5 | 0,0033 | 7 | 1,15E-07 | -1,98 |
| GUCA2A | -12,84 | 0,0134 | 18 | 8,25E-10 | -2,26 |
| GUCA2B | -3,08 | 0,0345 | 41 | 2,67E-08 | -2,18 |
| HAVCR1 | -12,55 | 3,60E-05 | 6 | 4,58E-12 | -2,28 |
| HMGCS2 | -13,13 | 6,56E-05 | 18 | 2,91E-08 | -2,41 |
| HSD17B2 | -2,69 | 0,0456 | 15 | 1,71E-23 | -2,11 |
| MEP1A | -11,49 | 0,009 | 18 | 1,76E-16 | -2,15 |
| OTC | -6,32 | 0,0359 | 28 | 7,09E-10 | -1,68 |
| PCK1 | -6,73 | 0,0212 | 18 | 1,52E-07 | -2,18 |
| PNLIPRP2 | -15,2 | 5,99E-05 | 23 | 1,63E-05 | -1,74 |
| PRAP1 | -1,32 | 0,0025 | 260 | 1,97E-09 | -2,49 |
| SLC17A8 | -2,89 | 0,0082 | 45 | 2,95E-06 | -1,6 |
| SLC22A4 | -0,35 | 0,0174 | 172 | 1,25E-27 | -1,72 |
| SLC25A34 | -12,01 | 0,0099 | 40 | 2,17E-15 | -1,76 |
| SLC30A10 | -7,67 | 0,0006 | 14 | 4,89E-10 | -1,89 |
| SLC3A1 | -15.79 | 0,0002 | 10 | 1,29E-17 | -2,43 |
| SLC6A19 | -12,17 | 0,0057 | 68 | 1,53E-09 | -2,82 |
| SULT1A2 | -11,71 | 0,003 | 13 | 7,56E-09 | -1,67 |
| TINCR | -0,66 | 0,0107 | 72 | 1,58E-12 | -1,98 |
| TMIGD1 | -9,19 | 0,0135 | 6 | 1,16E-09 | -2,32 |
| UGT1A10 | -8,16 | 0,0005 | 34 | 4,04E-15 | -1,79 |
| UGT1A8 | -5,96 | 0,0091 | 23 | 1,67E-10 | -1,85 |
#c indicates number of methylated cytosines. % methyl indicates % difference of DNA methylation normal [N]-UC.
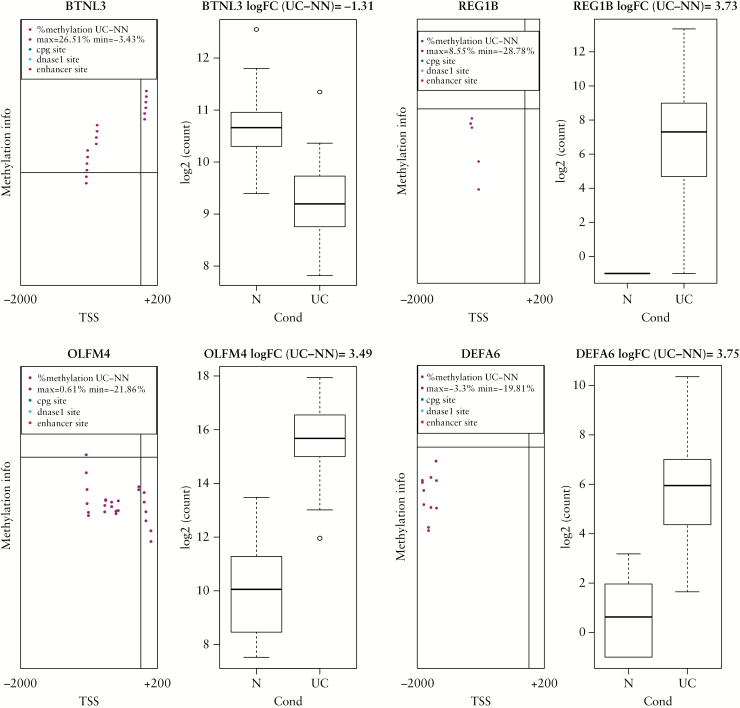
Hypo-methylated genes in treatment-naïve UC are listed in Table 4. DEGs with log2FC > 3.0 and P < 0.05 and their corresponding methylation status have been included in the list. The annotation of the hypo-methylated transcripts revealed genes involved in inflammatory responses, like chemokine receptors [CXCR1, CXCR2], chemokine ligands [CXCL5, CXCL6], interleukins [IL 17A, IL1B], defensins [REG3A, DEFA6], and genes involved in cytokine signalling [SAA1, SAA2, LCN2]. Other hypo-methylations relevant for UC are observed for transporters like aquaporin 9 [AQP9], members of the solute carrier family [SLC6A4, SLC6A14], oncostatin [OSM], and olfactomedin [OLFM4]. For four genes, the observed DNA methylation occurred in the absence of CpG islands or other well-known cis-acting regulatory domains; these genes are defensin A6 [DEFA6], olfactomedin 4 [OLFM4], regenerating protein beta 1 [REG1B], and butyrophilin like protein3 [BTNL3], as shown in Figure 2 and Table 5.
