Abstract
Unlike the WASP family of Arp2/3 complex activators, WISH/DIP/SPIN90 (WDS) family proteins activate actin filament nucleation by the Arp2/3 complex without the need for a preformed actin filament. This allows WDS proteins to initiate branched actin network assembly by providing seed filaments that activate WASP‐bound Arp2/3 complex. Despite their important role in actin network initiation, it is unclear how WDS proteins drive the activating steps that require both WASP and pre‐existing actin filaments during WASP‐mediated nucleation. Here, we show that SPIN90 folds into an armadillo repeat domain that binds a surface of Arp2/3 complex distinct from the two WASP sites, straddling a hinge point that may stimulate movement of the Arp2 subunit into the activated short‐pitch conformation. SPIN90 binds a surface on Arp2/3 complex that overlaps with actin filament binding, explaining how it could stimulate the same structural rearrangements in the complex as pre‐existing actin filaments. By revealing how WDS proteins activate the Arp2/3 complex, these data provide a molecular foundation to understand initiation of dendritic actin networks and regulation of Arp2/3 complex by its activators.
Keywords: actin, Arp2/3 complex, nucleation, SPIN90, WASP
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Structural Biology
Introduction
Arp2/3 complex is a seven‐subunit protein assembly that nucleates branched actin filaments in response to cellular signals. Dendritic actin networks assembled by Arp2/3 complex contribute to cellular processes like motility, endocytosis, and meiotic spindle positioning (Moreau et al, 1996; Yi et al, 2011; Suraneni et al, 2012; Wu et al, 2012; Liu et al, 2013). To orchestrate these processes, cells must tightly regulate Arp2/3 complex to control the timing and localization of dendritic actin network assembly. Cells rely on both inhibitory and activating regulators of Arp2/3 complex to accomplish this, and the molecular mechanisms by which inhibitors can block activation are becoming clearer, owing to structural, biochemical, and cell biological data (Humphries et al, 2002; Cai et al, 2008; Gandhi et al, 2010; Nakano et al, 2010; Liu et al, 2011; Maritzen et al, 2012; Dang et al, 2013; Luan & Nolen, 2013; Sokolova et al, 2017). On the other hand, how activating regulators of Arp2/3 complex (a.k.a. nucleation‐promoting factors, NPFs) influence the complex is less clear, both because their mechanisms are more complicated and because of insufficient structural information about how they bind Arp2/3 complex.
WASP proteins comprise the largest family of NPFs and are characterized by a C‐terminal region called VCA (Campellone & Welch, 2010). VCA binds and recruits actin monomers to Arp2/3 complex (Rohatgi et al, 1999; Marchand et al, 2001; Padrick et al, 2011; Boczkowska et al, 2014), making interactions that drive the two actin‐related subunits, Arp2 and Arp3 into filament‐like (short pitch) arrangement (Boczkowska et al, 2008; Xu et al, 2012; Hetrick et al, 2013; Rodnick‐Smith et al, 2016a,b). Adoption of the short‐pitch conformation is a critical activation step, and stimulating this structural change is the main function of WASP and actin monomers recruited by WASP (Rodnick‐Smith et al, 2016b). WASP‐mediated activation of Arp2/3 complex also requires a preformed actin filament (Machesky et al, 1999; Maul et al, 2003; Achard et al, 2010; Smith et al, 2013; Wagner et al, 2013); only once bound to the side of a pre‐existing filament can the WASP‐bound complex nucleate a new filament. This biochemical requirement ensures that WASP‐activated Arp2/3 complex creates branches, allowing it to function specifically in dendritic actin network assembly. Filaments were initially thought to be required, along with WASP, to stimulate the short‐pitch conformation, but recent data indicate WASP alone can stimulate this structural change (Xu et al, 2012; Rodnick‐Smith et al, 2016b). This suggests actin filaments may trigger a distinct set of structural changes to allow activation. While the nature of these changes is unknown, low‐resolution EM reconstructions have identified the actin filament‐binding site on Arp2/3 complex (Rouiller et al, 2008). Comparing this site to the two WASP‐binding sites on Arp2/3 complex shows that WASP and actin filaments bind distinct surfaces (Boczkowska et al, 2008; Padrick et al, 2011; Ti et al, 2011; Jurgenson & Pollard, 2015; Rodnick‐Smith et al, 2016a; Luan et al, 2018), consistent with the idea that they might stimulate distinct structural changes during activation.
The requirement for a preformed filament means WASP‐mediated assembly of branched actin networks must be primed with an initial seed filament (Achard et al, 2010). We recently showed that a class of Arp2/3 complex activators called WISH/DIP/SPIN90 (WDS) family proteins create seed filaments for WASP‐mediated activation (Wagner et al, 2013). Unlike WASP, WDS proteins can activate Arp2/3 complex without requiring a preformed actin filament, resulting in the nucleation of a linear actin filament. These linear filaments are thought to seed assembly of branched networks by WASP‐bound Arp2/3 complex and may be important initiators of branched actin networks in cells. For example, in fission yeast, deletion of the WDS protein Dip1 significantly decreases the rate of initiation of new branch actin networks at endocytic sites (Basu & Chang, 2011). Less is known about the biological function SPIN90, the mammalian WDS protein (also called WISH, DIP and NCKIPSD; Fukuoka et al, 2001; Lim et al, 2001; Meng et al, 2004), but existing studies show it plays a role in actin‐driven processes, such as lamellipodial protrusion, endocytosis, and neurite outgrowth, so the seeding function of WDS proteins may be also important in mammals (Kim et al, 2006, 2011; Oh et al, 2013).
To fulfill their role as seeders, WDS proteins must stimulate all activating steps in Arp2/3 complex that are normally triggered by the coordinated action of WASP and actin filaments. How WDS proteins could accomplish this is unknown. One early study predicted that WDS proteins might have WASP‐like features, explaining how they could stimulate activating steps triggered by WASP (Kim et al, 2006). However, our data showed that WDS proteins are biochemically distinct from WASP because they do not bind actin monomers and they lack the Arp2/3‐activating VCA segment conserved in WASP proteins (Wagner et al, 2013). Despite harboring structural properties distinct from WASP, we showed that the Schizosaccharomyces pombe WDS protein, Dip1, simulates the short‐pitch conformation as potently as WASP (Wagner et al, 2013). This activity resides in the conserved C‐terminal segment of Dip1, since this is the region responsible for Arp2/3 complex activation (Wagner et al, 2013). How this segment could stimulate the short‐pitch conformational change using a different structural moiety than WASP is unclear. In addition, because it is not understood how the WDS C‐terminal segment interacts with Arp2/3 complex, it is unknown how WDS proteins could stimulate the same activating changes as actin filaments.
Here, we present X‐ray crystal structures of SPIN90 on its own and bound to Arp2/3 complex. These structures, along with our biochemical data, reveal a mechanism by which SPIN90 can carry out the activating functions of both WASP proteins and actin filaments. Specifically, we show that SPIN90 folds into an armadillo repeat motif domain (ARM) that binds to the same surface on Arp2/3 complex as actin filaments. Binding to this site likely allows SPIN90 to trigger conformational changes normally stimulated by filaments, since it causes structural changes that move Arp3 toward a “flattened” conformation, an activating conformational change that occurs in actin upon incorporation into filaments (Oda et al, 2009). Further, while the SPIN90‐binding site is distinct from the two binding sites for WASP (Padrick et al, 2011; Ti et al, 2011; Boczkowska et al, 2014; Jurgenson & Pollard, 2015; Rodnick‐Smith et al, 2016a; Luan et al, 2018), it binds along a long alpha helix in the ARPC4 subunit that bends to rotate Arp2 into the short‐pitch conformation in some models for activation (Robinson et al, 2001; Dalhaimer & Pollard, 2010). Therefore, our data support a model in which SPIN90 binds to a different site on Arp2/3 complex than WASP to allosterically stimulate the same activating conformational change as WASP. Together, these data reveal a mechanism by which WDS proteins can activate Arp2/3 complex without requiring a preformed actin filament. These results have implications for understanding both how WDS proteins initiate branched actin networks and the molecular mechanisms by which different NPF proteins activate Arp2/3 complex.
Results
The C‐terminus of SPIN90 folds into an armadillo repeat (ARM) domain that harbors a conserved patch of surface residues near its C‐terminus
To understand how SPIN90 activates Arp2/3 complex, we attempted to solve its structure by x‐ray crystallography. SPIN90 is a 722 amino acid protein containing an N‐terminal SH3 domain, a long proline rich and hydrophilic stretch predicted to be unstructured, and a C‐terminal Arp2/3‐activating region that contains a conserved leucine‐rich segment (annotated as a “leucine‐rich domain”, LRD; Kim et al, 2006; Wagner et al, 2013; Fig 1A). In the S. pombe WDS protein, Dip1, a construct containing the full LRD plus 29 N‐terminal flanking residues activated Arp2/3 complex as potently as full‐length Dip1, suggesting the LRD is responsible for activation (Wagner et al, 2013). While less is known about the sequence requirements for SPIN90 activity, previous studies showed a C‐terminal construct containing the LRD plus an additional 277 residues N‐terminal to the LRD [SPIN90 (269–722)] activated Arp2/3 complex (Kim et al, 2006; Wagner et al, 2013). However, the SPIN90 (269–722) construct failed to crystallize, so we made several shorter N‐terminally truncated constructs and set up crystallization screens (Fig 1A). We found that a SPIN90 construct containing 195 residues N‐terminal to the LRD, SPIN90 (351–722), produced monoclinic crystals in space group P21 that diffracted to 2.2 Å (Table EV1). We solved this structure by SAD phasing and refined the structure to a final Rfree of 24.0% (Table EV1).
