Abstract
Numerous microRNAs (miRNA/miRs) have been reported to be associated with the initiation and progression of non-small cell lung cancer (NSCLC). The aim of the present study was to examine the expression and biological role of miR-939 in human NSCLC, in vitro. Reverse transcription-quantitative polymerase chain reaction analysis was used to evaluate the expression of miR-939 in NSCLC tissues. Cell Counting Kit-8, 5-ethynyl-29-deoxyuridine and Transwell assays were also used to determine the effects of miR-939 on tumor cell proliferation and invasion in two human NSCLC cell lines (H1299 and SPCA1). Furthermore, tissue inhibitor of metalloproteinases 2 (TIMP2) was confirmed to be a target of miR-939 by luciferase reporter assay, western blotting and bioinformatics analysis. Following downregulation of miR-939 expression, cell proliferative and invasive abilities were significantly suppressed. Collectively, these findings indicated that the knockdown of miR-939 may inhibit cell proliferation and invasion by regulating the expression of TIMP2 in NSCLC cells. Thus, miR-939 may be a potential target in the treatment of NSCLC, although this requires further investigation.
Keywords: microRNAs, non-small cell lung cancer, tissue inhibitor of metalloproteinases 2, invasion, proliferation
Introduction
Lung cancer is among the most prevalent types of cancer worldwide and represents a serious threat to human health (1). The most common subtype of lung cancer is non-small cell lung cancer (NSCLC), which comprises ~85% of all lung cancer cases (2). Due to a lack of early screening and diagnosis based on specific biological markers, and the unapparent clinical symptoms of early lung cancer, the majority of lung cancer cases are diagnosed at a late stage (3). The 5-year survival rate for lung cancer is <10% (4). Therefore, it is important to identify novel biomarkers to provide more accurate diagnosis and individualized treatment regimens for patients with lung cancer.
MicroRNAs (miRNA/miRs) are non-coding single-stranded RNAs of ~19–22 nucleotides in length. Numerous studies have demonstrated that miRNAs are involved in the processes of tumorigenesis and tumor development, and serve similar oncogenic roles in cell colonization, apoptosis, migration and other biological processes (5–9). Recent studies have indicated that a number of miRNAs, including miR-126 (10,11) and miR-16-1 (12), are involved in the initiation and progression of NSCLC through the regulation of target genes. miR-939 promotes the proliferation of human ovarian cancer cells by repressing adenomatous polyposis coli like-2 expression (13). Decreased expression of miR-939 contributes to the chemoresistance and metastasis of gastric cancer via dysregulation of sodium-dependent phosphate transport protein 2B and the RAF proto-oncogene serine/threonine-protein kinase/dual specificity mitogen-activated protein kinase kinase/extracellular signal-regulated kinase pathway. mir-939 is also involved in the regulation of breast cancer. Notably, when up taken in exosome-releasing triple negative breast cancer cells, researchers demonstrated that inhibiting miR-939 expression caused downregulation of vascular endothelial cadherin expression (14). Using a number of bioinformatics software packages and platforms, Ma et al (15) reported that miR-939 was the most connected miRNA that regulated a large number of genes, and was involved in lung cancer and the nervous system via functional annotation analysis.
The aim of the present study was to observe the function of miR-939 in NSCLC and its mechanism of action, in order to provide data to potentially aid in the early diagnosis and clinical treatment of NSCLC. Additionally, the present study aimed to provide a theoretical basis for the development of novel drugs against genes associated with NSCLC.
Materials and methods
Clinical samples
NSCLC samples and the adjacent tissues were obtained from 60 patients (27 males/33 females; age range, 40–70 years) who underwent surgery at the First Affiliated Hospital of Nanjing Medical University (Nanjing, China) from March 2014 to July 2016. Patients who had received radiotherapy and/or chemotherapy were excluded. The specimens were immediately frozen in liquid nitrogen and stored at −80°C until further analysis. Approval to conduct human experiments was obtained from the Ethical Committee at the First Affiliated Hospital of Nanjing Medical University and all patients enrolled in the study signed consent forms. All clinical procedures were conducted in accordance with the guidelines of the Ethical Committee of the First Affiliated Hospital of Nanjing Medical University.
