Abstract
The concept that inflammation serves a leading role in osteoclast-induced bone loss under pathological circumstances is now widely accepted. In the present study, it was observed that lipopolysaccharides (LPSs) demonstrated a synergic effect on receptor activator of nuclear factor κ-B ligand (RANKL)-induced osteoclast differentiation in Raw264.7 cells, with increasing levels of multiple pro-inflammatory cytokines including interleukin (IL)-6, tumor necrosis factor-α and IL-1β. Furthermore, microRNA (miR)-146a was highly induced by LPS and RANKL co-stimulation during the process of osteoclast differentiation. Overexpression of miR-146a inhibited osteoclast transformation by targeting the key regulators of nuclear factor (NF)-κβ signaling, TNF receptor-associated factor 6 and interleukin-1 receptor-associated kinase 1. The downstream activation of NF-κβ signaling was also inhibited by transfection with a miR-146a mimic. Altogether, the results of the present study demonstrated that miR-146a prevents osteoclast differentiation induced by LPS and RANKL co-stimulation, suggesting that miR-146a may be a promising therapeutic target for treatment of inflammation mediated bone loss.
Keywords: Raw264.7 cells, osteoclast, lipopolysaccharide, receptor activator of nuclear factor κ-B ligand, microRNA-146a
Introduction
Osteoclasts are multinucleated, exclusively bone-absorbing cells, which share the same precursor cells with macrophages and dendritic cells (1). Imbalance of osteoclastic bone resorption and osteoblastic bone formation are associated with multiple pathological disorders, including osteoporosis, osteopenia, Crohn's disease, rheumatoid arthritis (RA) and psoriatic arthritis (2–5). Immune cells profoundly influence the pathogenesis of these diseases in multiple processes, as macrophage precursors are able to differentiate into osteoclasts and multiple inflammatory cells provide cytokines for the progression of osteolysis (6,7). Since vital signaling factors including interleukin (IL)-1β, tumor necrosis factor-α (TNF-α), IL-6 and IL-17 have been revealed to give rise to osteoclast differentiation and activation (8,9), the strong association between inflammation and bone degradation at the molecular signaling level has received extensive attention.
Bacterial lipopolysaccharide (LPS), one of the key components of the outer membrane of Gram-negative bacteria, is able to stimulate the innate immune system to initiate inflammatory responses, exclusively through its pattern recognition receptor Toll-like receptor 4 (TLR4) (10). It is reported that LPS treatment substantially increases the mRNA and protein expression levels of the osteoclast-associated genes receptor activator of nuclear factor κ-B (RANK), TNF receptor-associated factor 6 (TRAF6) and cyclooxygenase-2 in Raw264.7 cells (11). Furthermore, the activation of TLRs is able to suppress expression of RANK in human primary cell cultures, therefore attenuating the activity of RANK ligand (RANKL) and macrophage colony-stimulating factor mediated pathways, consequently resulting in the inhibition of osteoclastogenesis (12). The intriguing results concerning the function of TLRs in addition to other inflammatory factors in osteoclastogenesis indicates a homeostasis of these molecules and their feedback regulation (13).
MicroRNAs (miRNAs/miRs) are a group of highly conserved, single-stranded, small noncoding RNA molecules that regulate gene expression in a post-transcriptional manner. Accumulating evidence has demonstrated that miRNAs serve a critical role in regulating immune responses and inflammation (14,15). Previously, a number of studies have revealed that miRNAs are involved in the development and differentiation of bone cells. For example, miR-223, which is almost exclusively expressed in mouse bone marrow, is regulated by the enhancer modulators PU.1 and CAAT (16). In addition, miR-125b, miR-26a, miR-133 and miR-135 have all been implicated in the differentiation of osteoblasts (17–19).
miR-146a is a typical multi-functional microRNA that serves an important role in regulating multiple biological processes in various types of cells (20,21). The aberrant expression of miR-146a is frequently observed in various diseases, for example rheumatoid arthritis (RA) (22). In the present study, the functions of LPS and miR-146a in the development of osteoclast induced by RANKL were investigated.
Materials and methods
Materials
LPS from Escherichia coli O55:B5 was purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). Recombinant murine RANKL was purchased from PeproTech EC Ltd., (London, UK). A tartrate-resistant acid phosphatase (TRAP) staining kit was obtained from Kamiya Biomedical Company (Seattle, WA, USA). Phosphorylated (Ser) p65, p65, phosphorylated (Ser) IKBα and TRAF6 antibodies were purchased from Cell Signaling Technology, Inc., (Danvers, MA, USA). IRAK1 and GAPDH antibodies were purchased from Abcam (Cambridge, UK). Primers were synthesized by Sangon Biotech Co., Ltd., (Shanghai, China) and the miR-146a mimic was purchased from Shanghai GenePharma, Co., Ltd., (Shanghai, China). TaqMan dye-labeled probes were purchased from Applied Biosystems (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Cell culture and transfection. The murine macrophage cell line Raw264.7 was obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat inactivated fetal calf serum (HIFCS) penicillin (100 U/ml) and streptomycin (100 µg/ml; all from Gibco; Thermo Fisher scientific, Inc.) at 37°C under 5% CO2. For osteoclast cultures, cells were cultured in α-Minimum Essential Medium (α-MEM; Gibco; Thermo Fisher Scientific, Inc.) with 10% HIFCS in the presence of RANKL (30 ng/ml), LPS (100 ng/ml), or together.
For transfection, the synthetic miR-146a mimics, miR-146a inhibitor, or negative control were transfected into cells using Lipofectamine® RNAiMAX (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Sequences of the miR-146a mimics were 5′-UGAGAACUGAAUUCCAUGGGUU-3′ and 5′-CCCAUGGAAUUCAGUUCUCAUU-3′ (antisense), negative control sequence were 5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGGAATT-3′ (antisense). miR-146a inhibitor was a 29-O methylated single-stranded miR-146a antisense oligonucleotides, the sequence was 5′-AACCCAUGGAAUUCAGUUCUCA-3′, and that of the negative control for the inhibitor was 5′-CAGUACUUUUGUGUAGUACAA-3′. In brief, 5×103 cells were seeded into a 48-well plate 24 h prior to transfection. The next day, 200 nM miRNA mimic, miRNA inhibitor, or negative control were transfected into cells. Cells were then cultured for an additional 3 days with RANKL (30 ng/ml) and LPS (100 ng/ml) to collect RNA, protein or observe osteoclast differentiation.
