Abstract
As one of the most common and aggressive cancer types, hepatocellular carcinoma (HCC) leads to a large number of fatalities every year. However, the pathogenesis of HCC remains largely unknown. In the present study, it was identified that FEZF1-AS1 was significantly upregulated in HCC cell lines and tissues, as determined by reverse transcription-quantitative polymerase chain reaction. Additionally, it was observed that higher expression of FEZF1-AS1 in patients with HCC indicated poorer prognosis. Furthermore, it was identified that knockdown of FEZF1-AS1 markedly inhibited the proliferation, colony formation, migration and invasion of Hep3B and Huh7 cells, as determined by Cell Counting Kit-8, colony formation and Transwell assays. In terms of mechanism, it was observed that FEZF1-AS1 acted as a sponge for microRNA (miR)-4443. The results of a luciferase reporter assay revealed that overexpression of miR-4443 significantly inhibited the luciferase activity in Hep3B and Huh7 cells. Additionally, miR-4443 overexpression markedly inhibited the expression of FEZF1-AS1, and vice versa. It was additionally identified that miR-4443 was downregulated in HCC tissues. There was an inverse correlation between the expression of miR-4443 and FEZF1-AS1 in HCC tissues. Furthermore, through functional experiments, it was identified that knockdown of FEZF1-AS1 significantly inhibited the proliferation, migration and invasion of HCC cells, whereas inhibition of miR-4443 reversed these effects. Collectively, the present results demonstrated that FEZF1-AS1 acts as an oncogene by acting as a sponge for miR-4443.
Keywords: hepatocellular carcinoma, FEZF1-AS1, proliferation, migration, micro RNA-4443
Introduction
Hepatocellular carcinoma (HCC) is derived from hepatocytes and has become one of the most prevalent and malignant cancer types worldwide (1). Every year, a large number of cancer-associated mortalities are induced by HCC (2). Despite certain advances in HCC therapy being achieved, the outcomes of patients with HCC remain unsatisfactory (3). A number of patients with HCC are diagnosed at the advanced stage, which makes curative treatment is no longer possible and increases the probability of cancer recurrence and metastasis (4). Therefore, in order to better treat HCC, it is vital to identify novel biomarkers for HCC diagnosis and prognosis, and therapeutic targets.
Long non-coding RNAs (lncRNAs) are a class of noncoding RNAs with a length >200 nucleotides, which possess no protein-coding potential (5). In recent years, lncRNA has attracted much attention in the field of biology. Increasing evidence demonstrated that lncRNA exerts pivot functions in the majority of biological processes, including cell survival, proliferation, migration and invasion (6–8). Dysregulation of lncRNAs is closely associated with cancer development and progression (9). For instance, the lncRNA Sox2ot is overexpressed in cholangiocarcinoma and promotes tumor cell proliferation and invasion (10). Zhang et al (11) reported that lncRNA HOXD-AS1 promotes epithelial ovarian cancer cell proliferation and invasion by targeting microRNA (miRNA/miR)-133a-3p and activating the Wnt/β-catenin signaling pathway. Therefore, it is crucial to determine the mechanism of lncRNAs in tumor progression.
FEZF1-AS1 has been reported to regulate tumor progression in a number of cancer types, including colorectal carcinoma (12), gastric cancer (13) and non-small cell lung cancer (14). However, whether FEZF1-AS1 serves a role in HCC requires investigation. In the present study, it was identified that FEZF1-AS1 was significantly upregulated in HCC tissues and predicted a poor prognosis for patients with HCC. It was demonstrated that knockdown of FEZF1-AS1 inhibited the proliferation, migration and invasion of HCC cells. Additionally, it was identified that FEZF1-AS1 acted as a sponge to miR-4443, which was significantly downregulated in HCC tissues. Furthermore, it was identified that inhibition of miR-4443 abolished the effects of FEZF1-AS1 on HCC cell proliferation, migration and invasion. Collectively, the present results demonstrated that FEZF1-AS1 serves as an oncogene in HCC via inhibition of miR-4443.
Materials and methods
Patient samples
A total of 116 specimens, including 58 tumor-adjacent tissues and 58 tumour tissues (female, 11 and male, 47; mean age, 49.16±13.42 years), were obtained from patients with HCC, who underwent surgical resection from August 2010 to October 2016 at The First College of Clinical Medical Science, China Three Gorges University (Yichang, China). Patients who received chemotherapy or radiotherapy prior to surgery were excluded. The final diagnosis was confirmed by pathological analysis. All the specimens were collected immediately following liver resection and stored in liquid nitrogen at −80°C until analysis. Written consent was obtained from every patient and the research protocol was approved by the Ethics Committee of The First College of Clinical Medical Science, China Three Gorges University.