Table 4.
TOP up-regulated and hypo-methylated genes in treatment-naïve ulcerative colitis [UC].
| Gene symbol | % methyl | p < 0.05 methyl | #c |
p < 0.05
transcript |
Log2 fc > 3.0 transcript |
|---|---|---|---|---|---|
| AQ9 | 6,39 | 0,0013 | 17 | 3,562E-32 | 5,37 |
| C2CD4A | 2,43 | 0,0007 | 64 | 1,99E-35 | 3,72 |
| CD300E | 13,58 | 0,0036 | 11 | 4,45E-22 | 3,44 |
| CHI3L2 | 11,96 | 6,26E-06 | 38 | 8,53E-17 | 3,32 |
| CHRDL2 | 10,62 | 0,0403 | 65 | 5,82E-20 | 3,34 |
| CXCL5 | 18,85 | 1,20E-05 | 30 | 7,17E-20 | 4,63 |
| CXCL6 | 11,03 | 0,0020 | 18 | 5,46E-26 | 4,06 |
| CXCR1 | 6,08 | 0,0013 | 6 | 3,92E-23 | 4,64 |
| CXCR2 | 6,03 | 0,0159 | 20 | 2,37E-17 | 3,44 |
| DEFA6 | 10,44 | 0,0002 | 12 | 1,74E-15 | 3,75 |
| DMBT1 | 11,24 | 5,84E-06 | 16 | 4,19E-17 | 3,49 |
| FCN1 | 8,55 | 0,0459 | 12 | 3,57E-28 | 3,25 |
| FFAR2 | 2,16 | 9,25E-05 | 54 | 1,91E-20 | 3,02 |
| GABRP | 7,43 | 0,0257 | 16 | 8,36E-35 | 3,72 |
| GZMB | 12,88 | 0,0005 | 6 | 3,37E-24 | 3,16 |
| HCAR2 | 9,48 | 0,0012 | 26 | 8,69E-22 | 3,58 |
| HCAR3 | 14,16 | 0,0010 | 18 | 5,01E-22 | 4,32 |
| IL17A | 9,08 | 7,82E-06 | 32 | 1,50E-33 | 5,03 |
| IL1B | 9,15 | 2,73E-08 | 13 | 1,09E-32 | 3,28 |
| LCN2 | 5,23 | 0,0243 | 29 | 2,24E-53 | 4,95 |
| LYPD5 | 3,93 | 0,0254 | 40 | 5,55E-23 | 3,07 |
| OLFM4 | 10,75 | 0,0251 | 24 | 1,02E-13 | 3,49 |
| OSM | 22,21 | 0,0008 | 36 | 2,01E-17 | 3,10 |
| PI3 | 2,26 | 0,0153 | 14 | 2,73E-31 | 4,11 |
| REG1B | 6,63 | 0,0283 | 7 | 3,76E-12 | 3,73 |
| REG3A | 13,27 | 0,0299 | 2 | 2,63E-15 | 4,19 |
| S100A9 | 8,26 | 0,0002 | 40 | 1,00E-17 | 3,46 |
| SAA1 | 10,55 | 5,19E-06 | 26 | 5,16E-46 | 6,40 |
| SAA2 | 11,89 | 0,0075 | 6 | 7,31E-43 | 6,23 |
| SLC26A4 | 5,22 | 8,27E-07 | 54 | 3,26E-15 | 3,28 |
| SLC6A14 | 3,99 | 0,0432 | 29 | 1,73E-48 | 5,66 |
#c indicates number of methylated cytosines. % methyl indicates % difference of DNA methylation normal [N]-UC..
Figure 2.
