Abstract
Small ubiquitin-like modifier proteins are involved in tumorigenesis; however, the potential effects and functions of the family member ubiquitin-like modifier-activating enzyme 2 (UBA2) on colorectal cancer are not clear. The present study aimed to examine the effects of UBA2 on the proliferation of colorectal cancer cells in vitro and in vivo. The mRNA and protein expression levels of UBA2 in patients with colorectal cancer were measured by reverse transcription-quantitative polymerase chain reaction and immunohistochemistry, respectively. UBA2 expression levels in colorectal cancer tissues were significantly increased compared with the paracancerous normal tissues. The expression of UBA2 was also associated with higher stage colorectal cancer and poor prognosis. MTT and colony formation assays were used to examine proliferation in colorectal cancer cell lines. Flow cytometry was performed to examine the effects of UBA2 on the cell cycle and apoptosis of colorectal cancer cell lines and protein expression levels were examined by western blotting. Athymic nude mice were used to examine the ability of transfected colorectal cancer cells to form tumors in vivo. Downregulation of UBA2 inhibited the proliferation of colorectal cancer cell lines in vitro and in vivo through the regulation of cell cycle associated protein expression and apoptosis. Furthermore, downregulation of UBA2 decreased the expression levels of cyclin B1, B-cell lymphoma-2, phosphorylated protein kinase B and E3 ubiquitin-protein ligase MDM2 in colorectal cancer cells, whereas the expression levels of p21 and p27 were increased. UBA2 was demonstrated to serve an essential role in the proliferation of colorectal cancer and may be used as a potential biomarker to predict prognosis and as a therapeutic target in colorectal cancer.
Keywords: ubiquitin-like modifier activating enzyme 2, colorectal cancer, cell proliferation, apoptosis
Introduction
Colorectal cancer is the third most common cancer worldwide (1); the lifetime risk of developing colorectal cancer is 4.7% for men and 4.4% for women. Although the mortality rate from colorectal cancer has been declining for several decades owing to the early diagnosis and improved treatment, >1 million novel cases are diagnosed each year. Therefore, it is crucial to identify novel biomarkers and therapeutic targets for colorectal cancer to improve the prognosis of the disease.
Sumoylation is a transient post-translational modification process that is highly regulated by the balance between enzyme-mediated conjugating and deconjugating activities. Small ubiquitin-like modifier (SUMO) conjugation involves an associated enzymatic cascade that includes an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme and an E3 ubiquitin ligase (2). It has been demonstrated that sumoylation is associated with a range of cellular processes, including cell cycle progression, apoptosis regulation, maintenance of genome integrity, DNA repair, cell survival, modulation of subcellular transport and transcription (3,4). A previous study reported that sumoylation regulates the cell cycle in glioblastoma by stabilizing cyclin-dependent kinase (CDK) 6 (5). Other studies have demonstrated the interaction between CDKs and sumoylation during cell cycle regulation (6,7). The imbalance between sumoylation and desumoylation is associated with a variety of diseases, such as cancer (8–10). For example, sumo-conjugating enzyme UBC9 is upregulated in a number of types of cancer, including prostate cancer, breast cancer, advanced melanomas and colon cancer (11,12). UBA2/SAE2 was reported to promote the growth of colon tumors in mice (13), and tumor aggressiveness was associated with increased MDM2 expression induced by the upregulation of SUMO1 in patients with oral squamous cell carcinoma (14).
Ubiquitin-like modifier-activating enzyme 2 (UBA2), also known as sumo-activating enzyme subunit 2 (SAE2), is a subcomponent of the sumoylated E1 enzyme. The human UBA2 gene is located on chromosome19q12, which is one of the most important enzymes that directly affect the levels of sumoylation in the body (3,15,16). A number of previous studies have implicated UBA2 in clinical diseases; for example, haploinsuffiency of the UBA2 gene is associated with cutis aplasia (17,18). In addition, UBA2 was reported to form a fusion protein with the DNA dC>dU-editing enzyme APOBEC3G to prevent the degradation of the APOBEC3G protein by human immunodeficiency virus (HIV)-viral infectivity factor (19). Therefore, UBA2 may serve an inhibitory role in HIV infectivity, which suggested that UBA2 may exert its biological function by stabilizing certain proteins. Recent studies have demonstrated that UBA2 was highly expressed in certain malignant diseases, including liver cancer (20), small cell lung cancer (21) and gastric cancer (22). Another study reported that the expression levels of UBA2 were different in metastatic colorectal cancer compared with the primary tumor (23). However, the function and clinical significance of UBA2 in colorectal cancer are not clear.
In the present study, the expression level of UBA2 was examined in patients with colorectal cancer, and UBA2 expression was associated cancer stage. Furthermore, the function of UBA2 on cell proliferation in vitro and in vivo was examined. The results of the present study indicated that UBA2 may serve important roles in tumorigenesis and may act as a potential therapeutic target in colorectal cancer.
Materials and methods
Patients
Colorectal cancer tissues were collected from 237 patients with colorectal cancer in Bethune First Hospital of Jilin University (Changchun, China) between January 2009 and December 2015; patient clinicopathological characteristics are presented in Table I. Patients who received preoperative radiotherapy or chemotherapy and patients with multiple tumors were excluded from the present study. The pathological stage of cancer was determined according to the American Joint Committee on Cancer stage. All tumors were confirmed by a pathologist and have been collected intraoperatively. The fresh specimens for molecular analysis were immediately frozen in liquid nitrogen and stored in −80°C. Specimens for immunohistochemistry (IHC) were fixed in formalin for 24 h at room temperature and embedded in paraffin for further experiments. The present study was approved by the Ethics Review Committee at Jilin University. All patients were informed their participation rights and signed the written informed consent.
Table I.
Clinicopathological characteristics of patients with colorectal cancer.
| UBA2 expression | ||||
|---|---|---|---|---|
| Clinicopathological feature | Negative | Positive | Total | P-value |
| Number of patients | 35 | 202 | 237 | |
| Age (years) | 0.110 | |||
| <61 | 20 | 86 | 106 | |
| ≥61 | 15 | 116 | 131 | |
| Sex | 0.058 | |||
| Male | 24 | 98 | 122 | |
| Female | 11 | 104 | 115 | |
| Tumor location | <0.001 | |||
| Ascending colon | 6 | 87 | 93 | |
| Descending colon | 25 | 112 | 137 | |
| Transverse colon | 4 | 3 | 7 | |
| Histological grade | <0.001 | |||
| Well and moderate | 14 | 177 | 191 | |
| Poor and other | 21 | 25 | 46 | |
| Depth of invasion | 0.546 | |||
| T1-T2 | 2 | 13 | 15 | |
| T3 | 23 | 148 | 171 | |
| T4 | 10 | 41 | 51 | |
| Lymph node metastasis | 0.056 | |||
| Absent | 14 | 116 | 130 | |
| Present | 21 | 86 | 107 | |
| TNM stage | 0.041 | |||
| I–II | 23 | 95 | 118 | |
| III–IV | 12 | 107 | 119 | |
TNM, tumor, node, metastasis; UBA2, ubiquitin like modifier activating enzyme 2.