Cell culture and transfection
HCC cell lines, Hep3B and Huh7, and normal hepatocyte LO2 were all purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). In addition, all the cell lines were cultured with their specified basic culture medium [Dulbecco's modified Eagle's medium (DMEM); Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA] supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin sulfate and maintained at 37°C in a humidified atmosphere containing 5% CO2.
FEZF1-AS1 small interfering (si)RNA (5′-GAAAGUGUUGUGUCAAUAACG-3′) and non-targeting siRNA [si negative control (siNC, 5′-AATTCTCCGAACGTGTCACGT-3′)], miR-4443 mimics (5′-UUGGAGGCGUGGGUUUU-3′), inhibitors (5′-AAAACCCACGCCUCCAA-3′) and controls (5′-ACAUCUGCGUAAGAUUCGAGUCUA-3′) were purchased from Shanghai Integrated Biotech Solutions Co., Ltd. (Shanghai, China). Transfection was performed with Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol, with plasmids or siRNAs transfected at a concentration of 50 nM. A total of 48 h post-transfection, efficiency was validated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturers' protocol, and subsequently converted into complementary DNA (cDNA) using a PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China), according to the manufacturer's protocol. The RNA expression levels were examined by real-time PCR using a SYBR Premix Dimmer Eraser kit (Takara Biotechnology Co., Ltd.). The thermocycling conditions of qPCR were as follows: 94°C for 15 min, followed by 45 cycles at 94°C for 10 sec, 60°C for 30 sec and 72°C for 30 sec. Gene expression in each sample was normalized to U6. The expression of miR-4443 was quantified using TaqMan MicroRNA Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.), and human U6 RNA, which was amplified as a control. Data are presented as the mean ± standard deviation from three independent experiments. The relative expression fold-change of mRNA was calculated using the 2−ΔΔCq method (15). The primer sequences were as follows: FEZF1-AS1, forward 5′-TTAGGAGGCTTGTTCTGTGT-3′, reverse 5′-GCGCAGGTACTTAAGAAAGA-3′; miR-4443, forward 5′-GTTGGAGGCGTGGGT-3′, reverse 5′-GGTCCAGTTTTTTTTTTTTTTTAAAACC-3′; and U6, forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′.
RNA-FISH assay
Fluorescence-conjugated FEZF1-AS1 probes for RNA-FISH were generated, according to protocols of LGC Biosearch Technologies (Petaluma, CA, USA). HCC cells were treated in a non-denaturing condition, followed by hybridization with DNA probe sets, as previously described (9). DAPI (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used to stain the nuclei at room temperature for 10 min. Treated samples were visualized by confocal microscopy at ×600 magnification (FV1000; Olympus Corporation, Tokyo, Japan).
Cellular proliferation assays
Cells were seeded into 96-well plates at a density of 1×103 cells/well. Following 24, 48, 72 and 96 h of incubation at 37°C in DMEM (Gibco; Thermo Fisher Scientific, Inc.), cellular viability was evaluated using a Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan), according to the manufacturer's protocol. The absorbance was measured at a wavelength of 450 nm using a Multiskan™ GO Microplate Spectrophotometer (Thermo Fisher Scientific, Inc.).
A colony formation assay was also performed. A total of 2×103 cells/well were seeded into 6-well plates with DMEM supplemented with 10% FBS and cultured at 37°C with 5% CO2 for 14 days. Colonies were fixed with 100% methanol at room temperature for 20 min and stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) at 25°C for 30 min. The total number of visible colonies was determined under an optical light microscope at ×40 magnification (Olympus Corporation). All experiments were repeated three times.
Cellular migration and invasion assays
Transwell migration and matrigel invasion assays were assessed using transwell membranes (8 µm pore size, 6.5 mm diameter; Corning Incorporated, Corning, NY, USA) in 24-well plates of 8 µm (BD Biosciences, Franklin Lakes, NY, USA) according to the manufacturer's protocol. For the transwell migration assay, cells were resuspended in serum-free 200 µl DMEM medium at a density of 5×104 cells/well. The bottom chamber of transwell plates was supplemented with 600 µl DMEM medium, containing 20% FBS as a chemoattractant. Cells were subsequently seeded into the upper chamber and incubated for another 20 h at 37°C with 5% CO2. For the invasion assay, matrigel matrix (1:8; 50 µl/well, BD Biosciences) was polymerized in the transwell membrane, according to the manufacturer's protocol. Cells (1×105 cells/well) were used and incubated for another 48 h at 37°C with 5% CO2. Following incubation, cells on the upper surface of the membrane were scraped off with cotton swabs. Cells that migrated to the lower surface were fixed with polyoxymethylene at 25°C for 30 min, stained with 0.1% Crystal Violet staining solution at 25°C for 30 min. The cells on the bottom of the membrane were calculated from five random light microscopic fields at ×40 magnification.