Figure 1. Structure and biochemical characterization of the C‐terminus of SPIN90.
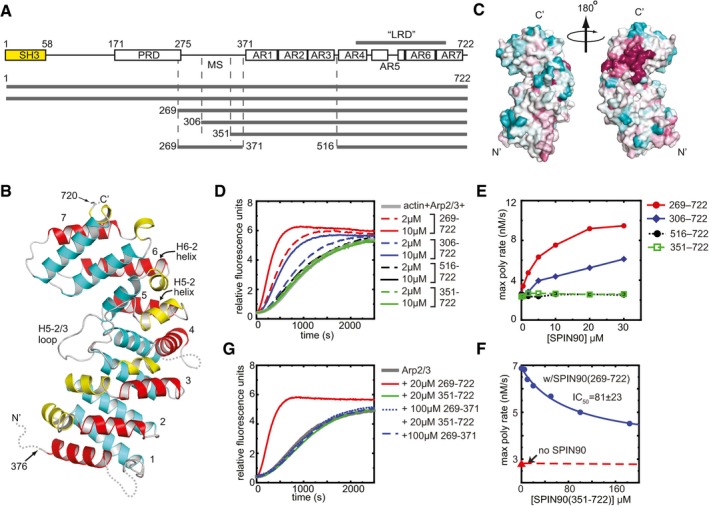
- Domain organization of SPIN90 and SPIN90 constructs used in this study (SH3, Src homology 3; PRD, proline‐rich domain; MS, middle segment; AR, armadillo repeat).
- Ribbon diagram of SPIN90 (351–722). Helices are colored according to their position within the repeat (1—yellow, 2—red, 3—cyan). Armadillo repeats are numbered 1–7. Disordered residues are indicated with dotted lines.
- Surface representation of SPIN90 (351–722) showing conservation scores derived from an alignment of WDS family proteins from 31 diverse species (see Materials and Methods). Conservation score is indicated by color, with dark red > pink > light pink > gray > light cyan > cyan, and higher scores meaning greater conservation.
- Time course of pyrene actin polymerization showing the influence of different SPIN90 constructs in reactions containing 50 nM Bos taurus Arp2/3 complex and 3 μM 15% pyrene‐labeled actin.
- Plot of maximum polymerization rate versus concentration of SPIN90 for reactions shown in (D).
- Plot of maximum polymerization rate versus concentration of SPIN90 for pyrene actin polymerization assays containing 2.5 μM SPIN90, 500 nM Arp2/3 complex, 3 μM 15% pyrene‐labeled actin and the indicated concentration of the SPIN90 ARM domain construct.
- Time courses of pyrene actin polymerization as described in panel (D), except the influence of the SPIN90 middle segment is tested on its own or in the presence of the 351–722 construct.
The SPIN90 (351–722) structure contains two molecules in the asymmetric unit (ASU). Because the two molecules bury only ~500 Å2 surface area at their interaction interface, the dimer interface is unlikely to be biologically relevant. The two molecules in the ASU are nearly identical (RMSD = 0.169 Å for 1,228 backbone atoms), so here we limit our description to chain A. The N‐terminal 25 residues of SPIN90 (351–722), two C‐terminal residues, and two short loop segments are disordered and missing from the density (Fig 1B). The 336 ordered residues form an armadillo repeat motif domain (ARM domain) containing 6 2/3 repeats (Figs 1B and EV1). Canonical ARM repeats contain one short (H1) and two long alpha helices (H2 and H3) that form a right‐handed supercoil (Groves & Barford, 1999). The SPIN90 ARM domain conforms to this pattern, except that helix H1 is missing in the first ARM repeat and helices H4‐1 and H7‐1 are 310 rather than alpha helices. In addition, the SPIN90 ARM domain contains a long (16 aa) insert between α‐helices 2 and 3 of repeat 5 that binds along the concave surface of the ARM domain (Fig 1B, H5‐2/3 loop). The conserved LRD sequence spans from helix H4‐3 in repeat 4 to helix H7‐2 in repeat 7, forming the C‐terminal half of the ARM domain (Figs 1A and B, and EV1). The fold of the SPIN90 ARM domain is similar to other ARM domain‐containing proteins, including importin‐α, β‐catenin, the adenomatous polyposis coli protein (APC), and the regulatory domains in some formin proteins (Huber et al, 1997; Kobe, 1999; Morishita et al, 2011). Of these, SPIN90 structurally aligns most closely with APC, and the backbone atoms of the two proteins superpose with an RMSD of 5.7 Å (Zhang et al, 2011; Appendix Fig S1). The backbone atoms in repeats 3–7 in SPIN90 superposed to repeat 2 with RMSD values ranging from 1.3 to 5.1 Å, indicating considerable structural diversity within ARM repeats (Appendix Fig S2). To identify residues of SPIN90 that might interact with Arp2/3 complex, we mapped sequence conservation onto the surface of the ARM domain. This analysis revealed a cluster of conserved solvent‐exposed residues in the LRD sequence near the C‐terminus that might be important for activation of Arp2/3 complex (Fig 1C). Most of the residues that constitute this surface patch are contributed by two alpha helices: H5‐2 and H6‐2, i.e., the second alpha helix from ARM repeats 5 and 6.
Figure EV1. Sequence alignment of SPIN90 from diverse species.
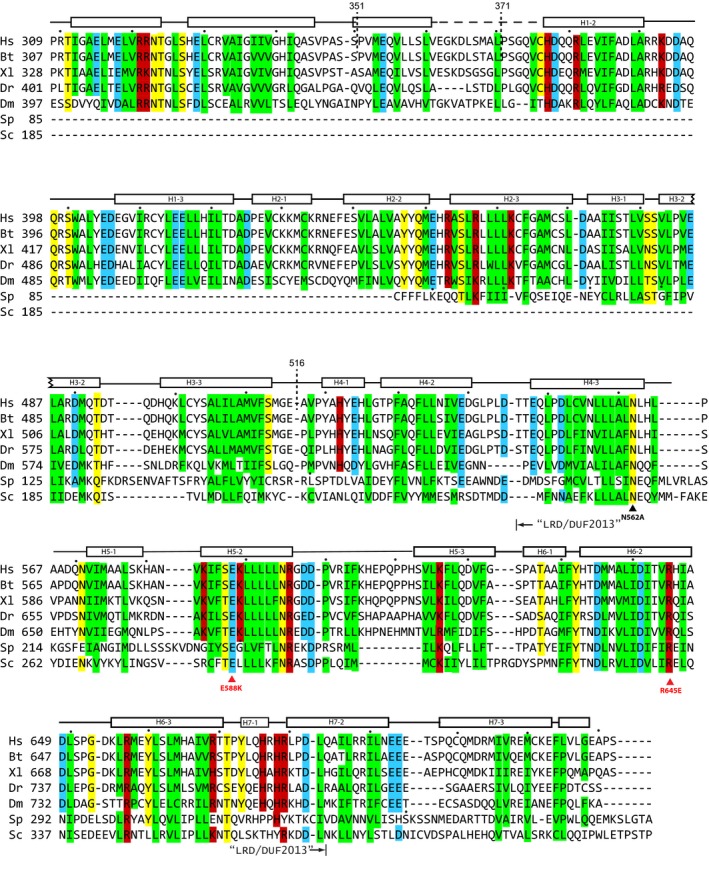
Conserved residues are indicated by colored boxes (green: hydrophobic; yellow: hydrophilic; red: basic; cyan: acidic). The highly conserved segment among WDS proteins, the “leucine‐rich domain” (LRD)/DUF2013 segment, is indicated below the sequences, along with residues mutated in this study. Truncation sites are indicated with dashed lines. Alpha helices in the SPIN90 structure are shown above the sequence as rectangles. Hs, H. sapiens; Sc, S. cerevisiae; Sp, S. pombe; Bt, B. taurus; Dm, D. melanogaster; Xl, X. laevis; Dr, D. rerio.
Both the ARM domain and its flexible flanking N‐terminal region are required for activation of Arp2/3 complex
In Dip1, the S. pombe WDS protein, a construct containing the LRD plus 29 additional N‐terminal residues [Dip1 (160–374) activated Arp2/3 complex as potently as full‐length Dip1 (Wagner et al, 2013], suggesting the LRD may be sufficient for activation. Our SPIN90 (351–722) structure shows that Dip1 160–374 is homologous to ARM repeats 4–7, so we wondered whether a SPIN90 construct containing repeats 4–7 could activate Arp2/3 complex. Unexpectedly, this construct, SPIN90 (516–722), had no activity, even at concentrations as high as 30 μM (Fig 1D and E). Perhaps more surprisingly, the construct we used for the structural analysis described above [SPIN90 (351–722)] also failed to activate the complex, despite the fact that it contains the entire LRD sequence encompassed within the stably folded full ARM domain (Fig 1D and E). These data indicate that residues from the sequence N‐terminal to the ARM domain, which we term the middle segment (MS), are also required for activity. Importantly, high concentrations of the 351–722 construct inhibited SPIN90 (269–722) from activating the complex, indicating that the full ARM domain in isolation can bind to Arp2/3 complex, albeit weakly (Fig 1F). The middle segment fragment (269–371) added in trans to the full ARM domain (351–722) had no influence on Arp2/3 complex activity, demonstrating MS and the ARM domain must be covalently linked for SPIN90 to activate Arp2/3 complex (Fig 1G). To zero in on the region of the MS required for activity, we made an N‐terminal truncation mutant that contained an additional 45 residues of the MS compared to SPIN90 (351–722). This fragment, SPIN90 (306–722), is predicted to harbor two alpha helices near the start of the ARM domain but otherwise is thought to be unstructured (Appendix Fig S3). SPIN90 (306–722) was active in pyrene actin polymerization assays, although its activity was about 50% lower than the SPIN90 (269–722) construct (Fig 1D and E).