TRAP staining assays
Using the K-ASSAY TRAP Staining kit, TRAP staining was performed according to the manufacturer's protocol. In brief, the medium was removed from the cells, which were then washed and fixed for 5 min at room temperature. Then, the chromogenic substrate was added to each well and cells were incubated at 37°C for 60 min, washed by ddH2O. Cells were observed using a Leica DM IRB inverted microscope. TRAP-positive multinuclear cells containing >3 nuclei were classified as osteoclasts. Cell images were obtained using LAS EZ software version 3.4.0.272 (Leica DMIL LED Inverted microscope; Leica Microsystems GmbH, Wetzlar, Germany). TRAP-positive cells were counted as the mean number of positive cells of 6 fields of view under the microscope.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
For RNA extraction, Raw264.7 cells were plated in a 6-well plate at a density of 5×104 cells/ml in α-MEM with 10% HIFCS. Cells were treated with 30 ng/ml RANKL, 100 ng/ml LPS, a combination or 200 nM miR-146a mimic. Total RNA was extracted using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. For TNF-α, IL-6 and IL-1β genes analysis, 300 ng total RNA was converted to cDNA using PrimeScript™ RT Master Mix (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's protocol. qPCR was performed for different genes in separate wells (Singleplex assay) of a 384-well plate in a reaction volume of 5 µl using TB Green™ Premix Ex Taq™ (Takara Biotechnology Co., Ltd., Dalian, China) and reaction condition as following: 95°C for 15 sec, 1 cycle; 95°C for 5 sec, 60°C for 30 sec, 40 cycles. GAPDH served as the endogenous control for all experiments. The primers used were as follows: GAPDH forward, 5′-TGGATTTGGACGCATTGGTC-3′ and reverse, 5′-TTTGCACTGGTACGTGTTGAT-3′; 5′-ATCTTTTGGGGTCCGTCAACT-3′; TNF-α forward, 5′-CCCTCACACTCAGATCATCTTCT-3′ and reverse, 5′-GCTACGACGTGGGCTACAG-3′; IL-6 forward, 5′-TAGTCCTTCCTACCCCAATTTCC-3′ and reverse, 5′-TTGGTCCTTAGCCACTCCTTC-3′; TRAF6 forward, 5′-AAAGCGAGAGATTCTTTCCCTG-3′ and reverse, 5′-ACTGGGGACAATTCACTAGAGC-3′; IRAK1 forward, 5′-AGCCGAGGTCTGCATTACATT-3′ and reverse, 5′-TGGCAGTCTGGATAACTGATGA-3′. For miRNA analysis, 50 ng total RNA was converted to miRNA using the TaqMan™ MicroRNA Reverse Transcription kit (Invitrogen; Thermo Fisher Scientific, USA) according to the manufacturer's protocol. qPCR was performed for different miRNA in separate wells (Singleplex assay) of a 384-well plate in a reaction volume of 5 µl. Sno202 served as the endogenous control for all experiments. TaqMan dye-labeled probes were purchased from Applied Biosystems (Thermo Fisher Scientific, Inc.). The PCR reaction mixture was performed in an Applied Biosystems ABI Prism 7300 sequence detection system instrument using reaction condition as follows: 50°C for 2 min, 95°C for 10 min, 1 cycle; 95°C for 15 sec, 60°C for 1 min, 40 cycles. Analysis of relative gene expression data was made using real-time quantitative PCR and the 2−ΔΔCq method (23).
Western blotting
Cells were lysed using RIPA buffer (Thermo Fisher Scientific, Inc.). Then, 20 µg lysate proteins were separated using 10% SDS-PAGE, transferred to polyvinylidene fluoride membranes, blocked with 5% BSA in TBS solution at room temperature for 1 h and probed with primary antibodies directed against phosphorylated (Ser) p65 (cat. no. 3033; 1:1,000), p65 cat. no. 8242; 1:1,000), phosphorylated (Ser) IKBα (cat. no. 2859; 1:1,000), TRAF6 (cat. no. 8028; 1:1,000; all Cell Signaling Technology, Inc.), IRAK1 (Abcam; cat. no. ab238; 1:1,000) and GAPDH (cat. no. ab9485; 1:5,000; Abcam) at 4°C overnight. The next day, the membranes were incubated with anti-mouse IgG (cat. no. 7076; 1:5,000) or anti-rabbit IgG, HRP-linked antibody (cat. no. 7074; 1:5,000; both Cell Signaling Technology, Inc.) at room temperature for 1 h and visualized with a SuperSignal West Pico kit (Thermo Fisher Scientific, Inc.). All results were normalized against GAPDH as loading controls.