Luciferase assays
The potential targets of FEZF1-AS1 were predicted using the miRDB tool (http://mirdb.org/miRDB/index.html). Subsequently, FEZF1-AS1 sequences containing the wild-type (WT) binding site or mutated-type (Mut) binding site for miR-4443 were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) and cloned into the pmiR-GLO vector (Promega Corporation, Madison, WI, USA). Luciferase reporter gene assays were performed using the Dual-Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer's protocol. Prior to transfection, cells were seeded in 24-well plates (5×103 cells/well) and cultured for 24 h. Subsequently, the WT or Mut of FEZF1-AS1 was transiently co-transfected with miR-4443 mimics or miR-NC (Guangzhou RiboBio Co., Ltd., Guangzhou, China) into logarithmic phase cells using Lipofectamine® 2000 transfection reagent. Luciferase assays were performed using the Dual-Luciferase Reporter Assay system (Promega Corporation), according to the manufacturer's protocol, after a further 48 h culture. The value of relative luciferase activity indicates the firefly luciferase activity normalized to that of Renilla for each assay.
Statistical analysis
The statistical significance of the differences between groups was assessed using Student's t-test for pair-wise comparisons or one-way analysis of variance followed by Fisher's least significant difference post hoc test for multiple comparisons. Pearson's correlation coefficient analysis was used to determine the correlations. Kaplan-Meier survival curve analysis and log-rank test were used for survival analysis. P<0.05 was considered to indicate a statistically significant difference. Data are presented as the mean ± standard deviation of three independent experiments. Statistical analysis was performedv using the SPSS software version 20.0 (IBM Corp., Armonk, NY, USA).
Results
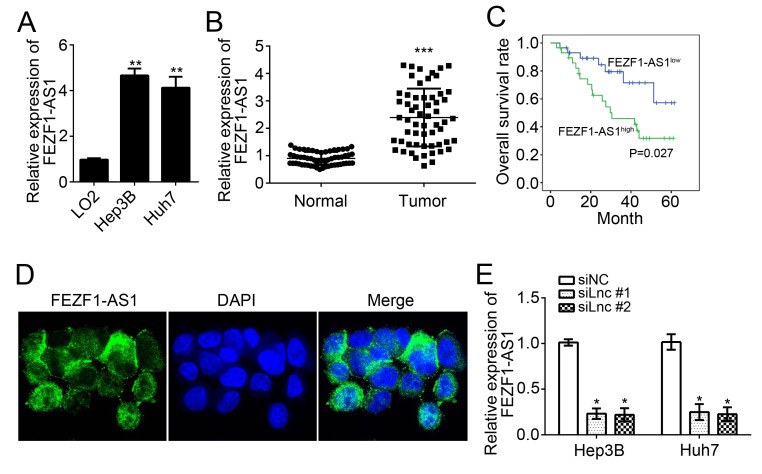
FEZF1-AS1 is upregulated in HCC tissues
To examine the expression patterns of FEZF1-AS1, RT-qPCR analysis was conducted using HCC cell lines and LO2 cells. It was identified that FEZF1-AS1 expression was significantly upregulated in HCC cell lines (Hep3B and Huh7) compared with normal LO2 hepatocytes (P<0.01; Fig. 1A). To further verify its overexpression in HCC cells, 58 HCC tissues and 58 adjacent normal tissues were obtained, and FEZF1-AS1 expression was analyzed using RT-qPCR. The results suggested that FEZF1-AS1 expression levels were significantly upregulated in HCC tissues compared with adjacent normal tissues (P<0.001; Fig. 1B). To determine whether FEZF1-AS1 is able to serve as a prognostic biomarker of patients with HCC, a Kaplan-Meier survival curve analysis was conducted. It was identified that patients with HCC with higher expression of FEZF1-AS1 exhibited a significantly lower survival rate (P<0.05; Fig. 1C). The location of FEZF1-AS1 in HCC cells was analyzed and it was identified that FEZF1-AS1 was located in the cytoplasm (Fig. 1D). FEZF1-AS1 expression was subsequently knocked down in Hep3B and Huh7 cells by transfection with specific RNAs. As presented, the expression levels of FEZF1-AS1 were significantly downregulated in Hep3B and Huh7 cells following transfection with the two siRNAs (P<0.05; Fig. 1E).
Figure 1.
