Abstract
Growing evidence suggests that long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) are involved in the occurrence and development of tumors and fibrotic diseases. However, the integrated analysis of lncRNA and circRNA expression, alongside associated co-expression and competing endogenous RNA (ceRNA) networks, has not yet been performed in human hypertrophic scars (HS). The present study compared the expression levels of lncRNAs, circRNAs and mRNAs in human HS and normal skin tissues by high-throughput RNA sequencing. Numerous differentially expressed lncRNAs, circRNAs and mRNAs were detected. Subsequently, five aberrantly expressed lncRNAs and mRNAs, and six circRNAs were measured to verify the RNA sequencing results by reverse transcription-quantitative polymerase chain reaction. Furthermore, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed for the dysregulated genes, in order to elucidate their principal functions. In addition, a coding-noncoding gene co-expression (CNC) network and ceRNA network were constructed for specific significantly altered genes. The CNC network analysis suggested that AC048380.1 and LINC00299 were associated with metastasis-related genes, including inhibin subunit βA (INHBA), SMAD family member 7 (SMAD7), collagen type I α1 chain (COL1A1), transforming growth factor β3 (TGFβ3) and MYC proto-oncogene, bHLH transcription factor (MYC). Inhibitor of DNA binding 2 was associated with the lncRNAs cancer susceptibility 11, TGFβ3-antisense RNA 1 (AS1), INHBA-AS1, AC048380.1, LINC00299 and LINC01969. Circ-Chr17:50187014_50195976_-, circ-Chr17:50189167_50194626_-, circ-Chr17:50189167_ 50198002_- and circ-Chr17:50189858_50195330_- were also associated with INHBA, SMAD7, COL1A1, TGFβ3 and MYC. COL1A1 and TGFβ3 were associated with circ-Chr9:125337017_125337591_+ and circ-Chr12:120782654_120784593_-. The ceRNA network indicated that INHBA-AS1 and circ-Chr9:125337017_125337591_+ were ceRNAs of microRNA-182-5p targeting potassium voltage-gated channel subfamily J member 6, ADAM metallopeptidase with thrombospondin type 1 motif 18, SRY-box 11, MAGE family member L2, matrix metallopeptidase 16, thrombospondin 2, phosphodiesterase 11A and collagen type V a1 chain. These findings suggested that lncRNAs and circRNAs may act as ceRNAs, which are implicated in the pathophysiology and development of human HS, and lay a foundation for further insight into the novel regulatory mechanism of lncRNAs and circRNAs in hypertrophic scarring.
Keywords: hypertrophic scars, circular RNA, long noncoding RNA, competing endogenous RNA, RNA sequencing
Introduction
Hypertrophic scars (HS) derive from the overproliferation of fibroblasts following surgery, inflammation, trauma or burns. They are often associated with functional impairments, and severe physiological and psychological problems (1). Despite numerous studies (2,3) being conducted on the etiology and pathogenesis of HS, the progression of fibrosis remains poorly understood. Recently, increasing evidence has suggested that noncoding RNAs (ncRNAs), including microRNAs (miRNAs/miRs) and long noncoding RNAs (lncRNAs), serve important roles in the formation of HS (4), and can regulate gene expression at the levels of transcription, RNA processing and translation (5).
MicroRNA is a class of small ncRNA, ranges from 18 to 25 nucleotides (nt). As we know it plays important roles in fibrosis processes including transforming growth factor (TGF)-beta signaling, ECM deposition, epithelial to mesenchymal transition (EMT), fibroblast proliferation and differentiation (6). With the study of scarring, more and more miRNAs and their function were found and identified. For instance, by affecting the collagen I and III synthesis and TGF-β1/α-SMA signalling, miR-200b could regulate the cell proliferation of human hypertrophic scar fibroblasts and affected hypertrophic scar (7).
Previous studies have shown that lncRNAs served a crucial role in regulation of gene expression and disease processes (8,9). It's reported that lncRNAs participated in cell proliferation, differentiation, migration and apoptosis (10). Notably, they can regulate the stages of wound healing, including re-epithelialization, angiogenesis, remodelling and scar formation (11). lncRNA has become a new research hotspot in wound healing.
In addition, a newly identified type of ncRNA, circRNA, is molecularly stable and can function as a miRNA sponge, thus affecting the functions of miRNAs (12). For example, circZNF91 contains 24 target sites for miR-23b-3p, which is known to play important roles in keratinocyte differentiation (13). This means that circRNAs bind to miRNAs and affect biological pathways of miRNAs and consequently repress their function. Such as, circRNA_100782 can regulate BxPC3 cell proliferation by acting as miR-124 sponge through the IL6-STAT3 pathway (14). Resistant to exonuclease, circRNAs are stable molecules in cells which make the possibility of circRNAs as biological markers in clinical samples. To the best of the authors' knowledge, there is no report about the functional roles of circRNA in HS.
LncRNAs and circRNAs can function as competing endogenous RNAs (ceRNAs) and regulate each other by competing for the same miRNA response elements (MREs) (15). ncRNAs have become a novel area of research in wound healing; however, the potential roles of lncRNAs and circRNAs as ceRNAs in human HS remain to be elucidated.
The present study aimed to investigate the differentially expressed patterns of lncRNAs, circRNAs and mRNAs in HS using high-throughput gene sequencing. Meanwhile, the functions of the lncRNAs and circRNAs were predicted by investigating the co-expression and ceRNA networks of these dysregulated RNAs, such as AC048380.1 and LINC00299 were associated with metastasis-related genes, including INHBA, SMAD7, COL1A1, TGF-β3, which might be associated with HS.
Patients and methods
Patients and samples
The present study was approved by the Ethics Committee of The First Affiliated Hospital of Nanchang University (Nanchang, China). Written informed consent was obtained from all subjects. A total of six pairs of HS and adjacent normal skin tissues were collected from patients (4 men and 2 women; age range, 20–56 years old) who lived in the first affiliated hospital of Nanchang University from April to September 2016. All 6 patients underwent surgical treatment without preoperative drug therapy or radiotherapy. Subsequently, the epidermis and subcutaneous tissue were removed from the samples in a bacteria-free operating environment within 15 min, and were stored in liquid nitrogen.
RNA isolation and quality control
Total RNAs were isolated from HS and normal skin tissues using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. Subsequently, total RNA was cleaned using the RNeasy Mini kit (Qiagen, Inc., Valencia, CA, USA), and RNA quantity was analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). RNA integrity and chromosomal DNA contamination were assessed by denaturing agarose gel electrophoresis (concentration of agarose gel is 1%, and 0.5 µg/ml ethidium bromide to help it visualized).
Library preparation for RNA sequencing (RNA-seq)
Ribosomal RNA depletion was performed using a 5′-phosphate-dependent exonuclease (GeneRead rRNA Depletion kit (Qiagen, Germany), which degraded transcripts with a 5′ monophosphate. Linear RNA depletion was performed using Ribonuclease R. The dosage ratio of Ribonuclease R to RNA is 3U:1 µg and the digestive conditions were 37°C for 30 min. According to experimental protocols, Rayscript cDNA Synthesis kit (GENEray, GK8030) was used for the reverse transcription. RNA fragments were used for sequential first-strand and second-strand cDNA synthesis, and the cDNA templates were enriched. Libraries were constructed using the TruSeq Stranded mRNA LT Sample Prep kit (Illumina, Inc., San Diego, CA, USA). Library quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.) prior to sequencing. Total concentrations were determined using a Qubit 12.0 Fluorometer (Thermo Fisher Scientific, Inc.). Libraries were sequenced on the Illumina NextSeq 500 platform (Illumina, Inc.) with 2×150 bp paired-end reads. The libraries were constructed and sequenced by Shanghai Personal Biotechnology Co., Ltd. (www.personalbio.cn/; Shanghai, China).
Data quality control
The quality of raw sequencing data was assessed by using FASTQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). All sequences were subjected to quality control to trim the adapter contaminants and filter out low-quality reads. Cutadapt (version 1.2.1; pypi.org/project/cutadapt/1.2.1/#files), which can perform various useful forms of trimming (16), was used for quality control of the raw data.
