Abstract
To identify the key genes and pathways in the development of hepatocellular carcinoma (HCC) from hepatitis B virus (HBV)-positive liver cirrhosis, differentially expressed genes (DEGs) between HCC and liver cirrhosis tissue samples from the GSE17548 gene expression profile dataset were screened. A total of 1,845 DEGs were identified, including 1,803 upregulated and 42 downregulated genes. Gene Ontology, Kyoto Encyclopedia of Genes and Genomes (KEGG) and protein-protein interaction (PPI) network analyses were performed. It was identified that the ‘cell cycle’ and ‘progesterone-mediated oocyte maturation’ KEGG pathways were significantly enriched in the DEGs. In addition, the high expression of the hub genes from the PPI network (including cyclin dependent kinase 1, cyclin B1, cyclin B2, mitotic arrest deficient 2 like 1, BUB1 mitotic checkpoint serine/threonine kinase and cyclin A2; P=0.00116, 0.00021, 0.04889, 0.00222, 0.00015 and 0.00647, respectively) was associated with a decrease in overall survival for patients with HCC as identified using survival and expression data from The Cancer Genome Atlas. The identified hub genes and pathways may help to elucidate the molecular mechanisms of HCC progression from HBV-positive liver cirrhosis. Additionally, they may be useful as therapeutic targets or serve as novel biomarkers for HCC prognosis prediction.
Keywords: bioinformatics analysis, differentially expressed genes, hepatocellular carcinoma, liver cirrhosis, hepatitis B virus
Introduction
Hepatocellular carcinoma (HCC) is the most common type of liver cancer and is a principal health problem worldwide (1). The majority of HCC cases are associated with viral infection; hepatitis B virus (HBV) is the principal cause of HCC in Asia (2–4). HBV is epidemiologically associated with the development of liver cirrhosis (5). HCC frequently develops in the background of cirrhosis following chronic HBV infection (6,7).
Bioinformatics analysis combined with microarray technology has provided a novel method for comprehensively studying the alterations in expression associated with disease at the molecular level. Critical tumor-associated genes and cellular signaling pathways have been identified in recent decades using these methods. Therefore, examining the differentially expressed genes (DEGs) between HCC and liver cirrhosis tissues is helpful for identifying the key genes and pathways leading to tumor progression.
However, previous studies have focused primarily on progression from liver cirrhosis to HCC without regard to the etiology of cirrhosis (8,9). Furthermore, analysis regarding the associations between gene expression and patient survival is lacking. At present, there is insufficient knowledge regarding the molecular mechanisms for HCC transformation from liver cirrhosis in patients with HBV infection.
In the present study, expression profiles from HCC and liver cirrhosis tissue samples in a Gene Expression Omnibus (GEO) dataset were compared to identify DEGs. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment and protein-protein interaction (PPI) network analyses were performed using the DEGs. The prognostic value of the hub genes from the PPI network was validated using survival and expression data from The Cancer Genome Atlas (TCGA) database. The results of the present study may help to identify the genes involved in the progression of HBV-positive liver cirrhosis to HCC, and provide information regarding biomarkers for verification and development in further studies.
Materials and methods
Microarray data
The GSE17548 gene expression profile dataset, based on the GPL570 platform [(HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array; Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA] and deposited by Yildiz et al (8), was downloaded from the GEO database (www.ncbi.nlm.nih.gov/geo/). In total, 21 HBV-positive samples, including 11 liver cirrhosis and 10 HCC tissues, were analyzed.
Identification of DEGs
The raw expression data were preprocessed using the robust multiarray average method (10) in the affy package (11) in Bioconductor. Following preprocessing, HCC tissue samples were compared with liver cirrhosis tissues using an unpaired t-test in the Bioconductor limma package (version 3.10.3; bioconductor.org/biocLite.R). An adjusted P<0.01 and |log fold change| >1 were set as thresholds for the identification of DEGs.
GO term and KEGG pathway enrichment analysis of the DEGs
Subsequent to obtaining the DEGs, GO term and KEGG pathway enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.ncifcrf.gov/). GO (12) analysis determines the potential functions of a list of genes, including the associated biological process (BP), molecular function (MF) and cellular component (CC) terms. KEGG (http://www.genome/ad.jp/kegg/) is a database of genomes, biological pathways, associated diseases and possible drug targets. P<0.05 was considered to indicate a statistically significant difference for the GO and KEGG enrichment analyses.
PPI network construction
A PPI network of DEGs was constructed using the Search Tool for the Retrieval of Interacting Genes (http://string.embl.de/) database and Cytoscape software (version 3.6.0; www.cytoscape.org). As a large number of DEGs was being analyzed, an interaction confidence score threshold of >0.90 was set to avoid uncertain PPIs. PPI networks were visualized with Cytoscape software. Modules from the PPI network were screened using the Molecular Complex Detection (MCODE) plugin (13) with the following criteria: Degree cutoff, 2; node score cutoff, 0.2; k-core, 2; and max depth, 100. P<0.05 was used as a threshold value. The top 10 genes, as ranked by degree, were considered the hub genes.