The SPIN90 (306–722) construct shows structural differences compared to the SPIN90 (351–722) ARM domain construct
To understand why the SPIN90 (306–722) construct is active, we solved its structure and compared it to the structure of the inactive 351–722 construct. We grew crystals of SPIN90 (306–722) that diffracted to 3.0 Å and refined the structure to an Rfree of 30.7% (Table EV1). We note that the refinement statics were worse than expected for a structure of this resolution for reasons that are not currently clear. The SPIN90 (306–722) structure shows density for residues 308–361 of the MS (Fig 2A). This short segment forms three alpha helices disconnected from the main density of the ARM domain that pack between two neighboring symmetry‐related molecules in the crystal and are disordered in another structure (see below), suggesting they are flexible in solution (Appendix Fig S4). The ARM domain of SPIN90 (306–722) adopts a similar conformation to the ARM domain in the SPIN90 (351–722) structure (RMSD of 0.613 Å for 1,187 backbone atoms) but shows a potentially important structural difference (Fig 2B). Specifically, the loop connecting helices H5‐1 and H5‐2 (H5‐1/2 loop) is flipped toward the C‐terminus of the ARM domain in the SPIN90 (306–722) structure compared to both SPIN90 molecules in the SPIN90 (315–722) structure (Fig 2B and C). This structural change, which is characterized by a backbone flip of His580, brings the N‐termini of helices H5‐2 and H6‐2 1.8 Å closer together than in the inactive structure (Fig 2B). We show below that helices H5‐2 and H6‐2 contribute most of the residues to the interaction with Arp2/3 complex. Therefore, the conformational differences in these helices comparing the inactive 351–722 and the active ARM domain may contribute to the observed activity differences. How the presence or absence of the residues N‐terminal to the ARM domain could influence the position of the H5‐1/2 loop and the H5‐2 and H6‐2 helices is unclear and is currently under investigation.
Figure 2. Comparison of active and inactive ARM domain structures.
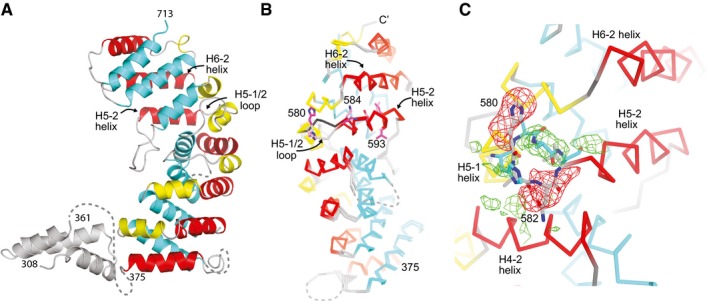
- Ribbon diagram of SPIN90 (306–722). Helices within the ARM domain are colored according to their position within the repeat (1—yellow, 2—red, 3—cyan). Structural features that are not part of the armadillo repeat domain are colored gray.
- Cα trace of SPIN90 (351–722) superposed onto the structure of SPIN90 (306–722). His580 is shown in stick representation, as are residues on the H5‐2 helix that contribute to the conserved surface patch shown in Fig 1C. H5‐1/2 is flipped away from the C‐terminus of the ARM domain in the inactive 351–722 structure.
- Zoomed in view from panel (B) showing the difference in conformation of the H5‐1/2 loop in the inactive (351–722, sticks with gray carbon atoms) versus active (306–722, sticks with cyan carbon atoms) SPIN90 structures. Positive (green) or negative (red) Fo−Fc electron density maps are contoured at ±2.5 σ and were generated by replacing H5‐1/2 loop residues (577–585) of the 306–722 structure with the same residues from chain A in the 351–722 structure.
The SPIN90 ARM domain binds to a surface on the front side of Arp2/3 complex distinct from WASP‐binding sites
To determine how SPIN90 binds to Arp2/3 complex, we solved a co‐crystal structure of SPIN90 bound to Arp2/3 complex. This co‐complex crystal contained the SPIN90 (269–722) construct and bovine Arp2/3 complex and diffracted to 4.6 Å. We solved this structure by molecular replacement using the GMF‐bound Arp2/3 complex structure (4jd2) and the ARM domain of the SPIN90 (306–722) structure (Luan & Nolen, 2013). The refined structure has an Rfree of 32.0% and contains two molecules of SPIN90 (269–722) and two Arp2/3 complexes per asymmetric unit (Table EV1). The electron density allowed us to model 4,080 of the 4,870 residues in the asymmetric unit, but only about 1% of the side chains could be modeled due to missing electron density (Table EV1, Appendix Figs S5 and S6). However, the backbone trajectories in both structures of unbound SPIN90 are very similar to Arp2/3 complex‐bound SPIN90, so the sequence register and the backbone trajectory of SPIN90 are unambiguous. Unlike the SPIN90 (306–722) structure, all residues N‐terminal to the ARM domain are disordered in both SPIN90 molecules in the ASU in the co‐complex structure. The possible to exception to this are two disconnected tubes of density near the N‐termini of the ARM domains that we modeled as 6 or 9 residue alpha helices. It is unclear whether this density is contributed by residues N‐terminal to the ARM domain or by a flexible segment from one of the Arp2/3 complex subunits.
The resulting model shows the ARM domain of SPIN90 binds to the front side of Arp2/3 complex, contacting the ARPC2 and ARPC4 subunits (Fig 3A). An electron density omit map verified the position of SPIN90 in the structure (Fig 3B). Importantly, the co‐complex structure shows that SPIN90 binds a distinct surface on the complex compared to WASP proteins, which use their conserved CA segments to bind to Arp3 on the back side of the complex and to Arp2/ARPC1 on the back/bottom side of the complex (Padrick et al, 2011; Ti et al, 2011; Boczkowska et al, 2014; Jurgenson & Pollard, 2015; Rodnick‐Smith et al, 2016a; Luan et al, 2018; Fig 3C and D).
Figure 3. SPIN90 binds to the front side of Arp2/3 complex.
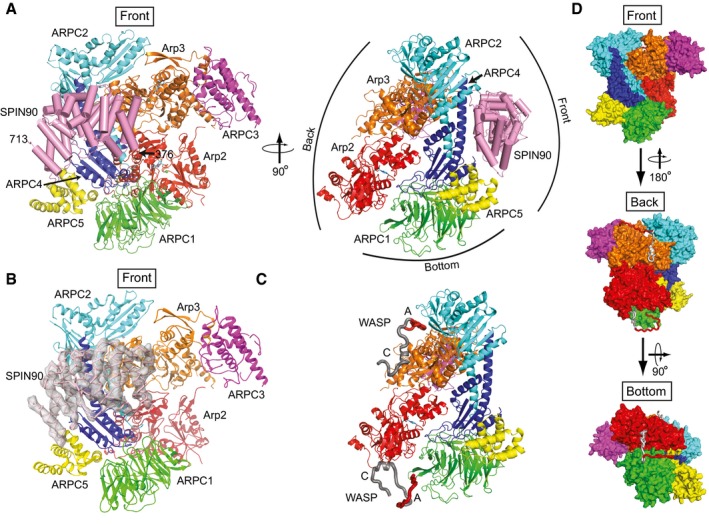
- Ribbon diagram of Arp2/3 complex with bound SPIN90 from the co‐complex structure. SPIN90 is shown in pink with cylindrical helices. Conventional designations of “Front”, “Back”, and “Bottom” views are indicated in right panel.
- Ribbon diagram showing front side of Arp2/3 complex and bound SPIN90 with Fo−Fc electron density map contoured at 2.1 σ calculated without phases from SPIN90 (chain M).
- Model of bovine Arp2/3 complex with bound WASP CA segments modeled based on crosslinking/mass‐spectrometry, homology modeling, x‐ray crystallography, and FRET (Padrick et al, 2011; Ti et al, 2011; Boczkowska et al, 2014; Jurgenson & Pollard, 2015; Rodnick‐Smith et al, 2016a; Luan et al, 2018). The complex is in an identical orientation to the right panel in (A).
- Surface representation of model described in (C) showing the convention for front, back, and bottom views of Arp2/3 complex.
Bound SPIN90 buries approximately 1,200 Å2 of surface area on Arp2/3 complex. The majority (~60%) of the buried area results from contacts with the ARPC4 subunit (Fig 4A). Helices H5‐2 and H6‐2 in SPIN90 form the main interface that interacts with Arp2/3 complex, packing against the long alpha helix (αD) in ARPC4 to make an anti‐parallel three helix bundle (Fig 4A). In addition to interactions with the ARPC4 αD helix, SPIN90 also makes minor contacts with APRC2 and ARPC5 and the globular base of ARPC4 (Appendix Fig S7). The N‐terminus of ARPC4 rearranges to accommodate this interaction (Appendix Fig S7). Together, these minor interactions account for an additional ~480 Å2 of buried surface area at the periphery of the main interaction with ARPC4. The residues in SPIN90 involved in contacting the ARPC4 αD helix are well conserved and form the highly conserved patch of surface‐exposed residues described above (Fig 1C). Much of the interaction surface for SPIN90 on Arp2/3 complex is also conserved (Fig EV2; Beltzner & Pollard, 2004). We note that the interaction between SPIN90 ARM domain and Arp2/3 complex is distinct from other ARM domains, which typically use their concave surface to accommodate extended polypeptides (Gul et al, 2017).
