Abstract
The discs large-associated protein (DLGAP) family has been implicated in psychological and neurological diseases. However, few studies have explored the association between the expression of DLGAPs and different types of cancer. Therefore, the present study analyzed the status of DLGAPs in gastric cancer (GC) using bioinformatic tools. Analyses of data obtained from The Cancer Genome Atlas and Gene Expression Omnibus databases revealed that there was selective upregulation of DLGAP4 and DLGAP5 expression in GC tissues when compared with normal gastric tissues. In addition, survival analysis using OncoLnc indicated that high expression of DLGAP4 was significantly correlated with shorter overall survival for all GC patients. However, Kaplan-Meier plots demonstrated that the expression of all DLGAPs, except for DLGAP3, was correlated with patient prognosis; DLGAP4 was consistently associated with GC. DLGAP4 mRNA and protein distributions were examined by reverse transcription-quantitative polymerase chain reaction and immunohistochemsitry. Furthermore, its mutation rate and associated biological processes and signaling pathways were assessed in GC with cBioPortal and FunRich analyses. Taken together, these results indicated that DLGAP4 may serve an oncogenic role in GC development and may be a monitoring target for GC prognosis.
Keywords: discs large associated protein, gastric cancer, The Cancer Genome Atlas, prognostic significance, bioinformatic analysis
Introduction
Gastric cancer (GC) is a highly aggressive disease, with an incidence that is ranked fifth among all types of cancers worldwide in recent years (1,2). There are nearly 1,000,000 new cases of GC diagnosed globally each year (3). The majority of patients succumb to GC due to late recurrence or distant metastasis, and the 5-year survival rate for GC patients is <5% (4). Thus, there is an urgent requirement to identify biomarkers associated with the prognosis of patients with GC. Although some targets are currently in clinical use, including human epidermal growth factor receptor 2 (5,6), vascular endothelial growth factor receptor-2 (7), excision repair cross complementing gene 1 (8), B-cell lymphoma 2, and Ki-67 (9), the heterogeneous nature of GC renders these as only weakly predictive. Therefore, the aim of the present study was to identify novel molecular markers associated with the prognosis of GC.
The discs large-associated protein (DLGAP) family includes five members, namely DLGAP1, 2, 3, 4 and 5, which are distributed on different chromosomes and generate transcript variants of varying lengths (10). They were originally detected in the rat (11,12), and then the structures and functions of their human homologues were described. All DLGAPs share three key domains, including a dynein light chain domain (13), a 14-amino-acid repeat domain (14–16), and a guanylate kinase-associated protein homology domain (17,18). These specific regions enable DLGAPs to interact with numerous other proteins, including SH3 and mutiple ankyrin repeat domain protein (19), DLG4 proteins (14–16), Stargazin proteins (20–23) and the Homer family proteins (24). To date, the role of DLGAPs in cancer remains unclear.
Until now, there have been no studies that have investigated the function of DLGAPs in GC. The present study evaluated the expression of DLGAPs in The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases, and investigated the correlation between prognostic significance and the expression of DLGAPs to identify which DLGAPs may be relevant for GC.
Materials and methods
Identification of DLGAPs from Oncomine
Data was obtained from the Oncomine™ database (www.oncomine.org), which includes 715 datasets and 86,733 samples. The present sutdy screened the obtained data for differentially expressed genes (DEGs) by comparing gastric adenocarcinoma tissues with normal gastric (NG) tissues. The judgement criteria were as follows: i) P<0.05 was considered to indicate a statistically significant difference; and ii) DEGs were accepted with a fold change >2.
Expression of DLGAPs from TCGA and GEO databases
To assess the expression of DLGAPs in GC, samples were collected from the TCGA and GEO databases. First, the expression of DLGAPs was copmared between NG and GC tissues, and then matched GC and adjacent para-cancer (APC) tissues were also compared and evaluated.
Prognostic significance of DLGAPs
The present study used Kaplan-Meier plotter (kmplot.com/) and OncoLnc (www.oncolnc.org/) online tools to evaluate the correlation between the expression of each DLGAP and the prognosis of patients with GC, respectively.