Data arrangement and analysis
Raw reads were subjected to quality control and the remaining ‘clean reads’ were mapped to the Ensembl database (www.ensembl.org/) using Tophat version 2 (ccb.jhu.edu/software/tophat/index.shtml). The reads per kilobase of transcript per million mapped reads was calculated for each gene and used to analyze the fold change. The differentially expressed genes (DEGs) were detected using DEGseq Software (version 1.18.0) (17). A fold-change >2, a P-value <0.05 and a false-discovery rate (FDR) <0.05 were considered the criteria for differential expression. The differentially expressed genes obtained from all groups were analyzed by bidirectional clustering through Pheatmap package. Euclidean method was used to calculate the distance, and the Complete Linkage was used for clustering. The gene functional annotation included: ENSEMBL ID, information on the chromosome distribution in each locus, gene name [Human Genome Variation Society Symbol, National Center for Biotechnology Information (NCBI) Gene ID, UniProtKB ID], gene classification [Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO), enzyme classification]. The Gene Ontology Consortium (www.geneontology.org/) was used for GO functional enrichment analysis. It aided the description of the biological function of the DEGs, including molecular functions, cellular components and biological processes. The analysis first mapped all DEGs to GO terms within the database and the gene numbers were calculated for each term. Subsequently, ultra-geometric tests were used to identify the GO terms markedly enriched among the DEGs relative to the genomic background. KEGG (www.kegg.jp/) includes KO and KEGG Pathway. KEGG Pathway was used to conduct a pathway enrichment analysis of the DEGs. It identified signal transduction pathways or metabolic pathways associated with the DEGs, which were significantly enriched. In the present study, significant pathways were identified as those with P-values <0.05 and FDR <0.05. The lower values indicated greater significance.
Validation with reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The RNA-seq data were validated by RT-qPCR. RNA extracted by using TRIzol™ Reagent (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). cDNA was generated from RNA using the Rayscript cDNA Synthesis kit (cat. no. GK8030; Shanghai Generay Biotech Co., Ltd., Shanghai, China). cDNA was subjected to RT-qPCR analysis using Power qPCR PreMix (cat. no. GK8020; Shanghai Generay Biotech Co., Ltd.). The primers were designed using NCBI-Primer BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast/) and primers were checked using OLIGO7.37 software in accordance with the sequences of the corresponding human RNAs in GenBank (www.ncbi.nlm.nih.gov/genbank/). A list of primers of the selected genes for RT-qPCR is provided in Tables I–III. GAPDH and Human β-actin (H-actin) were used as the control genes for normalization of cDNA loading differences. Cycling conditions were: 95°Cx10 min→95°C ×15 sec, total 45 cycles and it was repeated 3 times.
Table I.
Primer sequences of long noncoding RNAs used for reverse transcription-quantitative polymerase chain reaction.
| Primer ID | Primer sequences (5′-3′) | Annealing temperature (°C) | Length (base pairs) |
|---|---|---|---|
| ENSG00000258876 (TGFβ3-AS1) | F:TTGAGTGAATGAATGGAGGA | 60 | 91 |
| R:AAATGGAGGGAAGAGGACA | |||
| ENSG00000268262 (AC011445.1) | F:CTTTGACGCCTTCCTGTCC | 60 | 111 |
| R:AGCGTGAGGGTGTTTAGGTTC | |||
| ENSG00000223749 (STX17-AS1) | F:AATAACCAGGGTCACAGAAAA | 60 | 147 |
| R:AAAGCAGCATACAGTCATTCA | |||
| ENSG00000237548 (TTLL11-IT1) | F:TACAAGTCCCCAGAAACCA | 60 | 196 |
| R:AAAATCCACCCAGATAGCC | |||
| ENSG00000251085 (LINC01969) | F:CAGACGGTCTCCTTAAAGATTCTCC | 60 | 89 |
| R:GAACCCATCTTCACCCTTGCTAA | |||
| GAPDH | F:GGACCTGACCTGCCGTCTAG | 60 | 100 |
| R:GTAGCCCAGGATGCCCTTGA |
AS1, antisense RNA 1; F, forward; R, reverse; STX17, syntaxin 17; TGF-β3, transforming growth factor β3; TTLL11-IT1, TTLL11 intronic transcript 1.
Table III.
Primer sequences of mRNA used for reverse transcription-quantitative polymerase chain reaction.
| Primer ID | Primer sequences | Annealing temperature (°C) | Length (base pairs) |
|---|---|---|---|
| COL1A1 | F:AGACATCCCACCAATCACCT | 60 | 118 |
| R:GTCATCGCACAACACCTTG | |||
| ASXL1 | F:CCTACTGTCCTCCCAAACC | 60 | 118 |
| R:CTCCTCATCATCACTTTCCC | |||
| HIPK3 | F:GTTGTGATACGGTGGATGG | 60 | 135 |
| R:CTGGCTTGGTTTCTGTGTC | |||
| CTGF | F:TACCAATGACAACGCCTCCTG | 60 | 208 |
| R:GCCGTCGGTACATACTCCACA | |||
| TGFb3 | F:GCTACTATGCCAACTTCTGCTCA | 60 | 224 |
| R:GCTACATTTACAAGACTTCACCACC | |||
| GAPDH | F:GGACCTGACCTGCCGTCTAG | 60 | 100 |
| R:GTAGCCCAGGATGCCCTTGA |