Statistical analysis
GraphPad Prism version 5.0 (Graphpad Software, Inc., La Jolla, CA, USA) was used for data analysis. The reproducibility of each experiment was confirmed by three independent experiments. Bars represent the mean ± standard error of the mean. Differences between groups were analyzed for statistical significance using an unpaired Student's t-test or two-way analysis of variance followed by Bonferroni post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
LPS exerts a synergic function in promoting RANKL-induced osteoclast differentiation
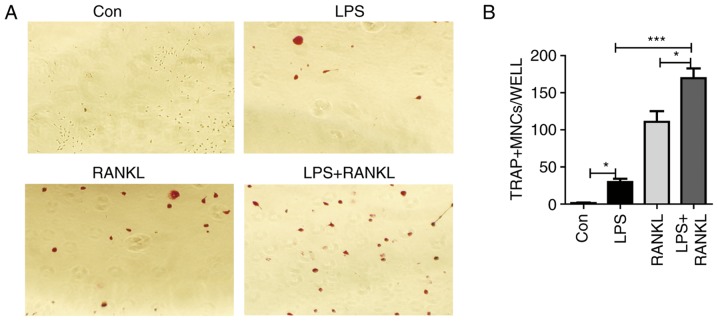
RANKL is an essential regulator of osteoclast differentiation and activation (24). LPS, the key component of the Gram-negative bacterial cell wall, is a well-known stimulator of inflammatory bone loss (25). To investigate whether there is a synergic effect of LPS on RANKL-induced osteoclast differentiation, murine macrophage Raw264.7 cells were treated with LPS and/or RANKL. Consistent with previous studies (11,26), Raw264.7 cells were able to form multinucleated cells in response to either RANKL or LPS stimulation (Fig. 1A). Results from histological staining with TRAP confirmed the presence of osteoclasts (Fig. 1A). In addition, the number of osteoclasts generated from LPS and RANKL combined treatment was significantly (LPS + RANKL vs. RANKL, P<0.05; LPS + RANKL vs. LPS, P<0.001) increased compared with stimulation with either treatment alone (Fig. 1B).
Figure 1.
Effect of LPS and RANKL on osteoclast formation in Raw264.7 cell cultures. (A) Raw264.7 cells were treated with 30 ng/ml RANKL or 100 ng/ml LPS or a combination of RANKL and LPS for 3 days. Cells were fixed and stained for TRAP. TRAP+ cells with >3 nuclei were classified as osteoclasts. Magnification, ×200. (B) Bars represent the mean ± standard error of the mean. *P<0.05 and ***P<0.001. LPS, lipopolysaccharide; RANKL, receptor activator of nuclear factor κβ ligand; TRAP, tartrate-resistant acid phosphatase; MNCs, multinuclear cells; Con, control.
LPS stimulation triggers pro-inflammatory cytokine production during osteoclastogenesis
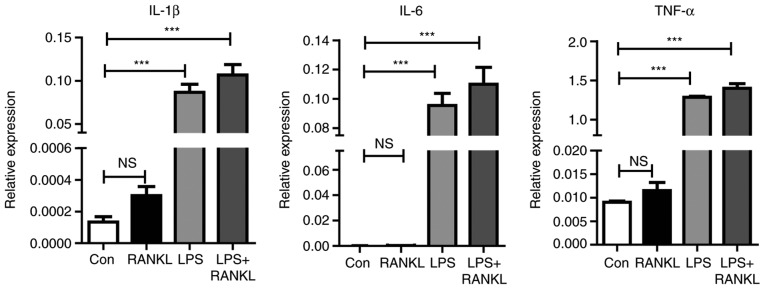
One previous study has revealed that pro-inflammatory cytokines serve a critical role in osteoclast development and bone loss (27). Therefore, to understand how LPS enhances osteoclast differentiation induced by RANKL, cytokine expression during osteoclastogenesis was assessed. As presented in Fig. 2, the expression levels of inflammatory mediators, including IL-6, TNF-α and IL-1β were significantly (P<0.001) increased following LPS stimulation compared with the control.
Figure 2.
LPS highly induces the cytokine production in osteoclastogenesis. Raw264.7 cells treated with 30 ng/ml RANKL and 100 ng/ml of LPS for 3 days. A reverse transcription-quantitative polymerase chain reaction was performed for the detection of IL-1β, IL-6 and TNF-α mRNA levels. The figure presents the relative expression levels of target mRNA normalized to endogenous GAPDH mRNA levels. Bars represent the mean ± standard error of the mean. ***P<0.001. LPS, lipopolysaccharide; RANKL, receptor activator of nuclear factor κβ ligand; IL, interleukin; TNF-α, tumor necrosis factor-α; NS, non-significant; Con, control.
miR-146a is highly induced during osteoclastogenesis
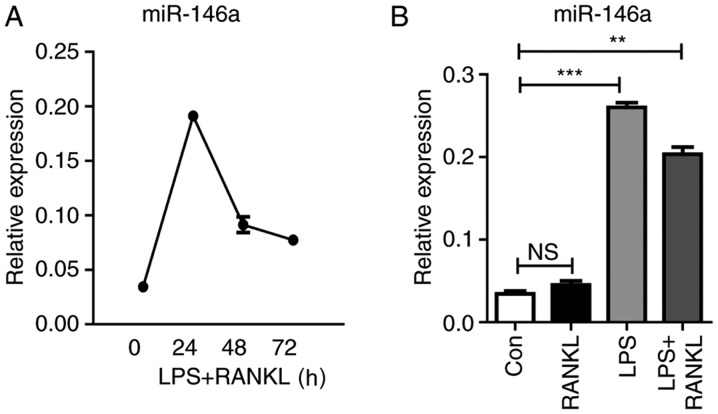
In response to LPS stimulation, Raw264.7 cells express various types of miRNAs. Among them, miR-146a is highly induced and serves a negative regulatory role in the TLR4 signaling pathway (28). To investigate the function of miR-146a in the development of osteoclasts, the present study analyzed the kinetic change of miR-146a expression during osteoclastogenesis. The expression levels of miR-146a were continuously elevated within 24 h following LPS and RANKL treatment, which then subsequently declined. In addition, it was revealed that the combined treatment of LPS and RANKL reduced miR-146a expression compared with LPS stimulation alone (Fig. 3).
Figure 3.
LPS highly induces miR-146a expression in osteoclastogenesis in Raw264.7 cells. (A) Cells were treated with 30 ng/ml RANKL and 100 ng/ml of LPS for 24, 48 and 72 h. (B) Cells were treated with 100 ng/ml LPS, or a combination of 30 ng/ml RANKL and 100 ng/ml LPS for 24 h. Total RNA was extracted for the detection of miR-146a expression using the Taqman method. Bars represent the mean ± standard error of the mean. **P<0.01, ***P<0.001. LPS, lipopolysaccharide; miR, microRNA; RANKL, receptor activator of nuclear factor κβ ligand; Con, control; NS, not significant.
miR-146a inhibits osteoclast differentiation
To further elucidate the role of miR-146a in osteoclast differentiation, the expression levels of miR-146a in Raw264.7 cells were manipulated to demonstrate whether it could affect the outcomes.