mRNA and protein expression of DLGAP4 in GC tissues
Based on these previous analyses, DLGAP4 was selected for further evaluation. The present study collected multiple pairs of clinical samples that contained GC and APC tissues, which were obtained during routine surgery, from the First Affiliated Hospital of Xi'an Jiaotong University (Shaanxi, China). A total of 19 patients (12 males and 7 females; median age, 65 years; age range, 48–80 years) were included, according to certain inclusion criteria (pathological diagnosis was clear; no distant metastasis; no history of cardiovascular disease; no history of radiotherapy or chemotherapy) and gave written informed consent. Our experiments were approved by the Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University. Then, the total RNA of each sample was extracted according to a published protocol (25) and the expression of DLGAP4 was detected using QuantiTect SYBR Green polymerase chain reaction (PCR) kits (Qiagen GmbH, Hilden, Germany) on a Bio-Rad CFX96 system (Bio-Rad Laboratories, Inc, Hercules, CA, USA). β-actin was applied as an internal standard. The thermocycling conditions for reverse transcription-quantitative (RT-q) PCR were as follows: 95°C for 30 sec, 39 cycles of 5 sec at 95°C and 30 sec at 58°C. The expression was calculated using the 2−ΔΔCq method (26). The sequences of the primers were as follows: β-actin forward, 5′-CCTTGCACATGCCGGAG-3′ and reverse, 5′-GCACAGAGCCTCGCCTT-3′; and DLGAP4 forward, 5′-GCTGTCTCTTTGTCTCTGCCC-3′ and reverse, 5′-TGGAAGGTGTTCTCAAGGGG-3′. At the same time, tissues were also fixed in 4% formaldehyde at room temperature for 48 h in preparation for immunohistochemistry (IHC) (27) with rabbit anti-DLGAP4 primary antibody (BIOSS, Beijing, China; cat. no. AE080301) in PBS (1:500) overnight at 4°C.
Bioinformatics analysis using multiple tools
Proteins coexpressed with DLGAP4 with a Pearson score ≥0.4 in the cBioPortal database (www.cbioportal.org) were selected for examination and their correlation was assessed using Cytoscape software (version 3.6.0; www.cytoscape.org). In addition, all DLGAP4-associated proteins were analyzed for their roles in biological processes and signaling pathways with the Functional Enrichment (FunRich; version: 2.1.2) tool (www.funrich.org).
Statistical analysis
All statistical analyses were performed in SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 5.01 (GraphPad Software, Inc., La Jolla, CA, USA). The expression of DLGAPs among GC, NG and APC tissues in the TCGA and GEO databases were compared with Student's t-tests. In addition, the association between each DLGAP and the prognosis of patients with GC was evaluated with Kaplan-Meier plotter survival curves and OncoLnc survival curves. Experimental data were presented as the mean ± SD and experimental repeats were performed three times. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of DLGAPs in GC
