Abstract
Familial adenomatous polyposis (FAP) is an autosomal dominant-inherited colorectal cancer. Recent advances in genetics have indicated that the majority of patients with FAP carry germline mutations of the adenomatous polyposis coli (APC) and mutY DNA glycosylase (MUTYH) genes. However, a large subset of families with a history of FAP have undetectable pathogenic alterations, termed APC/MUTYH mutation-negative FAP. To investigate the germline mutations in the APC and MUTYH genes in Chinese patients with FAP, 13 unrelated patients were enrolled. Through genetic sequencing, four known pathogenic alterations (Lys1061LysfsTer2, Glu1309AspfsTer4, Arg283Ter and Ser1196Ter) of APC and two novel disease-associated pathogenic mutations (Tyr152Ter and Ter522Gly) in MUTYH were identified in six individuals. For samples that did not present with pathogenic alterations, the functional effects of missense, synonymous and intronic mutations were analyzed using bioinformatics tools and databases. Bioinformatics prediction suggested that the synonymous mutation Tyr486Tyr in APC (APC∆486s) was likely a disease-causing polymorphism and may have induced the exon skipping of APC. A hybrid mini-gene assay was performed, which confirmed that the synonymous single nucleotide polymorphism APC∆486s induced major splicing defects with skipping of exon 12 in APC. The data of the present study suggested that the synonymous polymorphism APC∆486s was a potential pathogenic alteration that predisposed APC/MUTYH mutation-negative patients to FAP.
Keywords: familial adenomatous polyposis, adenomatous polyposis coli, mutY DNA glycosylase, synonymous single nucleotide polymorphism, bioinformatics, exon skipping
Introduction
Familial adenomatous polyposis (FAP) is an autosomal dominant-inherited colorectal cancer syndrome with poor prognosis (1,2). It is well established that FAP is linked to germline mutations of the adenomatous polyposis coli (APC; transcript ID, ENST00000257430.8; https://www.ensembl.org/Homo_sapiens/Transcript/Summary?g=ENSG00000134982; r=5:112737888-112846239;t=ENST00000257430) gene (3,4), and screening for APC germline mutations in families with FAP is a standard and effective tool for the predictive testing of at-risk individuals (3–6). However, through conventional screening methods, a number of patients with FAP have no detectable APC mutations, which is defined as APC mutation-negative FAP (7,8). Certain patients with FAP without detectable germline APC mutations were revealed to carry biallelic mutations in the base-excision-repair gene mutY DNA glycosylase (MUTYH; transcript ID, ENST00000354383.10), and investigators demonstrated that mutations in MUTYH may explain up to 25% of APC mutation-negative FAP cases (8,9). However, a large subset of families with a history of FAP have pathogenic alterations that are undetectable, and this is termed APC/MUTYH mutation-negative FAP (8–10).
In the present study, in order to investigate the germline mutations and the potential causes of APC/MUTYH mutation-negative FAP, a cohort of 13 patients with FAP were enrolled. First, whole-gene sequencing of APC and MUTYH was applied to determine the pathogenic alterations predisposing patients to FAP (11–13), and the results identified four known pathogenic mutations in APC and two novel disease-associated pathogenic mutations in six individuals. For the APC/MUTYH mutation-negative FAP cases, missense, synonymous and intronic mutations in APC and MUTYH were analyzed using bioinformatics assessment tools and databases; notably, a synonymous mutation of Tyr486Tyr in APC (APC∆486s) was predicted to be a disease-causing polymorphism. In-depth analysis of the results suggested that this mutation may have caused exon skipping in APC and that the truncated APC protein resulted in FAP.
It has been established that the majority of synonymous and intronic mutations are silent mutations; however, previous investigations have also revealed that certain synonymous substitutions are associated with a risk of disease and other complex traits including susceptibility to cancer (14,15). Increasing amounts of evidence indicate that synonymous mutations may induce major splicing defects at the mRNA level and consequently result in disease (16).
Combined with the above conclusions and the findings of the present study, it was hypothesized that APC∆486s which was previously considered to be non-pathogenic polymorphism was a potential pathogenic mutation for APC/MUTYH mutation-negative FAP. To confirm the splicing regulation of the synonymous mutation APC∆486s in vitro, an exon skipping assay was performed to verify whether the synonymous mutation APC∆486s leads to APC protein truncation. It is hoped that following further studies on the mechanism of exon skipping of the synonymous mutation APC∆486s, it could be applied as a potential pathogenic alteration in the diagnostic application for those APC/MUTYH mutation-negative patients to FAP.
Materials and methods
Ethics statement
The present study was approved by and performed in accordance with the Research Ethics Board of Kunming Medical University (Kunming, China). All participants provided written informed consent, and the ethics committees approved the consent procedure.
Patients
Between January 2013 and December 2017, 13 patients [male:female 8:5, mean age 31 years (range, 16–41] clinically diagnosed with FAP were recruited from the First Affiliated Hospital of Kunming Medical University (Kunming, China). The diagnosis of all patients referred for the present study was confirmed by surgery, and all patients had a family history suggestive of FAP. Blood samples were collected from all patients prior to surgery.