FEZF1-AS1 is upregulated in HCC tissues. (A) Relative expression levels of FEZF1-AS1 in HCC cell lines (Hep3B and Huh7) and normal hepatocyte LO2. *P<0.05 vs. LO2. (B) Relative expression levels of FEZF1-AS1 in HCC tissues (n=58) and adjacent normal tissues (n=58) were determined by reverse transcription-quantitative polymerase chain reaction assays. ***P<0.001 vs. normal tissue (C) Kaplan-Meier curve was determined, according to FEZF1-AS1 expression in HCC tissues. (D) RNA-fluorescence in situ hybridization indicated FEZF1-AS1 was located in the cytoplasm in HCC cells (×600 magnification). (E) Relative expression of FEZF1-AS1 in Hep3B and Huh7 cells transfected with siNC or siLnc (siFEZF1-AS1). *P<0.05 vs. respective siNC. HCC, hepatocellular carcinoma; si, small interfering; NC, negative control; lnc, long non-coding.
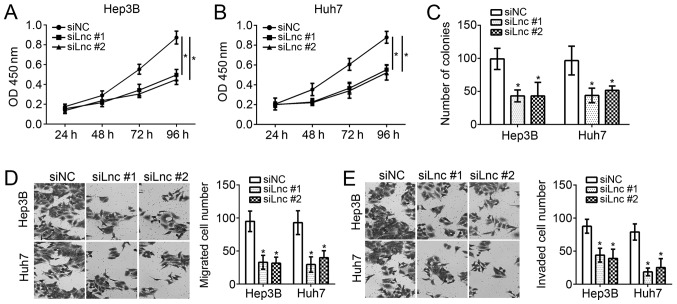
FEZF1-AS1 knockdown inhibits the proliferation, migration and invasion of Hep3B and Huh7 cells
To examine the function of FEZF1-AS1 in HCC cells, CCK-8, colony formation and transwell assays were performed. Through CCK-8 assays, it was observed that knockdown of FEZF1-AS1 significantly inhibited the proliferation of Hep3B and Huh7 cells (Fig. 2A and B) at 96 h after culture. The colony formation assays suggested that the downregulation of FEZF1-AS1 significantly decreased the number of colonies formed by Hep3B and Huh7 cells (P<0.05; Fig. 2C). Furthermore, it was identified that FEZF1-AS1 depletion significantly decreased the numbers of migrated and invaded Hep3B and Huh7 cells (P<0.05; Fig. 2D and E). Collectively, these data demonstrated that FEZF1-AS1 is important for the proliferation, migration and invasion of HCC cells.
Figure 2.
FEZF1-AS1 knockdown inhibits the proliferation, migration and invasion of Hep3B and Huh7 cells. Cell Counting Kit-8 assays were conducted to examine the proliferation of (A) Hep3B and (B) Huh7 cells. (C) Knockdown of FEZF1-AS1 reduces the number of colonies formed by Hep3B and Huh7 cells. Transwell assays were utilized to determine (D) the migration and (E) invasion potential of Hep3B and Huh7 cells (×200 magnification). *P<0.05 vs. the control group. OD, optical density; si, small interfering; NC, negative control; lnc, long non-coding.
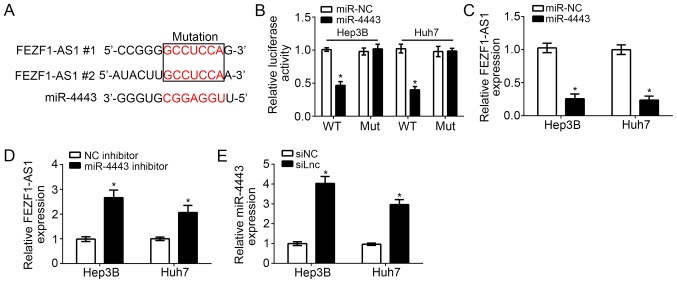
FEZF1-AS1 acts as a sponge for miR-4443
lncRNAs serve as competing endogenous RNAs (ceRNAs) to bind with miRs and regulate gene expression (16). In order to investigate the downstream mechanism of FEZF1-AS1, bioinformatics analysis was performed, which identified that numerous miRs exhibited potential binding sites for FEZF1-AS1. Among all targeting miRs, miR-4443 ranked top and has been reported to inhibit colon cancer progression (17). Therefore, miR-4443 was selected for further examination. It was identified that there were two potential binding sites of miR-4443 in FEZF1-AS1 (Fig. 3A). Through luciferase reporter assays, it was demonstrated that miR-4443 overexpression inhibited the luciferase activity in Hep3B and Huh7 cells transfected with FEZF1-AS1-WT, while mutation of the binding sites in FEZF1-AS1 abrogated this effect (Fig. 3B). Furthermore, it was identified that overexpression of miR-4443 significantly inhibited the expression levels of FEZF1-AS1 in Hep3B and Huh7 cells (P<0.01; Fig. 3C); whereas, inhibition of miR-4443 significantly increased the expression of FEZF1-AS1 (P<0.05; Fig. 3D). Consistently, knockdown of FEZF1-AS1 additionally significantly upregulated the expression levels of miR-4443 in Hep3B and Huh7 cells (P<0.05; Fig. 3E). Collectively, these data suggested that FEZF1-AS1 directly targets miR-4443 in HCC cells.