ASXL1, ASXL transcriptional regulator 1; COL1A1, collagen type I α1 chain; CTGF, connective tissue growth factor; F, forward; HIPK3, homeodomain interacting protein kinase 3; R, reverse; TGF β3, transforming growth factor β3.
All reactions were analyzed using the 2−ΔΔCq method (18). Student's t-test was applied to statistically analyze the data and P<0.05 was considered to indicate a statistically significant difference. The values are expressed as the mean ± standard deviation. The data was analyzed by SPSS19 (IBM Corp., Armonk, NY, USA).
Co-expression networks and ceRNA network analysis
The expression levels of the lncRNAs/circRNAs and mRNAs were analyzed to elucidate any meaningful association. The sequences of lncRNAs/circRNAs and mRNAs were searched by Cytoscape (www.cytoscape.org/) and the present study aimed to discover their potential miRNA response elements. The overlapping of the same miRNA-binding site on both lncRNA/circRNA and mRNA may aid the prediction of the lncRNA/circRNA-miRNA-mRNA interactions. The lncRNA-miRNA, circRNA-miRNA, miRNA-mRNA interactions were predicted using miRanda (www.microrna.org/), Targetscan (www.targetscan.org/) and psRobot (omicslab.genetics.ac.cn/psRobot/).
Results
Differential lncRNA, mRNA and circRNA expression profiles, as determined by RNA-seq
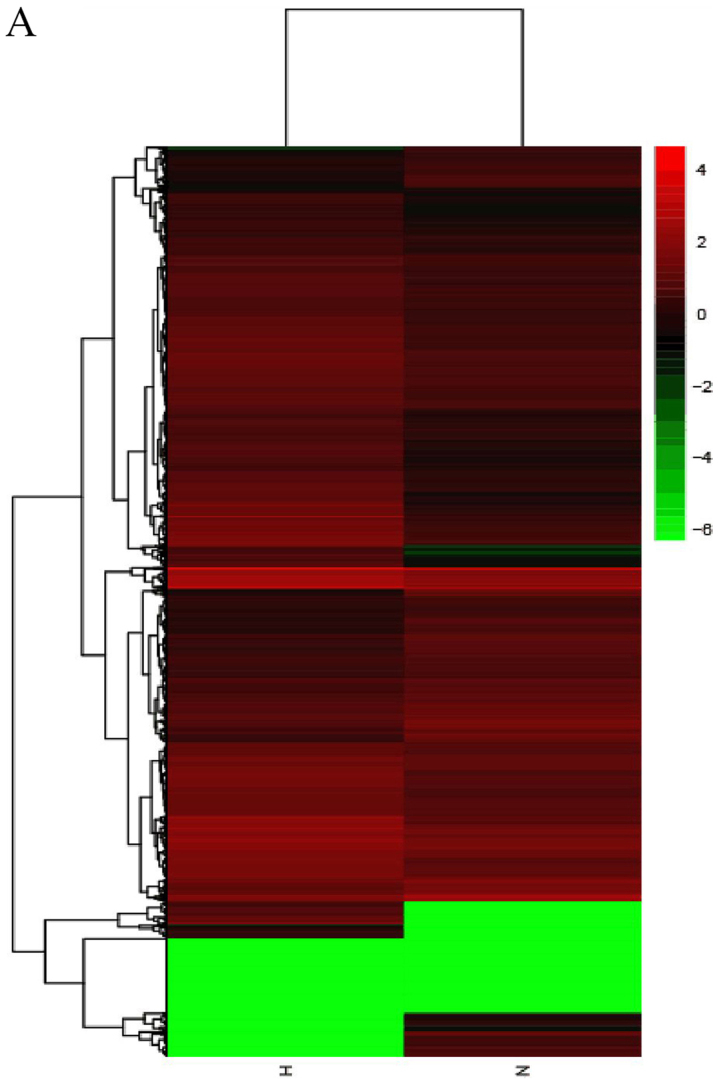
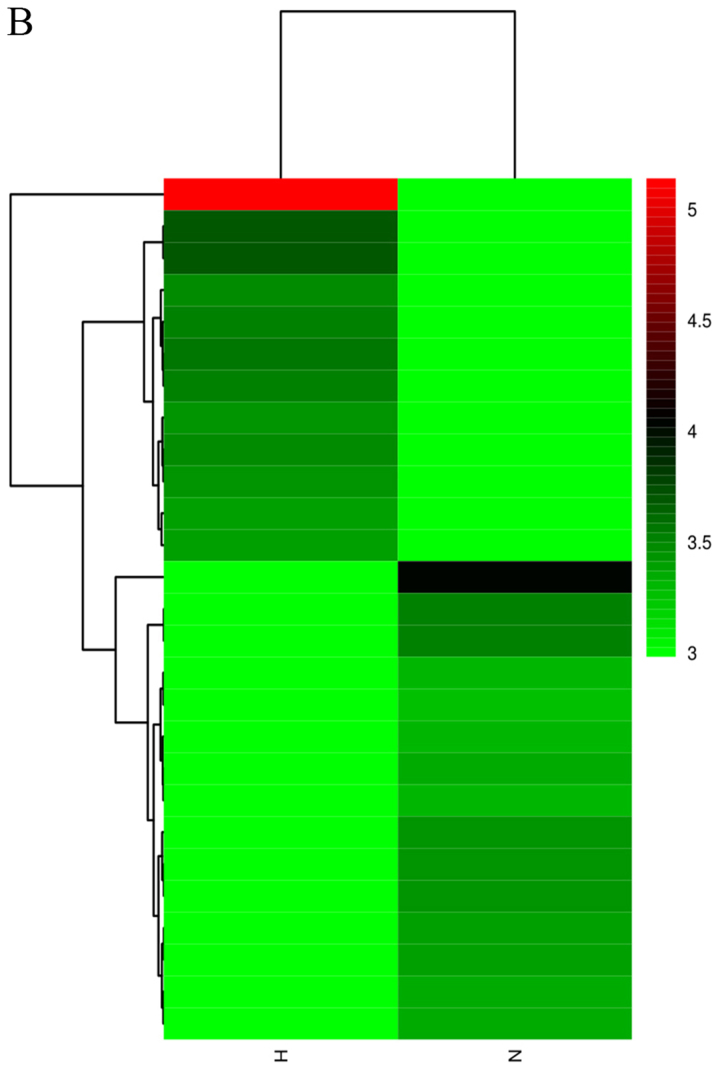
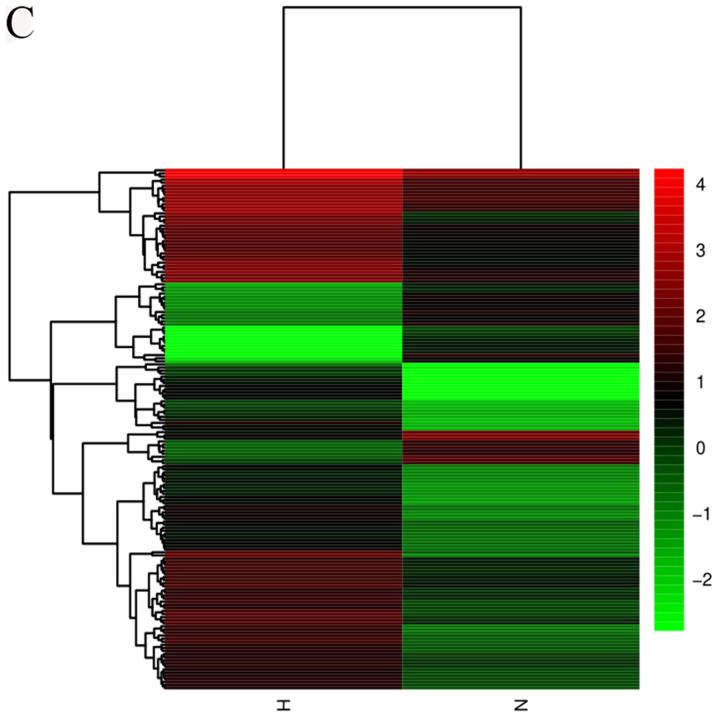
High throughput sequencing is an efficient method for studying the biological function of RNAs. Thousands of transcripts were detected in HS and normal tissues by RNA-seq. Among them, a total of 3,469 lncRNAs and 536 mRNAs were detected to be significant differentially expressed (|fold change|>2.0, P<0.05). Of these, 2,479 and 990 lncRNAs were upregulated and downregulated, respectively. In addition, 345 and 191 mRNAs were upregulated and downregulated in HS compared with in normal tissues. Furthermore, the expression profiles included 3,171 circRNAs in HS and 4,519 in normal tissues respectively. A total of 11 circRNAs (fold change ≥2.0, P<0.05) were differentially expressed between HS and normal tissues, 10 upregulated and 1 downregulated. In addition, a total of 644 lncRNAs exhibited a fold change of ≥10, 493 of which were upregulated and 151 were downregulated. AC022034.2 (fold change: 15,030,055) was the most upregulated lncRNA. Meanwhile, the coding gene profile suggested that 299 mRNAs had a fold change ≥10. Hierarchical clustering demonstrated the distinguishable gene expression profiles of lncRNA, circRNA and mRNA among the samples. These results suggested that the expression profiles of lncRNAs, circRNAs and mRNAs in HS tissues differed from those of normal skin tissues (Table IV; Fig. 1).
Table IV.
Significant differentially expressed RNAs in hypertrophic scar tissues compared with the control group (|fold change| >2.0, P<0.05).
| Category | Upregulated genes | Downregulated genes | Total differentially expressed genes |
|---|---|---|---|
| mRNA | 345 | 191 | 536 |
| Long noncoding RNA | 2,479 | 990 | 3,469 |
| Circular RNA | 10 | 1 | 11 |
Figure 1.
Heatmaps showing expression profiles of (A) lncRNAs, (B) circRNAs and (C) mRNAs. The maps were based on the expression values of all expressed lncRNAs, circRNAs and mRNAs detected by RNA sequencing with fold change ≥2.0, P<0.05 and false-discovery rate <0.05. The expression values are depicted according to the color scale. The intensity increases from green to red (Red indicates high expression and green indicates low expression). circRNAs, circular RNAs; H, hypertrophic scar tissues; lncRNAs, long noncoding RNAs; N, normal tissues.
All of these dysregulated RNAs were diffusely distributed in all chromosomes, including sex chromosomes X and Y. All forms of splicing transcripts were determined by StringTie (ccb.jhu.edu/software/stringtie/index.shtml?t=manual#input), mapped to the Ensembl database by gffcompare (ccb.jhu.edu/software/stringtie/gffcompare.shtml) and novel transcripts were identified (Table V).
Table V.
Information regarding transcripts.
| Group | Total transcripts | Known transcripts | Novel transcripts |
|---|---|---|---|
| H | 119,518 | 26,160 | 93,358 |
| N | 190,765 | 29,093 | 161,672 |
H, hypertrophic scar; N, normal skin.
Validation with RT-qPCR
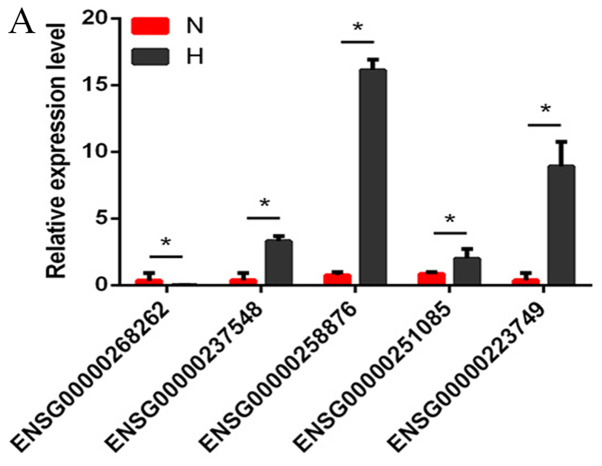
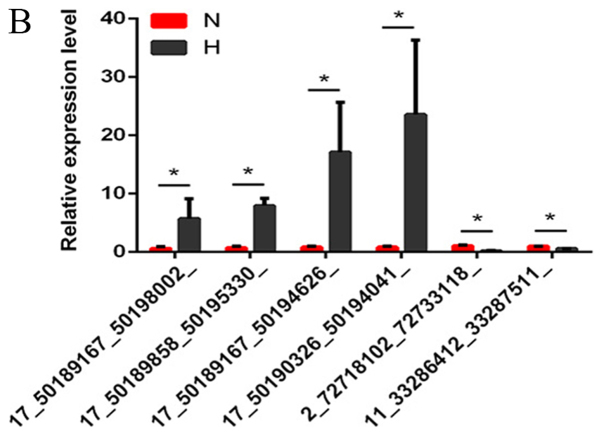
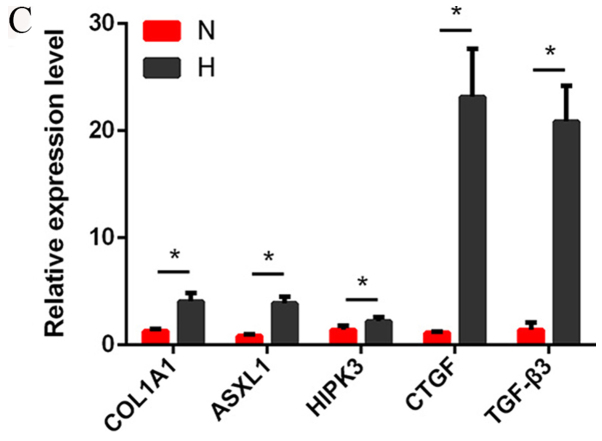
Several differentially expressed lncRNAs, circRNAs and mRNAs were randomly selected and RT-qPCR was used to verify the RNA-seq results. The results demonstrated that the fold changes observed in the sequencing analysis were consistent with the sequencing assay (Fig. 2A-C; Tables VI–VIII).