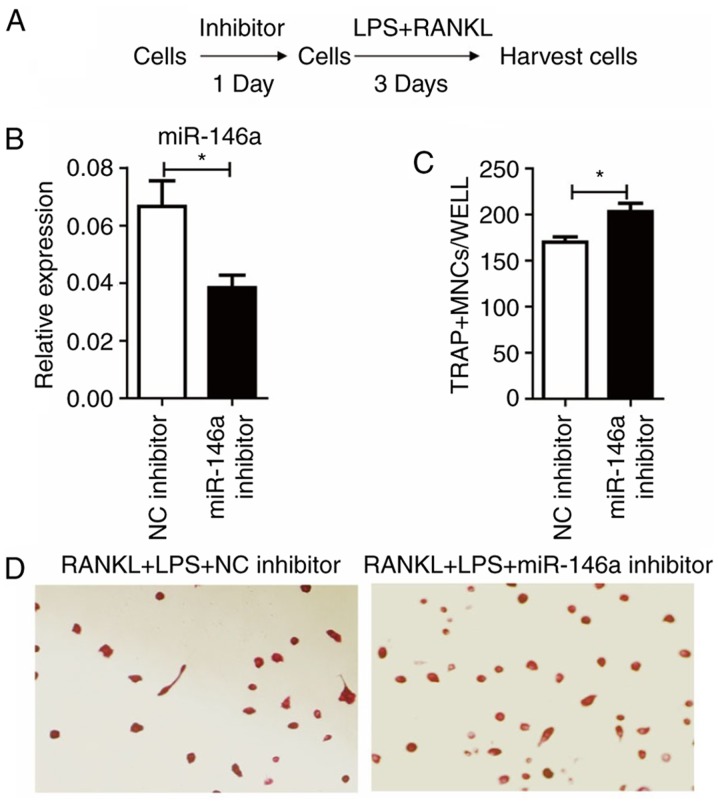
As miR-146a was upregulated following LPS and RANKL treatment, the effect of miR-146a inhibitor was tested (Fig. 4A). Treatment of miR-146a inhibitor 24 h prior to LPS and RANKL administration significantly (P<0.05) reduced the levels of miR-146a, compared with the NC inhibitor group (Fig. 4B). Notably, it was demonstrated that miR-146a inhibition significantly increased the number of TRAP+ cells induced by LPS and RANKL (P<0.05; Fig. 4C and D).
Figure 4.
Silencing of miR-146a promotes osteoclast differentiation in Raw264.7 cells. (A) Study design of the experiment. Cells were treated with 200 nM miR-146a inhibitor for 24 h, then 30 ng/ml RANKL and 100 ng/ml LPS were added and incubated for 3 days. (B) The expression levels of miR-146a. (C) Numbers of osteoclasts were determined by averaging the positive cell number of six fields of view under the microscope. Bars represent the mean ± standard error of the mean. (D) Subsequent to culturing, the cells were fixed and stained for TRAP. TRAP+ cells with >3 nuclei were classified as osteoclasts. Magnification, ×200. *P<0.05. miR, microRNA; LPS, lipopolysaccharide; RANKL, receptor activator of nuclear factor κβ ligand; TRAP, tartrate-resistant acid phosphatase; NC, negative control; MNCs, multinucleated cells.
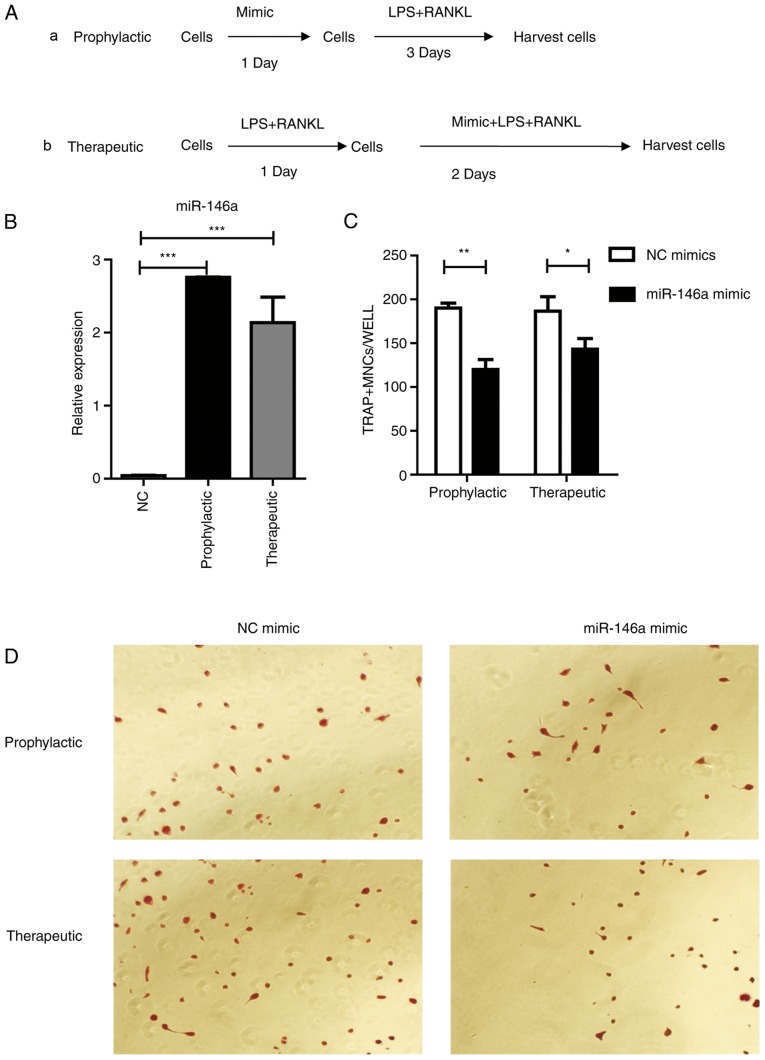
Next, if overexpression of miR-146a could prevent osteoclast differentiation was tested. Two strategies were designed, named ‘Prophylactic’ and ‘Therapeutic’ (Fig. 5A). In the ‘Prophylactic’ strategy, Raw264.7 cells were transfected with miR-146a mimics one day prior to LPS and RANKL treatment; in the ‘Therapeutic’ strategy, Raw264.7 cells received miR-146a mimic transfection one day following LPS and RANKL treatment. Then, cells were harvested 3 days following miR-146a mimic transfection, with LPS and RANKL stimulation. Data from RT-qPCR results revealed that miR-146a expression was significantly (P<0.001) elevated in both ‘Prophylactic’ and ‘Therapeutic’ groups, compared with the negative controls (Fig. 5B). Pre-treatment with the miR-146a mimic revealed a potent inhibitory role in osteoclast differentiation, with a 37% decrease compared with the control group (Fig. 5C and D). Furthermore, treatment with the miR-146a mimic during the process of osteoclast development was able to prevent osteoclast formation by 24% (Fig. 5C).