Cell culture
H1299, SPCA1, A549, H358 and H1650 cells were purchased from the Chinese Academy of Sciences (Shanghai, China). The 16HBE human normal lung cell line was obtained from our laboratory. All cell lines were maintained in Dulbecco's modified Eagle's medium or RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2 atmosphere.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA of both tissues and cell lines were extracted from the cell lines and clinical samples with a TRIzol kit (Thermo Fisher Scientific, Inc.), used according to the manufacturer's protocol. RNA quality was confirmed with a NanoDrop 300 spectrophotometer (Thermo Fisher Scientific, Inc.). RT was performed using a SuperScript II first-strand cDNA synthesis kit (Thermo Fisher Scientific, Inc.). RT was performed at 42°C for 40 min, followed by 70°C for 15 min. The following PCR cycling conditions were used: 95°C for 5 min; followed by 45 cycles of 95°C for 45 sec; 60°C for 60 sec; 72°C for 45 sec. PCR for the detection of miR-939 was performed with an ABI Prism 7900 detection system (Thermo Fisher Scientific, Inc.) using a TaqMan MicroRNA Assay (Thermo Fisher Scientific, Inc.). The primer sequences were as follows: miR-939 forward, TGGGGAGCTGAGGCTCTG and reverse, AGTGCAGGGTCCGAGGTATT; miR-939 RT Primer: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACCCC; GAPDH forward, AACTTTGGCATTGTGGAAGG and reverse, CACATTGGGGGTAGGAACAC. miR-939 expression was normalized to the expression level of GAPDH. Relative miR-939 expression was analyzed by the 2−ΔΔCq method (16).
Transfection
Human miR-939 inhibitor and inhibitor negative control (inhibitor NC) (miR-939 mimics sense, UGGGGAGCUGAGGCUCUGGGGGUG and mimics antisense, CCCCCAGAGCCUCAGCUCCCCAUU; mimics NC sense, UUCUCCGAACGUGUCACGUTT and mimics NC antisense, ACGUGACACGUUCGGAGAATT; inhibitor, CACCCCCAGAGCCUCAGCUCCCCA and inhibitor NC, CAGUACUUUUGUGUAGUACAA) oligonucleotides were synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The NSCLC cell lines, H1299 and SPCA1, were transfected with the miR-939 inhibitor and NC using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) at a final concentration of 100 nM. Between 2 and 5 days following transfection, NSCLC cells were harvested and RT-qPCR was performed to determine transfection efficiency.
Proliferation experiment
Cell Counting Kit-8 (CCK-8) experiments were used to detect cell proliferative ability. According to the kit instructions, CCK-8 reagent (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added to 4×103 transfected H1299 or SPCA1 cells/well in a 96-well plate, which were incubated at 37°C for 2 h. The optical density of the wells was evaluated at 450 nm with a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). According to the manufacturer's protocol, a 5-ethynyl-29-deoxyuridine (EdU) assay (Guangzhou RiboBio Co, Ltd., Guangzhou, China) was additionally performed to detect cell proliferation. In brief, 24 h after transfection, H1299 or SPCA1 cells were incubated with EdU (50 µM) for 2 h at 37°C. Apollo staining (Guangzhou RiboBio Co, Ltd., Guangzhou, China; 400 µl for 30 min at room temperature) and DAPI (Guangzhou RiboBio Co, Ltd.) staining (400 µl for 30 min at room temperature) were performed and the EdU positive cells were evaluated with a fluorescence microscope (×200; Nikon Corporation, Tokyo, Japan). The EdU incorporation rate was calculated as the ratio of EdU-positive cells, to the total number of DAPI-positive cells (blue).
Transwell assay
Cell invasion was assessed using a 24-well, 8-mm pore size (EMD Millipore, Billerica, MA, USA) and BioCoat Matrigel (BD Biosciences, San Jose, CA, USA). Briefly, transfected H1299 and SPCA1 cells (1×105 cells/well) were plated in the upper chamber in DMEM with 1% FBS. The lower chamber was filled with DMEM with 10% FBS as a chemoattractant. At 24 h following incubation at 37°C in a 5% CO2 atmosphere, the upper chamber was removed. Cells that had invaded into the lower chamber were fixed with 70% ethanol at 4°C for 15 min and stained with hematoxylin at room temperature for 15 min. In each well, cell numbers (5×104) in five random rectangular fields were counted under a light microscope (×200), and the average value was used to express the invasive ability of the cells, and then normalized to the cell numbers in control wells. Images were obtained using NIS-Elements Viewer software through a Nikon microscope (Nikon Corporation, Tokyo, Japan).
Western blot analysis
Total protein of transfected H1299 and SPCA1 were extracted and quantified using a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology, Haimen, China). Cells were lysed in ice-cold radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology) with 10 nM phenylmethylsulfonyl fluoride for 30 min at 4°C and collected to extract total protein. The proteins (10 µl) were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% fat-free milk at room temperature for 1 h, incubated with mouse monoclonal anti-tissue inhibitor of metalloproteinases 2 (TIMP2; 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA; cat. no. sc-21735) and anti-GAPDH antibodies (1:1,000; Santa Cruz Biotechnology, Inc.; cat. no. sc-32233). Mouse IgGk binding protein horseradish peroxidase-conjugated secondary antibody (1:1,000; Santa Cruz Biotechnology, Inc.; cat. no. sc-516102) was added for 2 h at room temperature. Bands were visualized with the Pierce ECL Western Blotting substrate (Thermo Fisher Scientific, Inc.) and exposed in a Molecular Imager ChemiDoc XRS system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). According to the manufacturer's protocol, the analysis was performed using Image Lab (version 5.2.1; Bio-Rad Laboratories, Inc.) and GAPDH was used as the normalization.