Gene-sequencing of APC and MUTYH
Blood samples from all patients with FAP underwent exon-specific DNA sequencing for APC and DNA was purified following the manufacturer's protocols of the DNA extraction kit (QIAamp DNA blood mini kit; Qiagen, Valencia, CA, USA). The extracted DNA was quantified by a Pearl nanophotometer (Implen, Munich, Germany). The promoter region, exons 1-14 and 21 segments of exon 15 in the APC gene were separately amplified by polymerase chain reaction (PCR) (11,13,17). Following the DNA sequencing of APC, whole-gene sequencing of MUTYH was performed for the APC mutation-negative FAP samples. The primer pairs used for MUTYH sequencing in the present study were designed as previously described (12,13).
In silico analysis and functional single nucleotide polymorphism (SNP) assessment
The sequencing data for each gene was initially analyzed using Mutation Surveyor (18) (www.softgenetics.com). Human Gene Mutation Database (19) (www.hgmd.org), International HapMap Project (20) (ftp.ncbi.nlm.nih.gov/hapmap/), dbSNP database (21) (www.ncbi.nlm.nih.gov/SNP), 1000 Genomes (http://www.internationalgenome.org/), and Ensembl (22) (www.ensembl.org) were also used.
For APC gene mutations, the Leiden Open Variation Database (LOVD; databases.lovd.nl/shared/genes/APC), BIPMed SNP Array (bipmed.iqm.unicamp.br/snparray/genes/APC), ClinVar at the National Center for Biotechnology Information [NCBI; www.ncbi.nlm.nih.gov/clinvar/?term=APC(gene)], the Universal Mutation Database (UMD)-APC mutations database (www.umd.be/APC/W_APC), Iran Variation Database (genet.ir/variome/genes/APC), International Society for Gastrointestinal Hereditary Tumours Database (www.insight-database.org/genes/APC), LOVD (proteomics.bio21.unimelb.edu.au/lovd/genes/APC), Spain Mutation Database (lovd3.isciii.es/genes/APC) and the Zhejiang University Center for Genetic and Genomic Medicine (23) (www.genomed.org/lovd2/home.php?select_db=APC) were applied.
For the mutation screening of the MUTYH gene, the LOVD (proteomics.bio21.unimelb.edu.au/lovd/genes/MUTYH), Shared database (databases.lovd.nl/shared/genes/MUTYH), UMD-MUTYH mutations database (www.umd.be/MUTYH/), Iran Variation Database (genet.ir/variome/genes/MUTYH), ClinVar at NCBI [www.ncbi.nlm.nih.gov/clinvar/?term=MUTYH(gene)], International Society for Gastrointestinal Hereditary Tumours Database (www.insight-database.org/genes/MUTYH) and PharmGKB (https://www.pharmgkb.org/gene/PA31328/variantAnnotation) were used.
Bioinformatics predictions of the missense, synonymous and intronic substitutions in APC/MUTYH mutation-negative FAP
To investigate the possible genetic causes in patients with APC/MUTYH mutation-negative FAP, bioinformatics prediction tools were used to study the function of any identified missense, synonymous and intron substitutions. Polymorphism phenotype version 2 (24), Sorting Intolerant From Tolerant (SIFT) (25) and SNPs&Gene Ontology (GO) (26) were used to predict deleterious missense substitutions. Exonic splicing enhancer (ESE) finder 3.0 (27) (krainer01.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home), RESCUE-ESE (28) (genes.mit.edu/burgelab/rescue-ese), exonic-splicing regulatory (ESR) search (26) and putative exonic splicing enhancers (PESX) (29) were used to determine the corresponding functional impacts of the synonymous and intronic substitutions.
Hybrid mini-gene assay of synonymous SNP APC∆486s
Generation of mini-gene constructs of synonymous SNP APC∆486s
Through bioinformatics analysis, the synonymous and intronic variants in MUTYH were predicted to have no effect on the disease. However, five synonymous SNPs in APC were predicted to be disease-associated mutations (Fig. 1), and among them, APC∆486s was predicted to be associated with exon skipping in APC. To confirm the splicing regulation of the synonymous mutation APC∆486s in vitro, a mini-gene system of APC∆486s was constructed to investigate its splicing impact on the APC gene. The nucleotide sequence of exon 12 with wild-type and mutant APC∆486s was synthesized, together with 264 bp of the 12th flanking intron, carrying the XhoI restriction site, and 210 bp of the 13th flanking intron, carrying the HindIII restriction site (Fig. 2), as described previously (30). The wild-type and mutant mini-gene were inserted into pEGFP-N1 to construct the recombinant plasmids pEGFP-N1-wt-mini-gene and pEGFP-N1-mt-mini-gene, respectively (Takara Bio USA, Inc., CA, USA).
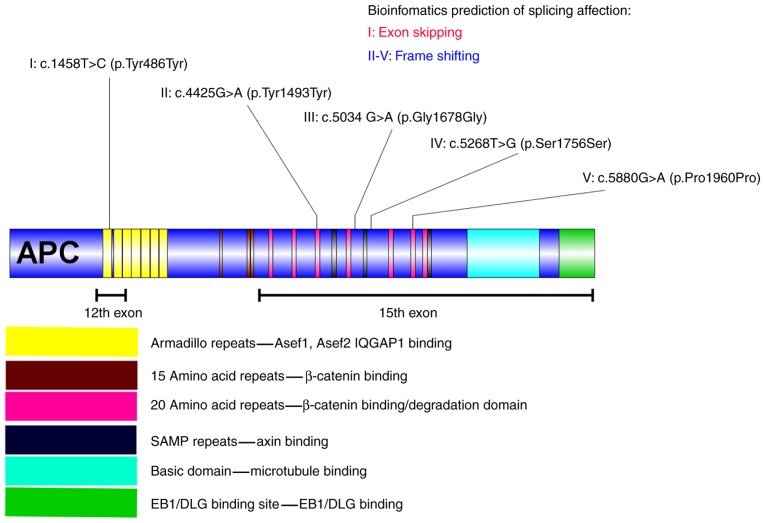
Figure 1.