Genes with novel DNA methylation features in treatment-naïve ulcerative colitis [UC]. The left side of individual illustrations shows the difference in relative methylation level between UC and normal samples [N]. Transcription start site [TSS] is indicated as a vertical line. The horizontal line shows where UC methylation equals N methylation. Each black circle represents a methylation event. Black circles over the horizontal line represent an increase in UC sample methylation compared with N methylation at that site. Black circles under the horizontal line represent an increase in N sample methylation compared with UC methylation at that site. The region between 2000 bp upstream and 200 bp downstream of the TSS is shown. The right side of individual illustrations shows boxplots of DESEQ2 log2 normalised values for gene of interest, normal control [N] versus ulcerative colitis [UC]. Genes are indicated: defensin A6 [DEFA6]; olfactomedin 4 [OLFM4]; butyrophilin like 3 [BTNL3]; regenerating protein 1B [REG1B].
Table 5.
Genes with novel DNA methylation features.
| Gene symbol | % methyl | p < 0.05 methyl | #c |
p < 0.05
transcript |
log2 FC > 1.0
transcript |
|---|---|---|---|---|---|
| BTNL3 | -13,20 | 0.0012 | 16 | 4,97E-07 | -1.31 |
| DEFA6 | 9,20 | 0.0002 | 12 | 4,56E-17 | +3.75 |
| OLFM4 | 9,95 | 0.0251 | 24 | 3,83E-15 | +3.49 |
| REG1B | 6,68 | 0.0282 | 7 | 1,95E-13 | +3.73 |
#c indicates number of methylated cytosines. % methyl indicates % difference of DNA methylation normal-ulcerative colitis [N-UC].
In order to explain if cell type populations in biopsies can explain some of the variation in DNA methylation profiles, previously reported cellndeconvolution results of the transcriptome33 and DNA methylation data were patient-matched and underwent PCA analysis. Further, partial least squares regression [PLSR]42 between the cell deconvolution PCA and the methylation PCA scores showed a strong correlation [Supplementary Data 4, available as Supplementary data at ECCO-JCC online].
To visualise co-variation between cell type contributions to methylation data, a biplot of the initial deconvolution PCA for the transcriptome was used in order to display information on both samples and variables [cell types] of the PCA result graphically. The procrustes algorithm43 was then used to overlay the methylation PCA sample scores onto the cell deconvolution biplot. The result shows that cell type is a significant determinant in methylation profile [Supplementary Data 5, available as Supplementary data at ECCO-JCC online].
Comparison of methylated genes [Supplementary data 2] showed significant overlaps with previous comparisons between methylation status peripheral blood mononuclear cells [PBMCs] from UC cases and controls,45 intestinal biopsies from controls and UC patients,46 and rectal biopsies22 [Supplementary Data 6, available as Supplementary data at ECCO-JCC online].
4. Discussion
It is generally accepted that epigenetic mechanisms like DNA methylation are contributing factors in the pathogenesis of IBD.13,45 The present study is the first comprehensive study giving a truly genome-wide description of DNA methylation of treatment-naïve UC using next-generation sequencing [NGS]-based bisulfite-sequencing. Furthermore, this study provides an interpretation of DNA methylation status in treatment-naïve UC with correlation to transcriptional levels of genes.33
Genome-wide DNA methylation changes in UC have been usually investigated by applying microarray technologies.19,45,49,50 A correlation between DNA methylation and gene transcription has not been established, except for two recent publication where a few gene candidates have been confirmed by pyro-sequencing.22,46 However, the evaluated degree of overlap between the present gene list [Supplementary Data 2] and previous genome-wide analyses of methylation using microarray-based technologies22,45,46 is much larger than those expected by chance [Supplementary Data 6, available as Supplementary data at ECCO-JCC online]. The use of microarray technology has several limitations, one of which is that attached array oligo probes might include single nucleotide polymorphisms [SNPs] or repetitive elements which can affect the outcome of the methylation analysis.19 In addition, pre-defined oligo probes do not cover all regions in the genome where methylation could occur,28,29 leaving possible methylation events undetected and/or resulting in compromised DNA methylation patterns. This may be the reason for contradictory results regarding the DNA methylation of neutrophil-activating peptide 78 [CXCL5] in UC, where hyper-methylation has been recently reported which is in contrast to the hypo-methylation observed in this study.20 All these limitations are bypassed with next-generation sequencing technology where methylation detection occurs at a single base-pair, thereby providing methylation profiles with full nucleotide level resolution.26
The use of a thoroughly stratified patient group representing only treatment-naïve UC for DNA methylation analysis offered a unique opportunity to investigate the DNA methylation state before prescription of any medication. This is of importance, since recent reports implied that medication, such as various non-prescription, over-the-counter non-steroidal anti-inflammatory drugs [NSAIDs] and immunosuppressive drugs can have short- and long-term effects on the immune response.51 For example, aspirin has been shown to result in hypo-methylation of cadherin 1 [CDH1] in the gastric mucosa.52 In addition, immunosuppressant therapy and long-standing disease might inaugurate unwanted bias in experiments aiming to investigate the treatment-naïve status of UC.53–55 In addition, age may affect DNA methylation.56,57 However, the results from the patient population of this study indicated that age does not seem to play a significant role [Supplementary Data 1, available as Supplementary data at ECCO-JCC online]. This might be due to the small number of patients aged over 60 in both control and patient groups.