Figure 5.
miR-146a inhibits osteoclast differentiation in Raw264.7 cells. (A) Study design of the experiment. (a) Cells were treated with 200 nm mimic for 24 h, then 30 ng/ml RANKL and 100 ng/ml LPS were added and incubated for 3 days, (b) or cells were treated with 30 ng/ml RANKL and 100 ng/ml LPS for 24 h followed by the addition of 200 nM mimic for 2 days. (B) miR-146a expression subsequent to treatment with mimic as aforementioned. (C) Numbers of osteoclasts were determined by averaging the positive cell number of six fields of view under a microscope. Bars represent the mean ± standard error of the mean. (D) Subsequent to culturing, the cells were fixed and stained for TRAP. TRAP+ cells with >3 nuclei were classified as osteoclasts. Magnification, ×200. *P<0.05, **P<0.01 and ***P<0.001. miR, microRNA; LPS, lipopolysaccharide; RANKL, receptor activator of nuclear factor κβ ligand; TRAP, tartrate-resistant acid phosphatase; NC, negative control.
Overexpression of miR-146a reduces NF-κβ activation and cytokine production
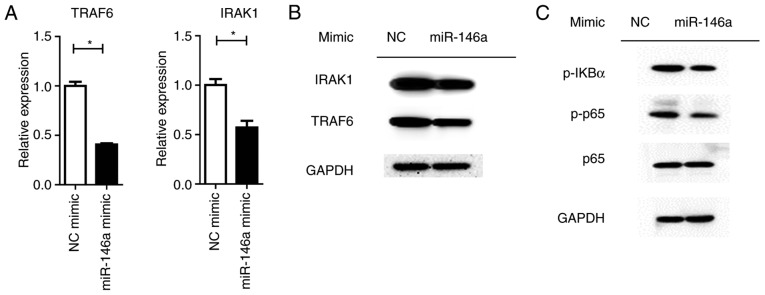
MiRNAs control a variety of biological processes by negatively regulating the expression of multiple genes. TRAF6 and IRAK1 are reported to be targets of miR-146a, which are the key components upstream of NF-κβ signaling (28). It was revealed that the overexpression of miR-146a significantly repressed TRAF6 and IRAK1 expression at the mRNA and protein levels (P<0.05; Fig. 6A). Downstream NF-κβ activation was dampened by the miR-146a mimic, as assayed by the phosphorylation levels of the p65 protein (Fig. 6B). Furthermore, the expression levels of pro-inflammatory cytokines were significantly (IL1-β and IL-6, P<0.01; TNF-α, P<0.05) reduced by miR-146a overexpression (Fig. 7).
Figure 6.
Overexpression of miR-146a reduces NF-κβ activation through targeting TRAF6 and IRAK1. Raw264.7 cells were treated with 30 ng/ml RANKL, 100 ng/ml LPS and 200 nM NC mimic or miR-146a mimic for 72 h. (A) mRNA levels of TRAF6 and IRAK1 were quantified, and (B) the protein levels of TRAF6 and IRAK1 were examined by western blotting. (C) Phosphorylated (Ser) p65 and p65 expression were examined by western blotting. GAPDH was used to confirm equal protein loading. Bars represent the mean ± standard error of the mean. *P<0.05. miR, microRNA; NF-κβ, nuclear factor-κβ; TRAF6, TNF receptor-associated factor 6; IRAK1, interleukin-1 receptor-associated kinase 1; NC, negative control.
Figure 7.
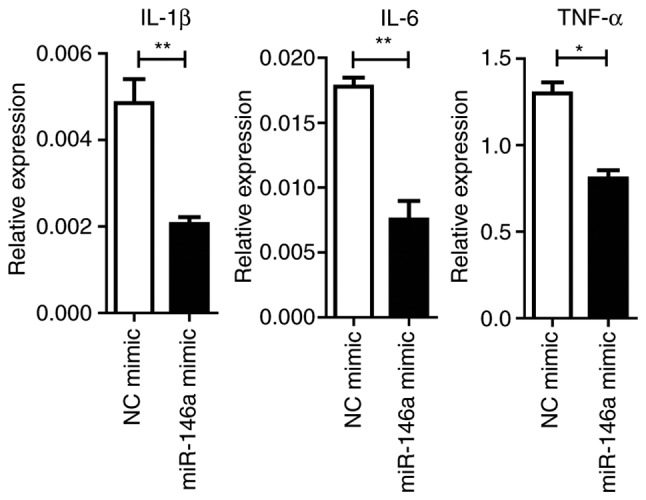
miR-146a mimic inhibits LPS-inducible cytokine production on osteoclastogenesis in Raw264.7 cells. Cells were treated with 30 ng/ml RANKL, 100 ng/ml LPS and 200 nM NC mimic or miR-146a mimic for 72 h. Total RNA was extracted to detect IL-1β, IL-6 and TNF-α expression. Bars represent the mean ± standard error of the mean. *P<0.05 and **P<0.01. miR, microRNA; LPS, lipopolysaccharide; NC, negative control; IL, interleukin; TNF-α, tumor necrosis factor-α.