Synonymous substitutions in the APC gene and the corresponding bioinformatics prediction of each mutant polymorphism. I indicates the synonymous substitution c.1458T>C, located in the first armadillo repeat of APC, which is in the Asef1, Asef2 and IQGAP binding domain. The splicing impact of c.1458T>C may have caused the skipping of exon 12 in APC. II indicates the synonymous substitution c.4425G>A located in the third 20 amino acid repeat of APC, which is located in the β-catenin binding/degradation domain. The splicing impact of c.4425G>A may have caused a frameshift mutation in APC. III indicates the synonymous substitution c.5034 G>A is located behind the fourth 20 amino acid repeat of APC, which is in the β-catenin binding/degradation domain. The splicing impact of c.5034 G>A may have resulted in a frameshift mutation. IV indicates the synonymous substitution c.5268T>G located behind the second SAMP repeat of APC. The splicing impact of c.5268T>G may also result in a frameshift mutation. V indicates the synonymous substitution c.5880G>A, located in the sixth 20 amino acid repeat of APC, which is in the β-catenin binding/degradation domain. The splicing impact of c. 5880G>A may have caused a frameshift mutation in APC. APC, adenomatous polyposis coli; Asef, APC-stimulated guanine nucleotide exchange factor; IQGAP1, IQ motif containing GTPase activating protein 1; EB1, microtubule associated protein RP/EB family member 1; DLG, discs large MAGUK scaffold protein 1.
Figure 2.
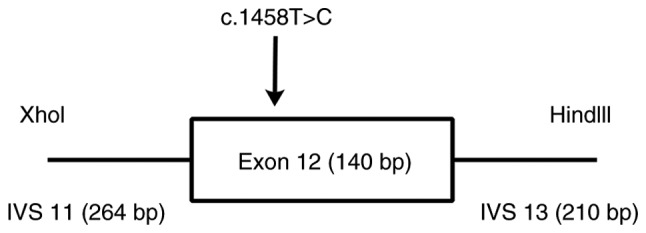
Mini-gene system encoding wild-type and mutant c.1458T>C. The nucleotide sequence of the exon 12 with wild-type and mutant APC∆486s, together with 264 bp of flanking intron 12, carrying the XhoI restriction site, and 210 bp of flanking intron 13, carrying the HindIII restriction site. APC, adenomatous polyposis coli; bp, base pairs; IVS, intervening sequence.
Transfection and reverse transcription (RT)-PCR
Following identification by plasmid sequencing (30), the wild-type and mutant mini-gene constructs were transiently transfected into HeLa cells using Lipofectamine® 2000 transfection reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to manufacturer's protocol: 1×105 HeLa cells were seeded in each 12-well plate and then cultured in DMEM (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) containing 100 ml/l fetal bovine serum (Gibco, Thermo Fisher Scientific, Inc.) and 50 ml/l CO2 at 37°C. 24 h following transfection, the cells were collected. Total RNA was extracted using the TriPure Isolation reagent (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and analyzed by RT-PCR (2XTSINGKE Master Mix, TSINGKE Biological Technology, Beijing, China). The resulting cDNA was amplified by PCR (30 cycles) using the following primers: APC forward, 5′-ATTATTTCGCTCAGCAAGATAAG-3′ and reverse, 5′-TTCCATCTGTAGATGTACCTTTGC-3′. GAPDH was selected as reference and the corresponding premiers were: GAPDH forward, 5′-TGAAGGTCGGAGTCAACGG-3′ and reverse, 5′-TCCTGGAAGATGGTGATGGG-3′. The PCR thermocycling conditions included an initial denaturation step at 95°C for 5 min, followed by 30 cycles of 95°C for 30 sec, 59°C for 30 sec and 72°C for 30 sec, and a final 5 min extension step at 72°C. PCR products were separated by electrophoresis on 2.5% agarose gels containing ethidium bromide, and visualized by exposure to ultraviolet light. The experiments were repeated 3 times.
Results
Micromutations of APC and MUTYH
Following DNA sequencing of APC and MUTYH, micromutations were analyzed using Mutation Surveyor (Table I). For APC, four disease-associated pathogenic alterations (Lys1061LysfsTer2, Glu1309AspfsTer4, Arg283Ter and Ser1196Ter) were identified, which had been previously reported to be pathogenic mutations (31–33,13). In addition to the pathogenic mutations mentioned above, seven missense mutations (Ser784Thr, Ile880Leu, Arg1171Gly, Ile1259Leu, Asp1519Glu, Glu1552Val and Pro2216Thr), five synonymous substitutions (Tyr486Tyr, Tyr1493Tyr, Gly1678Gly, Ser1756Ser and Pro1960Pro) and three intronic mutations (c.220+35T>A, c.645+129A>C and c.1548+133C>A) were identified.