Figure 2.
Verification of the expression of significantly dysregulated genes by reverse transcription-quantitative polymerase chain reaction. The relative expression levels of (A) five long noncoding RNAs, (B) six circular RNAs and (C) five mRNAs were detected in N and H groups. Data are presented as mean ± SD, *P<0.05. ASXL1, ASXL transcriptional regulator 1; COL1A1, collagen type I A1 chain; CTGF, connective tissue growth factor; H, hypertrophic scar tissues; HIPK3, homeodomain interacting protein kinase 3; N, normal tissues; TGF-β3, transforming growth factor β3.
Table VI.
Comparison of RT-qPCR data with RNA-seq data for lncRNAs.
| ENSG0000 | ENSG0000 | ENSG0000 | ENSG0000 | ENSG0000 | |
|---|---|---|---|---|---|
| lncRNA | 0251085 | 0223749 | 0258876 | 0237548 | 0268262 |
| RT-qPCR (H/N) | 2.20 | 8.14 | 15.42 | 6.78 | −6.84 |
| RNA-seq (H/N) | 5.48 | 192.80 | 21.11 | 54.27 | −13.05 |
lncRNA, long non-coding RNA; H, hypertrophic scar; N, normal skin; RNA-seq, RNA sequencing; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Table VIII.
Comparison of RT-qPCR data with RNA-seq data for mRNA.
| mRNA | COL1A1 | ASXL1 | CTGF | TGFβ3 | HIPK3 |
|---|---|---|---|---|---|
| RT-qPCR (H/N) | 8.65 | 12.86 | 20.39 | 15.99 | 4.32 |
| RNA-seq (H/N) | 69.23 | 2.17 | 21.19 | 17.19 | 0.95 |
ASXL1, ASXL transcriptional regulator 1; COL1A1, collagen type I α1 chain; CTGF, connective tissue growth factor; H, hypertrophic scar; HIPK3, homeodomain interacting protein kinase 3; N, normal skin; RNA-seq, RNA sequencing; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; TGFβ3, transforming growth factor β3.
GO and KEGG pathway analyses
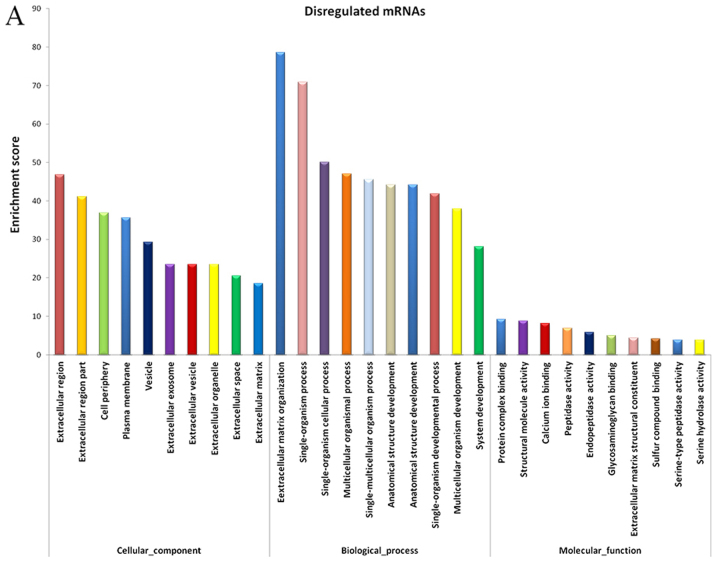
The evidently abnormal mRNAs were selected for functional enrichment analysis. The results demonstrated that certain mRNAs had important roles in various biological processes, including extracellular matrix (ECM) organization, collagen metabolic process, biological adhesion, negative regulation of cellular response to growth factor stimulus and skin development (Fig. 3A).
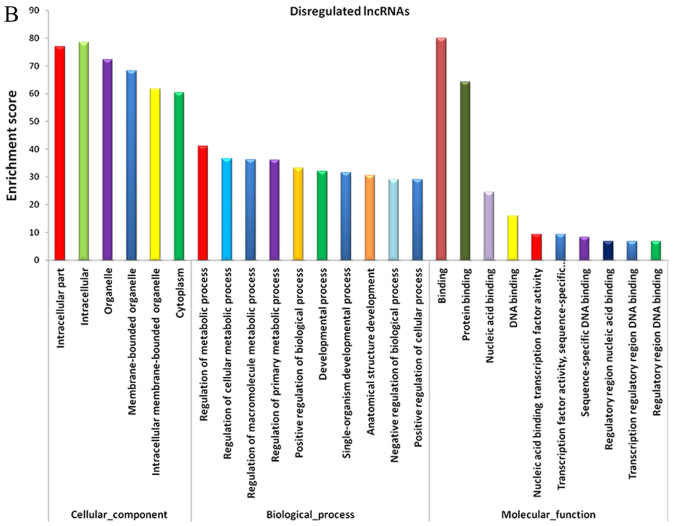
Figure 3.
GO analysis of dysregulated lncRNAs and mRNAs. GO annotation of dysregulated (A) mRNAs and (B) lncRNAs; top 10 enriched terms covering biological processes, cellular components and molecular functions are presented. GO, Gene Ontology; lncRNAs, long noncoding RNAs.
It is well known that lncRNAs influence DNA sequences by regulating the neighboring and overlapping coding gene expression levels (19). Therefore, the functions of lncRNAs may be reflected in nearby genes. GO enrichment analysis of the notably differentially expressed mRNAs in the vicinity of the lncRNAs may help to reveal the function of these lncRNAs. The data from the present study suggested that the dysregulated mRNAs and lncRNAs were associated with biological functions including regulation of collagen metabolic process, regulation of transcription and DNA-template, and ECM organization. The majority of these processes were associated with ECM deposition, cell proliferation and gene expression (Fig. 3B).
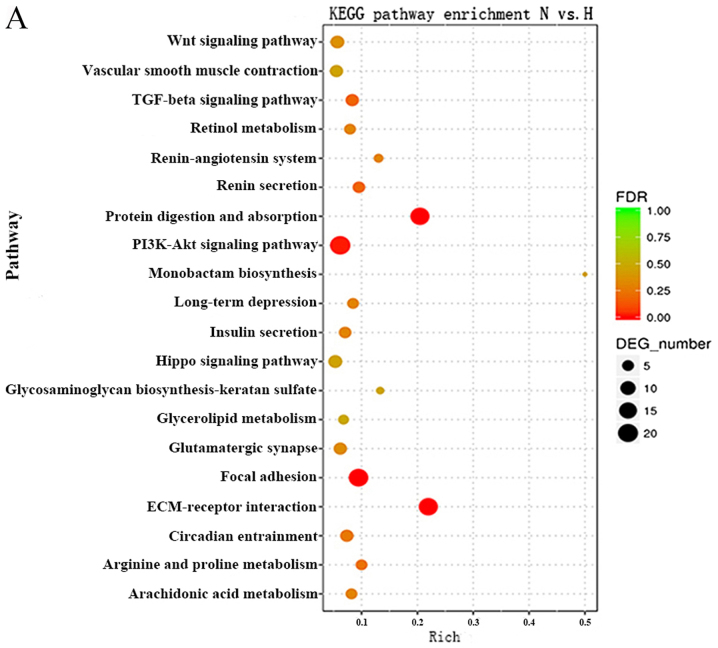
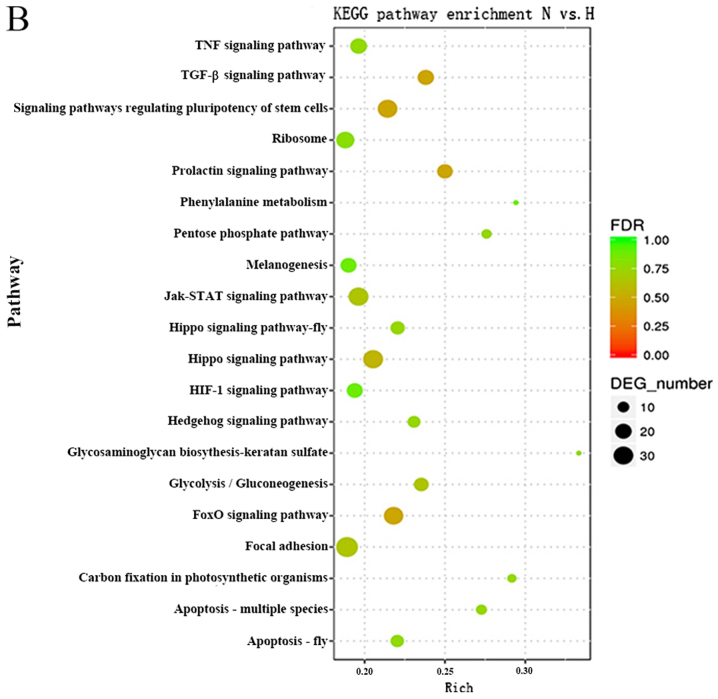
KEGG pathway analysis can reveal molecular interactions and the pathways associated with genes. Accordingly, the biological pathways associated with the significantly differentially expressed RNAs were investigated. The P-value cutoff was 0.05. The data demonstrated that the pathways associated with the mRNAs were mainly involved in protein digestion and absorption, the phosphoinositide 3-kinase (PI3K)-AKT serine/threonine kinase (Akt) signaling pathway, focal adhesion and the TGFβ signaling pathway (Fig. 4A). The Forkhead box (Fox)O signaling pathway, prolactin signaling pathway and the TGFβ signaling pathway were top pathways associated with the lncRNAs (Fig. 4B). These results suggested that these pathways may significantly contribute to the pathophysiology and development of HS.