Figure 3.
FEZF1-AS1 acts as a sponge for miR-4443. (A) Two potential binding sites of miR-4443 in FEZF1-AS1. (B) Luciferase reporter assay indicated overexpression of miR-4443 inhibits the luciferase activity in Hep3B and Huh7 cells transfected with WT FEZF1-AS1-WT. (C) Reverse transcription-quantitative polymerase chain reaction analysis demonstrated that overexpression of miR-4443 repressed the expression of FEZF1-AS1. P<0.05 vs. respective miR-NC. (D) Inhibition of miR-4443 upregulates the expression of FEZF1-AS1. P<0.05 vs. respective NC inhibitor. (E) Knockdown of FEZF1-AS1 promotes the expression level of miR-4443. *P<0.05 vs. respective siNC. miR, micro RNA; si, small interfering; NC, negative control; lnc, long non-coding; WT, wild-type; Mut, mutant.
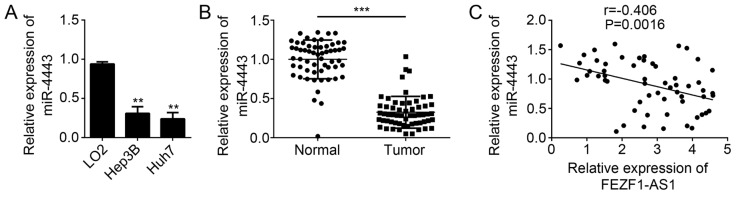
miR-4443 is downregulated in HCC
The function of miR-4443 remains unknown in HCC. To further determine whether miR-4443 is involved in HCC progression, the expression patterns of miR-4443 in HCC cell lines were first analyzed by RT-qPCR. The results suggested that miR-4443 was downregulated in HCC cell lines compared with LO2 cells (Fig. 4A). Similarly, the expression of miR-4443 was significantly downregulated in HCC tissues compared with adjacent normal tissues (P<0.001; Fig. 4B). Subsequently, the expression correlation between FEZF1-AS1 and miR-4443 was determined in HCC tissues by RT-qPCR. The results demonstrated that FEZF1-AS1 expression was negatively correlated with that of miR-4443 in HCC tissues (Fig. 4C). In summary, these data implied that miR-4443 may serve a function in HCC progression.
Figure 4.
miR-4443 is downregulated in HCC. (A) Relative expression of miR-4443 was determined by RT-qPCR in HCC cell lines. *P<0.05 vs. LO2. (B) Relative expression of miR-4443 was measured by RT-qPCR in HCC tissues. ***P<0.0001. (C) Expression correlation between FEZF1-AS1 and miR-4443 was determined by RT-qPCR in HCC tissues. **P<0.01 vs. the control group. miR, micro RNA; HCC, hepatocellular carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
FEZF1-AS1 promotes HCC cell proliferation, migration and invasion by regulating miR-4443
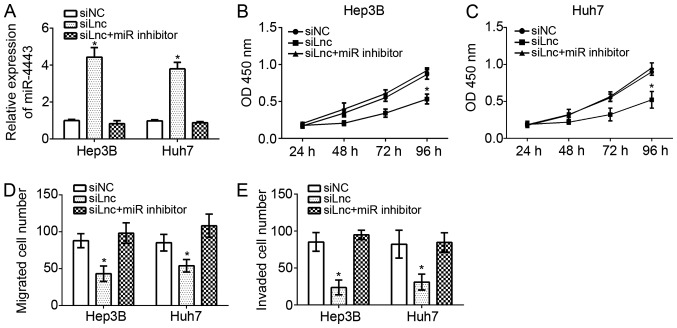
To verify whether miR-4443 was involved in FEZF1-AS1-mediated effects on HCC progression, the expression levels of miR-4443 were demonstrated to be downregulated by RT-qPCR in FEZF1-AS1-depleted Hep3B and Huh7 cells. The results suggested that the miR-4443 expression level was markedly downregulated in Hep3B and Huh7 cells transfected siFEZF1-AS1 plus miR-4443 inhibitor compared with the siFEZF1-AS1 group (Fig. 5A). CCK-8 assays were performed to evaluate the proliferation of Hep3B and Huh7 cells. It was identified that FEZF1-AS1 knockdown inhibited cellular proliferation while inhibition of miR-4443 significantly reversed this trend in Hep3B and Huh7 cells (P<0.05; Fig. 5B and C). Furthermore, transwell assays additionally demonstrated that FEZF1-AS1 depletion decreased the migration and invasion of Hep3B and Huh7 cells; whereas, knockdown of miR-4443 at the simultaneously promoted cell migration and invasion (Fig. 5D and E). FEZF1-AS1 promoted HCC cell proliferation, migration and invasion by acting as a sponge to miR-4443.