Figure 4. Contacts of SPIN90 with the long C‐terminal helix (αD) in ARPC4 are important for activation of Arp2/3 complex.
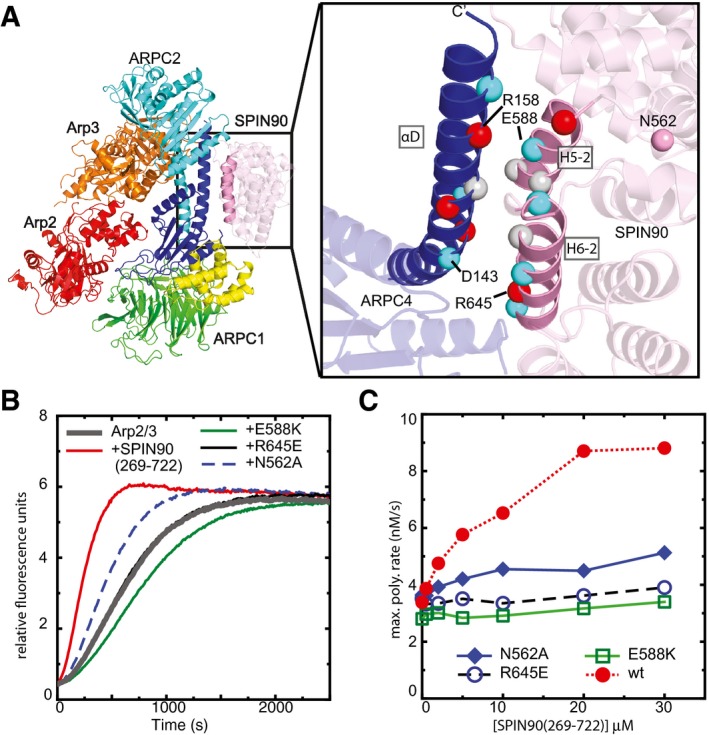
- Ribbon diagram of co‐complex structure with residues at the SPIN90‐Arp2/3 interface depicted as Cα atom spheres colored according to side chain chemistry (gray: hydrophobic, red: basic, blue: acidic). Only the ARPC4 subunit of Arp2/3 complex is shown in the zoomed in panel for clarity.
- Time courses of pyrene actin polymerization showing the influence of mutations on the ability of SPIN90 (269–722) to activate Arp2/3 complex. Reactions contained 3 μM 15% pyrene‐labeled actin, 50 nM Bos taurus Arp2/3 complex, and 20 μM wild‐type or mutant SPIN90 (269–722).
- Plot of maximum polymerization rate versus concentration of SPIN90 (269–722) for reactions as shown in panel (B).
Figure EV2. The SPIN90 contact surface overlaps with a conserved patch of residues on Arp2/3 complex.
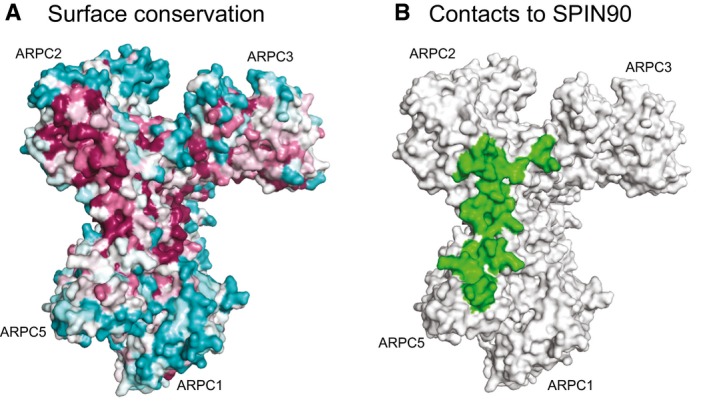
- Surface representation of the “front side” of Arp2/3 from the apo Arp2/3 complex structure (1k8k; Robinson et al, 2001) showing residue conservation based on analysis with the ConSurf server (Ashkenazy et al, 2016). Residues are colored according to conservation, with the following order, going from most to least conserved: dark magenta > pink > light pink > gray > light cyan > cyan.
- Surface representation of Arp2/3 complex (1k8k) showing residues (green) that are within 6 Å of SPIN90 in the co‐complex structure.
Comparing the structure of SPIN90 (269–722) to the isolated active [SPIN90 (306–722)] and inactive [SPIN90 (351–722)] structures revealed that the ARM domain of SPIN90 (269–722) adopts a conformation similar to the active SPIN90 structure. Specifically, as in the active structure of SPIN90 (306–722), the H5‐1/2 loop in Arp2/3‐bound SPIN90 is flipped toward the C‐terminus and the N‐termini of helices H5‐2 and H6‐2 are closer together than in the inactive structure (Fig EV3). These observations support the conclusion that these structural changes are important for SPIN90 to activate Arp2/3 complex.
Figure EV3. The H5‐1/2 loop is flipped toward the C‐terminus of the ARM domain in Arp2/3‐bound SPIN90.
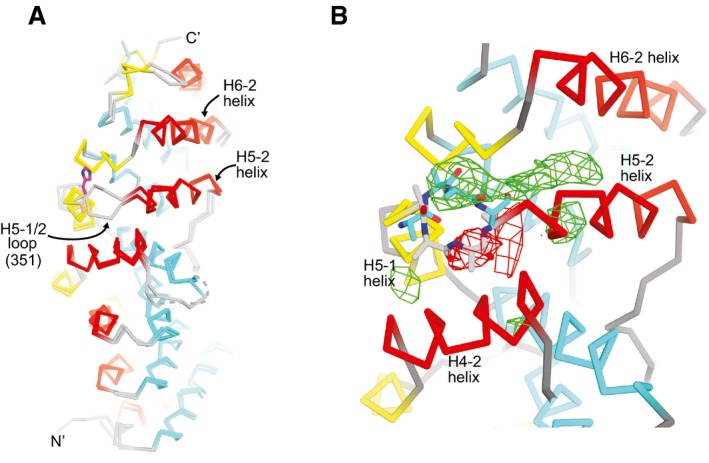
- Cα trace of SPIN90 (351–722) superposed onto the structure of SPIN90 (269–722) from the co‐complex structure. His580 is shown in stick representation. H5‐1/2 is flipped away from the C‐terminus in the inactive 351–722 structure and toward it in the co‐complex structure.
- Zoomed in view from panel (A) showing the difference in conformation of the H5‐1/2 loop in the inactive (351–722, sticks with gray carbon atoms) versus active (269–722, sticks with cyan carbon atoms) SPIN90 structures. Positive (green) or negative (red) Fo−Fc electron density maps are contoured at ±2.0 σ and were generated from the co‐complex reflection file plus a PDB file in which H5‐1/2 loop (residues 577–585) of the co‐complex structure was replaced with the same residues from chain A in the 351–722 structure.
SPIN90 interactions with ARPC4 are required for Arp2/3 complex activation
To test the importance of the observed interactions between SPIN90 and Arp2/3 complex, we mutated residues at the interface and tested their influence on activation of Arp2/3 complex by SPIN90. We focused on the three‐helix bundle formed between the long C‐terminal helix in ARPC4 and the H5‐2 and H6‐2 helices in SPIN90 because these contacts provide most of the binding surface for the interaction. Several complementary electrostatic interactions are provided by the H5‐2 and H6‐2 helices (Fig 4A). For instance, Arg645 and Glu588 in SPIN90 are in position to interact with Asp143 and Arg158 in ARPC4, respectively. Consistent with these observations, charge reversal mutations of SPIN90 Arg645 or Glu588 (R645E and E588K) reduced SPIN90‐mediated activation of Arp2/3 complex in a pyrene actin polymerization assay (Fig 4B and C). These experiments indicate that the interaction of SPIN90 with the ARPC4 helix of the complex is critical for SPIN90‐mediated activation of Arp2/3 complex.
We also tested the importance of N562, a residue conserved among WDS proteins, but on the opposite face of SPIN90 as the ARPC4‐interacting residues. N562 was previously reported to be important for the ability of SPIN90 to bind and inhibit mDia2, a linear actin filament nucleator from the formin protein family (Eisenmann et al, 2007). N562 may be conserved because of its formin‐regulating function and could thus mark a separate binding surface for an actin nucleator protein. However, we found that mutating N562 to alanine significantly decreased the ability of SPIN90 to activate Arp2/3 complex (Fig 4B and C). Examination of the 351–722 structure revealed that N562 hydrogen bonds to K616 and D620 in helix H5‐3 (Appendix Fig S8). We speculate that mutation of N562 may alter the position of helix H5‐3, thereby causing a shift in the ARPC4‐contacting helix H5‐2 and decreasing the affinity of the SPIN90‐Arp2/3 complex interaction (Appendix Fig S8).
SPIN90 binds at a putative hinge point for rotation of Arp2 into the short‐pitch position
Using a crosslinking assay with an engineered budding yeast Arp2/3 complex, we previously showed that movement of the complex into or toward the short‐pitch conformation is required for activation (Hetrick et al, 2013; Rodnick‐Smith et al, 2016a,b). The same assay also allowed us to demonstrate that Dip1 stimulates movement into or toward the short‐pitch conformation (Wagner et al, 2013). We could not use the same crosslinking assay to determine whether SPIN90 stimulates the short‐pitch conformation, because SPIN90 did not activate the budding yeast Arp2/3 complex (Appendix Fig S9). However, a small‐molecule inhibitor that blocks Arp2/3 complex from moving into or toward the short‐pitch conformation, CK666 (Hetrick et al, 2013), inhibits SPIN90‐mediated activation (Fig 5A). This experiment indicates that SPIN90‐mediated activation occurs through formation of the short‐pitch conformation. Because SPIN90 can activate the complex on its own (i.e., without WASP), we deduce that it must stimulate the short‐pitch conformation, as directly observed for Dip1.