Figure 4.
KEGG pathway analysis of dysregulated lncRNAs and mRNAs. KEGG pathway enrichment analysis of dysregulated (A) mRNAs and (B) lncRNAs; top 20 enriched pathways are presented. DEG, differentially expressed gene; FDR, false-discovery rate; H, hypertrophic scar tissues; KEGG, Kyoto Encyclopedia of Genes and Genomes; lncRNAs, long noncoding RNAs; N, normal tissues.
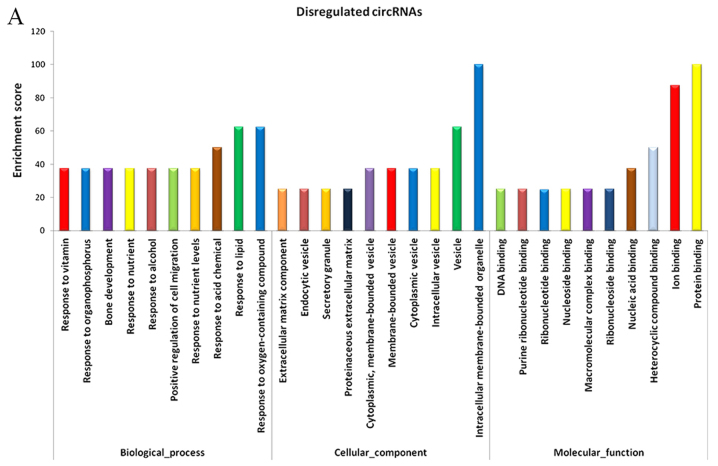
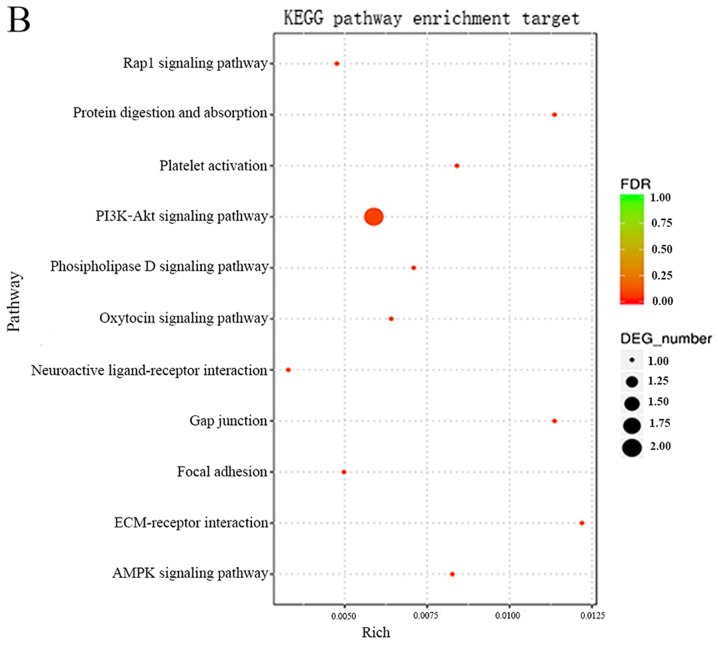
As miRNA sponges, circRNAs can regulate the function of miRNAs. Differentially expressed circRNAs, and the parental gene transcripts, were studied to determine their biological effects. Compared with the adjacent normal skin tissues, GO analysis revealed that the differentially overexpressed circRNAs in HS were associated with ECM component, response to nutrient and bone development, among others (Fig. 5A). Furthermore, KEGG analysis demonstrated that dysregulated circRNAs were associated with the PI3K-Akt signaling pathway, ECM-receptor interaction, and protein digestion and absorption (Fig. 5B). Based on these data, the biological functions of these circRNAs may contribute to the development of HS.
Figure 5.
GO and KEGG pathway analysis of circRNAs. (A) GO annotation of dysregulated circRNAs; top 10 enriched terms covering biological processes, cellular components and molecular functions are presented. (B) KEGG pathway enrichment analysis of dysregulated circRNAs; top 10 enriched pathways are presented. circRNAs, circulating RNAs; DEG, differentially expressed gene; FDR, false-discovery rate; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Co-expression networks of lncRNA-mRNA and circRNA-mRNA, and functional prediction
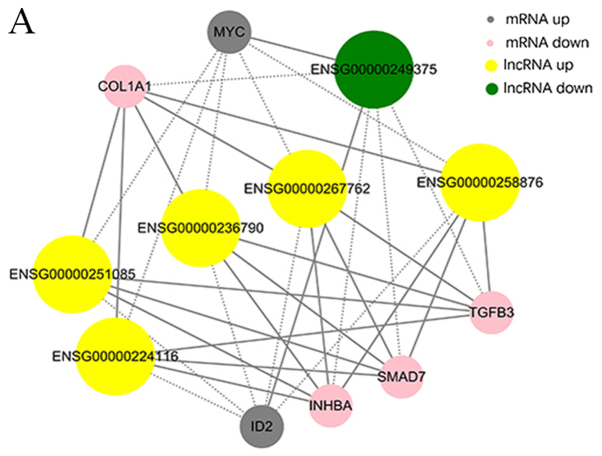
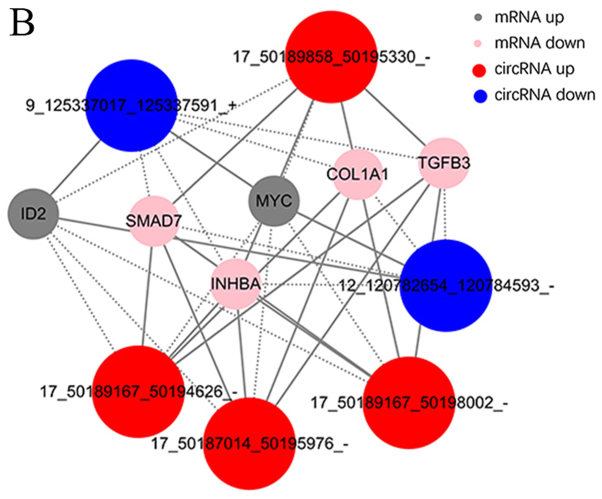
Thus far, the functions of lncRNAs and circRNAs have yet to be annotated; The prediction of lncRNAs or circRNAs function mainly depend on the annotation of the co-expressed mRNAs To obtain the core lncRNAs and circRNAs associated with fibroblast hyperplasia, significantly associated pairs of genes were selected to build the coding-noncoding gene co-expression (CNC) network, in order to identify the interactions and significance among the differentially expressed lncRNAs, circRNAs and mRNAs (Fig. 6A and B).
Figure 6.
Co-expression networks of six significantly dysregulated lncRNAs and circRNAs with their associated mRNAs. (A) Co-expression network of lncRNA-mRNA demonstrating the co-expression of lncRNAs and mRNAs in HS. (B) Co-expression network of circRNA-mRNA demonstrating the co-expression of circRNAs and mRNAs in HS. circRNAs, circular RNAs; HS, hypertrophic scar; lncRNAs, long noncoding RNAs.