Luciferase reporter assay
Using the prediction programs miRDB (mirdb.org) and TargetScan Human (www.targetscan.org), potential targets of miR-939 were predicted. The binding sites or mutant (mut) sequence of the TIMP2 3′-untranslated region (UTR) was inserted into the KpnI and SacI sites of the pGL3 promoter vector (GenScript, Piscataway, NJ, USA). H1299 cells (5×105 cells/well) were plated onto 6-well plates and were transfected with 100 ng pGL3-TIMP2 or pGL3-TIMP2-mut and miR-939 mimics (50 nM) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). The 3′-UTR of TIMP2 was determined to contain a binding site for miR-939. After 24 h, the luciferase activity of the constructs was evaluated according to the ratio of firefly fluorescence to Renilla fluorescence using a Luciferase Reporter Assay system (Promega Corporation, Madison, WI, USA).
Statistical analysis
Statistical analysis was performed with Stata software (version 11.0; StataCorp LP, College Station, TX, USA). The data were presented as mean ± standard deviation of three independent experiments. The Student's t-test, or two-way analysis of variance followed by Fisher's Least Significant Difference test were used to analyze statistical differences. Additionally, the analysis of statistical differences in the clinical factors of patients with NSCLC was conducted using the χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-939 is upregulated in NSCLC tissues
To investigate the underlying function of miR-939 expression in NSCLC progression, clinical specimens were collected from patients with NSCLC, and miR-939 expression was assessed by comparing cancer specimens with adjacent normal lung tissues. RT-qPCR analysis demonstrated that miR-939 was expressed at a higher level in the cancer specimens compared with its expression in the adjacent normal tissues (P<0.05; Fig. 1). In addition, in the analysis of the clinical characteristics of the patients with NSCLC (Table I), it was observed that increased miR-939 expression was significantly associated with higher tumor stage (stages III–IV; P=0.008), increased tumor size (>3 cm; P=0.015) and lymphatic metastasis (P=0.001). However, its abnormal expression was not associated with patient age, sex or smoking history. These findings indicated that miR-939 may be involved in the development of NSCLC, and thus subsequent experiments aimed to clarify its potential function.
Figure 1.
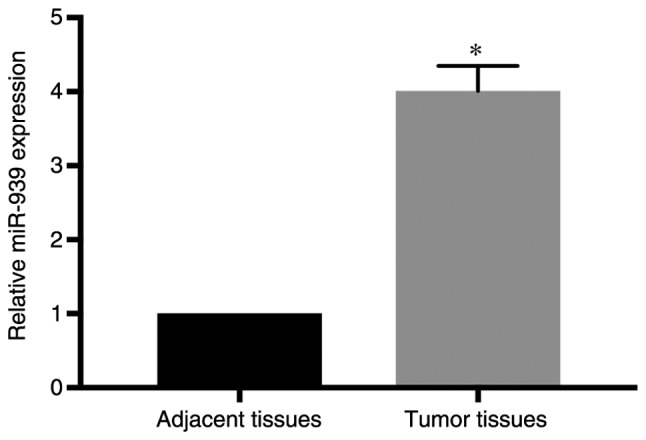
miR-939 expression is increased in tumor tissues. The relative expression of miR-939 was detected in tumor tissues and adjacent tissues by reverse transcription-quantitative polymerase chain reaction. *P<0.05 vs. adjacent tissues. miR, microRNA.
Table I.
Association between miR-939 expression and the clinicopathological characteristics of patients with non-small cell lung cancer.
| Expression of miR-939, n | ||||
|---|---|---|---|---|
| Characteristics | Patients, n | High | Low | P-value |
| Total | 60 | 30 | 30 | |
| Age, years | 0.793 | |||
| ≤50 | 25 | 12 | 13 | |
| >50 | 35 | 18 | 17 | |
| Sex | 0.436 | |||
| Male | 27 | 12 | 15 | |
| Female | 33 | 18 | 15 | |
| Smoke | 0.796 | |||
| Yes | 31 | 15 | 16 | |
| No | 29 | 15 | 14 | |
| TNM stage | 0.008a | |||
| I–II | 36 | 13 | 23 | |
| III–IV | 24 | 17 | 7 | |
| Tumor size | 0.015a | |||
| <3 cm | 39 | 15 | 24 | |
| >3 cm | 21 | 15 | 6 | |
| Lymphatic metastasis | 0.001a | |||
| No | 38 | 13 | 25 | |
| Yes | 22 | 17 | 5 | |
P<0.05. miR, microRNA; TNM, tumor, node, metastasis.
miR-939 expression is increased in NSCLC cell lines
The present study focused on investigating the potential function of miR-939 in vitro. miR-939 expression was detected in NSCLC cell lines (H1299, SPCA1, A549, H358 and H1650) and a normal lung cell line (16HBE). RT-qPCR analysis demonstrated that miR-939 expression was significantly increased in all NSCLC cell lines compared with that in the 16HBE cell line (P<0.05; Fig. 2).