Figure 5. Binding of the SPIN90 ARM domain to the ARPC4 long helix may stimulate movement of Arp2 into or toward the short‐pitch conformation.
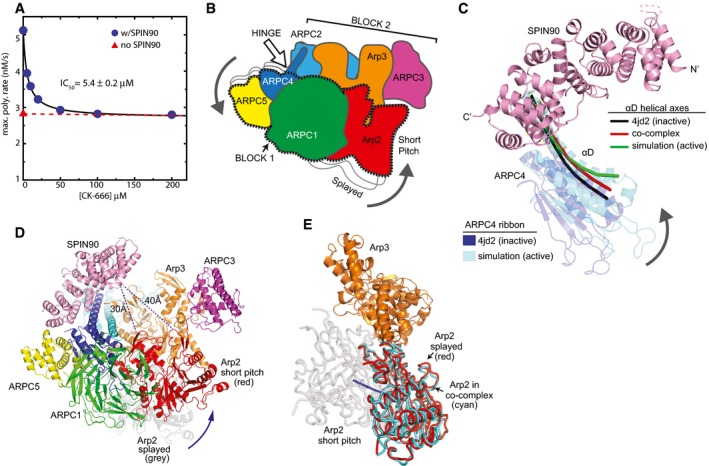
- Plot of maximum actin polymerization rate versus concentration of the small‐molecule Arp2/3 complex inhibitor CK‐666. Reactions contained 3 μM 15% pyrene‐labeled actin, 50 nM Bos taurus Arp2/3 complex, and the indicated concentrations of CK‐666.
- Cartoon of “rotation model” for movement of Arp2 into the short‐pitch conformation. White arrow shows the approximate position of a hinge point for the rotation.
- Structural superposition showing the proposed hinge movement in ARPC4 and the position of bound SPIN90 relative to the hinge. Residues 156–168 from ARPC4 and residues 247–260 from ARPC2 in an inactive crystal structure (4JD2) were overlaid with the same residues from the molecular dynamics (MD) short‐pitch model of Arp2/3 complex (Dalhaimer & Pollard, 2010). ARPC4 from 4JD2 (blue) and ARPC4 from the MD simulation (cyan) are shown. The helical axis of the long alpha helix (αD) in ARPC4 is shown as a black line in the 4JD2 structure and as a green line in the MD simulation structure. The red line represents the helical axis of helix αD in ARPC4 in the SPIN90‐bound co‐complex overlaid as described above. For clarity, the cartoon representation of the ARPC4 subunit in the co‐complex structure is not shown here, but is shown in Appendix Fig S10. The orientation of this panel is similar to that depicted in panels (B and D).
- Conceptual model of SPIN90‐bound Arp2/3 complex in the short‐pitch conformation. The model was made by superposing subdomains 1 and 2 of Arp3 from the co‐complex structure onto an actin subunit from a high‐resolution structure of an actin filament (3J8I; Galkin et al, 2015). Arp2 (red ribbon) was then moved into the short‐pitch position based on the filament structure. Arp2 in its original splayed position (as observed in the crystal structure) is shown in gray ribbon representation. Shortest Cα–Cα distances from SPIN90 to either subdomain 2 or subdomain 4 in Arp2 in the hypothetical short‐pitch conformation are indicated.
- Comparison of the position of Arp2 relative to Arp3 in the short‐pitch arrangement (gray Arp2), the inactive splayed arrangement (red Arp2), and the SPIN90 bound x‐ray crystal structure (cyan). The short‐pitch arrangement of Arp2‐Arp3 was modeled by overlaying subdomains 1 and 2 of Arp3 on the same subdomains in an actin subunit in the actin filament structure 3G37(Murakami et al, 2010) and then superposing Arp2 from the 4JD2 structure onto the actin subunit in the short‐pitch position. The SPIN90‐bound complex was then overlaid onto Arp3 subdomains 1 and 2 to determine the relative position of Arp2 in the SPIN90‐bound complex. Blue line shows a potential trajectory of the center of mass of Arp2 when it moves from the splayed to the short‐pitch position.
To investigate how SPIN90 stimulates the short‐pitch conformation, we first compared the SPIN90‐bound structure to a model of short‐pitch Arp2/3 complex from steered molecular dynamics simulations (Dalhaimer & Pollard, 2010). These simulations showed that adoption of the short‐pitch conformation occurs by rotation of a rigid body consisting of Arp2, ARPC1, ARPC5, and the globular portion of ARPC4 [block 1, (Fig 5B–D, Appendix Fig S10)]. This rotation requires bending of the long C‐terminal helices in ARPC4 and ARPC2, so that a block consisting of Arp3, ARPC3 and the globular portion of ARPC2 (block 2) remains fixed while block 1 rotates (Dalhaimer & Pollard, 2010). Importantly, SPIN90 binds near the putative hinge in the C‐terminal helix (αD) in ARPC4 (Fig 5C). Therefore, we speculate that SPIN90 allosterically stimulates movement of Arp2 into the short‐pitch conformation by triggering bending of the ARPC4 helix and subsequent rotation of a block of subunits containing Arp2. The ARPC4 αD helix is slightly more bent in the SPIN90‐bound structure compared to NPF‐free structures of Arp2/3 complex, but less bent than the MD‐derived short‐pitch model (Fig 5C). Unexpectedly, comparison to Arp2/3 complex structures without activator bound revealed that Arp2/3 complex is in the splayed (inactive) conformation in the SPIN90 co‐crystal structure (Fig 5E). Specifically, when Arp3 from each structure was superposed, the backbone atoms of Arp2 were in nearly identical positions (RMSD = 0.301 for 659 backbone atoms in Arp2). That Arp2/3 complex does not adopt the short‐pitch conformation in the crystal may mean that even with SPIN90 bound, the complex likely only weakly populates the short‐pitch conformation (see Discussion). However, we suspect that the partial bending of the ARPC4 αD helix may strain the complex, providing some destabilization of the inactive (splayed) conformation compared to Arp2/3 complexes without SPIN90 bound.
SPIN90 binds a surface that overlaps with the mother filament‐binding site
To investigate how WDS proteins can stimulate the activating changes caused by actin filaments during WASP‐mediated activation, we compared the SPIN90‐Arp2/3 co‐complex to the EM structure of Arp2/3 complex bound to the side of an actin filament at a branch junction (Rouiller et al, 2008). Importantly, this analysis showed that the SPIN90‐binding site on Arp2/3 complex overlaps with the site for preformed (mother) filaments of actin (Figs 6A and EV4). The long ARPC4 helix, which provides most of the binding interface for SPIN90, also provides a major portion (> 30%) of the overall interaction surface for actin filaments (Rouiller et al, 2008). These data indicate that SPIN90 and actin filaments compete for binding to Arp2/3 complex. To test this biochemically, we used an actin filament copelleting assay (Fig 6B and C). We found that 15 μM SPIN90 decreased the fraction of Arp2/3 complex that pelleted with filaments from 28 ± 2 to 8 ± 1%, demonstrating competition between actin filaments and SPIN90.
Figure 6. SPIN90 binds to the actin filament binding interface on Arp2/3 complex and blocks filament binding.
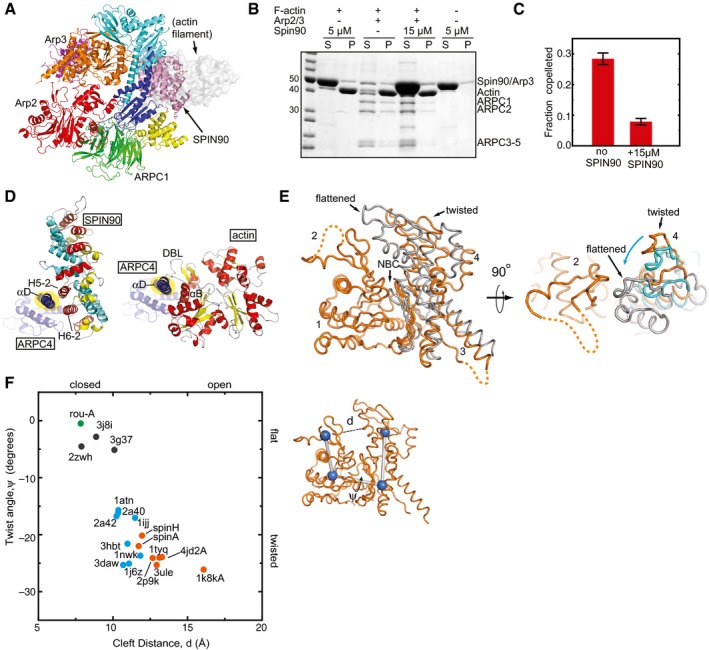
- Ribbon diagram of the co‐complex structure showing where an actin filament (gray semi‐transparent surface model) binds to Arp2/3 complex. To model the bound filament, ARPC2 and ARPC4 from the EM reconstruction of Arp2/3 complex at a branch junction were overlaid onto ARPC2 and ARPC4 from the co‐complex structure (Rouiller et al, 2008).
- SDS–PAGE gel showing results of actin filament copelleting assay. Actin filaments at 3.5 μM were used to co‐pellet 0.6 μM Bos taurus Arp2/3 complex in the presence or absence of the indicated concentrations of SPIN90 (269–722; S, soluble fraction; P, pellet)
- Plot of copelleting data as shown in (B), run in quadruplicate with error bars showing the standard deviation.