Survival validation in TCGA
The mRNA-sequencing expression profiles and clinical data from HCC samples (14) were downloaded from TCGA (https://cancergenome.nih.gov/). For the analysis of hub genes identified in the PPI network, patients with cancer were divided into different groups (high and low) based on the median expression of each hub gene. To investigate the association of each gene with overall survival, survival outcomes between the high and low gene expression groups were compared using the Kaplan-Meier method and log-rank test in the hash package in Bioconductor (https://CRAN.R-project.org/package=hash). P<0.05 was considered to indicate a statistically significant difference in survival.
Results
Data preprocessing and identification of DEGs
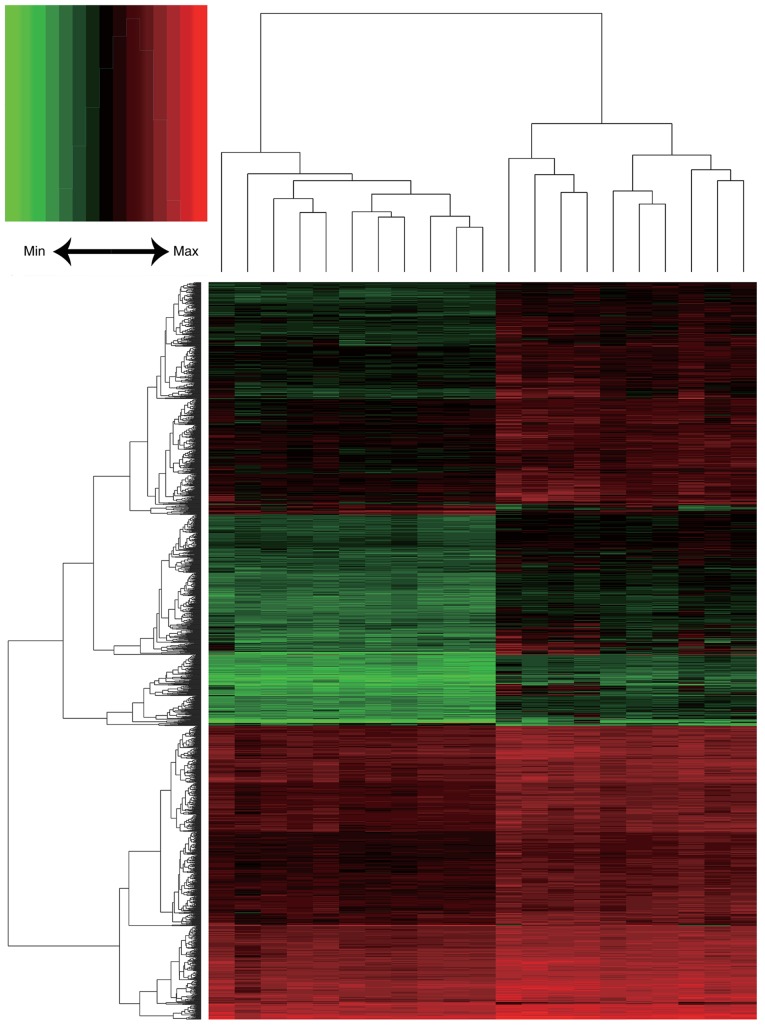
Following background correction and normalization, a total of 20,460 genes were annotated in the 21 HCC and liver cirrhosis expression profiles (Fig. 1). In total, 1,845 DEGs were identified, including 1,803 upregulated and 42 downregulated genes. A heat map of DEG expression is presented in Fig. 2.
Figure 1.
Boxplots of sample data. The × axis represents the samples and the y axis represents the alterations in gene expression. Blue boxes represent cirrhotic expression profiles and red boxes represent hepatocellular carcinoma tumor expression profiles.
Figure 2.
Heat maps of the differentially expressed genes. The × axis represents the samples and the y axis represents the genes. Red represents relatively upregulated genes and green represents relatively downregulated genes.
GO term and KEGG pathway enrichment analysis
Using the DAVID tool, the GO term and KEGG pathway enrichment of the identified DEGs was analyzed. GO BP analysis suggested that the DEGs were primarily associated with the ‘cell division’, ‘mitotic nuclear division’, ‘DNA replication’, ‘sister chromatid cohesion’, ‘DNA replication initiation’, ‘G1/S transition of mitotic cell cycle’, ‘telomere maintenance via recombination’, ‘DNA synthesis involved in DNA repair’, ‘chromosome segregation’ and ‘G2/M transition of mitotic cell cycle’ terms (Table I).
Table I.
Gene Ontology term enrichment analysis of the differentially expressed genes (top 10 terms selected according to P-value).