The co-expression networks were associated with numerous biological processes, including cell proliferation, adhesion, metastasis and differentiation. The network demonstrated that lncRNAs AC048380.1 (ENSG00000267762) and LINC00299 (ENSG00000236790) were associated with metastasis-related genes, including inhibin subunit βA (INHBA), SMAD family member 7 (SMAD7), collagen type I A1 chain (COL1A1), TGFβ3 and MYC proto-oncogene, bHLH transcription factor (MYC). Inhibitor of DNA binding 2 (ID2) was associated with lncRNA cancer susceptibility 11 (CASC11, ENSG00000249375), TGFβ3-antisense RNA 1 (AS1; ENSG00000258876), INHBA-AS1 (ENSG00000224116), AC048380.1 (ENSG00000267762), LINC00299 (ENSG00000236790) and LINC01969 (ENSG00000251085). In addition, the circRNA-mRNA co-expression network suggested that circ-Chr17:50187014_50195976_-, circ-Chr17:50189167_50194626_-, circ-Chr17:50189167_ 50198002_- and circ-Chr17:50189858_50195330_- were also associated with INHBA, SMAD7, COL1A1, TGFβ3 and MYC. COL1A1 and TGFβ3 were associated with circ-Chr9:125337017_125337591_+ and circ-Chr12:120782654_120784593_-. The co-expression networks demonstrated that one lncRNA, circRNA or mRNA may be associated with one or more mRNA, lncRNA or circRNA. This finding may aid the search for the regulation between lncRNAs, circRNAs and mRNAs implicated in HS.
Construction of a ceRNA network
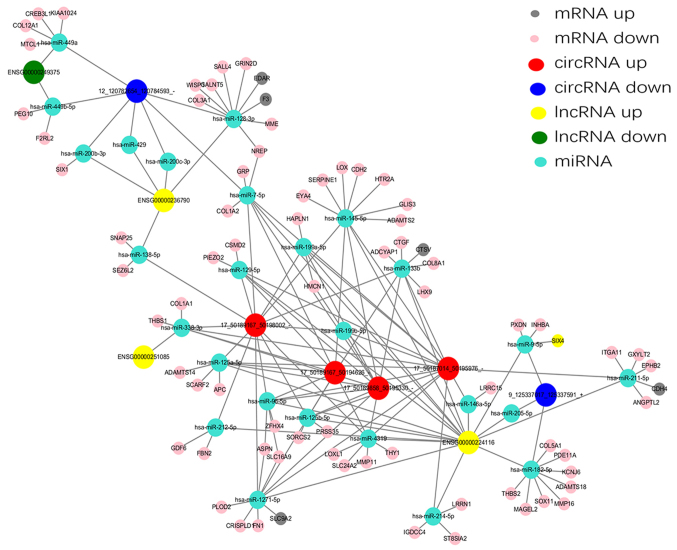
According to the ceRNA hypothesis, numerous lncRNAs, circRNAs and mRNAs may function as ceRNAs, which compete for the same MREs and regulate each other. Analysis of ceRNA interactions aids the functional characterization of such noncoding transcripts. A ceRNA network associated with HS was established in the present study using the high-throughput sequencing data (Fig. 7). Five differentially expressed lncRNAs and six differentially expressed circRNAs, sharing a common MRE site were selected. For example, INHBA-AS1 and circ-Chr9:125337017_125337591_+ were ceRNAs of miR-182-5p targeting potassium voltage-gated channel subfamily J member 6, ADAM metallopeptidase with thrombospondin type 1 motif 18, SRY-box 11, MAGE family member L2, matrix metallopeptidase 16, thrombospondin (THBS)2, phosphodiesterase 11A and collagen type V A1 chain. In addition, LINC01969 and circ-Chr17: 50187014_50195976_-, circ-Chr17:50189167_50194626_-, circ-Chr17: 50189167_50198002_- and circ-Chr17: 50189858_50195330_- were ceRNAs of miR-338-3p targeting THBS1 and COL1A1. These RNA interactions may supply novel insights into the mechanisms underlying HS.
Figure 7.
ceRNA network in hypertrophic scar. The ceRNA network was based on lncRNA/miRNA, circRNA/miRNA and mRNA/miRNA interactions. In this network, the lncRNAs or circRNAs expression linked to mRNAs via miRNAs. ceRNA, competing endogenous RNA; circRNA, circular RNA; lncRNA, long noncoding RNA; miRNA, microRNA.
Discussion
It has previously been reported that dysregulated lncRNAs serve an important role in tumorigenesis and tumor progression, and may be used as potential prognostic biomarkers (20). Recently, circRNAs have also been considered to possess this potential function in cancer (21). HS are characterized by excessive deposition of ECM molecules and the invasive growth of fibroblasts. Although it remains a benign disease, the fibroblasts express malignant features (22); therefore, it has been speculated that ncRNAs may serve an important role in HS. Existing studies have revealed that miRNAs serve a key role in HS (7,23). Previous studies reported that miR-21 promotes fibrosis via modulating the expression of TGFβ and SMAD7 at the post-transcriptional level (24,25). In addition, whether lncRNA8975-1 is overexpressed in HS tissues and dermal fibroblasts has been investigated, and it has been demonstrated that overexpression of lncRNA8975-1 inhibits cell proliferation and reduces the protein expression levels of collagen type I A2 chain, COL1A1, collagen type III A1 chain and α-smooth muscle actin in HS fibroblasts (26). These data suggested that the expression of ncRNAs undergoes significant alterations in HS, which may be closely associated with the development of HS. However, to the best of our knowledge, lncRNA and circRNA expression profiles, and their associated co-expression and ceRNA networks, have not been fully elucidated in HS.
In the present study, the expression profiles of lncRNAs, circRNAs and mRNAs were evaluated in HS and normal skin using RNA-seq, after which, CNC and ceRNA networks were constructed. The RNA-seq analysis demonstrated that a total of 3,649 lncRNAs, 536 mRNAs and 11 circRNAs were differentially expressed between the studied groups. Hierarchical clustering demonstrated that these differentially expressed genes were distinguishable. Functional annotation of the DEGs highly expressed in HS was shown. A total of 77 mRNAs were associated with ECM organization, 26 mRNAs were associated with collagen metabolic process; 693 lncRNAs were associated with the regulation of gene expression, and 591 lncRNAs were associated with nucleic acid-templated transcription; 4 circRNAs were associated with regulation of the RNA metabolic process and 2 circRNAs were associated with ECM organization.
KEGG pathway analysis demonstrated that the greatest number of genes was associated with pathways involved in the accumulation of ECM components, including the TGFβ signaling pathway, the PI3K-Akt signaling pathway and ECM receptor interaction. These genes may serve important roles during cell proliferation, differentiation, metastasis and apoptosis, all of which are associated with the stages of wound repair, inflammation, formation of new tissues, remodeling and formation of HS.
Previous studies have revealed that lncRNA H19 is aberrantly regulated in a wide range of cancers (27,28). The data from the present study demonstrated that H19 was markedly higher compared with in normal skin. According to previous studies, H19 mediates the expression of genes involved in invasion, promotion of epithelial-mesenchymal transition (EMT) and Wnt signaling (29), and promotes cell migration by acting as a sponge for miR-138 and miR-200a (30). In addition, the PI3K-Akt pathway has been reported to be of importance in EMT and wound healing (31,32), serving important roles in inflammation, angiogenesis, re-epithelialization and connective tissue regeneration.
The data from the present study also detected significant increases in INHBA-AS1, TGFβ3-AS1, LINC00299, AC048380.1 and AC037198.1 expression, and decreases in CASC11, AC012065.4 and AC016866.3 expression in HS tissues. These RNAs are associated with the TGFβ signaling pathway. Numerous studies (33,34) have focused on analysis of the expression patterns of lncRNAs and their potential crosstalk with adjacent protein-coding genes (PCGs). INHBA is a ligand belonging to the TGFβ superfamily, which promotes cell proliferation and metastasis (35,36). INHBA-AS1 has also been revealed to be increased in gastric cancer and chronic kidney disease (37,38). The data from the present study indicated that the expression of INHBA-AS1 was increased in HS. As an antisense lncRNA, INHBA-AS1 may influence the expression of its nearby PCGs through various mechanisms.
In the present study, the nearest PCGS was about 100 kb from LINC00299, and identified ID2 as a potential, corresponding cis-regulated target gene. Introduction of the ID2 gene results in Akt phosphorylation and negative regulation of cell differentiation. In terms of cell proliferation, the ID2 protein induces a malignant phenotype (39,40). Studies showed that ID2 is a negative regulator of EMT. Downregulated ID2 might associated with EMT such as cancer and fibrosis (41,42). To further investigate the mechanisms underlying LINC00299, a CNC co-expression network was constructed and 292 PCGs were identified to be associated with it. The majority of these genes were involved in cell proliferation, differentiation and metastasis. These findings suggested that LINC00299 may be an important regulator in HS via its co-expressed genes. In addition, certain major mRNAs were associated with different lncRNAs, including AC048380.1, LINC01969 and CASC11. Therefore, it was hypothesized that these lncRNAs may be associated with the pathophysiology of HS by regulating co-expressed genes.