- Comparison of the contacts made by SPIN90 (left panel) and actin filaments (Rouiller et al, 2008; right panel) with the αD helix in ARPC4 (ARPC4 is blue, with the αD helix highlighted in yellow). SPIN90 is colored according to the helix number within each repeat (helix 1—cyan, helix 2—yellow, helix 3—red). The actin subunit from the filament that contacts ARPC4 is colored according to secondary structure (helix—red, strand—yellow). Select elements of secondary structure are labeled, as is the DNase I‐binding loop of actin (DBL).
- Superposition of subdomains 1 and 2 of Arp3 from the inactive GMF‐bound structure (Luan & Nolen, 2013; 4JD2, orange) with Arp3 from the branch junction model (Rouiller et al, 2008; gray) and Arp3 from the SPIN90‐bound structure (cyan—shown only in right panel). Regions disordered in the structure are shown as dashed lines. Cyan arrow indicates the direction of motion in the transition from the twisted to flattened conformation.
- Plot of the nucleotide cleft width versus the twisting dihedral angle in Arp3 from x‐ray crystal structures, Arp3 from the branch junction EM reconstruction, or filamentous or monomeric actin structures. The PDB accession number is indicated for each data point. Orange: Arp3, x‐ray crystal structures; cyan: monomeric actin x‐ray crystal structures; black: actin filament fiber diffraction or cryoEM structures, green: Arp3 (rou‐A) from branch junction reconstruction. Twist angle was defined as the dihedral made by connecting the center of mass of each subdomain 1–4. Nucleotide cleft distance was defined as the distance between Cα atoms in residues 67 and 222 (Arp3) or 59 and 207 (actin).
Figure EV4. SPIN90 binds to the same surface of Arp2/3 complex as actin filaments.
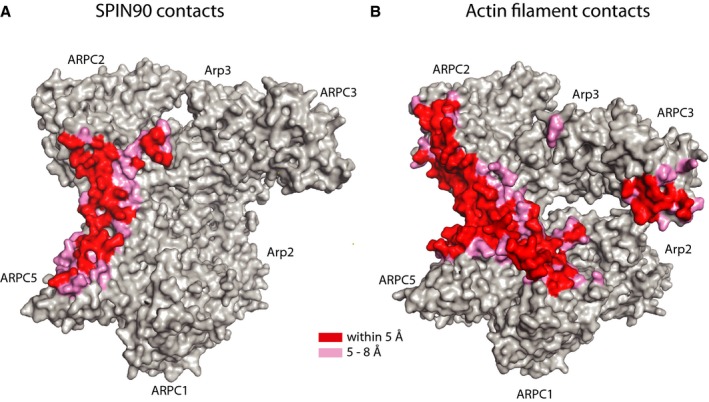
- Surface representation of the “front side” of Arp2/3 from SPIN90 co‐complex structure showing residues that are within 5 Å of SPIN90 (red) or between 5 and 8 Å (pink). Side chains were modeled onto the co‐complex structure before generating the surface.
- Surface representation of the “front side” of Arp2/3 from the branch junction model colored as described in (A) (Rouiller et al, 2008), except close contacts between the complex and actin filaments are colored.
We next asked whether SPIN90 structurally mimics part of an actin filament in its contacts with Arp2/3 complex. A comparison of the interfaces revealed the surfaces that engage Arp2/3 complex in actin filaments versus SPIN90 are distinct. For instance, while helices H5‐2 and H6‐2 of SPIN90 form an anti‐parallel three‐helix bundle with the αD helix in ARPC4, the same helix in ARPC4 packs between an obliquely aligned alpha helix (αB) and a loop segment (the DNase‐binding loop) in an actin subunit within a filament (Fig 6D). Therefore, SPIN90 and actin filaments use distinct structural modules to contact a common binding site along the long C‐terminal (αD) helix in ARPC4 in Arp2/3 complex.
The location of the SPIN90‐binding site on Arp2/3 complex suggests SPIN90 may stimulate the same activating conformational changes as actin filaments by engaging the actin filament‐binding site. The EM model of Arp2/3 complex at a branch junction, which represents the final activated conformation after all activating factors have acted on the complex, shows that in addition to adopting the short‐pitch conformation, both Arp2 and Arp3 undergo an intrasubunit scissoring motion (Rouiller et al, 2008). This motion changes the Arp subunits from a “twisted” to a “flattened” conformation, making them more closely resemble the structure of actin subunits within an actin filament (Oda et al, 2009). Because WASP and actin monomers stimulate movement of Arp2 and Arp3 into or toward the short‐pitch conformation (Boczkowska et al, 2008; Xu et al, 2012; Hetrick et al, 2013; Rodnick‐Smith et al, 2016b), one possibility is that actin filaments contribute to nucleation by stimulating the flattening conformational change. Recent FRET experiments support this model, showing that added actin filaments caused FRET probes on ARPC3 and ARPC1 to move closer together, as expected when Arp3 flattens (Espinoza Sanchez et al, 2018). To determine whether SPIN90 binding caused subunit flattening, we superposed subdomains 1 and 2 from Arp3 in the SPIN90‐bound complex to Arp3 in the branch junction model or in NPF‐free structures (Rouiller et al, 2008; Luan & Nolen, 2013). This analysis revealed that in the SPIN90‐bound complex, Arp3 rotates ~2° toward the flattened conformation relative to the conformation observed in the inactive crystal structures (Fig 6E). This slight “flattening” occurs in both Arp3 subunits in the asymmetric unit and is accompanied by a slight closing of the nucleotide cleft (Fig 6F). This structural change may represent an important SPIN90‐mediated shift toward the activated conformation, though we note that the twist angle varies from −15° to −25° for all nucleotide‐bound monomeric actin and inactive Arp3 subunits we analyzed, so it is not yet clear whether the small bias in Arp3 toward the flattened conformation is functionally relevant (Fig 6F).
Discussion
Here, we solved the structure of the ARM domain of SPIN90 bound to Arp2/3 complex. We show that SPIN90 binds to the front side of Arp2/3 complex, contacting the long C‐terminal helix (αD) in ARPC4. Our data indicate that by binding to this site, WDS proteins trigger the activating changes in the complex normally stimulated by the coordinated action of WASP and pre‐existing actin filaments (Fig 7). That SPIN90 binds to distinct sites on Arp2/3 complex compared to WASP was surprising, given that both classes of NPF stimulate the same conformational change: movement of Arp2 into or toward the short‐pitch conformation. Therefore, our results show that multiple interaction surfaces on the complex are allosterically linked to the relative arrangement of the Arp2 and Arp3 subunits.
Figure 7. Cartoon model showing the conformational pathway to activation of Arp2/3 complex by WASP or WDS family proteins.
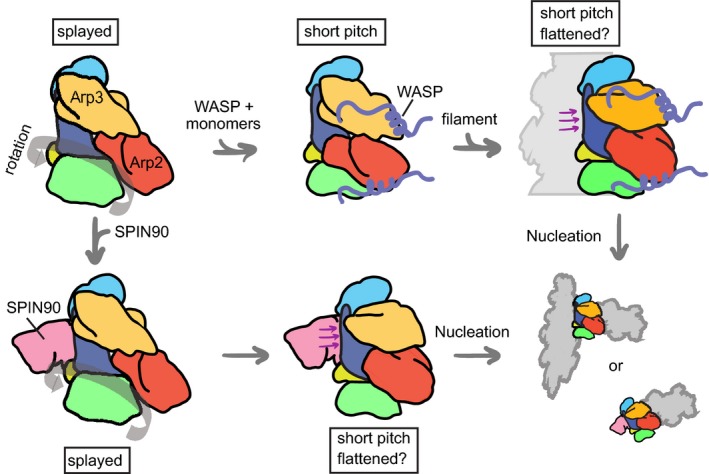
Arp2/3 complex subunits are colored as follows: Arp3, orange; Arp2, red; ARPC1, green; ARPC2, cyan; ARPC3 (not shown for clarity); ARPC4, blue; ARPC5, yellow. Actin monomers recruited by WASP are not shown. Purple arrows indicate contact between actin filaments or SPIN90 with ARPC4 that may stimulate a structural change required for activation. Thick gray arrow in left panels indicates potential rigid‐body rotation of Arp2, ARPC1, ARPC5, and the globular portion of ARPC4 hypothesized to move Arp2 into the short‐pitch arrangement (Dalhaimer & Pollard, 2010).