Bioinformatic analysis
Analysis of UBA2 mRNA expression levels was performed using datasets containing 334 colorectal cancer specimens and 28 normal colorectal specimens were obtained from the Cancer Genome Atlas via the starBase online database (http://starbase.sysu.edu.cn) (24,25).
Total RNA extraction
Total RNA from ~100 mg tissue and 5×106 cells of 7 colorectal cancer cell lines was extracted using TRIzol® Reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according the manufacturer's protocol. Briefly, the frozen specimens were placed in a microcentrifuge tube with 1 ml TRIzol and homogenized with a disposable plastic pestle. The specimens were incubated at room temperature for 5 min, and subsequently centrifuged at 12,000 × g for 10 min at 4°C. The supernatants were transferred to a fresh microcentrifuge tube and 200 µl chloroform was added for phase separation. RNA was precipitated by adding pre-chilled isopropanol. The specimens were centrifuged at 12,000 × g for 10 min at 4°C and the pellets were washed with 75% ethanol. The concentrations and purity of the extracted RNA were measured on a spectrophotometer at 260 and 280 nm wavelengths.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay
Total RNA from frozen colorectal cancer tissues or cell cultures was reverse transcribed into cDNA using a Reverse Transcriptase kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's protocol. UBA2 mRNA expression levels were measured by qPCR using 12.5 µl SYBR-Green (cat. no. DRR041B; Takara Biotechnology Co., Ltd., Dalian, China) 1 µl each primer, 2 µl cDNA and 8.5 µl diethyl pyrocarbonate-treated water. The thermocycling conditions were as follows: Initial denaturation at 95°C for 15 sec, followed by 40 cycles of 95°C (5 sec) and 60°C (30 sec). The primers used were as follows: GAPDH, forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′; UBA2, forward, 5′-CACAGGTTGCCAAGGAA-3′ and reverse, 5′-GACACTCATAACACTCGGTCA-3′; GAPDH was used as an internal control. UBA2 mRNA expression levels were quantified using the 2−ΔΔCq method and normalized to the expression levels of GAPDH (26). All experiments were performed three times in triplicate.
IHC analysis
IHC was performed to examine the protein expression of UBA2 in colorectal cancer specimens. The specimen sections (5 µm) were deparaffinized using xylene for 5 min at room temperature three times, which was followed by rehydration using 100% ethanol for 10 min, 95% ethanol for 10 min and water for 10 min. Antigen retrieval via steam treatment in citrate buffer was then performed at 120°C for 10 min. Endogenous peroxidase activity was blocked by 3% hydrogen peroxide at room temperature for 10 min. Non-specific protein binding was blocked by 10% normal sheep serum for 30 min at room temperature (Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China). Sections were incubated with anti-UBA2 primary antibody (1:100; Abcam, Cambridge, MA, USA; cat. no. ab185955) at 4°C overnight. Subsequently, the sections were incubated with a secondary antibody conjugated with HRP [cat. no. KIT-9706; UltraSensitive™ SP (Rabbit) IHC kit; Fuzhou Maixin Biotech Co., Ltd.] for 40 min at room temperature. The high-sensitivity 3,3-diaminobenzidine chromogenic substrate system was used for colorimetric visualization. In addition, specimen sections were also stained with 0.2% hematoxylin at room temperature for 5 min, 1% acid alcohol at room temperature for 30 sec and 0.5% eosin at room temperature for 1 min. The slides were examined using a light microscope (BX53F; Olympus Corporation, Tokyo, Japan). The paracancerous normal tissues were used as negative controls. The magnification was ×100 and 25 visual fields were analyzed.
Cell culture
The human colorectal cancer cell lines RKO, HCT116, HT-29, DLD-1, SW480, SW620 and LoVo were purchased from Shanghai GeneChem Co., Ltd., (Shanghai, China). The cells were routinely grown in RPMI-1640 (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal calf serum and 1% penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in a humidified incubator with 5% CO2.
Cell proliferation by MTT assay
RKO and HCT116 colorectal cancer cells at 70–80% confluency were transfected with UBA2-targeting small interfering (si)RNA (Invitrogen; Thermo Fisher Scientific, Inc.). The UBA2-siRNA sequence was 5′-GCCCGAAACCATGTTAATAGA-3′, and a scrambled siRNA (5′-GCCTAACTGTGTCAGAAGGAA-3′) was used as the negative control (NC-siRNA). siRNAs and Lipofectamine® 2000 (Gibco; Thermo Fisher Scientific, Inc.) were diluted in OPTI-MEM (Gibco; Thermo Fisher Scientific, Inc.) separately prior to being mixed. The siRNA-Lipofectamine 2000 mixture at a ratio of 1:0.1 (pmol:µl) was incubated for 30 min at room temperature, added to the colorectal cancer cell cultures for 6–8 h in a 37°C, 5% CO2 incubator and replaced with fresh culture medium. A total of 72 h post-transfection, proliferation was measured by MTT assay. Briefly, the transfected cancer cells were seeded in 96-well plates at a density of 1×104 cells/well. Following overnight incubation, MTT (5 mg/ml) was added to each well without replacing the medium. Following 4 h incubation, the medium was aspirated followed by the addition of 100 µl dimethyl sulfoxide into each well to dissolve the purple formazan crystals; the plates were agitated for 5–10 min. The optical density values were measured at 490 nm using a microplate reader. All experiments were performed three times in triplicate.
Determination of cell colony formation
To examine the effects of UBA2 on colony formation, 800 RKO and HCT116 colorectal cancer cells transfected with UBA2-siRNA or NC-siRNA were suspended in culture medium containing 10% FBS and seeded in 6-well plates; cells were cultured at 37°C in a humidified incubator with 5% CO2. Fresh complete culture medium was changed every 3 days, and the cells were grown for 14 days. Subsequently, the cells were washed with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature and stained with 500 µl clean, particle-free GIEMSA staining solution for 20 min at room temperature. Colonies were washed with PBS and air-dried. Images were captured with a digital camera. All experiments were performed three times in triplicate.