| A, Biological process | |||
|---|---|---|---|
| Term | Description | Count | P-value |
| GO:0051301 | Cell division | 96 | 1.40×10−26 |
| GO:0007067 | Mitotic nuclear division | 70 | 2.80×10−20 |
| GO:0006260 | DNA replication | 52 | 9.60×10−19 |
| GO:0007062 | Sister chromatid cohesion | 37 | 1.69×10−14 |
| GO:0006270 | DNA replication initiation | 18 | 5.43×10−11 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 32 | 7.00×10−11 |
| GO:0000722 | Telomere maintenance via recombination | 16 | 7.09×10−9 |
| GO:0000731 | DNA synthesis involved in DNA repair | 15 | 2.63×10−7 |
| GO:0007059 | Chromosome segregation | 21 | 2.99×10−7 |
| GO:0000086 | G2/M transition of mitotic cell cycle | 31 | 5.38×10−7 |
| B, Cellular component | |||
| Term | Description | Count | P-value |
| GO:0005654 | Nucleoplasm | 354 | 2.06×10−21 |
| GO:0005634 | Nucleus | 565 | 6.15×10−15 |
| GO:0005737 | Cytoplasm | 523 | 2.26×10−10 |
| GO:0000777 | Condensed chromosome kinetochore | 26 | 9.69×10−9 |
| GO:0000775 | Chromosome, centromeric region | 20 | 4.06×10−8 |
| GO:0005829 | Cytosol | 343 | 5.61×10−8 |
| GO:0000776 | Kinetochore | 23 | 2.22×10−7 |
| GO:0030496 | Midbody | 30 | 2.56×10−7 |
| GO:0005813 | Centrosome | 65 | 6.05×10−7 |
| GO:0005819 | Spindle | 26 | 8.38×10−6 |
| C, Molecular function | |||
| Term | Description | Count | P-value |
| GO:0005515 | Protein binding | 842 | 2.67×10−11 |
| GO:0019901 | Protein kinase binding | 60 | 1.10×10−6 |
| GO:0003677 | DNA binding | 180 | 1.19×10−4 |
| GO:0005524 | ATP binding | 163 | 1.33×10−4 |
| GO:0004674 | Protein serine/threonine kinase activity | 50 | 9.04×10−4 |
| GO:0004842 | Ubiquitin-protein transferase activity | 45 | 9.53×10−4 |
| GO:0003697 | Single-stranded DNA binding | 18 | 0.001422 |
| GO:0016874 | Ligase activity | 38 | 0.001512 |
| GO:0043130 | Ubiquitin binding | 16 | 0.00157 |
| GO:0042802 | Identical protein binding | 85 | 0.002182 |
ATP, adenosine triphosphate.
In the GO CC category, the ‘nucleoplasm’, ‘nucleus’, ‘cytoplasm’, ‘condensed chromosome kinetochore’, ‘chromosome, centromeric region’, ‘cytosol’, ‘kinetochore’, ‘midbody’, ‘centrosome’ and ‘spindle’ terms were enriched in the DEGs (Table I).
In the GO MF category, the ‘protein binding’, ‘protein kinase binding’, ‘DNA binding’, ‘ATP binding’, ‘protein serine/threonine kinase activity’, ‘ubiquitin-protein transferase activity’, ‘single-stranded DNA binding’, ‘ligase activity’, ‘ubiquitin binding’ and ‘identical protein binding’ terms were enriched (Table I).
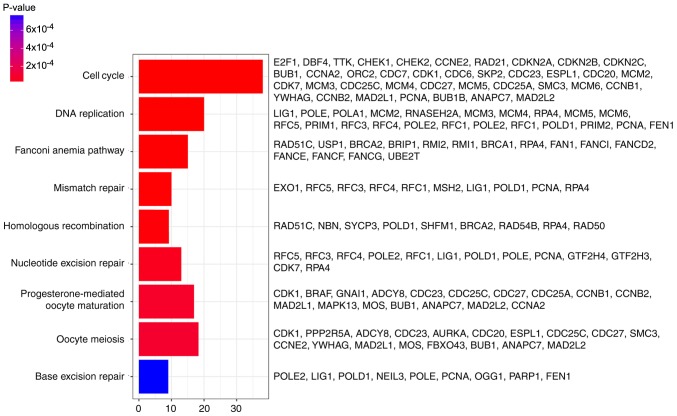
In addition, DEGs were primarily associated with the ‘cell cycle’, ‘DNA replication’, ‘fanconi anemia pathway’, ‘mismatch repair’, ‘homologous recombination’, ‘nucleotide excision repair’, ‘progesterone-mediated oocyte maturation’, ‘oocyte meiosis’ and ‘base excision repair’ pathways, determined by KEGG analysis (Fig. 3).
Figure 3.
Results of KEGG pathway enrichment analysis. The × axis represents the number of genes and the y axis represents the KEGG pathways significantly enriched by differentially expressed genes. KEGG, Kyoto Encyclopedia of Genes and Genomes.