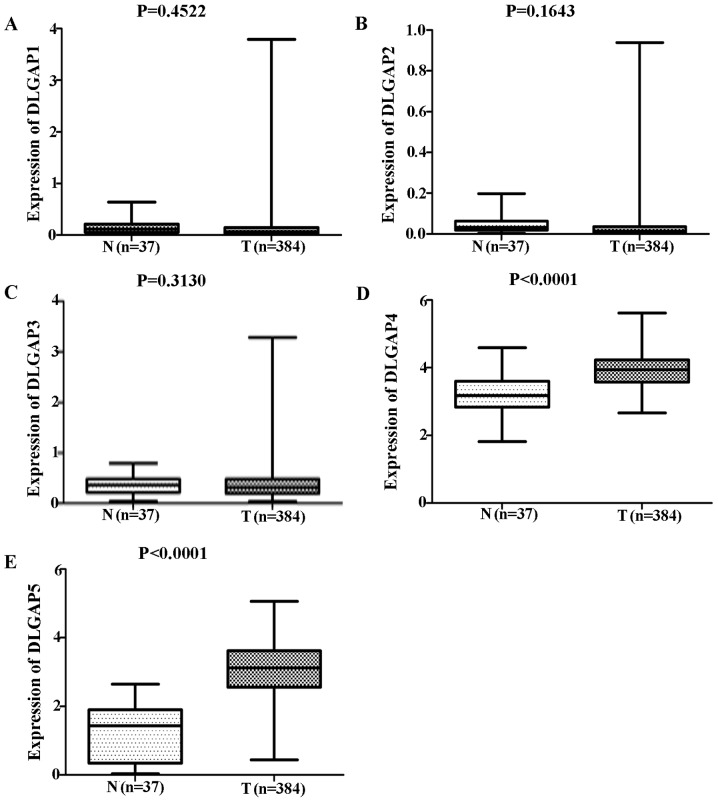
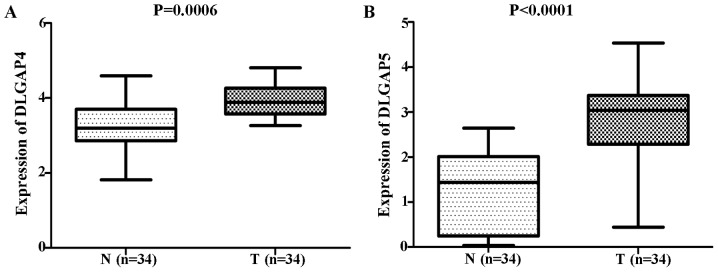
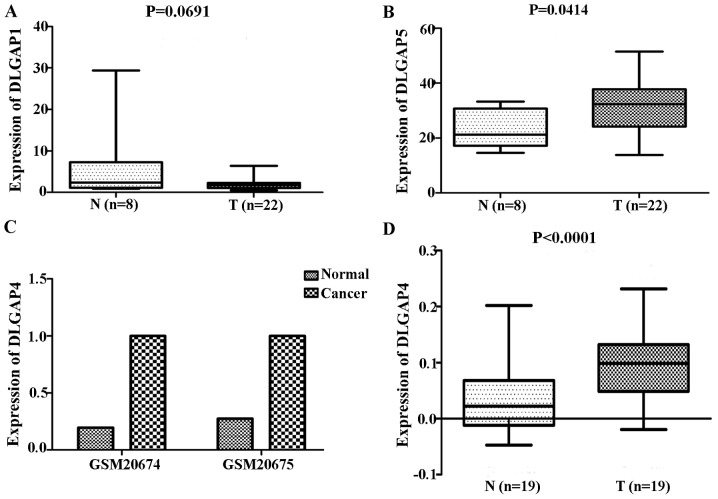
Comparisons of DLGAP expression data from TCGA database revealed no significant differences in the expression of DLGAP1 (P=0.4522; Fig. 1A), DLGAP2 (P=0.1643; Fig. 1B) or DLGAP3 (P=0.3130; Fig. 1C) between GC (n=384 cases) and NG (n=37 cases) tissues. However, DLGAP4 and DLGAP5 were significantly upregulated in GC tissues when compared with NG tissues (P<0.0001; Fig. 1D and E, respectively). Consistent results were obtained when the 34 matched pairs of GC and APC tissues were compared (DLGAP4: P=0.0006; Fig. 2A; and DLGAP5: P<0.0001; Fig. 2B). Similarly, repeating these analyses with GEO data demonstrated no significant differences in the expression of DLGAP1 (P=0.0691; Fig. 3A) and a significant difference in the expression of DLGAP5 (P=0.0414; Fig. 3B) between GC (n=22 cases) and NG tissues (n=8 cases). Due to the lack of expression data for DLGAP2 and DLGAP3, the present study was unable to evaluate their expression and perform statistical analysis. In addition, as only two samples were available for DLGAP4, a P-value was not able to be generated (Fig. 3C).
Figure 1.
DLGAP status between GC and NG tissues from TCGA database. The results showed that no significant differences were discovered in the expression of (A) DLGAP1, (B) DLGAP2 and (C) DLGAP3. However, (D) DLGAP4 and (E) DLGAP5 were significantly upregulated in GC tissues when compared with NG tissues. DLGAP, discs large-associated protein; GC, gastric cancer; NG, normal gastric.
Figure 2.
DLGAP status between matched GC and APC tissues from The Cancer Genome Atlas database. The results showed that consistent results for (A) DLGAP4 and (B) DLGAP5 were obtained when the 34 matched pairs of GC and APC tissues were compared. GC, gastric cancer; APC, adjacent para-cancer; DLGAP, discs large-associated protein.
Figure 3.
DLGAP status in GC from the Gene Expression Omnibus database and clinical samples. The results showed no significant difference in the expression of (A) DLGAP1 and a significant difference in the expression of (B) DLGAP5 between GC and normal gastric tissues. Due to the lack of expression data for DLGAP2 and DLGAP3, the present study was unable to evaluate its expression and perform statistical analysis. In addition, as only two samples were available for (C) DLGAP4, a P-value could not be generated. (D) Reverse transcription-quantitative polymerase chain reaction analysis of 19 pairs of clinical samples revealed a similar trend. DLGAP, discs large-associated protein; GC, gastric cancer.
Expression of DLGAP4 in clinical samples
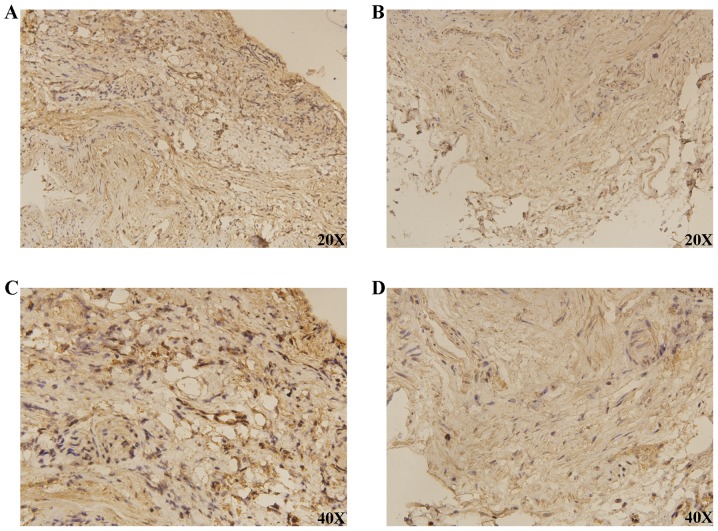
RT-qPCR analysis of 19 pairs of clinical samples revealed a similar trend to the above expression results from the TCGA and GEO databases (P<0.0001; Fig. 3D). IHC demonstrated that DLGAP4 was overexpressed in GC when compared with APC tissues (Fig. 4).
Figure 4.