Many studies have characterised DNA methylation in UC without relating the obtained data to transcriptional levels of genes.19,20,22 The correlation of DNA methylation status with transcription levels of genes are of importance in order to define physiological implications of the DNA methylation event. We have previously characterised the whole transcriptome of treatment-naïve UC and used these data in order to relate DNA methylation to gene expression.33 The results revealed that only 72% of DNA methylation events correspond with differential gene transcription levels [see Supplementary Data 2]. Annotations of DNA methylation sites covered regions of 2000 bp upstream and 200 bp downstream of the transcription start site [TSS] of genes, and were found to be correlated with transcription levels, thereby revealing possible disease-specific methylation patterns. It is noted that CpG islands are associated with the control of gene expression, and it would be expected that CpG islands might display tissue-specific patterns of DNA methylation.23,58,59 However, it has been shown that CpG islands associated with TSS rarely show tissue specific methylation patterns24,25,60–62 Instead, CpG regions located as far as 2 kb from CpG islands have highly conserved patterns of tissue-specific methylation, and methylation is highly correlated with reduced gene expression.63 Taking this into consideration, our data revealed 90 genes containing CpG sites in this region whereof 58 were at the transcription start site [TSS] of genes; 34 genes showed DNA methylation upstream TSS and might be considered as tissue-specific DNA methylation sites [Supplementary Data 3]. A number of hyper-methylations with corresponding gene expression levels have been found in this study [Table 3]. For example, bone morphogenic/retinoic acid inducible neural-specific protein 3 [BRINP3] has been reported to be usually under-expressed in UC.64 BRINP3 expression is influenced by DNA hyper-methylation within its promoter, as has been reported recently.65
Cell type populations present in tissue biopsies might also explain some of the variation observed in DNA methylation profiles [Supplementary Data 4 and 5]. During inflammation of the mucosa, the fraction of epithelial cells is diminished, which results in impaired intestinal permeability and a dysregulation of homeostasis.66,67 This might be reflected by the hyper-methylation and down-regulation of proline-rich acidic protein 1 [PRAP1] and members of the solute carrier protein family [SLC6A19 and SLC3A1] which are involved in the maintenance of homeostasis in epithelial cells.68–70 In concordance, hyper-methylation of genes that are involved in the gut mucosal defence system could also be detected, such as intestinal alkaline phosphatase 1 [ALPI] which is involved in the prevention of bacterial translocation in the gut,71 and defensin B1 [DEFB1] which is predominately expressed in neutrophils and is implicated in the resistance of epithelial surfaces to microbial colonisation.72 Members of the UDP glucuronosyltransferase family [UGT1A10 and UGT1A8] are located primarily in gastrointestinal [GI] mucosa from the duodenum to through the colon,73–75 and are involved in detoxification in order to restrict GI absorption of damaging chemicals via the cytochrome P450 system [CYP3A4]; all are hyper-methylated in UC [Table 3]. In addition, the reported down-regulation of guanylate cyclase activators GUCA2A and GUCA2B, which are involved in gastrointestinal fluid and electrolyte balance, is most likely linked to the hyper-methylation of both genes during inflammation observed here.76
On the other hand, hypo-methylation with corresponding up-regulation of genes relevant in UC has been observed. This include genes that are up-regulated due to the response to inflammation, and represent mostly genes with association to the innate immune system like chemokines, chemokine receptors, cytokines, interleukins, and transporters [Table 4].33
For further characterisation, the differentially methylated genes were related to currently known IBD susceptibility genes, as revealed by genome-wide association studies [GWAS].47,48 Of the significantly differentially DNA methylated genes, 25 are associated with IBD, [Table 2] of which six genes have CpG islands located at their transcription start site [TSS].