Table I.
Variants identified in the patient cohort by Mutation Surveyor.
| A, APC | |
|---|---|
| Variant | Functional class |
| c.3183_3187delACAAA (p.Lys1061LysfsTer2) | Frameshift mutations |
| c.3927_3931delAAAGA (p.Glu1309AspfsX4) | Frameshift mutations |
| c.847C>T (p.Arg283Ter) | Nonsense mutation |
| c.3587C>A (p.Ser1196Ter) | Nonsense mutation |
| c.2350T>A (p.Ser784Thr) | Missense mutation |
| c.2638A>C (p.Ile880Leu) | Missense mutation |
| c.3511C>G (p.Arg1171Gly) | Missense mutation |
| c.3775A>C (p.Ile1259Leu) | Missense mutation |
| c.4557T>A (p.Asp1519Glu) | Missense mutation |
| c.4655A>T (p.Glu1552Val) | Missense mutation |
| c.6646C>A (p.Pro2216Thr) | Missense mutation |
| c.220+35T>A | Intronic mutation |
| c.645+129A>C | Intronic mutation |
| c.1548+133C>A | Intronic mutation |
| c.1458T>C (p.Tyr486Tyr) | Synonymous mutation |
| c.4425G>A (p.Tyr1493Tyr) | Synonymous mutation |
| c.5034 G>A (p.Gly1678Gly) | Synonymous mutation |
| c.5268T>G (p.Ser1756Ser) | Synonymous mutation |
| c. 5880G>A (p.Pro1960Pro) | Synonymous mutation |
| B, MUTYH | |
| Variant | Functional class |
| c.456T>A (p.Tyr152Ter) | Nonsense mutation |
| c.1564T>G (p.Ter522Gly) | Nonstop mutation |
| c.553G>C (p.Glu185Gln) | Missense mutation |
| c.1232A>T (p.Gln411Leu) | Missense mutation |
| c.1268G>A (p.Arg423His) | Missense mutation |
| c.1483C>G (p.Gln495Glu) | Missense mutation |
| c.264+11G>A | Intronic mutation |
| c.304+23G>A | Intronic mutation |
| c.420+35A>G | Intronic mutation |
| c.1566+33_1566+35het_delTGT | Intronic mutation |
| c.423G>A (p.Glu141Glu) | Synonymous mutation |
| c.450C>A (p.Glu150Glu) | Synonymous mutation |
| c.1347G>C (p.Tyr449Tyr) | Synonymous mutation |
APC, adenomatous polyposis coli; FAP, familial adenomatous polyposis.
For MUTYH, one novel nonsense mutation (Tyr152Ter) and one nonstop mutation (Ter522Gly) were identified as pathogenic mutations and were predicted to cause premature protein truncation and the continued translation of an mRNA, respectively. In addition, four missense mutations (Glu185Gln, Gln411Leu, Arg423His and Gln495Glu), three synonymous substitutions (Glu141Glu, Glu150Glu and Tyr449Tyr) and four intronic mutations (c.264+11G>A, c.304+23G>A, c.420+35A>G and c.1566+33_1566+35het_delTGT) were identified.
Computational prediction of missense substitutions and ‘silent’ mutations
Computational prediction suggested that no missense substitutions in APC or MUTYH in the cohort of the present study were disease-associated variants (Table II). The synonymous and intronic substitutions of APC and MUTYH were further analyzed, and the in-silico analysis results indicated that the synonymous substitutions (Tyr486Tyr, Tyr1493Tyr, Gly1678Gly, Ser1756Ser and Pro1960Pro) of APC may have affected splicing regulation by creating or removing ESEs or exon splicing silencers. Through bioinformatics prediction, the synonymous mutations Tyr1493Tyr, Gly1678Gly, Ser1756Ser and Pro1960Pro were predicted to cause a frame shift in APC, and the synonymous mutation Tyr486Tyr (APC∆486s) was predicted to induce exon skipping in APC (Table III). No intron substitutions in APC or MUTYH were predicted to be disease-associated polymorphisms.
Table II.
Computational predictions of deleterious missense mutations.
| A, APC | ||||
|---|---|---|---|---|
| Variant | Functional class | PolyPhen-2 | SIFT | SNPs&GO |
| c.2350T>A (p.Ser784Thr) | Missense mutation | BENIGN (0.017) | Tolerated (0.41) | Neutral (RI: 5) |
| c.2638A>C (p.Ile880Leu) | Missense mutation | BENIGN (0.376) | Tolerated (0.92) | Disease (RI: 6) |
| c.3511C>G (p.Arg1171Gly) | Missense mutation | BENIGN (0.000) | Tolerated (0.11) | Disease (RI: 1) |
| c.3775A>C (p.Ile1259Leu) | Missense mutation | BENIGN (0.008) | Tolerated (0.58) | Disease (RI: 7) |
| c.4557T>A (p.Asp1519Glu) | Missense mutation | N/A | Tolerated (0.16) | Disease (RI: 6) |
| c.4655A>T (p.Glu1552Val) | Missense mutation | BENIGN (0.104) | Tolerated (0.24) | Neutral (RI: 1) |
| c.6646C>A (p.Pro2216Thr) | Missense mutation | BENIGN (0.000) | Tolerated (0.12) | Neutral (RI: 0) |
| B, MUTYH | ||||
| Variant | Functional class | PolyPhen-2 | SIFT | SNPs&GO |
| c.553G>C (p.Glu185Gln) | Missense mutation | BENIGN (0.020) | Tolerated (0.08) | Neutral (RI: 8) |
| c.1232A>T (p.Gln411Leu) | Missense mutation | BENIGN (0.000) | Tolerated (0.10) | Neutral (RI: 9) |
| c.1268G>A (p.Arg423His) | Missense mutation | BENIGN (0.001) | Tolerated (0.12) | Neutral (RI: 4) |
| c.1483C>G (p.Gln495Glu) | Missense mutation | BENIGN (0.146) | Tolerated (1.00) | Neutral (RI: 9) |
PolyPhen-2, >0.50 is predicted to be damaging; SIFT, <0.05 is predicted to be deleterious; RI, prediction reliability scored between 0 (unreliable) and 10 (reliable). Disease, disease-associated polymorphism; neutral, neutral polymorphism; PolyPhen, polymorphism phenotype; SIFT, Sorting Intolerant From Tolerant; SNPs&GO, Single Nucleotide Polymorphisms & Gene Ontology; RI, reliability index; APC, adenomatous polyposis coli; MUTYH, mutY DNA glycosylase.