Expression of DLGAP4 between GC and APC tissues using imunohistochemistry. The results demonstrated that DLGAP4 was overexpressed in GC tissues when compared with APC tissues. (A) DLGAP4; (B) APC; (C) DLGAP4 at higher magnification; (D) APC at higher magnification. GC, gastric cancer; APC, adjacent para-cancer; DLGAP, discs large-associated protein.
GC survival analysis in association with DLGAP expression
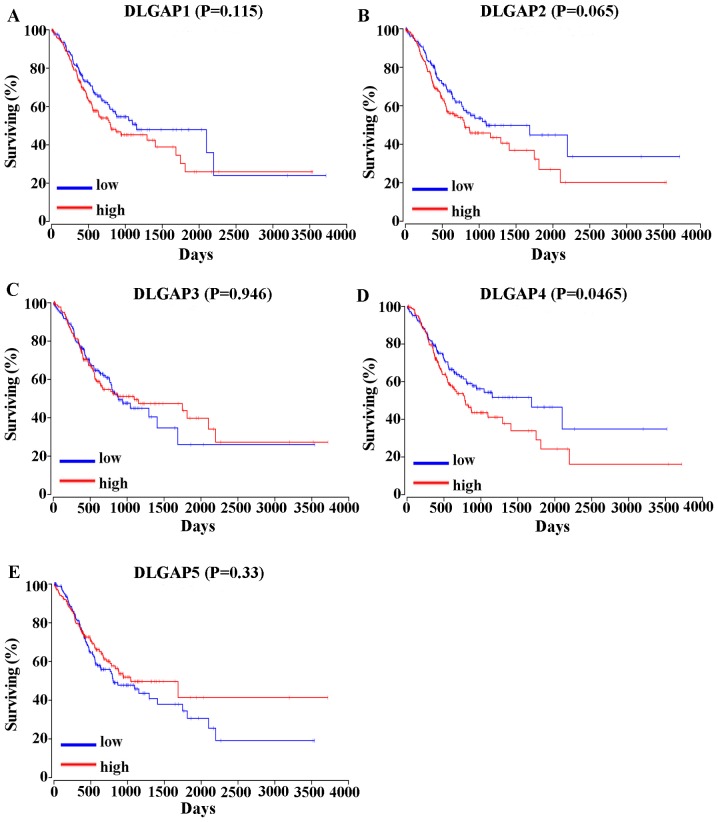
The correlation between the expression of DLGAPs and the prognosis of patients with GC was assessed in two different ways. Analysis of the overall survival (OS) of 378 GC patients in OncoLnc (www.oncolnc.org) revealed that high expression of DLGAP4 was correlated with a shorter OS in GC patients (P=0.0465; Fig. 5). However, the expression of other DLGAP family members was not statistically associated with the prognosis of patients with GC (P=0.115 for DLGAP1l P=0.065 for DLGAP2; P=0.946 for DLGAP3; and P=0.33 for DLGAP5; Fig. 5).
Figure 5.
Prognostic significance of DLGAPs in gastric cancer, assessed by OncoLnc. The results showed that out of all the DLGAP family members, only the overexpression of DLGAP4 was correlated with shorter overall survival. (A) DLPGAP1; (B) DLGAP2; (C) DLGAP3; (D) DLGAP4; (E) DLGAP5. DLGAP, discs large-associated protein.
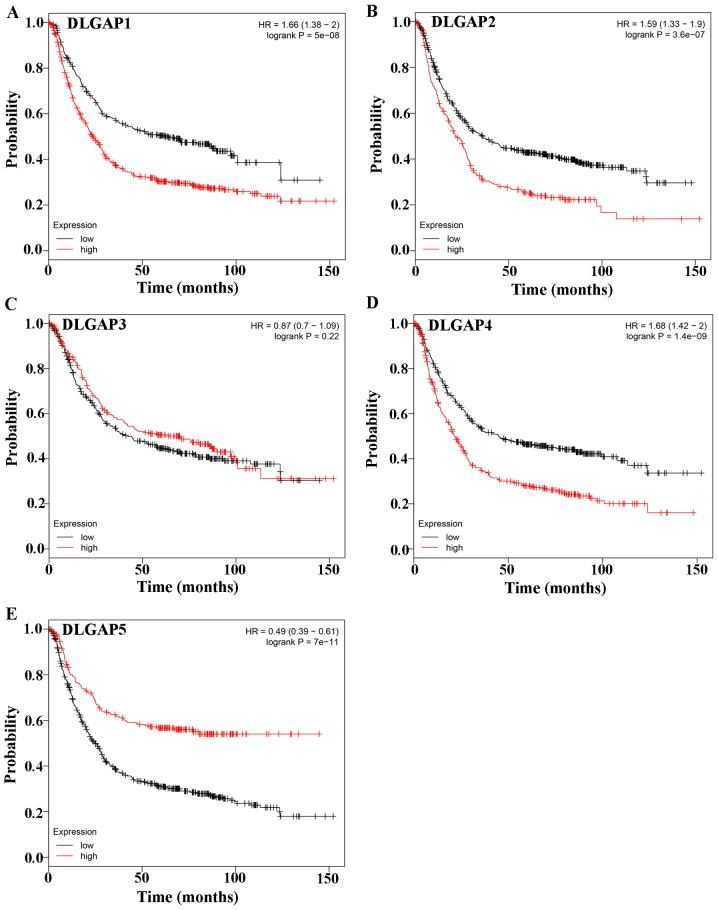
Evaluation of the prognostic significance of DLGAPs in GC with the Kaplan-Meier plotter, which included four types of cancers, revealed that the expression of all DLGAP family members, except DLGAP3, was correlated with the prognosis of patients with GC (P=5×10-8 for DLGAP1, P=3.6×10-7 for DLGAP2; P=0.22 for DLGAP3; P=1.4×10-9 for DLGAP4; and P=7×10-11 for DLGAP5; Fig. 6).