The majority of DNA methylation occurs on cytosines that precede a guanine nucleotide or CpG sites. However, this study reveals previously unknown DNA methylation patterns of genes in treatment-naïve UC, which are not dependent on CpG sites or known regulatory transcriptional cis-acting elements like DNAse1 and enhancers. This has been also reported in the tissue of adult mouse brain, where a significant percentage of methylated non-CpG sites have been identified.77 This phenomenon implies novel regulatory features of DNA methylation in UC, with genes involved in pro-inflammatory responses and possible antimicrobial activities. These would involve defensin A6 [DEFA6] and regenerating protein 1B [REG1B], facilitation of cell adhesion through interaction with lectins and cadherins (olfactomedin 4 [OLFM4]), and lipid metabolism (butyrophilin-like 3 [BTLN3]) [Figure 2 and Table 5].78 All four genes have been associated with colorectal cancer [CRC] and/or have shown to play a role in CRC progression and development.79–83 However, the role of non-CpG methylation is still unclear.
Regarding the heterogeneity of tissues, it is clear that the methylation events could occur in different cell subtypes present in the tissue samples from UC patients. This might be the situation for DEFA6, which is a Paneth cell-specific protein and which is predominantly abundant in the epithelia of the intestinal mucosal surface and in the granules of neutrophils.84 Epithelial cells are impaired and less abundant in the inflamed mucosa,66 and it is therefore believed that the observed hypo-methylation of DEFA6 most likely occurs in the neutrophils with elevated fractions in inflamed mucosal tissue.85 The same might be the situation for BTNL3 which modulates T-cell mediated immune response. The observed hyper-methylation of BTLN3 may take place in the increased fractions of T lymphocytes present in inflamed mucosa. However, with isolated cell fractions or single cell sequencing approaches, one would be able to confirm these results.
In conclusion, this comprehensive study shows for the first time that the use of well-stratified treatment- naïve UC patient samples in combination with genome-wide bisulfite-sequencing technology can reveal DNA methylation patterns of importance for UC pathogenesis. Potentially significant might be the differential DNA methylation patterns, with observed hyper-methylation of genes involved in homeostasis and defence, and hypo-methylation of genes involved in immune response with representative members of the innate immune system. Further investigation of such players may be useful for the development of epigenetic drugs and may allow new treatment strategies for UC patients in the future.
Funding
This work was supported by the Northern Norway Regional Health Authority [SPF-1209-14].
Conflict of Interest
The authors declare no conflict of interest regarding the publication of this paper.
Author Contributions
HT performed most of the experiments and wrote parts of the manuscript. CGF performed most statistical analyses and revised the manuscript. IVH performed a part of the experiments and revised the manuscript. EA performed a part of data analysis and revised the manuscript. JF was involved in evaluating and providing clinical samples from patients and healthy controls and revised the manuscript. RHP was involved in projects inception, design, analysis, supervision, manuscript writing, and revision.
Supplementary Material
Acknowledgments
The authors thank Ingrid Christiansen for the technical help performing the TNF-α levels measurements, and Odd-Sverre Moen for administrating the patient samples.
References
- 1. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011;474:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Head KA, Jurenka JS. Inflammatory bowel disease. Part 1: ulcerative colitis–pathophysiology and conventional and alternative treatment options. Altern Med Rev 2003;8:247–83. [PubMed] [Google Scholar]
- 3. Planell N, Lozano JJ, Mora-Buch R, et al. . Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut 2013;62:967–76. [DOI] [PubMed] [Google Scholar]
- 4. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium [IIBDGC] Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu JZ, van Sommeren S, Huang H, et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shaw KA, Cutler DJ, Okou D, et al. . Genetic variants and pathways implicated in a pediatric inflammatory bowel disease cohort. Genes Immun 2018, March 28. doi: 10.1038/s41435-018-0015-2. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Loddo I, Romano C. Inflammatory bowel disease: genetics, epigenetics, and pathogenesis. Front Immunol 2015;6:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karatzas PS, Gazouli M, Safioleas M, Mantzaris GJ. DNA methylation changes in inflammatory bowel disease. Ann Gastroenterol 2014;27:125–32. [PMC free article] [PubMed] [Google Scholar]