PPI network and module selection
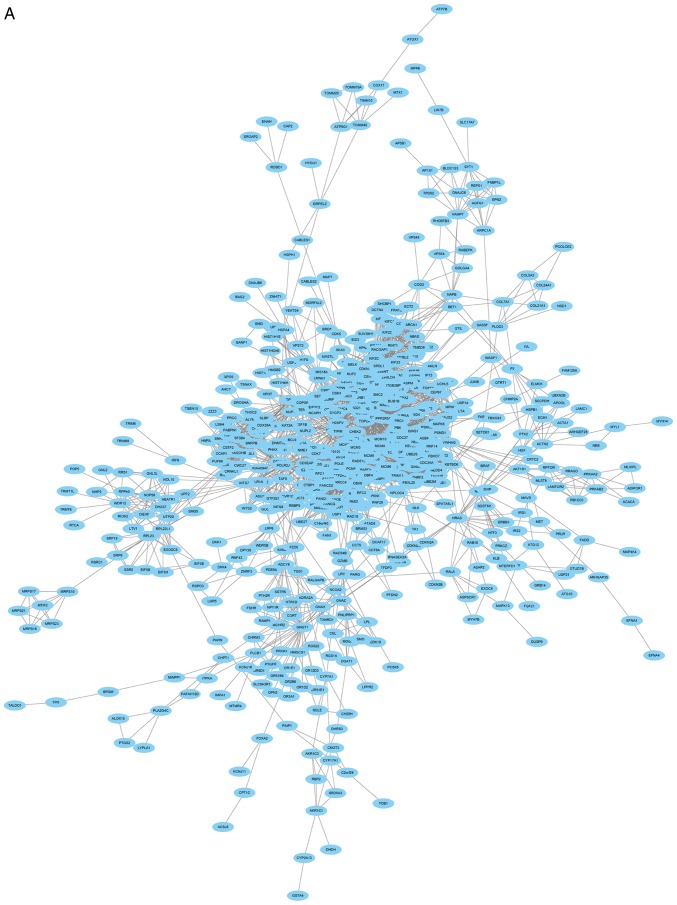
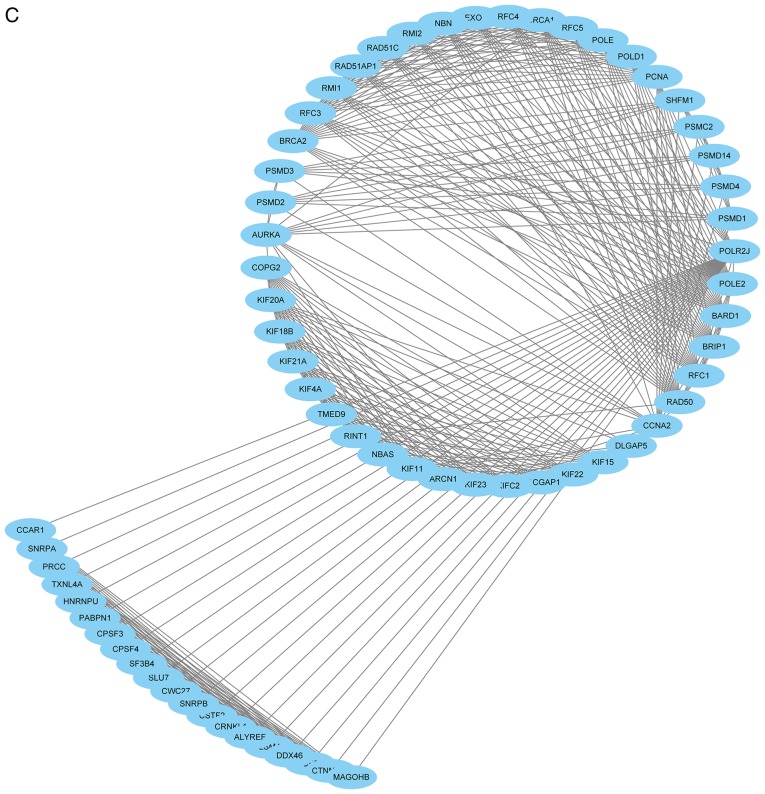
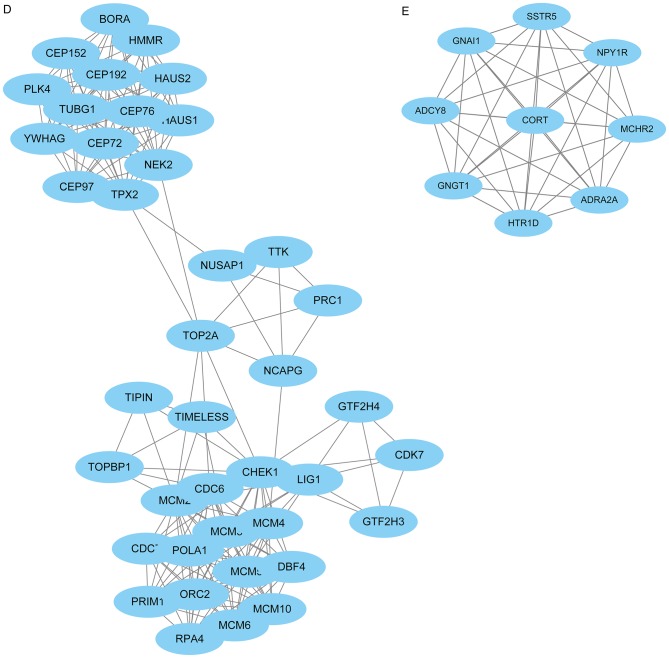
The PPI network consisted of 685 nodes and 4,603 edges (Fig. 4A). According to the degree for each gene, the top 10 hub genes were cyclin dependent kinase 1 (CDK1), cell division cycle 20, cyclin B1 (CCNB1), cyclin B2 (CCNB2), mitotic arrest deficient 2 like 1 (MAD2L1), aurora kinase B, BUB1 mitotic checkpoint serine/threonine kinase (BUB1), cyclin A2 (CCNA2), centromere protein E and kinesin family member 2C (Table II). Using the MCODE Cytoscape plugin, four significant modules were identified (Fig. 4B-E). An enrichment analysis of the genes involved in the top significant modules was performed. The results identified that the DEGs in modules were principally related to ubiquitin mediated proteolysis, cell cycle, oocyte meiosis, progesterone-mediated oocyte maturation, HTLV-I infection, p53 signaling pathway and Epstein-Barr virus infection.