Discussion
Inflammation is well known to be involved in osteoclast-induced bone loss under pathological circumstances. For instance, chronic infectious inflammation in the periodontium enhances local osteoclast activation and destroys the tooth-supporting alveolar bone, eventually resulting in periodontal disease (29). Systemic bone erosion occurs in RA as a consequence of distorted bone remodeling resulting from overactive osteoclastogenesis in the context of chronic inflammation (30). The mechanisms underlying the tight association between bone resorption and inflammation or infection have been studied for decades (31), yet the intricate and multi-factorial aspects involved in signal transduction have made it difficult to develop a comprehensive understanding of the different types of mediators, in addition to determining their active location in source cells, their upstream and downstream effectors, and the potential of how to control them. Further research is required to develop novel therapeutic strategies, therefore resulting in advancements in the treatment for osteoclast-induced dysfunctions.
The ability of LPS to augment cytokine synthesis and release by macrophages, fibroblasts and other cells has made LPS a potent inducer of osteoclast activation (32–34). Additionally, osteoclast differentiation, survival and fusion are also orchestrated by LPS-dependent cytokines IL-1, IL-6 and TNF-α in vivo and in vitro (35–38). Takami et al (36) once noted the mixed outcomes of LPS in osteoclastogenic assays in the presence or absence of stromal cells and osteoblasts. The induction of osteoclast differentiation by LPS requires the prior priming of RANKL, otherwise the inclusion of LPS may block the osteoclastogenic activity in primary bone marrow monocytes (39). In the present study, it was revealed that LPS exhibits a combined effect of enhancing RANKL-induced osteoclast differentiation. Additionally, in accordance with the previous reports (40–42), inflammatory mediators IL-6, TNF-α and IL-1β were elevated following LPS stimulation during the differentiation of osteoclasts.
miR-146a, which is located in the LOC285628 gene on human chromosome 5, was first discovered in the human acute monocytic leukemia cell line THP-1 (28). The elevated expression of miR-146a may be induced by LPS administration through a negative feedback loop involving the TLR and NF-κβ pathways (43,44). TRAF6 and IL-1 receptor-associated kinase 1 are directly downregulated by miR-146a in this mediation initiated following LPS treatment (17). Nakasa et al (45) revealed the suppressive role of miR-146a in the differentiation process of peripheral blood mononuclear cells into osteoclasts in a dose-dependent manner. In the present study, the high expression of miR-146a in response to LPS and RANKL stimulation in Raw264.7 cells was observed. miR-146a not only prevents osteoclast formation induced by either LPS or RANKL (data not shown), but also demonstrates a strong inhibition on LPS and RANKL co-stimulation. The inhibitory effect of miR-146a on osteoclastogenesis is further verified in the mouse primary bone marrow derived macrophages (data not shown). Therefore, these results suggest a protective role of miR-146a in inflammation-associated bone loss disorders. However, another study revealed that miR-146a facilitates osteoarthritis by regulating cartilage homeostasis (46). Further in vivo studies, particularly investigating cell type specific miR-146a knockout, should be conducted to elucidate this point.
The negative feedback loop of miR-146a and LPS stimulation through the TLR and NF-κβ pathway serves as an important regulatory mechanism in LPS-induced immune responses (47,48). The regulatory axis of miR-146a-NF-κβ has been observed in various kinds of diseases, including cancer (49,50). Considering that NF-κβ signaling is also highly responsive to osteoclastogenesis and osteoclast precursor activation, TRAF6 and IRAK1, two crucial upstream inducers of the NF-κβ pathway, were analyzed following the forced expression of miR-146a. It was revealed that the expression of TRAF6 and IRAK1 in response to the overexpression of miR-146a was decreased at the mRNA and protein levels. Furthermore, the activation of NF-κβ signaling was also inhibited by the miR-146a mimic. The aforementioned results demonstrated that miR-146a serves as a negative modulator in the process of osteoclast differentiation and therefore limits osteoclast number in a proper range and subsequently maintains bone homeostasis.