Table III.
Computational predictions of the effect on splicing of variants in each gene.
| A, APC | ||||||
|---|---|---|---|---|---|---|
| Variant | Functionalclass | ESE Finder | ESR Search | PESX | RESCUE_ESE | Splicingeffect |
| c.1458T>C(Tyr486Tyr) | Synonymousmutation | Changed | Changed | Changed | Notchanged | Exonskipping |
| c.4425G>A(Tyr1493Tyr) | Synonymousmutation | Changed | Changed | Changed | Notchanged | Frameshifting |
| c.5034G>A(Gly1678Gly) | Synonymousmutation | Changed | Changed | Changed | Changed | Frameshifting |
| c.5268T>G(Ser1756Ser) | Synonymousmutation | Changed | Changed | Changed | Changed | Frameshifting |
| c.5880G>A(Pro1960Pro) | Synonymousmutation | Changed | Changed | Changed | Changed | Frameshifting |
| c.220+35T>A | Intronmutation | N/A | N/A | N/A | N/A | Noeffect |
| c.645+129A>C | Intronmutation | N/A | N/A | N/A | N/A | Noeffect |
| c.1548+133C>A | Intronmutation | N/A | N/A | N/A | N/A | Noeffect |
| c.8532+99T>A | Intronmutation | N/A | N/A | N/A | N/A | Noeffect |
| B, MUTYH | ||||||
| Variant | Functionalclass | ESE Finder | ESR Search | PESX | RESCUE_ESE | Splicingeffect |
| c.423G>A(Glu141Glu) | Synonymousmutation | N/A | N/A | N/A | N/A | Noeffect |
| c.450C>A(Glu150Glu) | Synonymousmutation | N/A | N/A | N/A | N/A | Noeffect |
| c.1347G>C(Tyr449Tyr) | Synonymousmutation | N/A | N/A | N/A | N/A | Noeffect |
| c.264+11G>A | Intronmutation | N/A | N/A | N/A | N/A | Noeffect |
| c.304+23G>A | Intronmutation | N/A | N/A | N/A | N/A | Noeffect |
| c.420+35A>G | Intronmutation | N/A | N/A | N/A | N/A | Noeffect |
| c.1566+33_1566+35het_delTGT | Intronmutation | N/A | N/A | N/A | N/A | Noeffect |
APC, adenomatous polyposis coli; MUTYH, mutY DNA glycosylase; ESE, exonic splicing enhancer; ESR, exonic-splicing regulatory; PESX, putative exonic splicing enhancers.
Synonymous variant APC∆486s induces APC exon 12 skipping in a mini-gene splicing assay
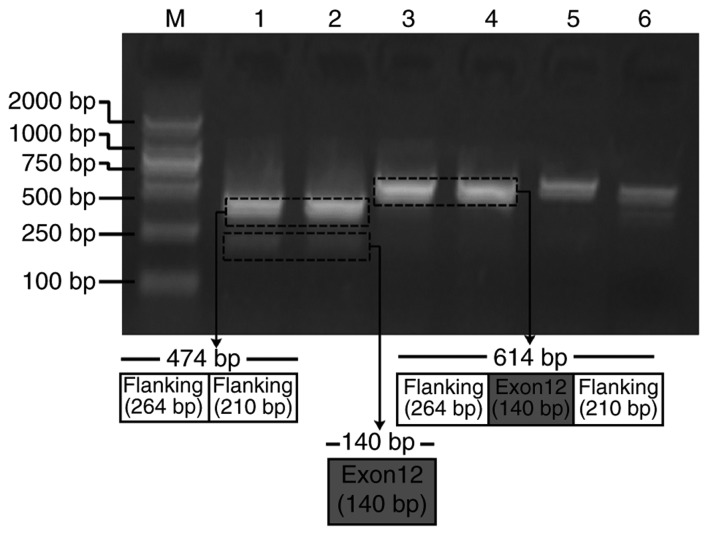
Due to the splicing impact predicted by bioinformatics analysis, APC∆486s was further studied to verify its exon splicing impact on the APC gene. Mini-gene systems encoding wild-type and mutant APC∆486s in APC exon 12 were synthesized and inserted into the pEGFP-N1 plasmid. The recombined constructs were subsequently transfected into HeLa cells, and the corresponding PCR products were separated by electrophoresis. The results revealed that the PCR product from the mutant mini-gene was smaller than that from the wild-type mini-gene (Fig. 3). Sequencing results of the two PCR products further confirmed the skipping of exon 12 in APC (data not shown). These results indicated that the mini-gene system encoding mutant APC∆486s induced the skipping of exon 12 in APC.