Figure 4.
PPI network of DEGs. (A) Constructed PPI network of DEGs. (B) Module 1 and (C) Module 2. (D) Module 3 and (E) Module 4. Nodes represent genes and lines represent interactions. PPI, protein-protein interaction; DEGs, differentially expressed genes.
Table II.
Core identified differentially expressed genes and their corresponding degrees.
| Gene | Degree |
|---|---|
| CDK1 | 129 |
| CDC20 | 101 |
| CCNB1 | 93 |
| CCNB2 | 81 |
| MAD2L1 | 77 |
| AURKB | 76 |
| BUB1 | 68 |
| CCNA2 | 67 |
| CENPE | 65 |
| KIF2C | 64 |
CDK1, cyclin dependent kinase 1; CDC20, cell division cycle 20; CCNB1, cyclin B1; CCNB2, cyclin B2; MAD2L1, mitotic arrest deficient 2 like 1; AURKB, aurora kinase B; BUB1, BUB1 mitotic checkpoint serine/threonine kinase; CCNA2, cyclin A2; CENPE, centromere protein E; KIF2C, kinesin family member 2C.
Validation with TCGA data
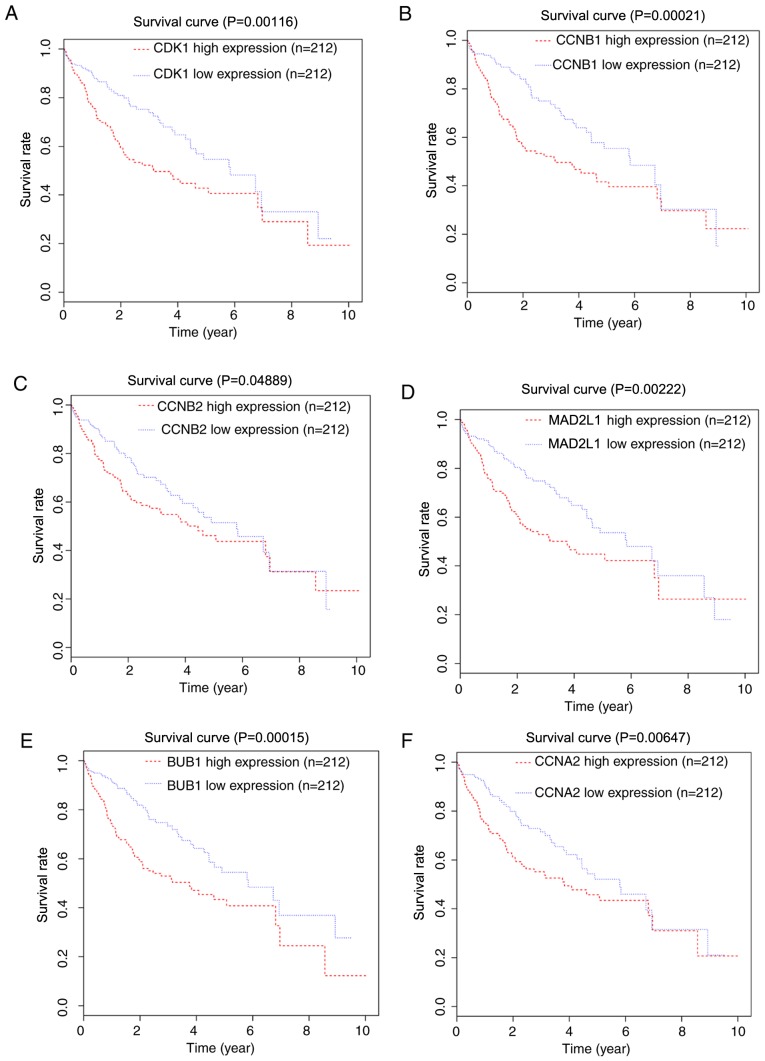
Information regarding 57,000 genes from 424 patients was included in the TCGA mRNA-sequencing expression data. Patients were sorted into high or low expression groups according to the median expression value. As CDK1, CCNB1, CCNB2, MAD2L1, BUB1 and CCNA2 were enriched in the ‘cell cycle’ and the ‘progesterone-mediated oocyte maturation’ pathways, the significance of these six hub genes in prognosis with TCGA data was subsequently validated with the Kaplan-Meier method. It was identified that the high expression of CDK1, CCNB1, CCNB2, MAD2L1, BUB1 and CCNA2 was associated with a decreased overall survival for patients with HCC (P=0.00116, 0.00021, 0.04889, 0.00222, 0.00015 and 0.00647, respectively; Fig. 5).
Figure 5.
Kaplan-Meier survival curves. Kaplan-Meier survival curves for patients stratified according to the median expression of (A) CDK1, (B) CCNB1, (C) CCNB2, (D) MAD2L1, (E) BUB1 and (F) CCNA2. CDK1, cyclin dependent kinase 1; CCNB1, cyclin B1; CCNB2, cyclin B2; MAD2L1, mitotic arrest deficient 2 like 1; BUB1, BUB1 mitotic checkpoint serine/threonine kinase; CCNA2, cyclin A2.