- 9. Saito S, Kato J, Hiraoka S, et al. . DNA methylation of colon mucosa in ulcerative colitis patients: correlation with inflammatory status. Inflamm Bowel Dis 2011;17:1955–65. [DOI] [PubMed] [Google Scholar]
- 10. Toyota M, Itoh F, Kikuchi T, et al. . DNA methylation changes in gastrointestinal disease. J Gastroenterol 2002;37[Suppl 14]:97–101. [DOI] [PubMed] [Google Scholar]
- 11. Mohn F, Weber M, Rebhan M, et al. . Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 2008;30:755–66. [DOI] [PubMed] [Google Scholar]
- 12. Murphy TM, Mill J. Epigenetics in health and disease: heralding the EWAS era. Lancet 2014;383:1952–4. [DOI] [PubMed] [Google Scholar]
- 13. Scarpa M, Stylianou E. Epigenetics: concepts and relevance to IBD pathogenesis. Inflamm Bowel Dis 2012;18:1982–96. [DOI] [PubMed] [Google Scholar]
- 14. Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods Mol Biol 2012;863:359–76. [DOI] [PubMed] [Google Scholar]
- 15. Dick KJ, Nelson CP, Tsaprouni L, et al. . DNA methylation and body-mass index: a genome-wide analysis. Lancet 2014;383:1990–8. [DOI] [PubMed] [Google Scholar]
- 16. Tsaprouni LG, Yang TP, Bell J, et al. . Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics 2014;9:1382–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mishkin S. Dairy sensitivity, lactose malabsorption, and elimination diets in inflammatory bowel disease. Am J Clin Nutr 1997;65:564–7. [DOI] [PubMed] [Google Scholar]
- 18. Rubin DT, Hanauer SB. Smoking and inflammatory bowel disease. Eur J Gastroenterol Hepatol 2000;12:855–62. [DOI] [PubMed] [Google Scholar]
- 19. Lin Z, Hegarty JP, Cappel JA, et al. . Identification of disease-associated DNA methylation in intestinal tissues from patients with inflammatory bowel disease. Clin Genet 2011;80:59–67. [DOI] [PubMed] [Google Scholar]
- 20. Karatzas PS, Mantzaris GJ, Safioleas M, Gazouli M. DNA methylation profile of genes involved in inflammation and autoimmunity in inflammatory bowel disease. Medicine [Baltimore] 2014;93:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nimmo ER, Prendergast JG, Aldhous MC, et al. . Genome-wide methylation profiling in Crohn’s disease identifies altered epigenetic regulation of key host defense mechanisms including the Th17 pathway. Inflamm Bowel Dis 2012;18:889–99. [DOI] [PubMed] [Google Scholar]
- 22. Cooke J, Zhang H, Greger L, et al. . Mucosal genome-wide methylation changes in inflammatory bowel disease. Inflamm Bowel Dis 2012;18:2128–37. [DOI] [PubMed] [Google Scholar]
- 23. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology 2013;38:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eckhardt F, Lewin J, Cortese R, et al. . DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet 2006;38:1378–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maunakea AK, Nagarajan RP, Bilenky M, et al. . Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010;466:253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Darst RP, Pardo CE, Ai L, Brown KD, Kladde MP. Bisulfite sequencing of DNA. Curr Protoc Mol Biol 2010;Chapter 7:Unit 7.9.1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Y, Tollefsbol TO. DNA methylation detection: bisulfite genomic sequencing analysis. Methods Mol Biol 2011;791:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Teh AL, Pan H, Lin X, et al. . Comparison of methyl-capture sequencing vs. infinium 450K methylation array for methylome analysis in clinical samples. Epigenetics 2016;11:36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harris RA, Wang T, Coarfa C, et al. . Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 2010;28:1097–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Magro F, Gionchetti P, Eliakim R, et al. ; European Crohn’s and Colitis Organisation [ECCO] Third European evidence-based consensus on diagnosis and management of ulcerative colitis. Part 1: definitions, diagnosis, extra-intestinal manifestations, pregnancy, cancer surveillance, surgery, and ileo-anal pouch disorders. J Crohns Colitis 2017;11:649–70. [DOI] [PubMed] [Google Scholar]
- 31. McNally PR. Literature review: Core I & II: colonic release budesonide for the induction of remission for mild-moderate ulcerative colitis. VHJOE 2014;13:1. [Google Scholar]
- 32. Olsen T, Goll R, Cui G, et al. . Tissue levels of tumor necrosis factor-alpha correlates with grade of inflammation in untreated ulcerative colitis. Scand J Gastroenterol 2007;42:1312–20. [DOI] [PubMed] [Google Scholar]