Figure 3.
Hybrid mini-gene assay results of the mini-gene system encoding wild-type and mutant APC∆486s in APC exon 12. Lane M indicates the DL2000 molecular marker ladder. Lanes 1 and 2 contain the electrophoresis results of the mutant-type mini-gene system encoding c.1458C. Lanes 3 and 4 contain the electrophoresis results of wild-type mini-gene system encoding c.1458T. APC, adenomatous polyposis coli; bp, base pairs.
Discussion
FAP has been linked to germline mutations in the APC gene. However, a number of patients with FAP have no APC mutations (1–3). Among the known genes, evidence has also implicated MUTYH mutations in the pathogenesis of FAP, which may explain up to 25% of all APC mutation-negative FAP cases (7–10). However, a large subset of families with FAP have undetectable pathogenic alterations in APC and MUTYH, and this condition is designated as APC/MUTYH mutation-negative FAP.
This apparent mutation negativity may suggest that APC has alterations that escape detection by routine techniques, and that other known or unknown genes are involved in FAP predisposition (8). Genetic counselling for these cases of FAP without an identified pathogenic alteration is frequently limited by a lack of knowledge about the pathogenic role of a large fraction of germline sequence variations (34).
In the present study, in order to investigate the germline mutations in Chinese patients with FAP, 13 patients with a classical clinical phenotype were examined by exon-specific DNA sequencing of APC and MUTYH to determine their micromutation types. Following the use of conventional analysis methods, only six types of clearly pathogenic mutations were identified in six individuals. Among the APC/MUTYH mutation-negative cases, seven missense mutations, five synonymous substitutions and three intron mutations were identified in APC, and four missense mutations, three synonymous substitutions and four intron mutations were identified in MUTYH.
Previous reports have indicated that certain missense mutations are deleterious (35). However, examining candidate missense mutations that may cause pathogenic alterations and identifying their relative effects on function is a time-consuming and labor-wasting process. Recently, computational prediction has been proposed as an efficient and economical strategy to screen for potential pathogenic missense mutations (36). By applying bioinformatics tools and databases in the present study, it was predicted that no identified missense mutations in APC or MUTYH were deleterious.
It was previously thought that all synonymous mutations were silent. However, further research into these mutations has demonstrated that silent mutations may alter protein expression, conformation and function (14). In 2007, it was demonstrated that synonymous mutations in transporter ATP binding cassette subfamily B member 1 may be implicated in drug resistance to chemotherapeutic agents, and it was further confirmed that synonymous SNPs (sSNPs) affect protein conformation and have functional and clinical consequences (37). Since then, a number of other synonymous mutations associated with human disease have been reported. For example, polymorphisms rs709816 and rs1061302 in gene NBN may be linked to smoking-associated cancer (38), and polymorphism rs11615 in ERCC1 aids in clinical outcome prediction following oxaliplatin-based chemotherapy in metastatic colorectal cancer (39). In addition, sSNPs rs2229069 and rs2227985 of the gene ABL1in the fusion protein BCR, RhoGEF and GTPase activating protein/ABL proto-oncogene 1, non-receptor tyrosine kinase, may contribute to primary, although not secondary, resistance to tyrosine kinase inhibitors (40). Furthermore, rs2293347 in gene EGFR may be a potential predictor of clinical outcome in patients with advanced non-small cell lung carcinoma when treated with gefitinib (41), and rs1045642 in gene ABCB1 has been associated with multidrug resistance to cancer chemotherapy (42).
Based on the above findings, the identified synonymous and intronic substitutions of APC and MUTYH were further analyzed by applying bioinformatics tools and databases, and the synonymous mutation Tyr486Tyr (APC∆486s) was predicted to have induced exon skipping in APC, which suggested that it may have caused pathogenic alterations that lead to FAP predisposition. Therefore, the synonymous substitution c.1458T>C was further studied to investigate its effect on exon splicing of the APC gene. A mini-gene system encoding wild-type or mutant c.1458T>C APC in exon 12 was synthesized and inserted into pEGFP-N1 plasmids. The synonymous substitution of APC∆486s was confirmed to induce a major splicing defect with skipping of exon 12 in APC.
In the present study, DNA sequencing of the APC and MUTYH genes in 13 patients with FAP was conducted. A total of four known pathogenic mutations in APC and two novel disease-associated pathogenic nonsense mutations in MUTYH were identified. For samples that did not present with obvious pathogenic alterations, the functional effects of the identified missense, synonymous and intronic mutations were analyzed with bioinformatics tools and databases. Bioinformatics analyses predicted that the synonymous mutation APC∆486s was likely a disease-causing polymorphism and that it may have induced exon skipping in APC. Using a hybrid mini-gene assay, synonymous SNP APC∆486s was demonstrated to induce a major splicing defect with the skipping of exon 12 in APC (Fig. 4).