Discussion
Different from the previous studies conducted by Yildiz et al (8) and He et al (9), which analyzed the key genes and pathways for HCC developing from liver cirrhosis regardless of etiology, the present study was conducted specifically with HBV-positive samples. Therefore, drugs targeting these identified key genes and pathways may be considered for HCC therapy.
The hub genes CDK1, CCNB1, CCNB2, MAD2L1, BUB1 and CCNA2 may be suitable for the diagnosis and treatment of HBV-associated HCC. Garnier et al (15) identified that CDK1 may enhance liver regeneration by promoting rapid and efficient hepatocyte proliferation in a rat model, which could promote mutations in the β-catenin gene, potentially promoting the progression to HCC (16). Zhang et al (17) suggested that miR-582-5p inhibits the proliferation of HCC cells by targeting CDK1. However, in a comparative study of HCC and cirrhosis, Masaki et al (18) observed that the expression of CDK1 did not differ between HCC and the adjacent cirrhotic tissues in all the tested patients with hepatitis C virus-RNA. Therefore, alterations of the role of CDK1 in HCC, depending on different viral types, require further evaluation. Wang et al (19) demonstrated that the increased expression of CCNB1 and CCNB2 was associated with the restoration of the young regenerating liver phenotype. Another previous study identified that CCNB1 was associated with G2/M phase cell cycle arrest-induced growth inhibition, suggesting that this gene may be suitable as an anti-cancer agent in future therapy (20). Sze et al (21) suggested that mitotic arrest deficient (MAD)1 interacted with MAD2, leading to its retention in the cytoplasm, which may oppose mitotic checkpoint control in hepatocarcinogenesis. Similarly, Sze et al (22) suggested that MAD2 deficiency may cause a mitotic checkpoint defect in hepatoma cells. Wills et al (23) suggested that BUB1, a loss-of-heterozygosity driver gene, may be implicated in the germline causes of genetic instability in polycystic liver disease. Furthermore, Wang et al (24) identified that BUB1B activated parathyroid hormone like hormone to promote the G-protein-coupled receptor-induced loss of cell adhesion. CCNA functions in the S and G2-M phases of the cell cycle, and CCNA2 overexpression is associated with carcinogenesis in the liver (25–27). Masaki et al (18) suggested that the activation of CCNA expression may serve an essential role in the transformation of cirrhosis to HCC. Wang et al (28) additionally identified that viral insertion was able to contribute to tumorigenesis in HBV-associated HCC by disrupting the control of the CCNA gene. Furthermore, Nault et al (29) identified that adeno-associated virus type 2 was associated with oncogenic insertional mutagenesis in human HCC, including in CCNA2. Yang et al (30) observed that bile acid-activated farnesoid X receptor stimulated the miR-22 silencing of CCNA2, exerting a protective effect in HCC cells.
It is noteworthy that CDK1, CCNB1, CCNB2 and MAD2L1 were enriched in the ‘p53 signaling pathway’. Cellular tumor antigen p53 (p53), discovered in 1979, suppresses cell growth and oncogenic transformation (31). The disruption of p53 protein expression is an important factor underlying tumorigenesis in humans. Reportedly, ≤50% of all malignant tumors exhibit p53 mutations (32). Furthermore, the p53 response pathway is frequently defective in HCC (33), and p53 status is associated with the prognosis of patients with HCC (34). This is consistent with the poor prognosis associated with the high expression of CDK1, CCNB1, CCNB2 and MAD2L1 in the present analysis.
The present study demonstrated that dysregulation of the cell cycle and progesterone-mediated oocyte maturation pathways was closely associated with the development and progression of HCC. It was established that the dysregulation of the cell cycle pathway is an initiating event in cancer (35). Cell cycle checkpoints ensure normal genetic stability, the dysregulation of which may cause deviant cell proliferation leading to tumorigenic mutations (36). Wong et al (37) identified that an abnormal cell cycle is crucial in the development of cancer, subsequent to analyzing cancer samples from early to late stage. Furthermore, the dysfunction of cell cycle regulators and checkpoint mediators in HCC was discussed in previous studies (38–40). Dituri et al (39) demonstrated that alterations in the expression of cell cycle checkpoint-associated proteins frequently occur during HCC development. Yang et al (41) identified that short hairpin RNAs were able to suppress HBV-associated HCC cell proliferation through an effect on the cell cycle pathway. Previous studies concerning the function of the progesterone-mediated oocyte maturation pathway in liver cancer development are limited. In agreement with the study conducted by Jin et al (42), the progesterone-mediated oocyte maturation pathway was significantly enriched in the DEGs in the present study. In addition, Duckworth et al (43) identified that protein kinase A negatively regulates cell division cycle 25C during oocyte maturation. Further studies investigating the function of the progesterone-mediated oocyte maturation pathway in liver cancer are required.