In conclusion, these results demonstrated that LPS enhances osteoclast differentiation from Raw264.7 cells induced by RANKL. miR-146a is highly induced by LPS and RANKL stimulation. The overexpression of miR-146a inhibits osteoclast transformation by targeting the key regulators of NF-κβ signaling, TRAF6 and IRAK1. Therefore, these results indicate that using an miR-146a mimic may be a promising therapeutic strategy for the prevention and treatment of inflammation mediated bone loss.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science Foundation Of China (grant no. 81773089).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WX conceived and designed the experiments. YG, BW and CS performed the experiments. YG produced the manuscript. BW and CS conducted the data analysis.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mizoguchi T, Muto A, Udagawa N, Arai A, Yamashita T, Hosoya A, Ninomiya T, Nakamura H, Yamamoto Y, Kinugawa S, et al. Identification of cell cycle-arrested quiescent osteoclast precursors in vivo. J Cell Biol. 2009;184:541–554. doi: 10.1083/jcb.200806139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldring SR, Gravallese EM. Mechanisms of bone loss in inflammatory arthritis: Diagnosis and therapeutic implications. Arthritis Res. 2000;2:33–37. doi: 10.1186/ar67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takayanagi H, Ogasawara K, Hida S, Chiba T, Murata S, Sato K, Takaoka A, Yokochi T, Oda H, Tanaka K, et al. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-gamma. Nature. 2000;408:600–605. doi: 10.1038/35046102. [DOI] [PubMed] [Google Scholar]
- 4.Oostlander AE, Everts V, Schoenmaker T, Bravenboer N, van Vliet SJ, van Bodegraven AA, Lips P, de Vries TJ. T cell-mediated increased osteoclast formation from peripheral blood as a mechanism for Crohn's disease-associated bone loss. J Cell Biochem. 2012;113:260–268. doi: 10.1002/jcb.23352. [DOI] [PubMed] [Google Scholar]
- 5.Horowitz MC, Xi Y, Pflugh DL, Hesslein DG, Schatz DG, Lorenzo JA, Bothwell AL. Pax5-deficient mice exhibit early onset osteopenia with increased osteoclast progenitors. J Immunol. 2004;173:6583–6591. doi: 10.4049/jimmunol.173.11.6583. [DOI] [PubMed] [Google Scholar]
- 6.Cappariello A, Maurizi A, Veeriah V, Teti A. The great beauty of the osteoclast. Arch Biochem Biophys. 2014;558:70–78. doi: 10.1016/j.abb.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 7.Gillespie MT. Impact of cytokines and T lymphocytes upon osteoclast differentiation and function. Arthritis Res Ther. 2007;9:103. doi: 10.1186/ar2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian J, Chen J, Gao J, Li L, Xie X. Resveratrol inhibits TNF-α-induced IL-1β, MMP-3 production in human rheumatoid arthritis fibroblast-like synoviocytes via modulation of PI3kinase/Akt pathway. Rheumatol Int. 2013;33:1829–1835. doi: 10.1007/s00296-012-2657-0. [DOI] [PubMed] [Google Scholar]
- 9.Tucci M, Stucci S, Savonarola A, Ciavarella S, Cafforio P, Dammacco F, Silvestris F. Immature dendritic cells in multiple myeloma are prone to osteoclast-like differentiation through interleukin-17A stimulation. Br J Haematol. 2013;161:821–831. doi: 10.1111/bjh.12333. [DOI] [PubMed] [Google Scholar]
- 10.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Hou GQ, Guo C, Song GH, Fang N, Fan WJ, Chen XD, Yuan L, Wang ZQ. Lipopolysaccharide (LPS) promotes osteoclast differentiation and activation by enhancing the MAPK pathway and COX-2 expression in RAW264.7 cells. Int J Mol Med. 2013;32:503–510. doi: 10.3892/ijmm.2013.1406. [DOI] [PubMed] [Google Scholar]
- 12.Ji JD, Park-Min KH, Shen Z, Fajardo RJ, Goldring SR, Mchugh KP, Ivashkiv LB. Inhibition of RANK expression and osteoclastogenesis by TLRs and IFN-gamma in human osteoclast precursors. J Immunol. 2009;183:7223–7233. doi: 10.4049/jimmunol.0900072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takayanagi H. Osteoimmunology: Shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol. 2007;7:292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- 14.Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157:163–179. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 16.Mandal P, Mcmullen MR, Park PH, Roge T, Nagy LE. Adiponectin decreases expression of TLR4 and MyD-88 independent signal transduction in RAW 264.7 macrophages. Cytokine. 2009;48:130. doi: 10.1016/j.cyto.2009.07.549. [DOI] [Google Scholar]
- 17.Tang P, Xiong Q, Wei G, Zhang L. The role of microRNAs in osteoclasts and osteoporosis. RNA Biol. 2014;11:1355–1363. doi: 10.1080/15476286.2014.996462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim K, Kim JH, Kim I, Lee J, Seong S, Park YW, Kim N. MicroRNA-26a regulates RANKL-induced osteoclast formation. Mol Cells. 2015;38:75–80. doi: 10.14348/molcells.2015.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Zhang W, Huang Y. MiRNA-133a is involved in the regulation of postmenopausal osteoporosis through promoting osteoclast differentiation. Acta Biochim Biophys Sin (Shanghai) 2018;50:273–280. doi: 10.1093/abbs/gmy006. [DOI] [PubMed] [Google Scholar]
- 20.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garciaflores Y, Luong M, Devrekanli A, Xu J, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Wan Y, Guo Q, Zou L, Zhang J, Fang Y, Zhang J, Zhang J, Fu X, Liu H, et al. Altered microRNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res Ther. 2010;12:R81. doi: 10.1186/ar3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi K, Takahashi N, Jimi E, Udagawa N, Takami M, Kotake S, Nakagawa N, Kinosaki M, Yamaguchi K, Shima N, et al. Tumor necrosis factor α stimulates osteoclast differentiation by a mechanism independent of the Odf/Rankl-Rank interaction. J Exp Med. 2000;191:275–286. doi: 10.1084/jem.191.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao Y, Grassi F, Ryan MR, Terauchi M, Page K, Yang X, Weitzmann MN, Pacifici R. IFN-gamma stimulates osteoclast formation and bone loss in vivo via antigen-driven T cell activation. J Clin Invest. 2007;117:122–132. doi: 10.1172/JCI30074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, Timms E, Tan HL, Elliott G, Kelley MJ, Sarosi I, et al. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci USA. 1999;96:3540–3545. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kudo O, Fujikawa Y, Itonaga I, Sabokbar A, Torisu T, Athanasou NA. Proinflammatory cytokine (TNFalpha/IL-1alpha) induction of human osteoclast formation. J Pathol. 2002;198:220–227. doi: 10.1002/path.1190. [DOI] [PubMed] [Google Scholar]