Cell cycle analysis by fluorescence-activated cell sorting
To determine the effects of UBA2 on the cell cycle, RKO and HCT116 colorectal cancer cells (1×106) transfected with UBA2-siRNA or NC-siRNA were harvested and washed in PBS (pH=7.2–7.4) followed by fixation in pre-chilled 70% ethanol for 1 h at −20°C. The cells were treated with RNase (2 U/ml) and stained with propidium iodide (50 µg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in the dark for 30 min at room temperature. The cell cycle was determined by flow cytometry analysis (NovoCyte Benchtop flow cytometer; ACEA Biosciences, San Diego, CA, USA) and NovoExpress software (version 1.2.1; ACEA Biosciences, San Diego, CA, USA). All experiments were performed three times in triplicate.
Apoptosis analysis by flow cytometry
To determine the effects of UBA2 on apoptosis, 5×105 RKO and HCT116 colorectal cancer cells transfected with UBA2-siRNA and NC-siRNA were harvested and washed with PBS. The cells were washed with binding buffer at room temperature and then incubated with Annexin V-APC (cat. no. 88-8007; eBioscience; Thermo Fisher Scientific, Inc.) in the dark at room temperature for 15 min prior to flow cytometric analysis. All experiments were performed three times in triplicate.
Western blotting
To detect the effects of UBA2 on target protein expression, western blotting was performed using the primary antibodies listed in Table II, as described previously (2). Cells (1×106) transfected with UBA2-siRNA and NC-siRNA were harvested and lysed with lysis buffer (cat. no. 9803; Cell Signaling Technology, Inc., Danvers, MA, USA). Protein concentration was analyzed using a protein assay kit (5000007) with bovine serum albumin standards according to the manufacturer's instructions (5000002) (both from Bio-Rad Laboratories, Inc., Hercules, CA, USA). Proteins (30 µg per lane) were separated by 10 or 15% SDS-PAGE gel and transferred to a polyvinylidene difluoride membrane. GAPDH was used as loading control. The densitometry of protein band was analyzed with ImageJ software (version 1.48v; National Institutes of Health, Bethesda, MD, USA).
Table II.
Antibodies used in western blotting.
| Protein | Vendor | Cat. no. | Dilution |
|---|---|---|---|
| UBA2 | Abcam (Cambridge, MA, USA) | ab185955 | 1:500 |
| p21 | Abcam | ab227443 | 1:300 |
| p27 | Abcam | ab137736 | 1:300 |
| Cyclin B1 | Cell Signaling Technology, Inc. (Danvers, MA, USA) | 4138 | 1:500 |
| Bcl-2 | Abcam | ab194583 | 1:300 |
| MDM2 | Abcam | ab226939 | 1:200 |
| p-AKT | Abcam | ab38449 | 1:300 |
| GAPDH | Cell Signaling Technology, Inc. | 2118 | 1:2,000 |
Bcl-2, B-cell lymphoma-2; MDM2, E3 ubiquitin-protein ligase MDM2; p-AKT, phosphorylated protein kinase B; UBA2, ubiquitin-like modifier-activating enzyme 2.
In vivo xenograft
All animal experiments were performed according to the approved protocols of the Animal Care and Use Committee at Jilin University. Female BALB/c nude mice (6 weeks; weight, 20–22 g; n=12) were purchased from Jilin University Animal Center and were housed under specific pathogen free conditions at 20–26°C, 40–70% humidity and a 12/12 h light/dark cycle. The animals were acclimated for 1 week prior to start of the experiment, could move freely and had free access to food and water. A total of 6 mice were subcutaneously injected with 2.0×106 RKO tumor cells transfected with UBA2-siRNA or NC-siRNA mixed with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) into left flank of the mice respectively to form a single tumor. Tumor growth and body weight were monitored every 2 days for 2 weeks, and tumor volume was measured in three dimensions using calipers on alternate days. The tumor volume (V) was calculated using the formula: V=π/6 × L × W × H; where W is width, L is length and H is height.
Statistical analysis
All data were analyzed by SPSS 21.0 (IBM Corp., Armonk, NY, USA). Clinicopathological features were evaluated using Pearson's χ2 test. The differences between groups were examined by the Student's t-test or the analysis of variance followed by a Tukey post-hoc test. For the IHC study, the Fisher's exact test was performed for statistical analysis. Overall survival rate was determined by the Kaplan-Meier method; log-rank test was used for statistical analysis of survival curves. P<0.05 was considered to indicate a statistically significant difference. All the data are presented as the mean ± standard deviation.
Results
UBA2 expression is increased and associated with prognosis in colorectal cancer
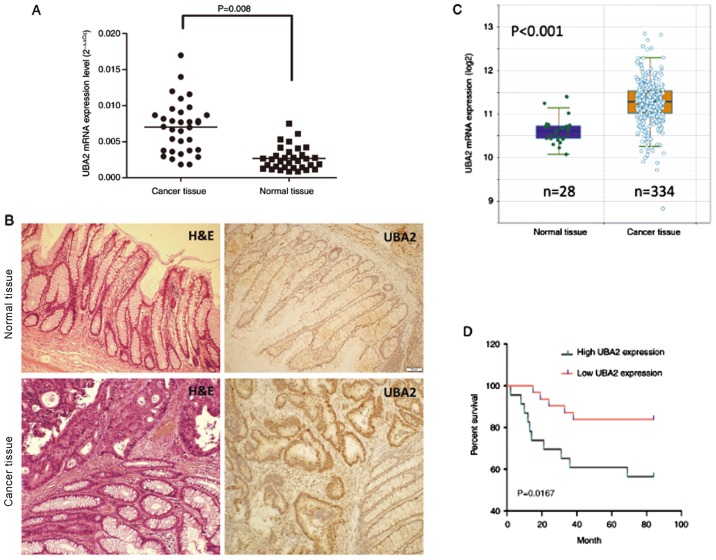
Colorectal cancer tissues and normal tissues were collected from patients with colorectal cancer, and RT-qPCR was performed to examine the UBA2 mRNA expression levels. UBA2 mRNA expression levels were significantly higher in cancer tissues compared with the normal tissues (P=0.008; Fig. 1A). UBA2 protein expression was also examined in the cancer tissues by IHC, which demonstrated that UBA2 expression was notably increased compared with the paracancerous tissues (Fig. 1B). UBA2 mRNA expression levels in 334 colorectal cancer specimens and 28 normal colorectal specimens from the Cancer Genome Atlas obtained through the starBase online database (http://starbase.sysu.edu.cn; Fig. 1C) were further analyzed; and the results suggested that UBA2 mRNA expression levels were significantly increased in cancerous tissues compared with normal tissues (P<0.001).