- 33. Taman H, Fenton CG, Hensel IV, et al. . Transcriptomic landscape of treatment - naive ulcerative colitis. J Crohns Colitis 2018;12:327–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dobin A, Davis CA, Schlesinger F, et al. . STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pearson K. On lines and planes of closest fit to systems of points in space. Philosophical Magazine 1901;2:14. [Google Scholar]
- 36. Ritchie ME, Phipson B, Wu D, et al. . limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc 1995;57:12. [Google Scholar]
- 38. Goeman JJ, Van de Geer SA, Van Houwelingen HC. Testing against a high dimensional alternative. J R Stat Soc B 2006;68:477–93. [Google Scholar]
- 39. Klein HU, Hebestreit K. An evaluation of methods to test predefined genomic regions for differential methylation in bisulfite sequencing data. Brief Bioinform 2016;17:796–807. [DOI] [PubMed] [Google Scholar]
- 40. MacArthur J, Bowler E, Cerezo M, et al. . The new NHGRI-EBI Catalog of published genome-wide association studies [GWAS Catalog]. Nucleic Acids Res 2017;45:D896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abbas AR, Baldwin D, Ma Y, et al. . Immune response in silico [IRIS]: immune-specific genes identified from a compendium of microarray expression data. Genes Immun 2005;6:319–31. [DOI] [PubMed] [Google Scholar]
- 42. Mevik B, Wehrens R. The pls package: principle component and partial least squares regression in r. J Stat Softw 2007;18:23. [Google Scholar]
- 43. Peres-Neto PR, Jackson DA. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia 2001;129:169–78. [DOI] [PubMed] [Google Scholar]
- 44. Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of p. J R Stat Soc 1922;85:7. [Google Scholar]
- 45. McDermott E, Ryan EJ, Tosetto M, et al. . DNA methylation profiling in inflammatory bowel disease provides new insights into disease pathogenesis. J Crohns Colitis 2016;10:77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Häsler R, Feng Z, Bäckdahl L, et al. . A functional methylome map of ulcerative colitis. Genome Res 2012;22:2130–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Anderson CA, Boucher G, Lees CW, et al. . Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 2011;43:246–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. de Lange KM, Moutsianas L, Lee JC, et al. . Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 2017;49:256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Harris RA, Nagy-Szakal D, Pedersen N, et al. . Genome-wide peripheral blood leukocyte DNA methylation microarrays identified a single association with inflammatory bowel diseases. Inflamm Bowel Dis 2012;18:2334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kang K, Bae JH, Han K, et al. . A genome-wide methylation approach identifies a new hypermethylated gene panel in ulcerative colitis. Int J Mol Sci 2016;17. doi: 10.3390/ijms17081291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tahara T, Shibata T, Yamashita H, et al. . Chronic nonsteroidal anti-inflammatory drug [NSAID] use suppresses multiple CpG islands hyper methylation [CIHM] of tumor suppressor genes in the human gastric mucosa. Cancer Sci 2009;100:1192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tahara T, Shibata T, Nakamura M, et al. . Chronic aspirin use suppresses CDH1 methylation in human gastric mucosa. Dig Dis Sci 2010;55:54–9. [DOI] [PubMed] [Google Scholar]
- 53. Harris RA, Nagy-Szakal D, Mir SA, et al. . DNA methylation-associated colonic mucosal immune and defense responses in treatment-naïve pediatric ulcerative colitis. Epigenetics 2014;9:1131–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Axelrad JE, Lichtiger S, Yajnik V. Inflammatory bowel disease and cancer: The role of inflammation, immunosuppression, and cancer treatment. World J Gastroenterol 2016;22:4794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Herfarth HH, Kappelman MD, Long MD, Isaacs KL. Use of methotrexate in the treatment of inflammatory bowel diseases. Inflamm Bowel Dis 2016;22:224–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Horvath S, Zhang Y, Langfelder P, et al. . Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol 2012;13:R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ciccarone F, Tagliatesta S, Caiafa P, Zampieri M. DNA methylation dynamics in aging: how far are we from understanding the mechanisms?Mech Ageing Dev 2018;174:3–17. [DOI] [PubMed] [Google Scholar]
- 58. Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet 2010;11:191–203. [DOI] [PubMed] [Google Scholar]
- 59. Zhang G, Pradhan S. Mammalian epigenetic mechanisms. IUBMB Life 2014;66:240–56. [DOI] [PubMed] [Google Scholar]