Figure 5.
FEZF1-AS1 promotes hepatocellular carcinoma cell proliferation, migration and invasion by regulating miR-4443. (A) Relative expression of miR-4443 in Hep3B and Huh7 cells transfected with siLnc in addition to miR-4443 inhibitor or not. Cell Counting Kit-8 assays were used to detect cell proliferation of (B) Hep3b and (C) Huh7 cells. Transwell assays were utilized to investigate (D) the migration and (E) invasion of Hep3B and Huh7 cells. *P<0.05 vs. respective siNC. miR, micro RNA; si, small interfering; NC, negative control; lnc, long non-coding.
Discussion
HCC contributes to a large number of cancer-associated mortalities around the world each year (2). Although HCC becomes a primary public health problem, there are no effective therapeutic methods for curative treatment of HCC. Therefore the examination of the underlying mechanism of hepatocarcinogenesis and the search for novel therapeutic targets for HCC treatment is urgently required. In the present study, it was identified that FEZF1-AS1 was highly expressed in HCC tissues and cell lines compared with adjacent normal tissues or hepatocytes. Additionally, it was demonstrated that FEZF1-AS1 expression serves as a prognostic marker for patients with HCC. Furthermore, the present results suggested that knockdown of FEZF1-AS1 suppressed the proliferation, migration and invasion of Hep3B and Huh7 cells in vitro. In terms of mechanism, it was observed that FEZF1-AS1 served as a ceRNA sponge to miR-4443 in HCC cells. Overexpression of miR-4443 repressed the expression of FEZF1-AS1 in HCC cells, and vice versa. Furthermore, it was demonstrated that miR-4443 was downregulated in HCC tissues and negatively correlated with the expression of FEZF1-AS1. Finally, through functional experiments, it was demonstrated that inhibition of miR-4443 abrogated the effects of FEZF1-AS1 on HCC cell proliferation, migration and invasion.
In recent years, an increasing number of studies document that lncRNAs are essential regulator in various biological processes (18–20). In human cancer, lncRNAs have been demonstrated to serve pivotal roles in regulating cellular proliferation, apoptosis, migration and invasion (19). Increasing evidence suggests that lncRNAs are involved in the development and progression of a diverse range of cancer types, including osteosarcoma (20), gastric cancer (21), non-small cell lung cancer (22), colorectal cancer (23), breast cancer (24), clear cell renal cell carcinoma (25) and hepatocellular carcinoma (26). However, the functions of the majority of lncRNAs in cancer remain unclear. As for FEZF1-AS1, a previous study suggested that FEZF1-AS1 facilitates cell proliferation and migration in colorectal carcinoma (12). Another previous study reported that FEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation (13). He et al (14) demonstrated that FEZF1-AS1 enhances epithelial-mesenchymal transition by suppressing epithelial-cadherin and regulating WNT pathway in non-small cell lung cancer. Whether FEZF1-AS1 regulates HCC progression remains largely unknown. In the present study, it was observed that FEZF1-AS1 was upregulated in HCC cells and promoted cancer cell proliferation, migration and invasion. The present results suggested that FEZF1-AS1 additionally serves as an oncogene in HCC.
In the past decades, the functions of miRNAs have been widely investigated. A large number of studies suggest that miRNAs are closely associated with tumorigenesis. Additionally, miRNAs are demonstrated to regulate tumor cell proliferation, migration and invasion by suppressing target gene expression post-transcriptionally (27). Abnormal expression of miRNAs is frequently observed in types of cancer, including breast cancer (28), colorectal cancer (29), non-small cell lung cancer (30) and hepatocellular carcinoma (31). Recently, a previous study suggested that miRNA expression levels may be regulated by lncRNAs (18). In the present study, FEZF1-AS1 was identified to act as a sponge of miR-4443 in HCC cells. miR-4443 has been demonstrated to decrease the invasiveness of human colon cancer cells (17). In contrast, another previous study suggested that miR-4443 induced malignancy of breast cancer (32). Whether miR-4443 is involved in HCC requires further investigation. Through functional experiments, it was observed that miR-4443 was downregulated in HCC tissues. Inhibition of miR-4443 abolished the effects of FEZF1-AS1 on HCC cell proliferation, migration and invasion, which suggested that miR-4443 suppressed HCC progression.