Two different models have been proposed to explain how Arp2/3 complex switches from the splayed to the short‐pitch conformation. In one model, supported by MD simulations, the complex bends at hinge points in the long alpha helices in ARPC2 and ARPC4 to move Arp2, along with a rigid block of other subunits, into the short‐pitch position (Robinson et al, 2001; Dalhaimer & Pollard, 2010; Fig 5B). The SPIN90 ARM domain binds at the proposed hinge point, so the rotation model provides a simple mechanism by which WDS family proteins could stimulate the short‐pitch conformation (Fig 7). In the second model, supported by low‐resolution EM reconstructions (Rouiller et al, 2008), Arp2 releases all contacts from the complex in the splayed conformation, then rebinds in the short‐pitch conformation. Our analysis of the co‐complex structure did not reveal any structural clues to how binding of the SPIN90 ARM could trigger release of Arp2. We also considered the possibility that SPIN90 could influence adoption of the short‐pitch conformation by stabilizing contacts made in the short‐pitch conformation. To address this, we overlaid short‐pitch Arp2/3 complex from the EM branch junction model onto the co‐complex structure (Fig 5D). Importantly, we found that while Arp2 moves toward SPIN90 when the complex adopts the short‐pitch position, the closest approach between short‐pitch Arp2 and SPIN90 is still ~30 Å, arguing against a model in which a direct contact between SPIN90 and Arp2 could stabilize the short‐pitch conformation. Therefore, our observations support a model in which SPIN90 binding along the ARPC4 αD helix stimulates rotation of Arp2 into the short‐pitch rearrangement. An important open question is why Arp2/3 complex adopts the splayed (inactive) conformation in our co‐crystal structure, even though SPIN90 is bound. The simplest explanation is that SPIN90 only weakly shifts the equilibrium toward the activated state, but this extends the time the complex inhabits the active state long enough to allow nucleation. Consistent with this hypothesis, we analyzed the number of filament ends nucleated at saturating concentrations of SPIN90 (269–722) and 50 nM Arp2/3 complex in the reactions shown in Fig 1E (Appendix Fig S11). We found that less than 0.1 nM ends were present at the beginning of the reaction and that new ends were initially created at 3 × 10−6 μM/s. We preincubated SPIN90 and Arp2/3 complex, so we assume 50 nM SPIN90‐Arp2/3 complex is present upon initiation of polymerization. These observations suggest additional steps must occur after SPIN90 binds Arp2/3 complex for the nucleus to be created. These steps might include an activating conformational switch, consistent with a heavily populated inactive state as suggested by the inactive conformation we observed in the SPIN90‐bound Arp2/3 complex structure, but could also be explained by other reactions, such as collision of SPIN90‐bound Arp2/3 complex with the first actin monomer(s).
Our data indicate that unlike Arp2/3 complex activators from the WASP family (Marchand et al, 2001), SPIN90 folds into a globular domain. The C‐terminal portion of this domain, which we show here is an armadillo repeat domain, is present in all WDS proteins and forms a conserved contact surface for SPIN90 with Arp2/3 complex. However, we note that WDS proteins from distinct species may differ in the way they activate Arp2/3 complex. For instance, in S. pombe Dip1, a fragment consisting of the conserved C‐terminus, which corresponds to repeats 4–7 in the ARM domain, is sufficient for full activity (Wagner et al, 2013). In contrast, we show here that in SPIN90, both the full ARM domain (repeats 1–7) and an additional flexible region (the middle segment, MS) are required for activation. While the function of the MS is not yet clear, our data suggest it influences the ability of the ARM domain to bind the complex (Fig 1). The MS is disordered in the co‐complex structure, with the potential exception of two short unassigned alpha helices that are disconnected from the ARM domain. We anticipate that understanding why the human but not the fungal WDS protein requires this additional segment to trigger nucleation activity will reveal important insights into the molecular basis for the regulation of WDS proteins.
A critical observation in this work is that the ARM domain of SPIN90 binds to a surface on Arp2/3 complex that overlaps with actin filament‐binding site. Binding to this site likely allows SPIN90 to stimulate the same activating step as actin filaments (Fig 7). While we outline evidence above that suggests that this step involves flattening of the Arp2 and Arp3 subunits, filaments may stimulate a different structural change for activation. For instance, some investigators have proposed a model in which WASP stimulates movement of Arp2 part way toward the short‐pitch conformation, and actin filaments trigger the transition from this intermediate state to the fully short‐pitch conformation (Espinoza Sanchez et al, 2018). There is no direct evidence for a stable intermediate conformation between the splayed and short‐pitch positions of Arp2, but recent FRET studies showed that both actin filaments and WASP‐binding move FRET probes on Arp2 and Arp3 closer together, indicating both ligands stimulate movement toward or into the short‐pitch conformation. Additional studies will be required to determine the exact nature of the filament‐induced activation step and precisely how SPIN90 could trigger it. Further, it will be important to determine why SPIN90 appears to be capable of triggering all the steps required for potent Arp2/3 complex activation by engaging the long ARPC4 helix, but actin filaments cannot. This will be critical for understanding Arp2/3 complex regulation, since removing this limitation would uncouple Arp2/3 complex from cellular signaling pathways. We anticipate that the distinct structural modules SPIN90 and filaments use to contact ARPC4 underpin these molecular differences (Fig 6D), but additional high‐resolution structural and biochemical information will be required to understand this issue.
We showed previously that SPIN90 not only activates Arp2/3 complex to generate linear filaments, but that these linear filaments may be blocked from annealing to the sides of pre‐existing filaments, since branches were never observed in TIRF microscopy experiments containing SPIN90 and Arp2/3 complex (Wagner et al, 2013). Our data here suggest that SPIN90 inhibits end‐to‐side annealing (branching without nucleation) by directly blocking Arp2/3 complex from binding to filament sides. Therefore, while WDS proteins likely play an important role in initiating the assembly of branched networks (Basu & Chang, 2011; Wagner et al, 2013), they nucleate linear filaments and prevent them from forming branches. This suggests that cells must finely tune WDS protein activity to create enough seed filaments to initiate branching without significantly altering the dendritic nature of Arp2/3‐generated networks. Understanding this regulation and how it balances the activity of WDS proteins relative to WASP will be important to address. The conformational change we observed in the H5‐1/2 loop comparing active to inactive SPIN90 structures provides an initial clue as to how the activity of SPIN90 might be switched on and off (Fig 2).
Finally, we note that because WDS and WASP proteins engage distinct sites on Arp2/3 complex, they could simultaneously bind and cooperatively trigger adoption of the short‐pitch conformation. While no previous data address whether WDS and WASP family proteins directly synergize, there is some precedence for cooperative activation of Arp2/3 complex by distinct families of NPFs. For instance, cortactin, an NPF that binds actin filaments instead of monomers, synergizes with dimeric WASP family proteins by interacting with one of the two WASP‐CA‐binding sites on the complex, accelerating WASP release from nascent branch junctions (Weaver et al, 2001, 2002; Helgeson & Nolen, 2013; Helgeson et al, 2014). Given that WASP and WDS family proteins are both present in multiple actin networks in cells (Kim et al, 2006; Basu & Chang, 2011), it will be important to understand how the coordinated activity of these NPFs influences the kinetics of nucleation and the architectures of networks assembled by Arp2/3 complex. We note that multiple Arp2/3 complex regulators harbor WASP‐like sequences, and are thought to engage one or both WASP‐CA‐binding sites on the complex (Goley & Welch, 2006; Liu et al, 2011; Padrick et al, 2011; Ti et al, 2011; Maritzen et al, 2012; Dang et al, 2013; Luan et al, 2018). These regulators include both activators and inhibitors of nucleation. Understanding whether and how they coordinately regulate Arp2/3 complex with WDS proteins will be important for understanding cellular actin assembly.
Materials and Methods
Cloning, protein expression, and purification
Bos taurus Arp2/3 complex and rabbit skeletal muscle actin were purified as previously described (Pollard, 1984; Robinson et al, 2001). A plasmid expressing Homo sapiens SPIN90 (269–722) was constructed previously (Wagner et al, 2013). Point mutations and truncations were generated from the SPIN90 (269–722) expression vector by PCR‐based mutagenesis and verified by sequencing. SPIN90 expression vectors were transformed into BL21(DE3)RIL Escherichia coli cells and grown in LB medium at 37°C to an OD595 of 0.6 before inducing with 0.4 mM IPTG overnight at 22°C. Cells were harvested and lysed by sonication, then clarified by centrifugation. The soluble fraction was loaded onto a Glutathione Sepharose 4B (GS4B, GE‐Healthcare) column equilibrated with GST‐binding buffer (20 mM Tris pH 8.0, 140 mM NaCl, 2 mM EDTA, 1 mM DTT), washed with GST‐binding buffer and eluted with GST‐binding buffer containing 50 mM reduced glutathione. TEV protease was added at a mass ratio of 1:100, and the solution was dialyzed against QA buffer (20 mM Tris pH 8.0, 50 mM NaCl, 1 mM DTT) at 4°C overnight. The dialysate was loaded on a 6‐ml source 30Q (GE‐Healthcare) column, washed with QA buffer, and eluted with a salt gradient from 50 to 500 mM. Pooled fractions were passed through a GS4B column to remove any remaining GST and then further purified on SuperDex 75 or 200 column (GE‐Healthcare) in 20 mM Tris pH 8.0, 100 mM NaCl, and 1 mM DTT. Pure fractions from the gel filtration column were concentrated in a Vivaspin Turbo 15 concentrator (Sartorius) and exchanged into storage buffer (20 mM Tris pH 8.0, 50 mM NaCl, 1 mM DTT) before flash freezing in liquid nitrogen and storing at −80°C. See Appendix Fig S12 for Coomassie‐stained SDS–PAGE gel of final purified protein preparations.
Selenomethionyl proteins were expressed as previously described (Doublié, 2007). Briefly, BL21(DE3)RIL E. coli cells transformed with the desired plasmid were grown in LB medium at 37°C overnight. Cells were harvested and resuspended in M9a medium (6 g/l Na2HPO4, 3 g/l KH2PO4, 0.5 g/l NaCl, 1 g/l (NH4)Cl, 0.4% glucose, 5 mg/l thiamine, 1 mM MgSO4) and grown at 37°C to OD595 of 0.6. Subsequently, 100 mg/l lysine, 100 mg/l threonine, 100 mg/l phenylalanine, 50 mg/l leucine, 50 mg/l isoleucine, and 50 mg/l valine were added, and the culture was incubated with shaking for 15 min. 50 mg/l selenomethionine was added, and the culture was incubated for an additional 15 min before adding 0.4 mM IPTG and growing at 22°C overnight. Cells were harvested, and protein was purified as described above.