Figure 6.
Prognostic significance of DLGAPs in GC, assessed by Kaplan-Meier plotter. The results revealed that the expression of all DLGAP family members except DLGAP3 was correlated with the prognosis of patients with GC. (A) DLPGAP1; (B) DLGAP2; (C) DLGAP3; (D) DLGAP4; (E) DLGAP5. DLGAP, discs large-associated protein; GC, gastric cancer.
Bioinformatic analysis of DLGAP4 in GC
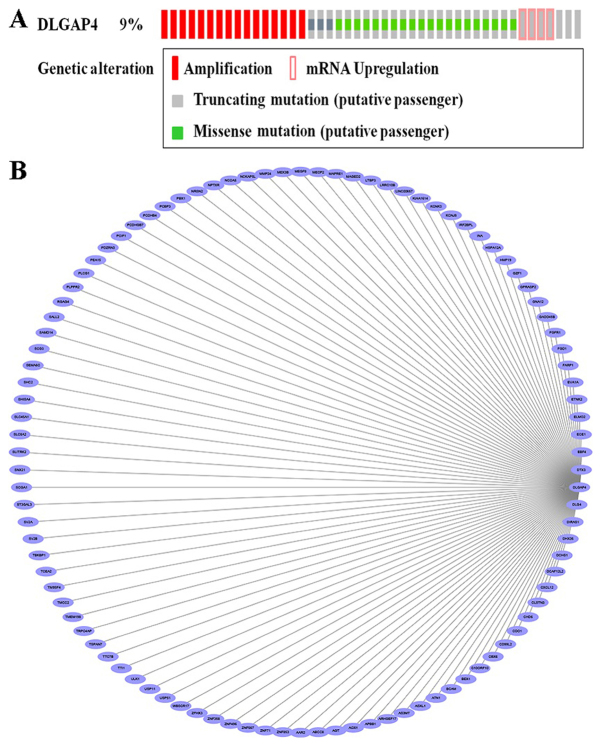
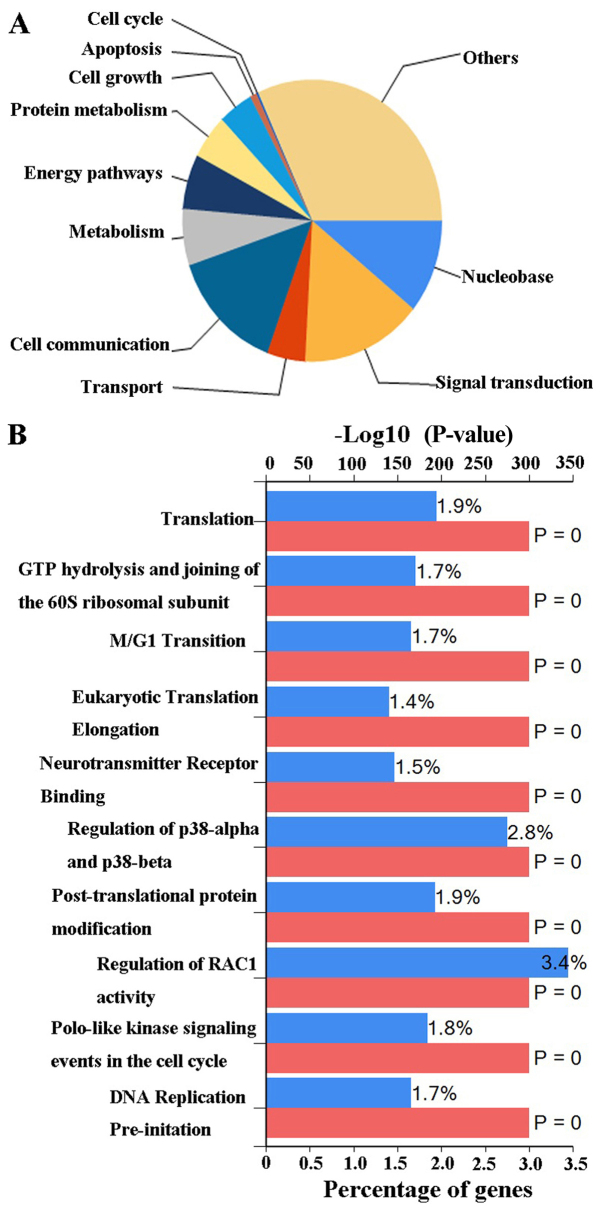
Based on the aforementioned analysis, the present study further investigated the potential DLGAP4-associated molecular mechanisms in the pathogenesis of GC. Analysis of the DLGAP4 variants in GC using data and tools provided by cBioPortal revealed that mutations, including missense mutations, amplification, mRNA upregulation and truncating mutations, were present in ~9% of GC tissue samples (Fig. 7A). When the coexpression data obtained from cBioPortal were evaluated, ~20,000 interacting proteins were observed in the DLGAP4 network. The proteins associated with DLGAP4 with a Pearson score ≥0.4 included, A1 α2 repressin protein splicing factor homolog (AAR2), matrix metalloproteinase 24 (MMP24), zinc finger protein 853 (ZNF853), cluster of differentiation 99 molecule like 2 (CD99L2), ZNF358 and ZNF396; the association network, as drawn in Cytoscape software, is presented in Fig. 7B. Finally, the possible biological processes and pathways of DLGAP4 were investigated using FunRich software (Fig. 8). The major biological processes of DLGAP4 were comprised of ‘signal transduction’ (15.1%) and ‘cell communication’ (14.2%). Biological pathways were dominated by ‘regulation of RAC1 activity’ (3.4%) and ‘regulation of p38-α and p38-β’ (2.8%).
Figure 7.
Alterations and coexpression network of DLGAP4. (A) DLGAP4 alterations, as determined by cBioPortal included (from high to low) missense mutations, amplification, mRNA upregulation and truncating mutations. (B) The coexpression network of DLGAP4 from Cytoscape; the top 100 genes were selected and included in the network. DLGAP4, discs large associated protein 4.
Figure 8.
FunRich analysis. The possible biological processes (A) and biological pathways (B) of DLGAP4 as determined by FunRich. FunRich, Functional Enrichment tool.
Discussion
GC is a common gastrointestinal tumor that threatens human health (28). As there are no typical, clear features of early GC, it is often not diagnosed until the late stages, leading to poor surgery prognosis (29) and a 5-year survival rate of only ~30% (30). The main cause of postoperative mortality is tumor metastasis or recurrence. Therefore, it is important to identify novel targets for monitoring patient prognosis with the aim of prolonging survival.