In conclusion, the results of the present study demonstrated that FEZF1-AS1 promoted the proliferation, migration and invasion of HCC cells by regulating miR-4443 expression levels. The present results highlight the importance of the FEZF1-AS1/miR-4443 axis during HCC development and progression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
JG and JW conceived and designed the study, and analyzed and interpreted the results. JZ performed experiments and wrote this manuscript. TL and JH conducted the experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
For the use of human samples, the protocol for the present study was approved by the Institutional Ethics Committee of China Three Gorges University (Yichang, China) and all enrolled patients signed a written informed consent document.
Patient consent for publication
All patients within the present study provide consent for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Parkin DM, Pisani P, Ferlay J. Global cancer statistics. CA Cancer J Clin. 1999;49:33–64. doi: 10.3322/canjclin.49.1.33. [DOI] [PubMed] [Google Scholar]
- 3.Colecchia A, Schiumerini R, Cucchetti A, Cescon M, Taddia M, Marasco G, Festi D. Prognostic factors for hepatocellular carcinoma recurrence. World J Gastroenterol. 2014;20:5935–5950. doi: 10.3748/wjg.v20.i20.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan Y, Qin T, Yin S, Zhang X, Gao X, Mu L. Long non-coding RNA UC001kfo promotes hepatocellular carcinoma proliferation and metastasis by targeting α-SMA. Biomed Pharmacother. 2017;87:669–677. doi: 10.1016/j.biopha.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 5.Lu L, Dai Z, Luo Q, Lv G. The long noncoding RNA cancer susceptibility candidate 2 inhibits tumor progression in osteosarcoma. Mol Med Rep. 2018;17:1947–1953. doi: 10.3892/mmr.2017.8080. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Wei G, Xia H, Tang Q, Bi F. Long noncoding RNA-ATB promotes cell proliferation, migration and invasion in gastric cancer. Mol Med Rep. 2018;17:1940–1946. doi: 10.3892/mmr.2017.8077. [DOI] [PubMed] [Google Scholar]
- 7.Tang WG, Hu B, Sun HX, Sun QM, Sun C, Fu PY, Yang ZF, Zhang X, Zhou CH, Fan J, et al. Long non-coding RNA00364 represses hepatocellular carcinoma cell proliferation via modulating p-STAT3-IFIT2 signaling axis. Oncotarget. 2017;8:102006–102019. doi: 10.18632/oncotarget.22039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J, Wang YJ, Sun CR, Qin B, Zhang Y, Chen G. Long noncoding RNA lncHERG promotes cell proliferation, migration and invasion in glioblastoma. Oncotarget. 2017;8:108031–108041. doi: 10.18632/oncotarget.22446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B, Du Y, Gao G, Tian Y, He L, Fan Z. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7:13608. doi: 10.1038/ncomms13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Li J, Ji D, Leng K, Xu Y, Huang L, Jiang X, Cui Y. Overexpressed long noncoding RNA Sox2ot predicts poor prognosis for cholangiocarcinoma and promotes cell proliferation and invasion. Gene. 2018;645:131–136. doi: 10.1016/j.gene.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Dun Y, Zhou S, Huang XH. LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells proliferation and invasion by targeting miR-133a-3p and activating Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2017;96:1216–1221. doi: 10.1016/j.biopha.2017.11.096. [DOI] [PubMed] [Google Scholar]
- 12.Chen N, Guo D, Xu Q, Yang M, Wang D, Peng M, Ding Y, Wang S, Zhou J. Long non-coding RNA FEZF1-AS1 facilitates cell proliferation and migration in colorectal carcinoma. Oncotarget. 2016;7:11271–11283. doi: 10.18632/oncotarget.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu YW, Xia R, Lu K, Xie M, Yang F, Sun M, De W, Wang C, Ji G. LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol Cancer. 2017;16:39. doi: 10.1186/s12943-017-0588-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC) Biomed Pharmacother. 2017;95:331–338. doi: 10.1016/j.biopha.2017.08.057. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Cao C, Zhang T, Zhang D, Xie L, Zou X, Lei L, Wu D, Liu L. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene. 2017;36:1112–1122. doi: 10.1038/onc.2016.278. [DOI] [PubMed] [Google Scholar]