Figure 1.
UBA2 expression in colorectal cancer. (A) UBA2 mRNA expression levels were examined in cancerous and paracancerous tissue by reverse transcription-quantitative polymerase chain reaction. (B) H&E staining and UBA2 immunohistochemistry of normal colorectal tissue and colorectal cancer tissue (magnification, ×100). (C) Analysis of UBA2 expression in colorectal cancer tissue and normal colorectal tissue using data obtained from the starBase database. (D) Survival rates of patients with colorectal cancer with different expression levels of UBA2. UBA2, ubiquitin-like modifier-activating enzyme 2; H&E, hematoxylin and eosin.
To determine the effects of UBA2 expression on the survival rate of patients with colorectal cancer, 237 colorectal cancer patients were followed up for 84 months. Patients with higher UBA2 protein expression levels exhibited a significantly decreased survival rate compared with those patients expressing lower UBA2 levels (P<0.05; Fig. 1D). In addition, UBA2 protein expression was revealed to be associated with the location of tumor, histological grade and tumor node and metastasis stage (Table I). In addition, UBA2 protein expression is enhanced in the ascending and descending colon compared with the transverse colon, and also enhanced in tumors without lymph node metastasis compared with tumors with lymph node metastasis (Table I). No significant associations were made between UBA2 expression and age, sex, lymph node and depth of invasion (Table I).
Downregulation of UBA2 inhibits colorectal cancer cell proliferation and colony formation
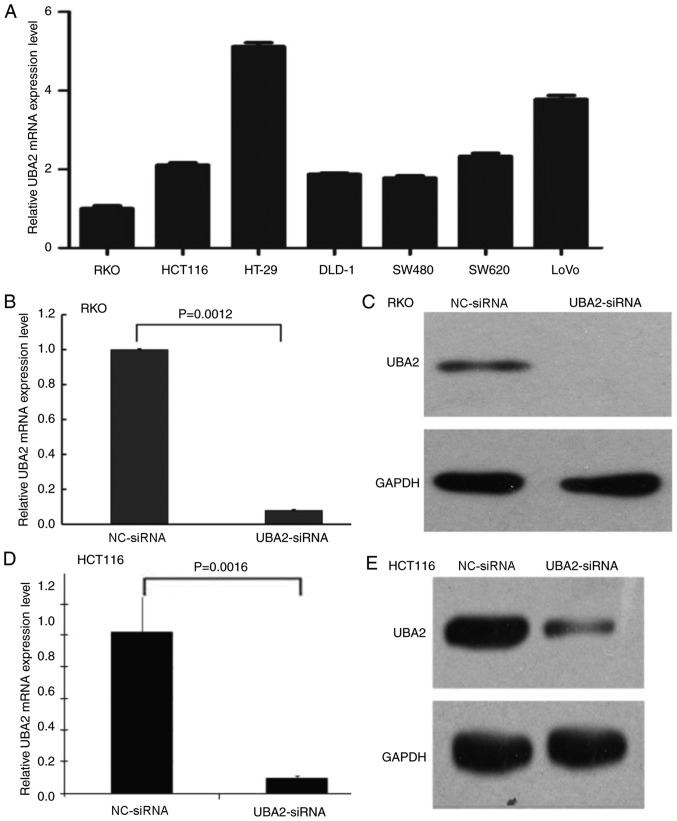
To investigate the effects of UBA2 on colorectal cancer cell proliferation, MTT and colony formation assays were performed using colorectal cancer cells transfected with UBA2-siRNA and NC-siRNA. First, the mRNA expression levels of UBA2 in different colorectal cancer cell lines were examined (Fig. 2A); UBA2 was expressed in all seven selected cell lines, including RKO, HCT116, HT-29, DLD-1, SW480, SW620 and LoVo cells. In order to compare the data generated in the present study with previous studies (27,28), RKO and HCT116 cells were selected to determine the role of UBA2 on cell proliferation. UBA2-siRNA transfection notably reduced the mRNA and protein expression levels of UBA2 in RKO cells and HCT116 cells (Fig. 2B-E).
Figure 2.
UBA2 expression levels in colorectal cancer cell lines. (A) UBA2 mRNA expression levels in colon cancer cell lines were detected by RT-qPCR. (B) UBA2 mRNA expression levels in RKO cells transfected with UBA2-siRNA or NC-siRNA. (C) UBA2 protein expression levels in RKO cells transfected with UBA2-siRNA or NC-siRNA were detected by western blotting. (D) UBA2 mRNA expression levels in HCT116 cells transfected with UBA2-siRNA or NC-siRNA were detected by RT-qPCR. (E) UBA2 protein expression levels in HCT116 cells transfected with UBA2-siRNA or NC-siRNA were detected by western blotting. All experiments were performed three times in triplicate. NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; siRNA, small interfering; UBA2, ubiquitin like modifier activating enzyme 2.
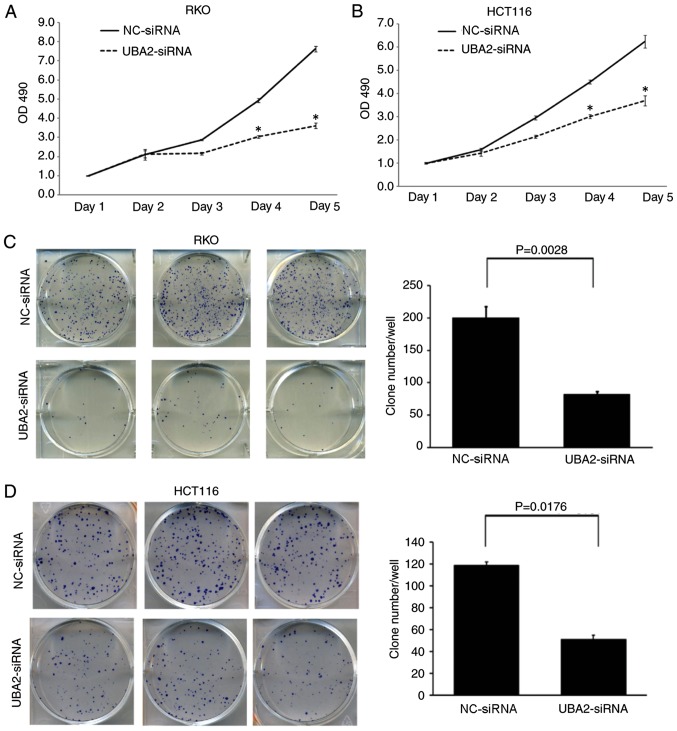
Downregulation of UBA2 significantly inhibited the proliferation of RKO and HCT116 cells compared with NC-siRNA-transfected cells at day 4 and day 5 time points (P<0.05; Fig. 3A and B). Furthermore, downregulation of UBA2 in RKO and HCT116 cells significantly decreased the colony formation capacity compared with the control cells (P<0.05; Fig. 3C and D).