In summary, the present study provided a comprehensive analysis of the DEGs between HCC and cirrhotic liver tissue samples through bioinformatics analysis to identify the potential genes and pathways involved in the transformation of HBV-positive liver cirrhosis to HCC. The identified genes and pathways may be suitable for use as therapeutic targets or novel biomarkers for prognosis. However, further studies are required to confirm these results and determine the underlying mechanisms.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and Technology Planning Project of Guangdong (Guangdong, China; grant no. 2014A020212532).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
QC, JX and PW conceived and designed the study. QC, JX, WL, LS and TH analyzed the data. TH performed literature searches. QC, JX and TH wrote the paper. QC, WL, LS, TH and PW reviewed and edited the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen QF, Jia ZY, Yang ZQ, Fan WL, Shi HB. Transarterial chemoembolization monotherapy versus combined transarterial chemoembolization-microwave ablation therapy for hepatocellular carcinoma tumors ≤5 cm: A propensity analysis at a single center. Cardiovasc Intervent Radiol. 2017;40:1748–1755. doi: 10.1007/s00270-017-1736-8. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273. doi: 10.1053/j.gastro.2011.12.061. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64(1 Suppl):S84–S101. doi: 10.1016/j.jhep.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 4.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 5.Zhou HY, Luo Y, Chen WD, Gong GZ. Hepatitis B virus mutation may play a role in hepatocellular carcinoma recurrence: A systematic review and meta-regression analysis. J Gastroenterol Hepatol. 2015;30:977–983. doi: 10.1111/jgh.12917. [DOI] [PubMed] [Google Scholar]
- 6.Intaraprasong P, Siramolpiwat S, Vilaichone RK. Advances in management of hepatocellular carcinoma. Asian Pac J Cancer Prev. 2016;17:3697–3703. [PubMed] [Google Scholar]
- 7.Sherman M. Hepatocellular carcinoma: Epidemiology, surveillance, and diagnosis. Semin Liver Dis. 2010;30:3–16. doi: 10.1055/s-0030-1247128. [DOI] [PubMed] [Google Scholar]
- 8.Yildiz G, Arslan-Ergul A, Bagislar S, Konu O, Yuzugullu H, Gursoy-Yuzugullu O, Ozturk N, Ozen C, Ozdag H, Erdal E, et al. Genome-wide transcriptional reorganization associated with senescence-to-immortality switch during human hepatocellular carcinogenesis. PLoS One. 2013;8:e64016. doi: 10.1371/journal.pone.0064016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He B, Yin J, Gong S, Gu J, Xiao J, Shi W, Ding W, He Y. Bioinformatics analysis of key genes and pathways for hepatocellular carcinoma transformed from cirrhosis. Medicine (Baltimore) 2017;96:e6938. doi: 10.1097/MD.0000000000006938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 11.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, Qiu P, Ji Y. TCGA-assembler: Open-source software for retrieving and processing TCGA data. Nat Methods. 2014;11:599–600. doi: 10.1038/nmeth.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garnier D, Loyer P, Ribault C, Guguen-Guillouzo C, Corlu A. Cyclin-dependent kinase 1 plays a critical role in DNA replication control during rat liver regeneration. Hepatology. 2009;50:1946–1956. doi: 10.1002/hep.23225. [DOI] [PubMed] [Google Scholar]
- 16.Nhieu JT, Renard CA, Wei Y, Cherqui D, Zafrani ES, Buendia MA. Nuclear accumulation of mutated beta-catenin in hepatocellular carcinoma is associated with increased cell proliferation. Am J Pathol. 1999;155:703–710. doi: 10.1016/S0002-9440(10)65168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Huang W, Ran Y, Xiong Y, Zhong Z, Fan X, Wang Z, Ye Q. miR-582-5p inhibits proliferation of hepatocellular carcinoma by targeting CDK1 and AKT3. Tumour Biol. 2015;36:8309–8316. doi: 10.1007/s13277-015-3582-0. [DOI] [PubMed] [Google Scholar]
- 18.Masaki T, Shiratori Y, Rengifo W, Igarashi K, Yamagata M, Kurokohchi K, Uchida N, Miyauchi Y, Yoshiji H, Watanabe S, et al. Cyclins and cyclin-dependent kinases: Comparative study of hepatocellular carcinoma versus cirrhosis. Hepatology. 2003;37:534–543. doi: 10.1053/jhep.2003.50112. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Quail E, Hung NJ, Tan Y, Ye H, Costa RH. Increased levels of forkhead box M1B transcription factor in transgenic mouse hepatocytes prevent age-related proliferation defects in regenerating liver. Proc Natl Acad Sci USA. 2001;98:11468–11473. doi: 10.1073/pnas.201360898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang G, Chen H, Huang M, Wang N, Zhang J, Zhang Y, Bai G, Fong WF, Yang M, Yao X. Methyl protodioscin induces G2/M cell cycle arrest and apoptosis in HepG2 liver cancer cells. Cancer Lett. 2006;241:102–109. doi: 10.1016/j.canlet.2005.10.050. [DOI] [PubMed] [Google Scholar]
- 21.Sze KM, Ching YP, Jin DY, Ng IO. Role of a novel splice variant of mitotic arrest deficient 1 (MAD1), MAD1beta, in mitotic checkpoint control in liver cancer. Cancer Res. 2008;68:9194–9201. doi: 10.1158/0008-5472.CAN-08-2600. [DOI] [PubMed] [Google Scholar]