The cellular biological role of DLGAPs as scaffold proteins suggests that they are able to bind to other substances and therefore, are potenitally involved in cancer progression and tumor metastasis. Until now, DLGAPs, particularly DLGAP1, DLGAP2 and DLGAP3, have been investigated largely in the context of psychological and neurological conditions, including schizophrenia (31), autism spectrum disorder (32), trichotillomania (33), obsessive compulsive disorder (33–35) and cerebellar ataxia (36). The results of the present study indicated that DLGAP4 and DLGAP5 had significantly higher expression levels in GC tissues than in NG tissues, as well as upregulated expression in GC tissues when compared with APC tissues.
In addition, the present evaluation of the assocations between the expression of DLGAPs and the prognosis of patients with GC supported the notion that DLGAPs may serve a role in GC. The analysis of the OncoLnc data (37) demonstrated that there was a significant association between the expression of DLGAP4 and the prognosis of patients with GC. Furthermore, the Kaplan-Meier plotter analysis (38) revealed that the expression of DLGAP1, DLGAP2, DLGAP4 and DLGAP5 were correlated with the prognosis of GC patients. Therefore, DLGAP4, in particular, was strongly implicated as a putative oncogene contributing to GC tumorigenesis. The RT-qPCR and IHC results supported this hypothesis. More mechanistic experiments, in vivo and in vitro, are required to determine whether DLGAP4 serves a causative role in GC.
The present cBioPortal analyses (39,40) demonstrated that there were GC-associated DLGAP mutations, with missense mutations being the most common type of alteration, followed by amplification, mRNA upregulation and truncating mutations. It was speculated that multiple DLGAP4 mutations may be associated with DLGAP4 protein expression changes. Any mutation that disrupts the CpG island located in the promoter region would have the potential to result in epigenetic changes. Thus, such alterations may facilitate the development of GC by increasing the expression of DLGAP4 transcript variants. Minocherhomji et al (36) obtained evidence that indicated that there was a similar mechanism in the cerebellar ataxia; however, more studies are required to confirm this.
The present gene coexpression analysis with cBioPortal data revealed that the genes that were the most likely to interact with DLGAP4 included AAR2, MMP24, ZNF853, CD99L2, ZNF358 and ZNF396. This result suggested that DLGAP4′s involvement in GC may potentially require interactions with other genes. The present examination of biological processes with the FunRich tool (41,42) revealed that DLGAP4, acting as a transcription factor, can participate in a series of signal pathways, and it was also demonstrated that DLGAP4 can function in a variety of biological processes, including signal transduction and cell communication. This information will be utilized to design our next in-depth study.