Figure 2.
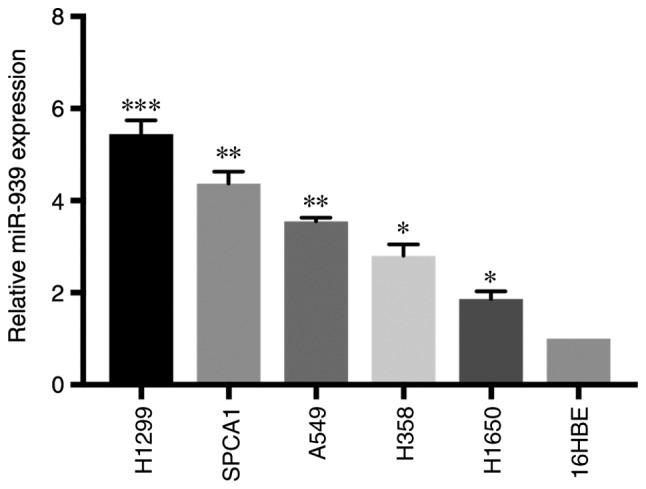
miR-939 is increased in tumor cell lines. The relative expression of miR-939 was detected in tumor cell lines (H1299, SPCA1, A549, H358 and H1650) and was compared with a normal cell line (16HBE) via reverse transcription-quantitative polymerase chain reaction. *P<0.05, **P<0.01 and ***P<0.001 vs. 16HBE. miR, microRNA.
miR-939 downregulation inhibits NSCLC cell line proliferation
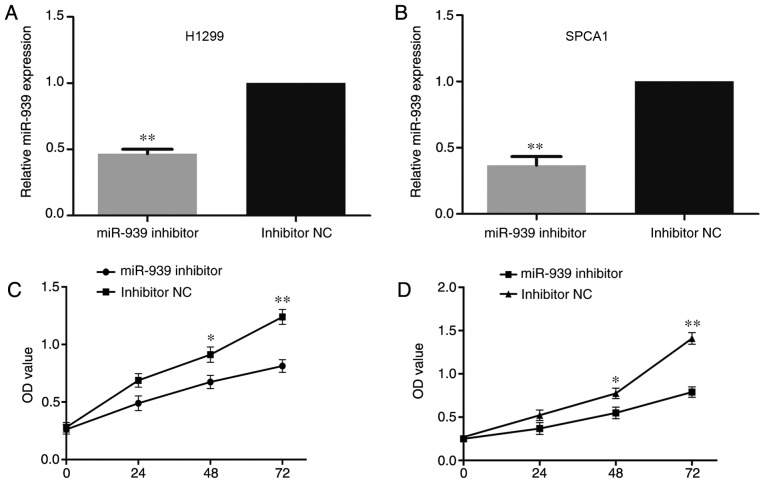
Subsequently, based on the abnormal expression of miR-939 among the NSCLC cell lines, H1299 and SPCA1 cells that demonstrated the highest miR-939 expression were transfected with an miR-939 inhibitor to detect the regulatory mechanisms affected by miR-939 knockdown in NSCLC development in vitro, with inhibitor negative control (NC)-transfected cells as the control. Following transfection of the cells with miR-939 inhibitor or inhibitor NC, it was identified via RT-qPCR that miR-939 was significantly downregulated in the miR-939-silenced cells compared with its expression in the inhibitor NC group (P<0.01; Fig. 3A and B).
Figure 3.
Knockdown of miR-939 inhibits cell proliferation. miR-939 expression was detected in (A) H1299 and (B) SPCA1 cells following transfection with an miR-939 inhibitor by reverse transcription-quantitative polymerase chain reaction, with inhibitor NC-transfected cells as the control group. Cell proliferation was also detected in the (C) H1299 and (D) SPCA1 cells by a Cell Counting Kit-8 assay at 0, 24, 48 and 72 h. *P<0.05 and **P<0.01 vs. respective inhibitor NC group. miR, microRNA; NC, negative control; OD, optical density.
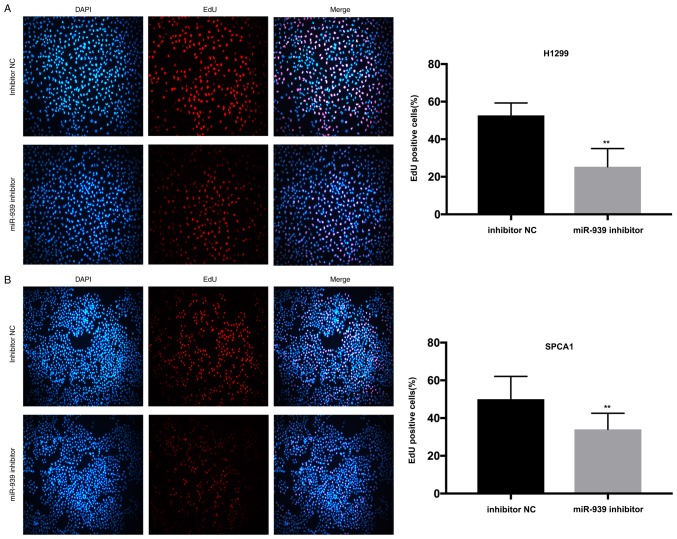
Additionally, to investigate the influence of miR-939 on the cell proliferative ability, CCK-8 and EdU assays were performed. In the H1299 and SPCA1 cell lines, the CCK-8 (Fig. 3C and D, respectively) and EdU (Fig. 4A and B, respectively) assays demonstrated that cell proliferative ability was markedly decreased by the silencing of miR-939 expression.