- 28.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Benedetto A, Gigante I, Colucci S, Grano M. Periodontal disease: Linking the primary inflammation to bone loss. Clin Dev Immunol. 2013;2013:503754. doi: 10.1155/2013/503754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romas E, Gillespie MT. Inflammation-induced bone loss: Can it be prevented? Rheum Dis Clin North Am. 2006;32:759–773. doi: 10.1016/j.rdc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Graves DT, Li J, Cochran DL. Inflammation and uncoupling as mechanisms of periodontal bone loss. J Dent Res. 2011;90:143–153. doi: 10.1177/0022034510385236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Yan M, Yu QF, Yang PF, Zhang HD, Sun YH, Zhang ZF, Gao YF. Puerarin prevents LPS-induced osteoclast formation and bone loss via inhibition of Akt activation. Biol Pharm Bull. 2016;39:2028–2035. doi: 10.1248/bpb.b16-00522. [DOI] [PubMed] [Google Scholar]
- 33.Kim DY, Jun JH, Lee HL, Woo KM, Ryoo HM, Kim GS, Baek JH, Han SB. N-acetylcysteine prevents LPS-induced pro-inflammatory cytokines and MMP2 production in gingival fibroblasts. Arch Pharm Res. 2007;30:1283–1292. doi: 10.1007/BF02980269. [DOI] [PubMed] [Google Scholar]
- 34.Rossol M, Heine H, Meusch U, Quandt D, Klein C, Sweet MJ, Hauschildt S. LPS-induced cytokine production in human monocytes and macrophages. Crit Rev Immunol. 2011;31:379–446. doi: 10.1615/CritRevImmunol.v31.i5.20. [DOI] [PubMed] [Google Scholar]
- 35.Islam S, Hassan F, Tumurkhuu G, Dagvadorj J, Koide N, Naiki Y, Mori I, Yoshida T, Yokochi T. Bacterial lipopolysaccharide induces osteoclast formation in RAW 264.7 macrophage cells. Biochem Biophys Res Commun. 2007;360:346–351. doi: 10.1016/j.bbrc.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 36.Takami M, Kim N, Rho J, Choi Y. Stimulation by toll-like receptors inhibits osteoclast differentiation. J Immunol. 2002;169:1516–1523. doi: 10.4049/jimmunol.169.3.1516. [DOI] [PubMed] [Google Scholar]
- 37.Mörmann M, Thederan M, Nackchbandi I, Giese T, Wagner C, Hänsch GM. Lipopolysaccharides (LPS) induce the differentiation of human monocytes to osteoclasts in a tumour necrosis factor (TNF) alpha-dependent manner: A link between infection and pathological bone resorption. Mol Immunol. 2008;45:3330–3337. doi: 10.1016/j.molimm.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 38.Baek JM, Kim JY, Yoon KH, Oh J, Lee MS. Ebselen is a potential anti-osteoporosis agent by suppressing receptor activator of nuclear factor Kappa-B ligand-induced osteoclast differentiation in vitro and lipopolysaccharide-induced Inflammatory bone destruction in vivo. Int J Biol Sci. 2016;12:478–488. doi: 10.7150/ijbs.13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou W, Bar-Shavit Z. Dual modulation of osteoclast differentiation by lipopolysaccharide. J Bone Miner Res. 2002;17:1211–1218. doi: 10.1359/jbmr.2002.17.7.1211. [DOI] [PubMed] [Google Scholar]
- 40.Itoh K, Udagawa N, Kobayashi K, Suda K, Li X, Takami M, Okahashi N, Nishihara T, Takahashi N. Lipopolysaccharide promotes the survival of osteoclasts via Toll-like receptor 4, but cytokine production of osteoclasts in response to lipopolysaccharide is different from that of macrophages. J Immunol. 2003;170:3688–3695. doi: 10.4049/jimmunol.170.7.3688. [DOI] [PubMed] [Google Scholar]
- 41.Kikuchi T, Matsuguchi T, Tsuboi N, Mitani A, Tanaka S, Matsuoka M, Yamamoto G, Hishikawa T, Noguchi T, Yoshikai Y. Gene expression of osteoclast differentiation factor is induced by lipopolysaccharide in mouse osteoblasts via Toll-like receptors. J Immunol. 2001;166:3574–3579. doi: 10.4049/jimmunol.166.5.3574. [DOI] [PubMed] [Google Scholar]
- 42.Kwan Tat S, Padrines M, Théoleyre S, Heymann D, Fortun Y. IL-6, RANKL, TNF-alpha/IL-1: Interrelations in bone resorption pathophysiology. Cytokine Growth Factor Rev. 2004;15:49–60. doi: 10.1016/j.cytogfr.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Liu R, Liu C, Chen D, Yang WH, Liu X, Liu CG, Dugas CM, Tang F, Zheng P, Liu Y, Wang L. FOXP3 controls an miR-146/NF-κB negative feedback loop that inhibits apoptosis in breast cancer cells. Cancer Res. 2015;75:1703–1713. doi: 10.1158/0008-5472.CAN-14-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yousefzadeh N, Alipour MR, Soufi FG. Deregulation of NF-кB-miR-146a negative feedback loop may be involved in the pathogenesis of diabetic neuropathy. J Physiol Biochem. 2015;71:51–58. doi: 10.1007/s13105-014-0378-4. [DOI] [PubMed] [Google Scholar]
- 45.Nakasa T, Shibuya H, Nagata Y, Niimoto T, Ochi M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. 2011;63:1582–1590. doi: 10.1002/art.30321. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Wang C, Zhao J, Xu J, Geng Y, Dai L, Huang Y, Fu SC, Dai K, Zhang X. miR-146a facilitates osteoarthritis by regulating cartilage homeostasis via targeting Camk2d and Ppp3r2. Cell Death Dis. 2017;8:e2734. doi: 10.1038/cddis.2017.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang L, Boldin MP, Yu Y, Liu CS, Ea CK, Ramakrishnan P, Taganov KD, Zhao JL, Baltimore D. miR-146a controls the resolution of T cell responses in mice. J Exp Med. 2012;209:1655–1670. doi: 10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng Y, Kuang W, Hao Y, Zhang D, Lei M, Du L, Jiao H, Zhang X, Wang F. Downregulation of miR-27a* and miR-532-5p and upregulation of miR-146a and miR-155 in LPS-induced RAW264.7 macrophage cells. Inflammation. 2012;35:1308–1313. doi: 10.1007/s10753-012-9443-8. [DOI] [PubMed] [Google Scholar]
- 49.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Grève J. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russo A, Saide A, Cagliani R, Cantile M, Botti G, Russo G. rpL3 promotes the apoptosis of p53 mutated lung cancer cells by down-regulating CBS and NFκB upon 5-FU treatment. Sci Rep. 2016;6:38369. doi: 10.1038/srep38369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.