Figure 3.
Downregulation of UBA2 inhibited colorectal cancer cell proliferation and colony formation. (A) Proliferation of RKO cells transfected with UBA2-siRNA or NC-siRNA. (B) Proliferation of HCT116 cells transfected with UBA2-siRNA or NC-siRNA. *P<0.05 vs. the NC-siRNA. (C) Colony formation capacity of RKO cells transfected with UBA2-siRNA or NC-siRNA. (D) Colony formation capacity of HCT116 cells transfected with UBA2-siRNA or NC-siRNA. All experiments were performed three times in triplicate.
Downregulation of UBA2 induces cell cycle arrest and apoptosis in colorectal cancer cells
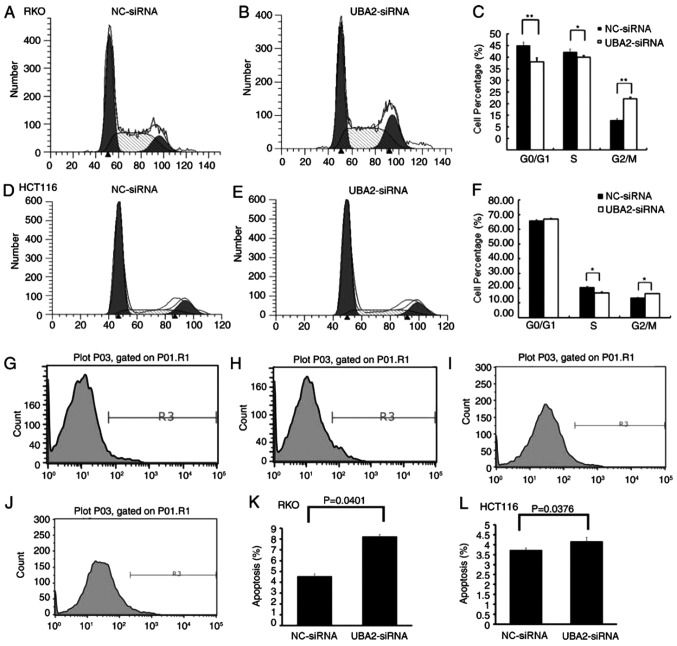
The effects of UBA2 on the cell cycle and apoptosis in colorectal cancer cells were examined by flow cytometry. Compared with NC-siRNA-treated cells, RKO and HCT116 cells transfected with UBA2-siRNA exhibited significant reductions in the number of cells in S phase, whereas the number of G2 phase cells was significantly increased (P<0.05; Fig. 4A-F). These results indicated that UBA2 induced G2/M arrest in these colorectal cancer cell lines. Downregulation of UBA2 significantly increased the apoptotic rates in RKO cells and HCT116 cells compared with NC-siRNA-treated cells (P<0.05; Fig. 4G-L). These results suggested that UBA2 might inhibit apoptosis in colorectal cells.
Figure 4.
Effects of UBA2 on the cell cycle and apoptosis. (A and B) Cell cycle analysis by flow cytometry in RKO cells transfected with (A) NC-siRNA and (B) UBA2-siRNA. (C) Relative quantity of cells in G0/G1, S and G2/M phases in the RKO cells transfected with UBA2-siRNA or NC-siRNA. (D and E) Cell cycle analysis by flow cytometry in HCT116 cells transfected with (D) NC-siRNA and (E) UBA2-siRNA. (F) Relative quantity of cells in G1/G1, S and G2/M phases in the HCT116 cells transfected with UBA2-siRNA or NC-siRNA. (G-J) Apoptosis analysis by flow cytometry in (G) RKO cells transfected with NC-siRNA, (H) RKO cells transfected with UBA2-siRNA, (I) HCT116 cells transfected with NC-siRNA, and (J) HCT116 cells transfected with UBA2-siRNA. (K and L) Apoptotic rates calculated from data in RKO cells and HCT116 cells. *P<0.05, **P<0.01 vs. the NC-siRNA. All experiments were performed three times in triplicate.
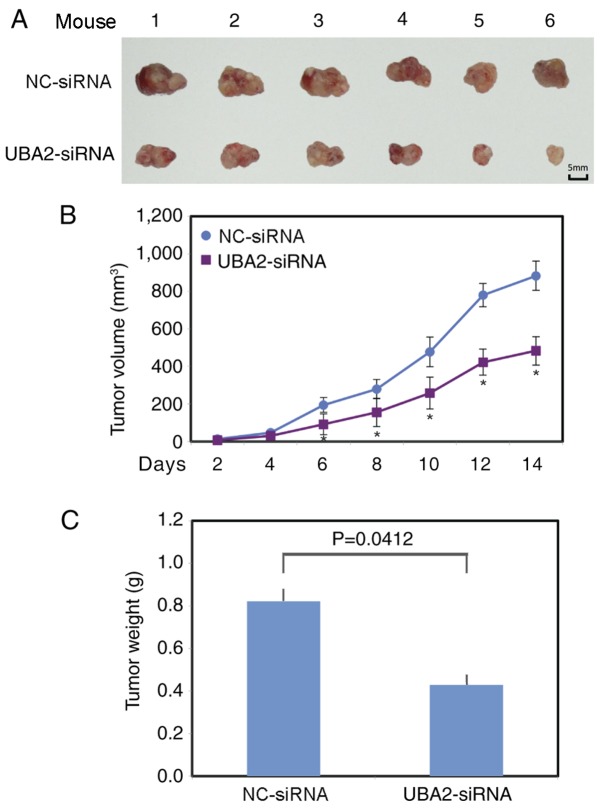
UBA2 promotes tumor growth in vivo
To further examine the function of UBA2 on colorectal cancer growth, UBA2-siRNA or NC-siRNA transfected RKO cells were injected subcutaneously into the left flank of the athymic nude mice. Tumor volume was measured every other day. The largest tumor diameter was ~14.9 mm in the control group and ~13 mm in the UBA2 downregulation group. A total of 2 weeks following xenotransplantation, the mice (weight, 25–28 g) were sacrificed and the tumors from each animal were collected (Fig. 5A). The tumors grew significantly more slowly in the UBA2-siRNA RKO cell group, compared with the NC-siRNA RKO cell group (P<0.05; Fig. 5B). The average tumor weight was also significantly lower in the UBA2-siRNA RKO cell group compared with the NC-siRNA RKO cell group (P<0.05; Fig. 5C).