Figure 4.
Downregulation of miR-939 represses cell proliferation. Cell proliferation in (A) H1299 and (B) SPCA1 cells, was detected by EdU assay (magnification, ×200). **P<0.01 vs. inhibitor NC group. miR, microRNA; NC, negative control.
miR-939 downregulation inhibits NSCLC invasion
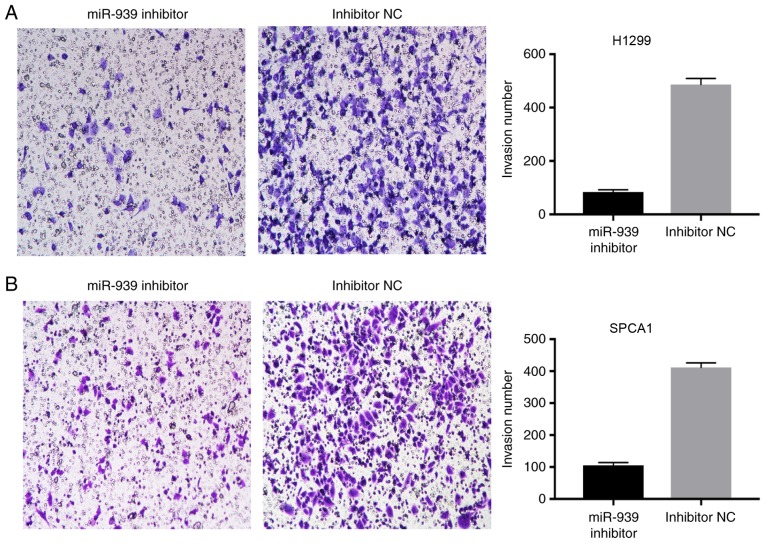
The effect of miR-939 downregulation on the invasion of H1299 and SPCA1 cells was also examined. A Transwell assay demonstrated that compared with the cells transfected with inhibitor NC, a reduced number of cells invaded into the lower chambers in the miR-939 inhibitor group (Fig. 5A and B). This data suggested that the knockdown of miR-939 markedly reduced NSCLC invasion.
Figure 5.
Downregulation of miR-939 inhibits cell invasion. Cell invasion in (A) H1299 and (B) SPCA1 cells was investigated by Transwell assay (magnification, ×40). miR, microRNA; NC, negative control.
miR-939 regulates TIMP2 by binding to its 3′-UTR
In order to define whether TIMP2 had a binding site for miR-939, cells were transfected with miR-939 mimics (Fig. 6A) and, using the prediction programs miRDB and TargetScan Human, the underlying target genes of miR-939 were verified. The 3′-UTR of TIMP2 was identified to have a binding site for miR-939 (Fig. 6B). To determine whether miR-939 directly targets TIMP2 in NSCLC cells, a luciferase reporter assay was conducted. The results demonstrated a reduction in luciferase activity following co-transfection with miR-939 mimics (P<0.01), whereas co-transfection with NC had no significant effect on luciferase activity (Fig. 6C), which indicated that miR-939 regulated TIMP2 by binding to its 3′-UTR.
Figure 6.