Functional pathway enrichment analysis was performed on the co-expressed genes of lncRNAs. The results indicated that these lncRNAs were involved in signaling pathways including FoxO, TGFβ and Hippo, all of which serve pivotal roles in regulating cell growth, proliferation, differentiation and apoptosis, and mediate the regulatory effects of cells in response to ECM adhesion (43–45). These results provided further insight into the molecular mechanisms of action underlying these lncRNAs.
circRNAs have been reported to be involved in transcriptional and post-transcriptional gene expression regulation. They are enriched with functional miRNA binding sites and work as miRNA sponges. For example, circZNF91 contains 24 target sites for miR-23b-3p, which is known to serve important roles in keratinocyte differentiation (13). Furthermore, circRNA_100782 can regulate BxPC3 cell proliferation by acting as a miR-124 sponge via the interleukin 6-signal transducer and activator of transcription 3 pathway (14). The present study performed a GO analysis on the dysregulated circRNAs in HS to analyze their functions. The results suggested that certain biological processes and molecular functions may be involved in the development of HS. KEGG pathway analysis for the differentially expressed circRNAs identified six pathways including PI3K-Akt signaling pathway, protein digestion and absorption, gap junction, platelet activation, AMPK signaling pathway, which may serve important roles during the wound repair stages, inflammation, formation of new tissues and remodeling, and may be strongly associated with the mechanisms underlying HS.
The concept of ceRNA indicates that RNA molecules harboring MREs can communicate with each other by competing for the same miRNA. Understanding this novel RNA crosstalk may provide a novel perspective to account for the function of uncharacterized ncRNAs involved in various types of disease (46). Prediction of ceRNA crosstalk is dependent on the identification of MREs on the relevant transcripts of interest. Previous studies (15,47) have reported that ceRNAs are widely implicated in various biological processes and associated with various diseases, including cancer. However, to the best of our knowledge, there are currently no studies regarding the functional roles of this novel RNA crosstalk in HS. In the present study, a ceRNA network of HS was constructed based on RNA-seq data. This network indicated the lncRNA-miRNA-circRNA-mRNA associations in HS, and included four lncRNAs, six circRNAs, 72 mRNAs and 24 miRNAs. It has previously been reported that miR-145 serves an important role in the differentiation and function of skin myofibroblasts (48), which may participate in the pathophysiology of HS.
With the development of RNA-seq, it has been revealed that ncRNAs serve a key role in the occurrence and development of disease. To date, the functions of the majority of lncRNAs and circRNAs are not well understood. The present study demonstrated that numerous ncRNAs, including lncRNAs and circRNAs, were markedly differentially expressed in HS compared with in normal skin. Although the results of the present study require further experimental verification, the profile of these dysregulated lncRNAs and circRNAs may aid identification of prospective clinical markers, and contribute to the understanding of the pathophysiology and development of HS.
Table II.
Primer sequences of circular RNAs used for reverse transcription-quantitative polymerase chain reaction.
| Primer ID | Primer sequences | Annealing temperature (°C) | Length (base pairs) |
|---|---|---|---|
| 17_50189167_50198002_- | F:CTGGACAGCGTGGTGTGGT | 60 | 187 |
| R:TCCCCATCATCTCCATTCTTTC | |||
| 17_50190326_50194041_- | F:TGGCAAGAGTGGTGATCGTG | 60 | 134 |
| R:GCTCTCCAGCAGCACCTTTAG | |||
| 17_50189167_50194626_- | F:GAGAGAGAGGCTTCCCTGGTC | 60 | 138 |
| R:ATCCTTTGGGGCCAGCAG | |||
| 17_50189858_50195330_- | F:CAAAGGTGCTCGCGGCAG | 60 | 71 |
| R:CACCAGCAATACCAGGAGCA | |||
| 2_72718102_72733118_- | F:ATGAAGCAAAATCAAGTGACGGA | 60 | 71 |
| R:ATTGCTATTACCAGTTCCTTTCCCT | |||
| 11_33286412_33287511_+ | F:CAATCTCGGTACTACAGGTATGGC | 60 | 87 |
| R:ACACTACAAAAGGCACTTGACTGAG | |||
| Human β-Actin | F:AGCACAGAGCCTCGCCTTTG | 60 | 208 |
| R:CTTCTGACCCATGCCCACCA |
F, forward; R, reverse.
Table VII.
Comparison of RT-qPCR data with RNA-seq data for circular RNA.
| circRNA | 17_50190326_50194041_- | 17_50189167_50194626_- | 17_50189858_50195330_- | 17_50189167_50198002_- | 11_33286412_33287511_+ | 2_72718102_72733118_- |
|---|---|---|---|---|---|---|
| RT-qPCR (H/N) | 25.86 | 18.49 | 11.12 | 8.49 | −1.75 | −5.10 |
| RNA-seq (H/N) | 63.80 | 76.86 | 94.26 | 136.31 | −184.80 | −62.36 |
CircRNA, circular RNA; H, hypertrophic scar; N, normal skin; RNA-seq, RNA sequencing; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Acknowledgements
Authors would like to thank Dr Shangfeng Fu from the First Affiliated Hospital of Nanchang University for cell culture support, and Professor Jianhua Fu from the First Affiliated Hospital of Nanchang University for providing samples of hypertrophic scar and normal skin tissue for the present study.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 81060323 and 81460293).
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ML and DL designed the experiments. ML, JW and HH performed the experimental work. JW also participated in the data analysis. ML wrote the manuscript with help from JW and DL. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The study procedures were approved by the Ethics Committee of the First Affiliated Hospital Nanchang University. Written informed consent was obtained from all subjects.
Patient consent for publication
Identifying information, including names, initials, date of birth or hospital numbers, images or statements did not be included in this manuscript. The patients consented for the publication of the associated data and accompanying images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhu Z, Ding J, Shankowsky HA, Tredget EE. The molecular mechanism of hypertrophic scar. J Cell Commun Signal. 2013;7:239–252. doi: 10.1007/s12079-013-0195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curran TA, Ghahary A. Evidence of a role for fibrocyte and keratinocyte-like cells in the formation of hypertrophic scars. J Burn Care Res. 2013;34:227–231. doi: 10.1097/BCR.0b013e318254d1f9. [DOI] [PubMed] [Google Scholar]
- 3.Butzelaar L, Soykan EA, Garre Galindo F, Beelen RH, Ulrich MM, Niessen FB, van der Molen Mink AB. Going into surgery: Risk factors for hypertrophic scarring. Wound Repair Regen. 2015;23:531–537. doi: 10.1111/wrr.12302. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Li J, Li Q, Yan H, Zhou B, Gao Y, Li J. Non-coding RNAs: The new insight on hypertrophic scar. J Cell Biochem. 2017;118:1965–1968. doi: 10.1002/jcb.25873. [DOI] [PubMed] [Google Scholar]
- 5.Arnvig KB, Cortes T, Young DB. Noncoding RNA in Mycobacteria. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.MGM2-0029-2013. [DOI] [PubMed] [Google Scholar]
- 6.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 7.Li P, He QY, Luo CQ. Overexpression of miR-200b inhibits the cell proliferation and promotes apoptosis of human hypertrophic scar fibroblasts in vitro. J Dermatol. 2014;41:903–911. doi: 10.1111/1346-8138.12600. [DOI] [PubMed] [Google Scholar]
- 8.Mehra M, Chauhan R. Long noncoding RNAs as a key player in hepatocellular carcinoma. Biomark Cancer. 2017;9:1179299X–17737301X. doi: 10.1177/1179299X17737301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rühle F, Stoll M. Long non-coding RNA databases in cardiovascular research. Genomics Proteomics Bioinformatics. 2016;14:191–199. doi: 10.1016/j.gpb.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Wu Z, Fu X, Han W. lncRNAs: Insights into their function and mechanics in underlying disorders. Mutat Res Rev Mutat Res. 2014;762:1–21. doi: 10.1016/j.mrrev.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Herter EK, Xu Landén N. Non-coding RNAs: New players in skin wound healing. Adv Wound Care (New Rochelle) 2017;6:93–107. doi: 10.1089/wound.2016.0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haque S, Harries LW. Circular RNAs (circRNAs) in health and disease. Genes (Basel) 2017;8:E353. doi: 10.3390/genes8120353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristensen LS, Okholm TLH, Venø MT, Kjems J. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2018;15:280–291. doi: 10.1080/15476286.2017.1409931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G, Shi Y, Zhang Y, Sun J. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco Targets Ther. 2017;10:5783–5794. doi: 10.2147/OTT.S150678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Chen X, Liu X, Yu Y, Pan H, Haak R, Schmidt J, Ziebolz D, Schmalz G. Complex integrated analysis of lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol. 2017;73:1–9. doi: 10.1016/j.oraloncology.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 16.Didion JP, Martin M, Collins FS. Atropos: Specific, sensitive, and speedy trimming of sequencing reads. Peer J. 2017;5:e3720. doi: 10.7717/peerj.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Feng Z, Wang X, Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 18.Adnan M, Morton G, Hadi S. Analysis of rpoS and bolA gene expression under various stress-induced environments in planktonic and biofilm phase using 2(−∆∆CT) method. Mol Cell Biochem. 2011;357:275–282. doi: 10.1007/s11010-011-0898-y. [DOI] [PubMed] [Google Scholar]