In conclusion, based on the present comprehensive analysis, the results of the present study revealed that there was a robust association between DLGAP4 expression and the prognosis of patients with GC. The significant overexpression of DLGAP4 in GC suggests that DLGAP4 may be a promising potential prognostic marker for GC. The actual mechanisms of DLGAP4 in GC remain unknown and require future investigation. The bioinformatic method employed in the present study is a useful tool for predicting and screening new features; however, it lacks mechanistic power. Therefore, further in vitro and in vivo validation is required to assess the role of DLGAP4 in GC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JL and TG conceived and designed the experiments. ZL performed the experiments. XZ analyzed the data. In addition, DY provided the gastric cancer specimens, gave final approval of the version to be published and agreed to be accountable for all aspects of the study.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the First Affiliated Hospital of Xi'an Jiaotong University; all patients provided written informed consent.
Patient consent for publication
All patients consented to the publication of data and any associated images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of caner in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Piazuelo MB, Correa P. Gastric cancer: overview. Colomb Med (Cali) 2013;44:192–201. [PMC free article] [PubMed] [Google Scholar]
- 3.Herrero R, Park JY, Forman D. The fight against gastric cancer-the IARC Working Group report. Best Pract Res Clin Gastroenterol. 2014;28:1107–1114. doi: 10.1016/j.bpg.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto M, Rashid OM, Wong J. Surgical management of gastric cancer: The East vs. West perspective. J Gastrointest Oncol. 2015;6:79–88. doi: 10.3978/j.issn.2078-6891.2014.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gravalos C, Jimeno A. HER2 in gastric cancer: A new prognostic factor and a novel therapeutic target. Ann Oncol. 2008;19:1523–1529. doi: 10.1093/annonc/mdn169. [DOI] [PubMed] [Google Scholar]
- 6.Qiu MZ, Li Q, Wang ZQ, Liu TS, Liu Q, Wei XL, Jin Y, Wang DS, Ren C, Bai L, et al. HER2-positive patients receiving trastuzumab treatment have a comparable prognosis with HER2-negative advanced gastric cancer patients: A prospective cohort observation. Int J Cancer. 2014;134:2468–2477. doi: 10.1002/ijc.28559. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs CS, Tomasek J, Yong CJ, Dumitru F, Passalacqua R, Goswami C, Safran H, Santos LV, Aprile G, Ferry DR, et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet. 2014;383:31–39. doi: 10.1016/S0140-6736(13)61719-5. [DOI] [PubMed] [Google Scholar]
- 8.Yamada Y, Boku N, Nishina T, Yamaguchi K, Denda T, Tsuji A, Hamamoto Y, Konishi K, Tsuji Y, Amagai K, et al. Impact of excision repair cross complementing gene 1 (ERCC1) on the outcomes of patients with advanced gastric cancer: Correlative study in Japan Clinical Oncology Group Trial JCOG9912. Ann Oncol. 2013;24:2560–2565. doi: 10.1093/annonc/mdt238. [DOI] [PubMed] [Google Scholar]
- 9.Tsamandas AC, Kardamakis D, Tsiamalos P, Liava A, Tzelepi V, Vassiliou V, Petsas T, Vagenas K, Zolota V, Scopa CD. The potential role of Bcl-2 expression, apoptosis and cell proliferation (Ki-67 expression) in cases of gastric carcinoma and correlation with classic prognostic factors and patient outcome. Anticancer Res. 2009;29:703–709. [PubMed] [Google Scholar]
- 10.Rasmussen AH, Rasmussen HB, Silahtaroglu A. The DLGAP family: Neuronal expression, function and role in brain disorders. Mol Brain. 2017;10:43. doi: 10.1186/s13041-017-0324-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim E, Naisbitt S, Hsueh YP, Rao A, Rothschild A, Craig AM, Sheng M. GKAP, a novel synaptic protein that interacts with the guanylate Kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeuchi M, Hata Y, Hirao K, Toyoda A, Irie M, Takai Y. SAPAPs. A family of PSD-95/SAP90-associated proteins localized at postsynaptic density. J Biol Chem. 1997;272:11943–11951. doi: 10.1074/jbc.272.18.11943. [DOI] [PubMed] [Google Scholar]
- 13.Naisbitt S, Valtschanoff J, Allison DW, Sala C, Kim E, Craig AM, Weinberg RJ, Sheng M. Interaction of the postsynaptic density-95/guanylate kinase domain-associated protein complex with a light chain of myosin-V and dynein. J Neurosci. 2000;20:4524–4534. doi: 10.1523/JNEUROSCI.20-12-04524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, Reissner C, Kuhlendahl S, Coblentz B, Reuver S, Kindler S, Gundelfinger ED, Garner CC. Intramolecular interactions regulate SAP97 binding to GKAP. EMBO J. 2000;19:5740–5751. doi: 10.1093/emboj/19.21.