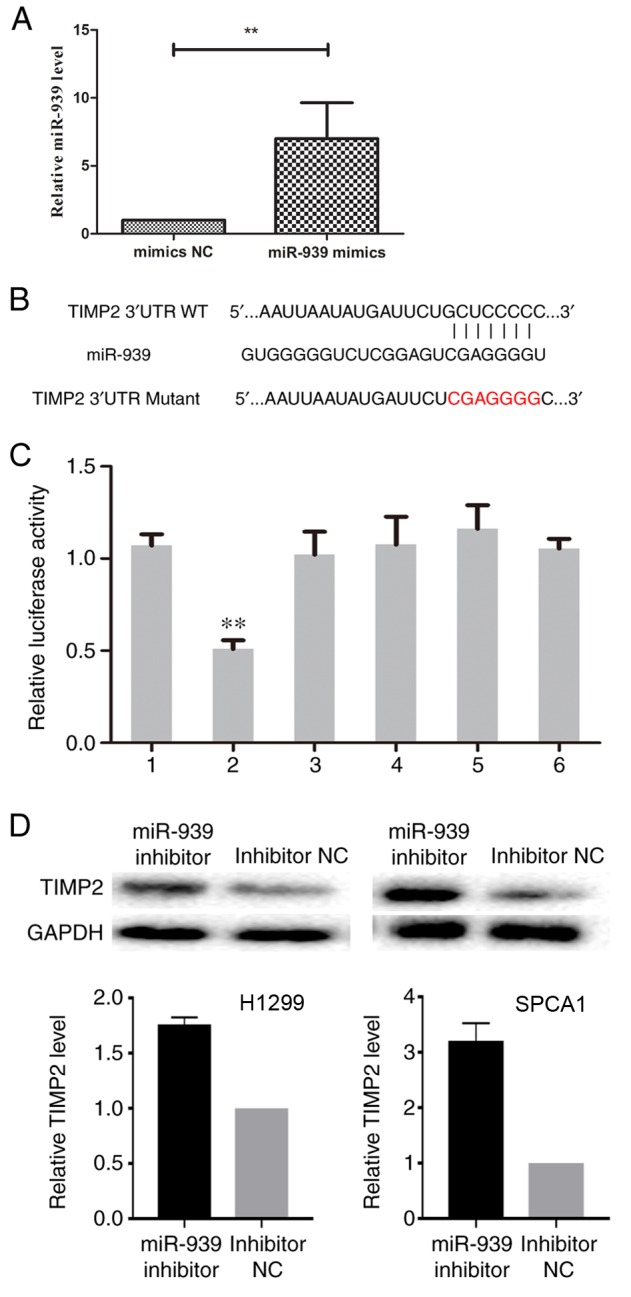
TIMP2 is negatively regulated by miR-939. (A) Successful transfection efficiency was determined by reverse transcription-quantitative polymerase chain reaction. **P<0.01. (B) In the prediction programs miRDB and TargetScan Human, the 3′-UTR of TIMP2 was indicated to have a binding site for miR-939. (C) Relative luciferase activity was evaluated. 1, pGL3-TIMP2; 2, pGL3-TIMP2+miR-939 mimics; 3, pGL3-TIMP2+NC; 4, pGL3-TIMP2 Mutant; 5, pGL3-TIMP2 Mutant+miR-939 mimics; 6, pGL3-TIMP2 Mutant+NC. **P<0.01 vs. group 1. (D) The relative protein expression of TIMP2 in H1299 (left) and SPCA1 (right) cells was detected by western blot analysis. miR, microRNA; NC, negative control; TIMP2, TIMP metallopeptidase inhibitor 2; UTR, untranslated region; WT, wild-type.
Subsequently, the effect of miR-939 downregulation on TIMP2 expression was assessed. Western blot analysis indicated that the silencing of miR-939 increased the protein expression level of TIMP2 in H1299 and SPCA1 cells (Fig. 6D). Collectively, these findings demonstrated that miR-939 was able to negatively regulate TIMP2 by binding to its 3′-UTR, although this requires further investigation.
Discussion
Numerous studies have reported that miRNAs function as oncogenes or tumor suppressors in the progression of different types of cancer. Lu et al (17) reported that miRNA-221 expression was increased in renal cell carcinoma, and promoted cell proliferation, migration and invasion by regulating TIMP2. In gastric cancer, miR-29a repressed cell migration and invasion by modulating roundabout homolog 1 (18). Low-level expression of miR-329 has been implicated in gastric cancer invasion and growth by targeting T cell lymphoma invasion and metastasis 1 (19). In addition, a number of key miRNAs have been established as biomarkers for early diagnosis and prognosis in a range of malignancy types (5). For instance, the downregulation of miR-197 predicted a poor prognosis for patients with esophageal cancer (20), whereas miR-126 and miR-200c dysregulation has been implicated in the prognosis of patients with NSCLC (21).
In the present study, RT-qPCR analysis demonstrated that the upregulation of miR-939 occurred in NSCLC cell lines and clinical specimens, thus indicating a potential prognostic marker for patients with NSCLC. Furthermore, RT-qPCR analysis identified that differential expression of miR-939 in NSCLC was significantly associated with the patient clinical factors of tumor stage, tumor size and lymphatic metastasis status. In addition, the functional role of miR-939 was investigated in vitro. On silencing of miR-939 in the NSCLC cell lines H1299 and SPCA1, inhibition of cell proliferation and invasion was observed. Collectively, these data demonstrated that miR-939 may be involved in the development of NSCLC, although this requires further investigation.
It is established that miRNAs regulate certain target genes by binding to their 3′-UTRs (22–24). miR-124 suppressed gastric cancer development by directly targeting enhancer of zeste homolog 2 (25); whereas, miR-26a, serving as a tumor suppressor, repressed cell proliferation by directly binding the high-mobility group AT-hook 2 3′-UTR in gallbladder cancer (26). Additionally, using the prediction programs miRDB and TargetScan Human, the underlying target genes of miR-939 were verified in the present study. A luciferase reporter assay identified a reduction in luciferase activity following co-transfection with miR-939 mimics, while co-transfection with the negative control did not significantly alter the luciferase activity, indicating that miR-939 regulated TIMP2 by binding to its 3′UTR. Subsequently, the effect of miR-939 downregulation on TIMP2 expression was examined. Western blot analysis indicated that the silencing of miR-939 increased the protein expression level of TIMP2.