Figure 5.
Downregulation of UBA2 inhibits tumor growth in vivo. (A) Tumors in the NC-siRNA and UBA2-siRNA groups were isolated from mice at the endpoint of experiments. (B) Tumor volume was monitored in the NC-siRNA and UBA2-siRNA groups for 2 weeks. *P<0.05 vs. the NC-siRNA. (C) Tumor weight was monitored in the NC-siRNA group and UBA2-siRNA group at the endpoint of experiments.
Target protein expression
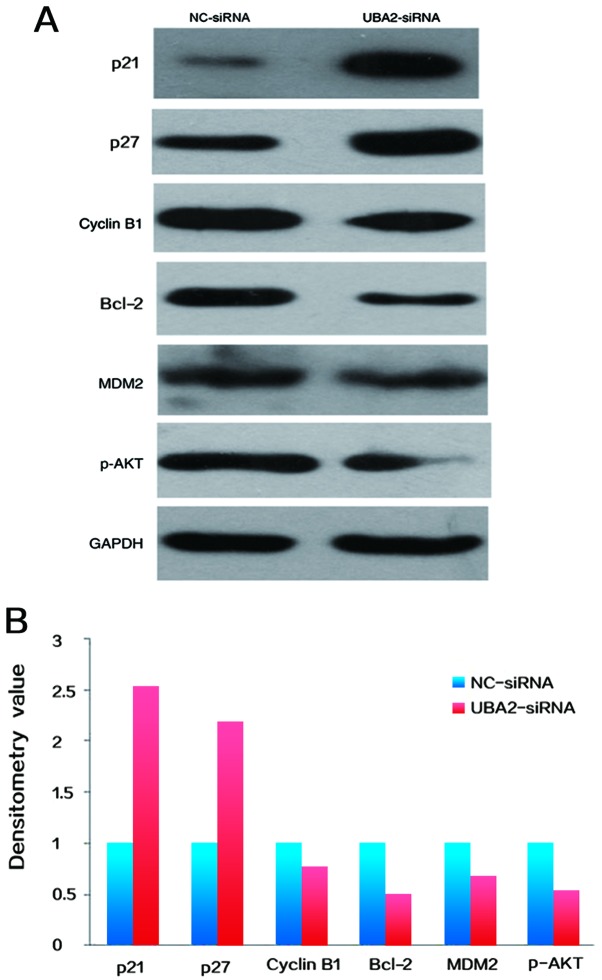
To determine the potential mechanisms underlying UBA2-mediated cell proliferation and apoptosis, the expression levels of cell cycle associated proteins in RKO colorectal cancer cells were examined by western blotting. As demonstrated in Fig. 6, the expression of cyclin B1, Bcl-2, phosphorylated protein kinase B (p-AKT) and E3 ubiquitin-protein ligase MDM2 were decreased, whereas the expression levels of p21 and p27 were increased in colorectal cancer cells following downregulation of UBA2, compared with NC-siRNA transfected cells (Fig. 6A and B).
Figure 6.
(A) Expression levels of cell cycle associated proteins (p21, p27 and cyclin B1) and apoptosis associated proteins (Bcl-2 and p-AKT) were determined by western blotting. (B) Densitometry analysis was performed to determine the protein expression. Data was normalized to GAPDH and then normalized to NC-siRNA. Bcl-2, B-cell lymphoma-2; MDM2, E3 ubiquitin-protein ligase MDM2; p-AKT, phosphorylated protein kinase B; NC, negative control; siRNA, small interfering RNA; UBA2, ubiquitin-like modifier-activating enzyme 2.
Discussion
UBA2 forms a heterodimer with SAE1, which acts as an E1-activating enzyme for the sumoylation of proteins (16). Adenosine triphosphate-dependent activation is mediated by the heterodimer through a thioester bond between sumo and a conserved active site cysteine residue on UBA2/SAE2 (29). Previous studies indicated that UBA2 may be a metastasis suppressor and prognostic marker in colorectal cancer. UBA2 expression levels are decreased in metastatic colorectal cancer cells (21–23). Another study demonstrated that the ubiquitin-associated protein 2-like (UBAP2L) gene, located on human chromosome 1q21.3, is associated with tumorigenesis of several types of human cancer, including multiple myeloma (30), liver cancer (31) and ovarian cancer (32). Chai et al demonstrated that UBAP2L serves important roles in colorectal cancer cell growth and survival (33), and that knockdown of UBAP2L in colorectal cancer cells led to suppression of proliferation, cell cycle arrest and apoptosis through the inhibition of p38 phosphorylation, and activation of proline-rich AKT1 substrate 1, Bcl-2-associated agonist of cell death, Bax, cleavage of poly[ADP-ribose] polymerase-4 and caspase-3.
Results from the present study demonstrated that the expression levels of UBA2 were increased in colorectal cancer tissues, and UBA2 expression was associated with higher stage in colorectal cancer and poor prognosis, which are consistent with a previous study (23). The results also demonstrated that UBA2 affected colorectal cancer cell proliferation in vitro and in vivo. In addition, downregulation of UBA2 expression induced apoptosis in colorectal cancer cells. These results indicated that UBA2 might serve important roles in colorectal cancer. However, normal colon cell lines were not included as a control in the present study, which limited our understanding of the expression level of UBA2 in non-cancerous colon cell lines.