- 17.Meerson A, Yehuda H. Leptin and insulin up-regulate miR-4443 to suppress NCOA1 and TRAF4, and decrease the invasiveness of human colon cancer cells. BMC Cancer. 2016;16:882. doi: 10.1186/s12885-016-2938-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng P, Yin Z, Wu Y, Xu Y, Luo Y, Zhang TC. LncRNA HOTAIR promotes cell migration and invasion by regulating MKL1 via inhibition miR206 expression in HeLa cells. Cell Commun Signal. 2018;16:5. doi: 10.1186/s12964-018-0216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, Zhao W, Lu Z, Zhang W, Yang X. Long noncoding RNA GAS5 promotes proliferation, migration, and invasion by regulation of miR-301a in esophageal cancer. Oncol Res. 2018;26:1285–1294. doi: 10.3727/096504018X15166193231711. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Yang C, Wu K, Wang S, Wei G. Long non-coding RNA XIST promotes osteosarcoma progression by targeting YAP via miR-195-5p. J Cell Biochem. 2018;119:5646–5656. doi: 10.1002/jcb.26743. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Wang Y, Sun L, Min J, Liu J, Chen D, Zhang H, Zhang H, Zhang H, Zhou Y, Liu L. Long noncoding RNA BC005927 upregulates EPHB4 and promotes gastric cancer metastasis under hypoxia. Cancer Sci. 2018;109:988–1000. doi: 10.1111/cas.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Y, Ma Y, Li L, Shen X, Xin T, Zhao Y, Ma R. Effect of long non-coding RNA LINC01116 on biological behaviors of non-small cell lung cancer cells via the hippo signaling pathway. J Cell Biochem. 2018;119:6310. doi: 10.1002/jcb.26711. [DOI] [Google Scholar]
- 23.Zhang Y, Tao Y, Li Y, Zhao J, Zhang L, Zhang X, Dong C, Xie Y, Dai X, Zhang X, Liao Q. The regulatory network analysis of long noncoding RNAs in human colorectal cancer. Funct Integr Genomics. 2018;18:261–275. doi: 10.1007/s10142-017-0588-2. [DOI] [PubMed] [Google Scholar]
- 24.Zhang S, Wang J, Ghoshal T, Wilkins D, Mo YY, Chen Y, Zhou Y. lncRNA gene signatures for prediction of breast cancer intrinsic subtypes and prognosis. Genes (Basel) 2018;9 doi: 10.3390/genes9020065. pii: E65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Tan C, Weng WW, Deng Y, Zhang QY, Yang XQ, Gan HL, Wang T, Zhang PP, Xu MD, et al. Long non-coding RNA Linc00152 is a positive prognostic factor for and demonstrates malignant biological behavior in clear cell renal cell carcinoma. Am J Cancer Res. 2016;6:285–299. [PMC free article] [PubMed] [Google Scholar]
- 26.Ma X, Liu H, Murphy JT, Foyil SR, Godar RJ, Abuirqeba H, Weinheimer CJ, Barger PM, Diwan A. Regulation of the transcription factor EB-PGC1α axis by beclin-1 controls mitochondrial quality and cardiomyocyte death under stress. Mol Cell Biol. 2015;35:956–976. doi: 10.1128/MCB.01091-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Wang W, Huang K, Wang Y, Li J, Yang X. MicroRNA-34a inhibits cells proliferation and invasion by downregulating Notch1 in endometrial cancer. Oncotarget. 2017;8:111258–111270. doi: 10.18632/oncotarget.22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong H, Yan T, Zhang W, Shi F, Jiang X, Wang X, Li S, Chen Y, Chen C, Zhu Y. miR-613 inhibits cell migration and invasion by downregulating Daam1 in triple-negative breast cancer. Cell Signal. 2018;44:33–42. doi: 10.1016/j.cellsig.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 29.Tong F, Ying Y, Pan H, Zhao W, Li H, Zhan X. MicroRNA-466 (miR-466) functions as a tumor suppressor and prognostic factor in colorectal cancer (CRC) Bosn J Basic Med Sci. 2018;18:252–259. doi: 10.17305/bjbms.2018.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang H, Zhang H, Hu X, Li W. Knockdown of long non-coding RNA XIST inhibits cell viability and invasion by regulating miR-137/PXN axis in non-small cell lung cancer. Int J Biol Macromol. 2018;111:623–631. doi: 10.1016/j.ijbiomac.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 31.Cui S, Qian Z, Chen Y, Li L, Li P, Ding H. Screening of up- and downregulation of circRNAs in HBV-related hepatocellular carcinoma by microarray. Oncol Lett. 2018;15:423–432. doi: 10.3892/ol.2017.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Zhong SL, Lu P, Wang DD, Zhou SY, Yang SJ, Shen HY, Zhang L, Zhang XH, Zhao JH, Tang JH. miR-4443 participates in the malignancy of breast cancer. PLoS One. 2016;11:e0160780. doi: 10.1371/journal.pone.0160780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.