Lang et al (27) reported miR-429 downregulates TIMP2 expression to promote the proliferation and metastasis of NSCLC cells. Wang et al (28) reported that microRNA-15b promotes proliferation and invasion of NSCLC by directly targeting TIMP2. Finally, another study reported that miRNA-221 serves an oncogenic role by directly targeting TIMP2 in NSCLC (29).
The development and progression of lung cancer is not determined by a single or a few factors; rather, it is affected by a number of genes and signaling pathways (30,31). The present study was limited due to the sample size and cell line type, and therefore a larger sample size is required. Further experiments are required in order to examine the association between miR-939 and invasion or migration in order to discover any potential regulatory role of miR-939 in the development of lung cancer.
In conclusion, the present study demonstrated that miR-939 was able to regulate NSCLC cell proliferation and invasion. Additionally, miR-939 negatively regulated TIMP2 by binding to its 3′UTR. Therefore, miR-939 may be a potential target in the treatment of NSCLC, although this required further investigation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author, on reasonable request.
Authors' contributions
YC and LW designed the experiments. AC and SL performed the experiments. XL analyzed the data. All authors have read and approved the manuscript.
Ethics approval and consent to participate
Approval to conduct human experiments was obtained from the Ethical Committee at the First Affiliated Hospital of Nanjing Medical University (Nanjing, China) and all patients enrolled in the study signed consent forms. All clinical procedures were conducted in accordance with the guidelines of the Ethical Committee of the First Affiliated Hospital of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Gompelmann D, Eberhardt R, Herth FJ. Advanced malignant lung disease: What the specialist can offer. Respiration. 2011;82:111–123. doi: 10.1159/000329703. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Sui J, Shen X, Li C, Yao W, Hong W, Peng H, Pu Y, Yin L, Liang G. Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of lung cancer. Oncol Rep. 2017;37:3543–3553. doi: 10.3892/or.2017.5612. [DOI] [PubMed] [Google Scholar]
- 4.de Cos Sanchez J, Sojo Gonzalez MA, Montero MV, Calvo Perez MC, Vicente MJ, Valle MH. Non-small cell lung cancer and silent brain metastasis. Survival and prognostic factors. Lung Cancer. 2009;63:140–145. doi: 10.1016/j.lungcan.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Kang SM, Lee HJ. MicroRNAs in human lung cancer. Exp Biol Med (Maywood) 2014;239:1505–1513. doi: 10.1177/1535370214533887. [DOI] [PubMed] [Google Scholar]
- 7.Ding XM. MicroRNAs: Regulators of cancer metastasis and epithelial-mesenchymal transition (EMT) Chin J Cancer. 2014;33:140–147. doi: 10.5732/cjc.013.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM, Yang PC. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat Commun. 2013;4:1877. doi: 10.1038/ncomms2876. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Huang R, Wang L, Hao J, Zhang Q, Ling R, Yun J. microRNA-762 promotes breast cancer cell proliferation and invasion by targeting IRF7 expression. Cell Prolif. 2015;48:643–649. doi: 10.1111/cpr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang CZ, Yuan P, Li Y. MiR-126 regulated breast cancer cell invasion by targeting ADAM9. Int J Clin Exp Pathol. 2015;8:6547–6553. [PMC free article] [PubMed] [Google Scholar]
- 11.Kong R, Ma Y, Feng J, Li S, Zhang W, Jiang J, Zhang J, Qiao Z, Yang X, Zhou B. The crucial role of miR-126 on suppressing progression of esophageal cancer by targeting VEGF-A. Cell Mol Biol Lett. 2016;21:3. doi: 10.1186/s11658-016-0004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang W, Tong JH, Lung RW, Dong Y, Zhao J, Liang Q, Zhang L, Pan Y, Yang W, Pang JC, et al. Targeting of YAP1 by microRNA-15a and microRNA-16-1 exerts tumor suppressor function in gastric adenocarcinoma. Mol Cancer. 2015;14:52. doi: 10.1186/s12943-015-0323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ying X, L-Ya Q, Feng Z, Yin W, Ji-Hong L. MiR-939 promotes the proliferation of human ovarian cancer cells by repressing APC2 expression. Biomed Pharmacother. 2015;71:64–69. doi: 10.1016/j.biopha.