- 22.Sze KM, Ching YP, Jin DY, Ng IO. Association of MAD2 expression with mitotic checkpoint competence in hepatoma cells. J Biomed Sci. 2004;11:920–927. doi: 10.1007/BF02254377. [DOI] [PubMed] [Google Scholar]
- 23.Wills ES, Cnossen WR, Veltman JA, Woestenenk R, Steehouwer M, Salomon J, Morsche Te RH, Huch M, Hehir-Kwa JY, Banning MJ, et al. Chromosomal abnormalities in hepatic cysts point to novel polycystic liver disease genes. Eur J Hum Genet. 2016;24:1707–1714. doi: 10.1038/ejhg.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Huang J, Jiang M, Lin H, Qi L, Diao H. Activated PTHLH coupling feedback phosphoinositide to G-protein receptor signal-induced cell adhesion network in human hepatocellular carcinoma by systems-theoretic analysis. ScientificWorldJournal. 2012;2012:428979. doi: 10.1100/2012/428979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinaudo Spiewak JA, Thorgeirsson SS. Detection of a tyrosine-phosphorylated form of cyclin A during liver regeneration. Cell Growth Differ. 1997;8:301–309. [PubMed] [Google Scholar]
- 26.Kim DH, Park SE, Kim M, Ji YI, Kang MY, Jung EH, Ko E, Kim Y, Kim S, Shim YM, Park J. A functional single nucleotide polymorphism at the promoter region of cyclin A2 is associated with increased risk of colon, liver, and lung cancers. Cancer. 2011;117:4080–4091. doi: 10.1002/cncr.25930. [DOI] [PubMed] [Google Scholar]
- 27.Simile MM, De Miglio MR, Muroni MR, Frau M, Asara G, Serra S, Muntoni MD, Seddaiu MA, Daino L, Feo F, Pascale RM. Down-regulation of c-myc and Cyclin D1 genes by antisense oligodeoxy nucleotides inhibits the expression of E2F1 and in vitro growth of HepG2 and Morris 5123 liver cancer cells. Carcinogenesis. 2004;25:333–341. doi: 10.1093/carcin/bgh014. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Chenivesse X, Henglein B, Brechot C. Hepatitis B virus integration in a cyclin A gene in a hepatocellular carcinoma. Nature. 1990;343:555–557. doi: 10.1038/343555a0. [DOI] [PubMed] [Google Scholar]
- 29.Nault JC, Datta S, Imbeaud S, Franconi A, Mallet M, Couchy G, Letouzé E, Pilati C, Verret B, Blanc JF, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet. 2015;47:1187–1193. doi: 10.1038/ng.3389. [DOI] [PubMed] [Google Scholar]
- 30.Yang F, Hu Y, Liu HX, Wan YJ. MiR-22-silenced cyclin A expression in colon and liver cancer cells is regulated by bile acid receptor. J Biol Chem. 2015;290:6507–6515. doi: 10.1074/jbc.M114.620369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson DC, Levine AJ. The properties of p53 proteins selected for the loss of suppression of transformation. Cell Growth Differ. 1994;5:61–71. [PubMed] [Google Scholar]
- 32.Vaughan C, Pearsall I, Yeudall A, Deb SP, Deb S. p53: Its mutations and their impact on transcription. Subcell Biochem. 2014;85:71–90. doi: 10.1007/978-94-017-9211-0_4. [DOI] [PubMed] [Google Scholar]
- 33.Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: Insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166–2176. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- 34.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong YH, Chen RH, Chen BS. Core and specific network markers of carcinogenesis from multiple cancer samples. J Theor Biol. 2014;362:17–34. doi: 10.1016/j.jtbi.2014.05.045. [DOI] [PubMed] [Google Scholar]
- 36.Elledge SJ. Cell cycle checkpoints: Preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 37.Wong YH, Li CW, Chen BS. Evolution of network biomarkers from early to late stage bladder cancer samples. Biomed Res Int. 2014;2014:159078. doi: 10.1155/2014/159078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheung KF, Zhao J, Hao Y, Li X, Lowe AW, Cheng AS, Sung JJ, Yu J. CITED2 is a novel direct effector of peroxisome proliferator-activated receptor γ in suppressing hepatocellular carcinoma cell growth. Cancer. 2013;119:1217–1226. doi: 10.1002/cncr.27865. [DOI] [PubMed] [Google Scholar]
- 39.Dituri F, Mazzocca A, Lupo L, Edling CE, Azzariti A, Antonaci S, Falasca M, Giannelli G. PI3K class IB controls the cell cycle checkpoint promoting cell proliferation in hepatocellular carcinoma. Int J Cancer. 2012;130:2505–2513. doi: 10.1002/ijc.26319. [DOI] [PubMed] [Google Scholar]
- 40.Furuta M, Kozaki K, Tanimoto K, Tanaka S, Arii S, Shimamura T, Niida A, Miyano S, Inazawa J. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013;8:e60155. doi: 10.1371/journal.pone.0060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Zheng B, Han Q, Zhang C, Tian Z, Zhang J. Targeting blockage of STAT3 inhibits hepatitis B virus-related hepatocellular carcinoma. Cancer Biol. 2016;17:449–456. doi: 10.1080/15384047.2016.1156257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin B, Wang W, Du G, Huang GZ, Han LT, Tang ZY, Fan DG, Li J, Zhang SZ. Identifying hub genes and dysregulated pathways in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2015;19:592–601. [PubMed] [Google Scholar]
- 43.Duckworth BC, Weaver JS, Ruderman JV. G2 arrest in Xenopus oocytes depends on phosphorylation of cdc25 by protein kinase A. Proc Natl Acad Sci USA. 2002;99:16794–16799. doi: 10.1073/pnas.222661299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.