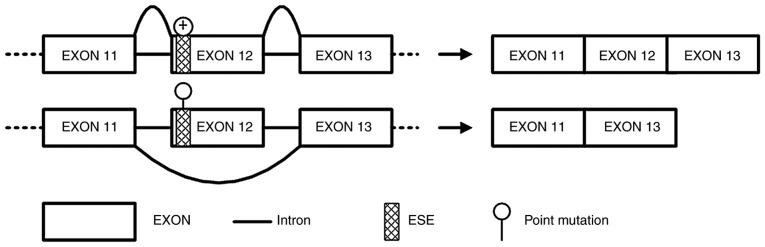
Figure 4.
Exon skipping impact induced by APC∆486s. Synonymous mutation of c.1458T>C induced a major splicing defect with skipping of exon 12 in APC, and the truncated APC protein results in FAP. APC, adenomatous polyposis coli; ESE, exon splicing enhancer; FAP, familial adenomatous polyposis.
Therefore, it was concluded that the synonymous mutation APC∆486s that affected exon splicing in APC was a pathogenic alteration that predisposed patients to APC/MUTYH mutation-negative FAP. However, a limitation to the present study was that the exon skipping effect of synonymous mutation of APC∆486s was predicted by bioinformatics analysis. This was the main focus of the present study; therefore, further ESE-dependent experiments and protein truncation tests are required to investigate the mechanism of exon skipping and to verify that this occurs in patients with FAP presenting with APC∆486s.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National Natural Science Foundation of China (grant nos. 81660472 and 81760511), the Scientific Research Foundation of Yunnan Province (grant no. 2015Y177), Yunnan Applied Basic Research Projects-Union Foundation (grant no. 2015FB035), the Scientific Research Project of Internal Research Institute in Yunnan Province (grant no. 2016NS009) and the Medical Candidate of Yunnan Province (grant no. H-201608).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
WQL, JY and JD conceived the present study. WQL, JY and YP carried out the bioinformatics prediction work and analyzed the data. WQL, JY, WLL and JD performed the experiments of hybrid mini-gene assay. All authors discussed the results and contributed to the final manuscript.
Ethics approval and consent to participate
The present study was approved by and performed in accordance with the Research Ethics Board of Kunming Medical University (Kunming, China). All participants provided written informed consent, and the ethics committees approved the consent procedure.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gardner EJ, Burt RW, Freston JW. Gastrointestinal polyposis: Syndromesand genetic mechanisms. West J Med. 1980;132:488–499. [PMC free article] [PubMed] [Google Scholar]
- 2.Kanth P, Grimmett J, Champine M, Burt R, Samadder NJ. Hereditary colorectal polyposis and cancer syndromes: A primer on diagnosis and management. Am J Gastroenterol. 2017;112:1509–1525. doi: 10.1038/ajg.2017.212. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler KW, Nilbert MC, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hamilton SR, Hedge P, Markham A, et al. Identification of a gene located at chromosome 5q21 that is mutated in colorectal cancers. Science. 1991;251:1366–1370. doi: 10.1126/science.1848370. [DOI] [PubMed] [Google Scholar]
- 4.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Identification and characterization of the familial adenomatous polyposis coligene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 5.Syngal S, Brand RE, Church JM, Giardiello FM, Hampel HL, Burt RW, American College of Gastroenterology ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol. 2015;110:223–262. doi: 10.1038/ajg.2014.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boman BM, Fields JZ. An APC: WNT counter-current-like mechanism regulates cell division along the human colonic crypt Axis: A mechanism that explains how APC mutations induce proliferative abnormalities that drive colon cancer development. Front Oncol. 2013;3:244. doi: 10.3389/fonc.2013.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C->T: A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 8.Renkonen ET, Nieminen P, Abdel-Rahman WM, Moisio AL, Järvelä I, Arte S, Järvinen HJ, Peltomäki P. Adenomatous polyposis families that screen APC mutation-negative by conventional methods are genetically heterogeneous. J Clin Oncol. 2005;23:5651–5659. doi: 10.1200/JCO.2005.14.712. [DOI] [PubMed] [Google Scholar]
- 9.Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RK, Bisgaard ML, Orntoft TF, Aaltonen LA, Hodgson SV, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 10.Ricci MT, Salemme M, Villanacci V, Varesco L. The genetics of inherited predispositions to colorectal polyps: A quick guide for clinicians. Colorectal Dis. 2015;17(Suppl 1):S3–S9. doi: 10.1111/codi.12814. [DOI] [PubMed] [Google Scholar]
- 11.Wei SC, Su YN, Tsai-Wu JJ, Wu CH, Huang YL, Sheu JC, Wang CY, Wong JM. Genetic analysis of the APC gene in Taiwanese familial adenomatous polyposis. J Biomed Sci. 2004;11:260–265. doi: 10.1007/BF02256569. [DOI] [PubMed] [Google Scholar]