- 60. Rakyan VK, Hildmann T, Novik KL, et al. . DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol 2004;2:e405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Meissner A, Mikkelsen TS, Gu H, et al. . Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008;454:766–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Illingworth RS, Gruenewald-Schneider U, Webb S, et al. . Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet 2010;6:e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Irizarry RA, Ladd-Acosta C, Wen B, et al. . The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 2009;41:178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen L, Su L, Li J, et al. . Hypermethylated FAM5C and MYLK in serum as diagnosis and pre-warning markers for gastric cancer. Dis Markers 2012;32:195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smith PJ, Levine AP, Dunne J, et al. . Mucosal transcriptomics implicates under expression of BRINP3 in the pathogenesis of ulcerative colitis. Inflamm Bowel Dis 2014;20:1802–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mankertz J, Schulzke JD. Altered permeability in inflammatory bowel disease: pathophysiology and clinical implications. Curr Opin Gastroenterol 2007;23:379–83. [DOI] [PubMed] [Google Scholar]
- 67. He F, Peng J, Deng XL, et al. . Mechanisms of tumor necrosis factor-alpha-induced leaks in intestine epithelial barrier. Cytokine 2012;59:264–72. [DOI] [PubMed] [Google Scholar]
- 68. Zhang J, Wong H, Ramanan S, Cheong D, Leong A, Hooi SC. The proline-rich acidic protein is epigenetically regulated and inhibits growth of cancer cell lines. Cancer Res 2003;63:6658–65. [PubMed] [Google Scholar]
- 69. Ghishan FK, Kiela PR. Epithelial transport in inflammatory bowel diseases. Inflamm Bowel Dis 2014;20:1099–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pérez-Torras S, Iglesias I, Llopis M, et al. . Transportome profiling identifies profound alterations in Crohn’s disease partially restored by commensal bacteria. J Crohns Colitis 2016;10:850–9. [DOI] [PubMed] [Google Scholar]
- 71. Fawley J, Gourlay DM. Intestinal alkaline phosphatase: a summary of its role in clinical disease. J Surg Res 2016;202:225–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Müller CA, Autenrieth IB, Peschel A. Innate defenses of the intestinal epithelial barrier. Cell Mol Life Sci 2005;62:1297–307. [DOI] [PubMed] [Google Scholar]
- 73. Basu NK, Kole L, Kubota S, Owens IS. Human UDP-glucuronosyltransferases show atypical metabolism of mycophenolic acid and inhibition by curcumin. Drug Metab Dispos 2004;32:768–73. [DOI] [PubMed] [Google Scholar]
- 74. Basu NK, Kubota S, Meselhy MR, et al. . Gastrointestinally distributed UDP-glucuronosyltransferase 1A10, which metabolizes estrogens and nonsteroidal anti-inflammatory drugs, depends upon phosphorylation. J Biol Chem 2004;279:28320–9. [DOI] [PubMed] [Google Scholar]
- 75. Dellinger RW, Fang JL, Chen G, Weinberg R, Lazarus P. Importance of UDP-glucuronosyltransferase 1A10 [UGT1A10] in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the UGT1A10139Lys isoform. Drug Metab Dispos 2006;34:943–9. [DOI] [PubMed] [Google Scholar]
- 76. Brenna Ø, Bruland T, Furnes MW, et al. . The guanylate cyclase-C signaling pathway is down-regulated in inflammatory bowel disease. Scand J Gastroenterol 2015;50:1241–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Xie W, Barr CL, Kim A, et al. . Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 2012;148:816–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Paulssen RH, Taman H, Fenton CG, et al. . Novel regulatory DNA methylation features of relevance for ulcerative colitis. Am J Gastroenterol 2018;113:P-050. [Google Scholar]
- 79. Park JM, Han NY, Han YM, et al. . Predictive proteomic biomarkers for inflammatory bowel disease-associated cancer: where are we now in the era of the next generation proteomics?World J Gastroenterol 2014;20:13466–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 2009;137:15–7. [DOI] [PubMed] [Google Scholar]
- 81. Lebrero-Fernández C, Wenzel UA, Akeus P, et al. . Altered expression of Butyrophilin [BTN] and BTN-like [BTNL] genes in intestinal inflammation and colon cancer. Immun Inflamm Dis 2016;4:191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. van Beelen Granlund A, Østvik AE, Brenna Ø, Torp SH, Gustafsson BI, Sandvik AK. REG gene expression in inflamed and healthy colon mucosa explored by in situ hybridisation. Cell Tissue Res 2013;352:639–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gersemann M, Becker S, Nuding S, et al. . Olfactomedin-4 is a glycoprotein secreted into mucus in active IBD. J Crohns Colitis 2012;6:425–34. [DOI] [PubMed] [Google Scholar]
- 84. Elphick DA, Mahida YR. Paneth cells: their role in innate immunity and inflammatory disease. Gut 2005;54:1802–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Arijs I, De Hertogh G, Lemaire K, et al. . Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One 2009;4:e7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.