Recent studies have demonstrated that knockdown sumo-conjugating enzyme UBC9 expression suppressed proliferation of RKO and HCT116 colorectal cancer cell lines (27,28). Similarly, knockdown UBA2 expression also suppressed proliferation of RKO and HCT116 cells, indicating SUMO pathway serves important roles in carcinogenesis of colorectal cancer. Knockdown of UBA2 decreased expression of cyclin B1, Bcl-2, MDM2 and p-AKT. Since sumoylation promotes cell cycle progress in glioblastoma by stabilizing CDK6 (5), the data from the present study indicated that expression reduction of cyclin B1, Bcl-2, MDM2 and p-AKT may be due to desumoylation by UBA2 inhibition, which is required to be studied in future. To further investigate the molecular mechanisms underlying UBA2-mediated cell proliferation and apoptosis in colorectal cancer, p53/MDM2/p21 signaling pathway was examined in the present study Knockdown of UBA2 by siRNA decreased MDM2, but increased p21 and p27 protein expression in colorectal cancer cells. Cyclin B1 and cell division control 2 form a complex that promotes the transition of cells from G2 to M phase (34). p21 and p27, downstream target proteins of p53, act as inhibitors of cell cycle progression at the G1, and S phase, and increased expression of such factors induces cell cycle arrest (35,36). Consistent with the alterations in cyclin B1, p21 and p27 expression levels, cells transfected with UBA2-siRNA were observed to be present in the G2/M cell cycle stage. Bcl-2 is an important member of the Bcl-2 family of regulator proteins that regulate apoptosis, and Bcl-2 downregulation leads to the induction of apoptosis (37). The results of the present study revealed that UBA2 silencing suppressed Bcl-2 expression and apoptosis levels, which indicated that downregulation of Bcl-2 is associated with UBA2-siRNA-induced apoptosis. The PTEN/PI3K/AKT signaling pathway is an essential pathway in the regulation of multiple biological processes, including apoptosis, cell proliferation and metabolism (38), in which both PTEN and AKT are part of the sumoylation pathway. The amino (N)-terminal domain of MDM2 interacts directly with the N-terminal transactivation domain of p53 and negatively regulates p53 transcriptional activation (39). The results suggested that UBA2 promoted cell proliferation and inhibited apoptosis by increasing cyclin B1, Bcl-2, p-AKT, MDM2, but decreasing p21 and p27 expression and involving PTEN/PI3K/AKT and p53/MDM2/p21 signaling pathways in colorectal cancer cells. These data suggest that elevated expression of UBA2 is an important contributor to the development of colorectal cancer.
In conclusion, results from the present study indicated that UBA2 expression may enhance colorectal cancer cell proliferation and inhibit apoptosis. Furthermore, the expression of UBA2 may be associated with p53/MDM2/p21 and PI3K/AKT signaling pathways. Although UBA2 is an ubiquitin-like modifier-activating enzyme, the present study did not establish a mechanistic link between UBA2 and SUMO modification of proteins. Further studies are required to determine how UBA2 silencing downregulates cyclin B1, Bcl-2 and p-AKT levels associated with SUMO modification. UBA2 may serve an essential role in proliferation of colorectal cancer and may be used as a potential biomarker to predict prognosis and a therapeutic target in colorectal cancer.
Acknowledgements
Not applicable.
Funding
The project is supported by the Young Scientists Award in Jilin Province (grant no. 20150204006YY).
Availability of data and materials
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PH, YZ, WL and YH conducted genetic analyses and cell proliferation experiments; PH and XM designed the study and drafted the manuscript; XS, QW, CZ and HC performed the western blotting experiments and analyzed the results. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Review Committee at Jilin University. All patients were informed their participation rights and provided written informed consent (Jilin, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Pichler A, Fatouros C, Lee H, Eisenhardt N. SUMO conjugation-a mechanistic view. Biomol Concepts. 2017;8:13–36. doi: 10.1515/bmc-2016-0030. [DOI] [PubMed] [Google Scholar]
- 3.Hay RT. SUMO: A history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Lee JS, Choi HJ, Baek SH. Sumoylation and Its contribution to cancer. Adv Exp Med Biol. 2017;963:283–298. doi: 10.1007/978-3-319-50044-7_17. [DOI] [PubMed] [Google Scholar]
- 5.Bellail AC, Olson JJ, Hao C. SUMO1 modification stabilizes CDK6 protein and drives the cell cycle and glioblastoma progression. Nat Commun. 2014;5:4234. doi: 10.1038/ncomms5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonne-Andrea C, Kahli M, Mechali F, Lemaitre JM, Bossis G, Coux O. SUMO2/3 modification of cyclin E contributes to the control of replication origin firing. Nat Commun. 2013;4:1850. doi: 10.1038/ncomms2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendriks IA, D'Souza RC, Yang B, Verlaan-de Vries M, Mann M, Vertegaal AC. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol. 2014;21:927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eifler K, Vertegaal ACO. SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem Sci. 2015;40:779–793. doi: 10.1016/j.tibs.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim KI, Baek SH. SUMOylation code in cancer development and metastasis. Mol Cells. 2006;22:247–253. [PubMed] [Google Scholar]
- 10.Zhou Z, Wang M, Li J, Xiao M, Chin YE, Cheng J, Yeh ET, Yang J, Yi J. SUMOylation and SENP3 regulate STAT3 activation in head and neck cancer. Oncogene. 2016;35:5826–5838. doi: 10.1038/onc.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moschos SJ, Jukic DM, Athanassiou C, Bhargava R, Dacic S, Wang X, Kuan SF, Fayewicz SL, Galambos C, Acquafondata M, et al. Expression analysis of Ubc9, the single small ubiquitin-like modifier (SUMO) E2 conjugating enzyme, in normal and malignant tissues. Hum Pathol. 2010;41:1286–1298. doi: 10.1016/j.humpath.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Zhu S, Sachdeva M, Wu F, Lu Z, Mo YY. Ubc9 promotes breast cell invasion and metastasis in a sumoylation-independent manner. Oncogene. 2010;29:1763–1772. doi: 10.1038/onc.2009.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He X, Riceberg J, Pulukuri SM, Grossman S, Shinde V, Shah P, Brownell JE, Dick L, Newcomb J, Bence N. Characterization of the loss of SUMO pathway function on cancer cells and tumor proliferation. PLoS One. 2015;10:e0123882. doi: 10.1371/journal.pone.0123882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katayama A, Ogino T, Bandoh N, Takahara M, Kishibe K, Nonaka S, Harabuchi Y. Overexpression of small ubiquitin-related modifier-1 and sumoylated Mdm2 in oral squamous cell carcinoma: Possible involvement in tumor proliferation and prognosis. Int J Oncol. 2007;31:517–524. [PubMed] [Google Scholar]