- 12.Prior TW, Bridgeman SJ. Identifying mutations for MYH-associated polyposis. Curr Protoc Hum Genet Chapter. 2010;10 doi: 10.1002/0471142905.hg1013s64. Unit 10.13. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Liu WQ, Li WL, Chen C, Zhu Z, Hong M, Wang ZQ, Dong J. Investigating polymorphisms by bioinformatics is a potential cost-effective method to screen for germline mutations in Chinese familial adenomatous polyposis patients. Oncol Lett. 2016;12:421–428. doi: 10.3892/ol.2016.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 15.Plotkin JB, Kudla G. Synonymous but not the same: The causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156:1324–1335. doi: 10.1016/j.cell.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 17.Chiang JM, Chen HW, Tang RP, Chen JS, Changchien CR, Hsieh PS, Wang JY. Mutation analysis of the APC gene in Taiwanese FAP families: Low incidence of APC germline mutation in a distinct subgroup of FAP families. Fam Cancer. 2010;9:117–124. doi: 10.1007/s10689-009-9292-2. [DOI] [PubMed] [Google Scholar]
- 18.Minton JA, Flanagan SE, Ellard S. Mutation surveyor: Software for DNA sequence analysis. Methods Mol Biol. 2011;688:143–153. doi: 10.1007/978-1-60761-947-5_10. [DOI] [PubMed] [Google Scholar]
- 19.Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The human gene mutation database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap project web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan M, Cong P, Wang Y, Lin C, Yuan Y, Dong J, Banerjee S, Zhang T, Chen Y, Zhang T, et al. Novel LOVD databases for hereditary breast cancer and colorectal cancer genes in the Chinese population. Hum Mutat. 2011;32:1335–1340. doi: 10.1002/humu.21588. [DOI] [PubMed] [Google Scholar]
- 24.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capriotti E, Calabrese R, Fariselli P, Martelli PL, Altman RB, Casadio R. WS-SNPs&GO: A web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genomics. 2013;14(Suppl 3):S6. doi: 10.1186/1471-2164-14-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeo G, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. In the Proceeding of Proc. Natl Acad Sci. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 30.Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods. 2005;37:331–340. doi: 10.1016/j.ymeth.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 31.Gómez-Fernández N, Castellví-Bel S, Fernández-Rozadilla C, Balaguer F, Muñoz J, Madrigal I, Milà M, Graña B, Vega A, Castells A, et al. Molecular analysis of the APC and MUTYH genes in Galician and Catalonian FAP families: A different spectrum of mutations? BMC Med Genet. 2009;10:57. doi: 10.1186/1471-2350-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kashfi SM, Farahbakhsh Behboudi F, Golmohammadi M, Mojarad Nazemalhosseini E, Azimzadeh P, Aghdaie Asadzadeh H. Frameshift mutations (deletion at codon 1309 and codon 849) in the APC gene in iranian FAP patients: A c series and review of the literature. Int J Mol Cell Med. 2014;3:196–202. [PMC free article] [PubMed] [Google Scholar]
- 33.Mohamed Z, Ahmad R, Yoke NS, Zakaria Z, Ahmad H, Yew TH. A nonsense mutation in exon 8 of the APC gene (Arg283Ter) causes clinically variable FAP in a Malaysian Chinese family. Cancer Sci. 2003;94:725–728. doi: 10.1111/j.1349-7006.2003.tb01509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Liu QW, Li LW, Wang QZ, Hong M, Dong J. Familial adenomatous polyposis in China. Oncol Lett. 2016;12:4877–4882. doi: 10.3892/ol.2016.5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menéndez M, González S, Obrador-Hevia A, Domínguez A, Pujol MJ, Valls J, Canela N, Blanco I, Torres A, Pineda-Lucena A, et al. Functional characterization of the novel APC N1026S variant associated with attenuated familial adenomatous polyposis. Gastroenterology. 2008;134:56–64. doi: 10.1053/j.gastro.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 36.Sucularli C, Arslantas M. Computational prediction and analysis of deleterious cancer associated missense mutations in DYNC1H1. Mol Cell Probes. 2017;34:21–29. doi: 10.1016/j.mcp.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 38.Park SL, Bastani D, Goldstein BY, Chang SC, Cozen W, Cai L, Cordon-Cardo C, Ding B, Greenland S, He N, et al. Associations between NBS1 polymorphisms, haplotypes and smoking-related cancers. Carcinogenesis. 2010;31:1264–1271. doi: 10.1093/carcin/bgq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang J, Jiang T, Yao RY, Liu ZM, Lv HY, Qi WW. The combination of ERCC1 and XRCC1 gene polymorphisms better predicts clinical outcome to oxaliplatin-based chemotherapy in metastatic colorectal cancer. Cancer Chemother Pharmacol. 2010;66:493–500. doi: 10.1007/s00280-009-1186-3. [DOI] [PubMed] [Google Scholar]
- 40.Ernst T, Hoffmann J, Erben P, Hanfstein B, Leitner A, Hehlmann R, Hochhaus A, Müller MC. ABL single nucleotide polymorphisms may masquerade as BCR-ABL mutations associated with resistance to tyrosine kinase inhibitors in patients with chronic myeloid leukemia. Haematologica. 2008;93:1389–1393. doi: 10.3324/haematol.12964. [DOI] [PubMed] [Google Scholar]
- 41.Ma F, Sun T, Shi Y, Yu D, Tan W, Yang M, Wu C, Chu D, Sun Y, Xu B, Lin D. Polymorphisms of EGFR predict clinical outcome in advanced non-small-cell lung cancer patients treated with Gefitinib. Lung Cancer. 2009;66:114–119. doi: 10.1016/j.lungcan.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 42.Fung KL, Gottesman MM. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim Biophys Acta. 2009;1794:860–871. doi: 10.1016/j.bbapap.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.