- 19.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong Y, Chen Z, Guo S, Liao X, Xie H, Zheng Y, Cai B, Huang P, Liu Y, Zhou Q, et al. TUG1, SPRY4-IT1, and HULC as valuable prognostic biomarkers of survival in cancer: A PRISMA-compliant meta-analysis. Medicine (Baltimore) 2017;96:e8583. doi: 10.1097/MD.0000000000008583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han YN, Xia SQ, Zhang YY, Zheng JH, Li W. Circular RNAs: A novel type of biomarker and genetic tools in cancer. Oncotarget. 2017;8:64551–64563. doi: 10.18632/oncotarget.18350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu BH, Chen L, Li SR, Wang ZX, Cheng WG. Smac/DIABLO regulates the apoptosis of hypertrophic scar fibroblasts. Int J Mol Med. 2013;32:615–622. doi: 10.3892/ijmm.2013.1442. [DOI] [PubMed] [Google Scholar]
- 23.Kwan P, Ding J, Tredget EE. MicroRNA 181b regulates decorin production by dermal fibroblasts and may be a potential therapy for hypertrophic scar. PLoS One. 2015;10:e0123054. doi: 10.1371/journal.pone.0123054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li G, Zhou R, Zhang Q, Jiang B, Wu Q, Wang C. Fibroproliferative effect of microRNA-21 in hypertrophic scar derived fibroblasts. Exp Cell Res. 2016;345:93–99. doi: 10.1016/j.yexcr.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 25.Zhou R, Zhang Q, Zhang Y, Fu S, Wang C. Aberrant miR-21 and miR-200b expression and its pro-fibrotic potential in hypertrophic scars. Exp Cell Res. 2015;339:360–366. doi: 10.1016/j.yexcr.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Chen L, Cao C, Yan H, Zhou B, Gao Y, Li Q, Li J. The long non-coding RNA LncRNA8975-1 is upregulated in hypertrophic scar fibroblasts and controls collagen expression. Cell Physiol Biochem. 2016;40:326–334. doi: 10.1159/000452548. [DOI] [PubMed] [Google Scholar]
- 27.Huang C, Cao L, Qiu L, Dai X, Ma L, Zhou Y, Li H, Gao M, Li W, Zhang Q, et al. Upregulation of H19 promotes invasion and induces epithelial-to-mesenchymal transition in esophageal cancer. Oncol Lett. 2015;10:291–296. doi: 10.3892/ol.2015.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weidle UH, Birzele F, Kollmorgen G, Ruger R. Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics. 2017;14:143–160. doi: 10.21873/cgp.20027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh YH, Wang SW, Yeh YC, Hsiao HF, Li TK. Rhapontigenin inhibits TGF-β-mediated epithelialmesenchymal transition via the PI3K/AKT/mTOR pathway and is not associated with HIF-1alpha degradation. Oncol Rep. 2016;35:2887–2895. doi: 10.3892/or.2016.4664. [DOI] [PubMed] [Google Scholar]
- 32.Baek SH, Ko JH, Lee JH, Kim C, Lee H, Nam D, Lee J, Lee SG, Yang WM, Um JY, et al. Ginkgolic acid inhibits invasion and migration and TGF-beta-induced EMT of lung cancer cells through PI3K/Akt/mTOR inactivation. J Cell Physiol. 2017;232:346–354. doi: 10.1002/jcp.25426. [DOI] [PubMed] [Google Scholar]
- 33.Liu T, Zhang X, Yang YM, Du LT, Wang CX. Increased expression of the long noncoding RNA CRNDE-h indicates a poor prognosis in colorectal cancer, and is positively correlated with IRX5 mRNA expression. Onco Targets Ther. 2016;9:1437–1448. doi: 10.2147/OTT.S98268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding LJ, Li Y, Wang SD, Wang XS, Fang F, Wang WY, Lv P, Zhao DH, Wei F, Qi L. Long noncoding RNA lncCAMTA1 promotes proliferation and cancer stem cell-like properties of liver cancer by inhibiting CAMTA1. Int J Mol Sci. 2016;17:E1617. doi: 10.3390/ijms17101617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HY, Li CC, Huang CN, Li WM, Yeh HC, Ke HL, Yang KF, Liang PI, Li CF, Wu WJ. INHBA overexpression indicates poor prognosis in urothelial carcinoma of urinary bladder and upper tract. J Surg Oncol. 2015;111:414–422. doi: 10.1002/jso.23836. [DOI] [PubMed] [Google Scholar]
- 36.Katayama Y, Oshima T, Sakamaki K, Aoyama T, Sato T, Masudo K, Shiozawa M, Yoshikawa T, Rino Y, Imada T, Masuda M. Clinical significance of INHBA gene expression in patients with gastric cancer who receive curative resection followed by adjuvant S-1 chemotherapy. In Vivo. 2017;31:565–571. doi: 10.21873/invivo.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smyth LJ, McKay GJ, Maxwell AP, McKnight AJ. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics. 2014;9:366–376. doi: 10.4161/epi.27161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ke D, Li H, Zhang Y, An Y, Fu H, Fang X, Zheng X. The combination of circulating long noncoding RNAs AK001058, INHBA-AS1, MIR4435-2HG, and CEBPA-AS1 fragments in plasma serve as diagnostic markers for gastric cancer. Oncotarget. 2017;8:21516–21525. doi: 10.18632/oncotarget.15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talkowski ME, Maussion G, Crapper L, Rosenfeld JA, Blumenthal I, Hanscom C, Chiang C, Lindgren A, Pereira S, Ruderfer D, et al. Disruption of a large intergenic noncoding RNA in subjects with neurodevelopmental disabilities. Am J Hum Genet. 2012;91:1128–1134. doi: 10.1016/j.ajhg.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamata YU, Sumida T, Kobayashi Y, Ishikawa A, Kumamaru W, Mori Y. Introduction of ID2 enhances invasiveness in ID2-null oral squamous cell carcinoma cells via the SNAIL axis. Cancer Genomics Proteomics. 2016;13:493–497. doi: 10.21873/cgp.20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen XF, Chen M, Wu Y, Chen MN, Glogowska A, Klonisch T, Zhang GJ. Inhibitor of DNA binding 2 inhibits epithelial-mesenchymal transition via up-regulation of Notch3 in breast cancer. Transl Oncol. 2018;11:1259–1270. doi: 10.1016/j.tranon.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gervasi M, Bianchi-Smiraglia A, Cummings M, Zheng Q, Wang D, Liu S, Bakin AV. JunB contributes to Id2 repression and the epithelial-mesenchymal transition in response to transforming growth factor-β. J Cell Biol. 2012;196:589–603. doi: 10.1083/jcb.201109045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahajan SG, Fender AC, Meyer-Kirchrath J, Kurt M, Barth M, Sagban TA, Fischer JW, Schrör K, Hohlfeld T, Rauch BH. A novel function of FoxO transcription factors in thrombin-stimulated vascular smooth muscle cell proliferation. Thromb Haemost. 2012;108:148–158. doi: 10.1160/TH11-11-0756. [DOI] [PubMed] [Google Scholar]
- 44.Wennmann DO, Vollenbröker B, Eckart AK, Bonse J, Erdmann F, Wolters DA, Schenk LK, Schulze U, Kremerskothen J, Weide T, Pavenstädt H. The Hippo pathway is controlled by Angiotensin II signaling and its reactivation induces apoptosis in podocytes. Cell Death Dis. 2014;5:e1519. doi: 10.1038/cddis.2014.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Zhang L, Yin X, Gao Z, Zhang H, Liu X, Pan X, Li N, Yu Z. Retinoic acid remodels extracellular matrix (ECM) of cultured human fetal palate mesenchymal cells (hFPMCs) through down-regulation of TGF-β/Smad signaling. Toxicol Lett. 2014;225:208–215. doi: 10.1016/j.toxlet.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 46.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sui J, Li YH, Zhang YQ, Li CY, Shen X, Yao WZ, Peng H, Hong WW, Yin LH, Pu YP, Liang GY. Integrated analysis of long non-coding RNA associated ceRNA network reveals potential lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol. 2016;49:2023–2036. doi: 10.3892/ijo.2016.3716. [DOI] [PubMed] [Google Scholar]
- 48.Gras C, Ratuszny D, Hadamitzky C, Zhang H, Blasczyk R, Figueiredo C. miR-145 contributes to hypertrophic scarring of the skin by inducing myofibroblast activity. Mol Med. 2015;21:296–304. doi: 10.2119/molmed.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the current study are available from the corresponding author on reasonable request.