- 15.Zhang D, Raasi S, Fushman D. Affinity makes the difference: Nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377:162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Truong K, Lee TD, Li B, Chen Y. Sumoylation of SAE2 C terminus regulates SAE nuclear localization. J Biol Chem. 2012;287:42611–42619. doi: 10.1074/jbc.M112.420877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melo JB, Estevinho A, Saraiva J, Ramos L, Carreira IM. Cutis Aplasia as a clinical hallmark for the syndrome associated with 19q13.11 deletion: The possible role for UBA2 gene. Mol Cytogenet. 2015;8:21. doi: 10.1186/s13039-015-0123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venegas-Vega C, Nieto-Martinez K, Martinez-Herrera A, Gómez-Laguna L, Berumen J, Cervantes A, Kofman S, Fernández-Ramírez F. 19q13.11 microdeletion concomitant with ins(2;19) (p25.3;q13.1q13.4)dn in a boy: Potential role of UBA2 in the associated phenotype. Mol Cytogenet. 2014;7:61. doi: 10.1186/s13039-014-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Liang D, Li JY, Zhao RY. APOBEC3G-UBA2 fusion as a potential strategy for stable expression of APOBEC3G and inhibition of HIV-1 replication. Retrovirology. 2008;5:72. doi: 10.1186/1742-4690-5-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu J, Chen Y, Cai L, Xu C, Zhang Y, Chen Y, Zhang C, Zhao J, Cheng J, Xie H, et al. Functional proteomics study reveals SUMOylation of TFII-I is involved in liver cancer cell proliferation. J Proteome Res. 2015;14:2385–2397. doi: 10.1021/acs.jproteome.5b00062. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Xu Y, Pang Z, Guo F, Qin Q, Yin T, Sang Y, Feng C, Li X, Jiang L, et al. Knockdown of SUMO-activating enzyme subunit 2 (SAE2) suppresses cancer malignancy and enhances chemotherapy sensitivity in small cell lung cancer. J Hematol Oncol. 2015;8:67. doi: 10.1186/s13045-015-0164-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao DF, Wang XH, Li ZY, Xing XF, Cheng XJ, Guo T, Du H, Hu Y, Dong B, Ding N, et al. High-level SAE2 promotes malignant phenotype and predicts outcome in gastric cancer. Am J Cancer Res. 2014;5:140–154. [PMC free article] [PubMed] [Google Scholar]
- 23.Torres S, Garcia-Palmero I, Bartolomé RA, Fernandez-Aceñero MJ, Molina E, Calviño E, Segura MF, Casal JI. Combined miRNA profiling and proteomics demonstrates that different miRNAs target a common set of proteins to promote colorectal cancer metastasis. J Pathol. 2017;242:39–51. doi: 10.1002/path.4874. [DOI] [PubMed] [Google Scholar]
- 24.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: A database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Bogachek MV, Park JM, De Andrade JP, Lorenzen AW, Kulak MV, White JR, Gu VW, Wu VT, Weigel RJ. Inhibiting the SUMO pathway represses the cancer stem cell population in breast and colorectal carcinomas. Stem Cell Reports. 2016;7:1140–1151. doi: 10.1016/j.stemcr.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomasi ML, Ryoo M, Ramani K, Tomasi I, Giordano P, Mato JM, Lu SC. Methionine adenosyltransferase α2 sumoylation positively regulate Bcl-2 expression in human colon and liver cancer cells. Oncotarget. 2015;6:37706–37723. doi: 10.18632/oncotarget.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem Biophys Res Commun. 1999;254:693–698. doi: 10.1006/bbrc.1998.9995. [DOI] [PubMed] [Google Scholar]
- 30.Sawyer JR. The prognostic significance of cytogenetics and molecular profiling in multiple myeloma. Cancer Genet. 2011;204:3–12. doi: 10.1016/j.cancergencyto.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Zhu ZZ, Wang D, Cong WM, Jiang H, Yu Y, Wen BJ, Dong H, Zhang X, Liu SF, Wang AZ, et al. Sex-related differences in DNA copy number alterations in hepatitis B virus-associated hepatocellular carcinoma. Asian Pac J Cancer Prev. 2012;13:225–229. doi: 10.7314/APJCP.2012.13.1.225. [DOI] [PubMed] [Google Scholar]
- 32.Naz RK, Dhandapani L. Identification of human sperm proteins that interact with human zona pellucida3 (ZP3) using yeast two-hybrid system. J Reprod Immunol. 2010;84:24–31. doi: 10.1016/j.jri.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chai R, Yu X, Tu S, Zheng B. Depletion of UBA protein 2-like protein inhibits growth and induces apoptosis of human colorectal carcinoma cells. Tumour Biol. 2016;37:13225–3522. doi: 10.1007/s13277-016-5159-y. [DOI] [PubMed] [Google Scholar]
- 34.Atherton-Fessler S, Liu F, Gabrielli B, Lee MS, Peng CY, Piwnica-Worms H. Cell cycle regulation of the p34cdc2 inhibitory kinases. Mol Biol Cell. 1994;5:989–1001. doi: 10.1091/mbc.5.9.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pestell RG, Albanese C, Reutens AT, Segall JE, Lee RJ, Arnold A. The cyclins and cyclin-dependent kinase inhibitors in hormonal regulation of proliferation and differentiation. Endocr Rev. 1999;20:501–534. doi: 10.1210/edrv.20.4.0373. [DOI] [PubMed] [Google Scholar]
- 36.Lloyd RV, Erickson LA, Jin L, Kulig E, Qian X, Cheville JC, Scheithauer BW. p27kip1: A multifunctional cyclin-dependent kinase inhibitor with prognostic significance in human cancers. Am J Pathol. 1999;154:313–323. doi: 10.1016/S0002-9440(10)65277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kontos CK, Christodoulou MI, Scorilas A. Apoptosis-related BCL2-family members: Key players in chemotherapy. Anticancer Agents Med Chem. 2014;14:353–374. doi: 10.2174/18715206113139990091. [DOI] [PubMed] [Google Scholar]
- 38.Carnero A, Blanco-Aparicio C, Renner O, Link W, Leal JF. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets. 2008;8:187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 39.Pant V, Xiong S, Iwakuma T, Quintás-Cardama A, Lozano G. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci USA. 2011;108:11995–12000. doi: 10.1073/pnas.1102241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.