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 14.Di Modica M, Regondi V, Sandri M, Iorio MV, Zanetti A, Tagliabue E, Casalini P, Triulzi T. Breast cancer-secreted miR-939 downregulates VE-cadherin and destroys the barrier function of endothelial monolayers. Cancer Lett. 2017;384:94–100. doi: 10.1016/j.canlet.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Ma R, Wang C, Wang J, Wang D, Xu J. miRNA-mRNA interaction network in non-small-cell lung cancer. Interdiscip Sci. 2015 Apr 11; doi: 10.1007/s12539-015-0117-8. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Lu GJ, Dong YQ, Zhang QM, Di WY, Jiao LY, Gao QZ, Zhang CG. miRNA-221 promotes proliferation, migration and invasion by targeting TIMP2 in renal cell carcinoma. Int J Clin Exp Pathol. 2015;8:5224–5229. [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Cai J, Sun Y, Gong R, Sun D, Zhong X, Jiang S, He X, Bao E, Yang L, Li Y. MicroRNA-29a inhibits cell migration and invasion via targeting Roundabout homolog 1 in gastric cancer cells. Mol Med Rep. 2015;12:3944–3950. doi: 10.3892/mmr.2015.3817. [DOI] [PubMed] [Google Scholar]
- 19.Li Z, Yu X, Wang Y, Shen J, Wu WK, Liang J, Feng F. By downregulating TIAM1 expression, microRNA-329 suppresses gastric cancer invasion and growth. Oncotarget. 2015;6:17559–17569. doi: 10.18632/oncotarget.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang TY, Liu SG, Zhao BS, Qi B, Qin XG, Yao WJ. Implications of microRNA-197 downregulated expression in esophageal cancer with poor prognosis. Genet Mol Res. 2014;13:5574–5581. doi: 10.4238/2014.July.25.12. [DOI] [PubMed] [Google Scholar]
- 21.Kim MK, Jung SB, Kim JS, Roh MS, Lee JH, Lee EH, Lee HW. Expression of microRNA miR-126 and miR-200c is associated with prognosis in patients with non-small cell lung cancer. Virchows Archiv. 2014;465:463–471. doi: 10.1007/s00428-014-1640-4. [DOI] [PubMed] [Google Scholar]
- 22.Xia Y, Gao Y. MicroRNA-181b promotes ovarian cancer cell growth and invasion by targeting LATS2. Biochem Biophys Res Commun. 2014;447:446–451. doi: 10.1016/j.bbrc.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 23.Fan D, Wang Y, Qi P, Chen Y, Xu P, Yang X, Jin X, Tian X. MicroRNA-183 functions as the tumor suppressor via inhibiting cellular invasion and metastasis by targeting MMP-9 in cervical cancer. Gynecol Oncol. 2016;141:166–174. doi: 10.1016/j.ygyno.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Lin Y, Liu AY, Fan C, Zheng H, Li Y, Zhang C, Wu S, Yu D, Huang Z, Liu F, Luo Q, et al. MicroRNA-33b inhibits breast cancer metastasis by targeting HMGA2, SALL4 and Twist1. Sci Rep. 2015;5:9995. doi: 10.1038/srep09995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie L, Zhang Z, Tan Z, He R, Zeng X, Xie Y, Li S, Tang G, Tang H, He X. MicroRNA-124 inhibits proliferation and induces apoptosis by directly repressing EZH2 in gastric cancer. Mol Cell Biochem. 2014;392:153–159. doi: 10.1007/s11010-014-2028-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhou H, Guo W, Zhao Y, Wang Y, Zha R, Ding J, Liang L, Hu J, Shen H, Chen Z, et al. MicroRNA-26a acts as a tumor suppressor inhibiting gallbladder cancer cell proliferation by directly targeting HMGA2. Int J Oncol. 2014;44:2050–2058. doi: 10.3892/ijo.2014.2360. [DOI] [PubMed] [Google Scholar]
- 27.Lang Y, Xu S, Ma J, Wu J, Jin S, Cao S, Yu Y. MicroRNA-429 induces tumorigenesis of human non-small cell lung cancer cells and targets multiple tumor suppressor genes. Biochem Biophys Res Commun. 2014;450:154–159. doi: 10.1016/j.bbrc.2014.05.084. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Zhan Y, Jin J, Zhang C, Li W. MicroRNA-15b promotes proliferation and invasion of nonsmall cell lung carcinoma cells by directly targeting TIMP2. Oncol Rep. 2017;37:3305–3312. doi: 10.3892/or.2017.5604. [DOI] [PubMed] [Google Scholar]
- 29.Yin Z, Xu M, Li P. miRNA-221 acts as an oncogenic role by directly targeting TIMP2 in non-small-cell lung carcinoma. Gene. 2017;620:46–53. doi: 10.1016/j.gene.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Ai X, Mao F, Shen S, Shentu Y, Wang J, Lu S. Bexarotene inhibits the viability of non-small cell lung cancer cells via slc10a2/PPARγ/PTEN/mTOR signaling pathway. BMC Cancer. 2018;18:407. doi: 10.1186/s12885-018-4224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lv M, Wang L. Comprehensive analysis of genes, pathways, and TFs in nonsmoking Taiwan females with lung cancer. Exp Lung Res. 2015;41:74–83. doi: 10.3109/01902148.2014.971472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author, on reasonable request.