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabio G, Arthur JS, Kuma Y, Peggie M, Carr J, Murray-Tait V, Centeno F, Goedert M, Morrice NA, Cuenda A. p38gamma regulates the localisation of SAP97 in the cytoskeleton by modulating its interaction with GKAP. EMBO J. 2005;24:1134–1145. doi: 10.1038/sj.emboj.7600578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manneville JB, Jehanno M, Etienne-Manneville S. Dlg1 binds GKAP to control dynein association with microtubules, centrosome positioning, and cell polarity. J Cell Biol. 2010;191:585–598. doi: 10.1083/jcb.201002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naisbitt S, Kim E, Weinberg RJ, Rao A, Yang FC, Craig AM, Sheng M. Characterization of guanylate kinase-associated protein, a postsynaptic density protein at excitatory synapses that interacts directly with postsynaptic density-95/synapse-associated protein 90. J Neurosci. 1997;17:5687–5696. doi: 10.1523/JNEUROSCI.17-15-05687.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong J, Yang H, Eom SH, Chum C, Im YJ. Structure of the GH1 domain of guanylate kinase-associated protein from Rattus norvegicus. Biochem Biophys Res Commun. 2014;452:130–135. doi: 10.1016/j.bbrc.2014.08.073. [DOI] [PubMed] [Google Scholar]
- 19.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/S0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, El-Husseini A, Tomita S, Bredt DS, Nicoll RA. Stargazin differentially controls the trafficking of alpha-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate and kainate receptors. Mol Pharmacol. 2003;64:703–706. doi: 10.1124/mol.64.3.703. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- 22.Bats C, Groc L, Choquet D. The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron. 2007;53:719–734. doi: 10.1016/j.neuron.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 23.Matsuda S, Kakegawa W, Budisantoso T, Nomura T, Kohda K, Yuzaki M. Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long-term depression. Nat Commun Nat Res. 2013;4:2759. doi: 10.1038/ncomms3759. [DOI] [PubMed] [Google Scholar]
- 24.Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/S0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 25.Rio DC, Ares M, Jr, Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent) Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5439. pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–208. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Goodpaster T, Randolph-Habecker J. A flexible mouse-on-mouse immunohistochemical staining technique adaptable to biotin-free reagents, immunofluorescence and multiple antibody staining. J Histochem Cytochem. 2014;62:197–204. doi: 10.1369/0022155413511620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 29.Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014;55:621–628. doi: 10.11622/smedj.2014174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 31.Kajimoto Y, Shirakawa O, Lin XH, Hashimoto T, Kitamura N, Murakami N, Takumi T, Maeda K. Synapse-associated protein 90/postsynaptic density-95-associated protein (SAPAP) is expressed differentially in phencyclidine-treated rats and is increased in the nucleus accumbens of patients with schizophrenia. Neuropsychopharmacology. 2003;28:1831–1839. doi: 10.1038/sj.npp.1300212. [DOI] [PubMed] [Google Scholar]
- 32.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welch JM, Lu J, Rodriguiz RM, Trotta NC, Peca J, Ding JD, Feliciano C, Chen M, Adams JP, Luo J, et al. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature. 2007;448:894–900. doi: 10.1038/nature06104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bienvenu OJ, Wang Y, Shugart YY, Welch JM, Grados MA, Fyer AJ, Rauch SL, McCracken JT, Rasmussen SA, Murphy DL, et al. Sapap3 and pathological grooming in humans: Results from the OCD collaborative genetics study. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:710–720. doi: 10.1002/ajmg.b.30897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryu S, Oh S, Cho EY, Nam HJ, Yoo JH, Park T, Joo YH, Kwon JS, Hong KS. Interaction between genetic variants of DLGAP3 and SLC1A1 affecting the risk of atypical antipsychotics-induced obsessive-compulsive symptoms. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:949–959. doi: 10.1002/ajmg.b.31242. [DOI] [PubMed] [Google Scholar]
- 36.Minocherhomji S, Hansen C, Kim HG, Mang Y, Bak M, Guldberg P, Papadopoulos N, Eiberg H, Doh GD, Møllgård K, et al. Epigenetic remodelling and dysregulation of DLGAP4 is linked with early onset cerebellar ataxia. Hum Mol Genet. 2014;23:6163–6176. doi: 10.1093/hmg/ddu337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anaya J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peerj Comput Sci. 2016;2:e67. doi: 10.7717/peerj-cs.67. [DOI] [Google Scholar]
- 38.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pathan M, Keerthikumar S, Chisanga D, Alessandro R, Ang CS, Askenase P, Batagov AO, Benito-Martin A, Camussi G, Clayton A, et al. A novel community driven software for functional enrichment analysis of extracellular vesicles data. J Extracell Vesicles. 2017;6:1321455. doi: 10.1080/20013078.2017.1321455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A, et al. FunRich: An open access standalone functional enrichmentand interaction network analysis tool. Proteomics. 2015;15:2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.