Pyrene actin polymerization assays
Time courses of pyrene actin polymerization were carried by adding 2 μl of MgCl2 (50 μM)/EGTA (0.2 mM) to 20 μl of a solution of 15 μM 15% pyrene‐labeled actin and incubating for 30 s. A polymerization initiation solution (78 μl) was added to this solution to make a final reaction mix containing 3 μM 15% pyrene‐labeled actin in 1× KMEI buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole, pH 7.0). Polymerization was monitored by the fluorescence at 407 nm (excitation = 365 nm) in a Tecan Safire plate reader. The maximum rate of actin polymerization was determined by multiplying the slope of polymerization curve at each time point (RFU/s) by the concentration of polymer per fluorescence unit. The ratio of polymer to fluorescence was determined by assuming that the polymer concentration at equilibrium was the total actin concentration minus 0.1 μM, the critical concentration (Pollard, 1986).
Actin binding/copelleting assays
Actin monomers in G‐buffer (2 mM Tris‐Cl pH 8.0, 0.2 mM ATP, 0.5 mM DTT, 0.1 mM CaCl2) were added to a polymerization solution to make a final solution containing 3.5 μM actin, 50 mM KCl, 10 mM imidazole pH 7.0, 1 mM MgCl2, and 1 mM EGTA. This solution was incubated at room temperature for 1 h before adding 0.6 μM Bos taurus Arp2/3 complex with or without 5 or 15 μM SPIN90 (269–722). After incubation for 30 min at room temperature, the solution was spun at 85 K in a TLA100 rotor at 22°C for 20 min. The supernatant was removed and the pellet resuspended in boiling SDS–PAGE loading buffer. Both the supernatant and the pellet fractions were analyzed by SDS–PAGE.
Crystal growth, data collection, refinement, and structural analysis
Crystals of SPIN90 (351–722) were grown by mixing 0.5 μl of a 470 μM protein solution with 0.5 μl 50 mM Tris pH 7.5 and 700 mM sodium potassium tartrate and equilibrating by vapor diffusion from a hanging drop at room temperature. Crushed crystals were used as micro seeds in various dilutions to improve the quality of the crystals. Micro‐seeded crystals grew to ~50 × 75 × 200 μm within 2 days. Upon harvest, the crystals were sequentially dipped in three drops of crystallization solution containing 40% glycerol as cryoprotectant and flash frozen in liquid nitrogen. Data were collected at 100 K at wavelength of 0.979423 Å at beam 19‐BM at Argonne National Laboratory and processed with HKL3000 (Minor et al, 2006). To solve the phases, selenomethionyl SPIN90 (351–722) was crystallized and prepared for data collection using the same protocol, except that 40% ethylene glycol was used as cryoprotectant. Crystals grew to ~30 × 50 × 150 μm in 2 days. Anomalous data were collected at 100 K at wavelength of 0.9797418 Å at beam 19‐BM and indexed, integrated, and scaled with HKL3000 (Minor et al, 2006; Table EV1). Phases were solved using program Phenix Autosol (Adams et al, 2010), which generated an interpretable electron density map with an overall figure of merit of 0.405 and two molecules in the asymmetric unit. The model generated from Autosol was used as a search model in a Phaser molecular replacement run against the native dataset (McCoy et al, 2007). This model was expanded with Phenix Autobuild (Adams et al, 2010) and manually rebuilt using Coot (Emsley et al, 2010). The XYZ coordinates, TLS parameters, and individual B‐factors of the model were refined in Phenix and Refmac using weighted NCS restrains (Murshudov et al, 1997). The Ramachandran statistics of the final model were as follows: 99% favored, 1% allowed, and 0% outlier. The clash score was 0.93. The model has been deposited in the Protein Data Bank with accession code 6DED.pdb.
SPIN90 (306–722) was crystallized by mixing 0.5 μl of 320 μM protein with 0.5 μl 50 mM MES pH 6.0, 100 mM MgSO4, and 1.5% PEG6000 and equilibrating by vapor diffusion from a hanging drop at room temperature. Crystals grew to ~50 × 50 × 50 μm within 3 days. Upon harvest, the crystals were sequentially dipped in three drops of crystallization solution containing 45% glycerol as cryoprotectant and flash frozen in liquid nitrogen. Data were collected at 100 K at wavelength of 0.9791829 Å at beam 19‐ID at Argonne National Laboratory and processed with HKL3000 (Minor et al, 2006). Molecular replacement was carried out with Phaser, using the SPIN90 (351–722) structure with side chains removed from three regions (389–407, 516–611 and 648–712; McCoy et al, 2007). The N‐terminal sequence outside of the ARM domain was built with Phenix Autobuild, and the sequence registry was assigned based on the anomalous signal from a selenomethionyl SPIN90 (306–722) dataset (Table S1). Side chains of the amino acid residues were included or excluded from the model based on the electron density coverage and R‐free changes upon refinement. Geometry and B‐factor refinements were carried out in Refmac and Phenix with secondary structure restraints. Manual rebuilding was carried out iteratively in Coot (Emsley et al, 2010). The Ramachandran statistics of the final model were as follows: 98% favored, 2% allowed, and 0% outlier. The clash score was 2.36. The model has been deposited in the Protein Data Bank with accession code 6DEE.pdb.
To crystallize the co‐complex, a solution containing 25 μM bovine Arp2/3 complex, 50 μM SPIN90 (269–722), 0.5 mM ATP, 0.5 mM CaCl2 and 1 mM DTT was mixed 1:1 with 50 mM Hepes pH 7.5, 5% PEG3350, and 50 mM L‐proline and equilibrated by vapor diffusion from a hanging drop at room temperature. Crushed crystals were used as micro seeds in various dilutions to improve the quality of the crystals. Crystals grew to ~40 × 40 × 120 μm in about a week. Upon harvest, the crystals were sequentially dipped in three drops of crystallization solution containing 40% ethylene glycol as cryoprotectant and flash frozen in liquid nitrogen. Data were collected at 100 K at wavelength of 0.9791829 Å at beam 19‐ID at Argonne National Laboratory and processed with HKL3000. Poly‐alanine versions of Arp2/3 complex from 4jd2 (Luan & Nolen, 2013) and the ARM domain of the SPIN90 (306–722) structure were used as search models for molecular replacement with Phaser. The molecular replacement solution was first minimized by rigid‐body refinement allowing individual subunits of Arp2/3 complex, SPIN90, and subdomains 1 and 2 or 3 and 4 of the Arp2 and Arp3 subunits to move independently. Geometry, TLS parameters, and group B‐factors were refined in Refmac with weighted NCS and secondary structure restraints. Manual adjustments to the model were made using Coot and validated based on the electron density coverage and R‐free changes upon refinement. The Ramachandran statistics of the final model were as follows: 98% favored, 2% allowed, and 0% outlier. The clash score was 2. The model has been deposited in the Protein Data Bank with accession code 6DEC.pdb).
The conservation of surface residues in the ARM domain was analyzed using the ConSurf server, using 31 sequences of WDS proteins from diverse species (Ashkenazy et al, 2016). The sequences included were from the following species: S. cerevisiae, S. pombe, X. tropicalis, D. discoideum, B. taurus, C. porcellus, D. melanogaster, L. thermotolerans, C. elegans, P. alecto, H. sapiens, A. aegypti, O. latipes, C. albicans, C. glabrata, E. caballus, C. jacchus, T. thermophla, B. impatiens, G. gallus, T. nigroviridis, T. guttata, C. quinquefasciatus, N. vitripennis, A. terreus, D. rerio, A. florea, M. musculus, T. rubripes, A. gossypi, and X. laevis.
Data availability
The three x‐ray crystal structures reported here have been deposited in the Protein Data Bank under accession codes 6DEC,6DEE, and 6DED (http://www.rcsb.org/pdb/explore/explore.do?structureId=6DEC, http://www.rcsb.org/pdb/explore/explore.do?structureId=6DEE, and http://www.rcsb.org/pdb/explore/explore.do?structureId=6DED, respectively).
Author contributions
QL, S‐LL, and LAH performed the experiments, BJN and QL designed the experiments and interpreted the results, BJN wrote the manuscript with input from all authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Review Process File
Acknowledgements
We thank Jim Remington for his advice and assistance with solving the phases and Christian La France for assistance with cloning SPIN90 point mutations. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers R01 GM092917 and R01 GM127440 (B.J.N) and T32 GM007759 (L.A.H.). Results are derived from work performed at Argonne National Laboratory, Structural Biology Center (SBC) at the Advanced Photon Source. SBC‐CAT is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE‐AC02‐06CH11357.
The EMBO Journal (2018) 37: e100005
References
- Achard V, Martiel J‐L, Michelot A, Guérin C, Reymann A‐C, Blanchoin L, Boujemaa‐Paterski R (2010) A ‘primer’‐based mechanism underlies branched actin filament network formation and motility. Curr Biol 20: 423–428 [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L‐W, Kapral GJ, Grosse‐Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben‐Tal N (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44: W344–W350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R, Chang F (2011) Characterization of dip1p reveals a switch in Arp2/3‐dependent actin assembly for fission yeast endocytosis. Curr Biol 21: 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltzner CC, Pollard TD (2004) Identification of functionally important residues of Arp2/3 complex by analysis of homology models from diverse species. J Mol Biol 336: 551–565 [DOI] [PubMed] [Google Scholar]
- Boczkowska M, Rebowski G, Petoukhov MV, Hayes DB, Svergun DI, Dominguez R (2008) X‐ray scattering study of activated Arp2/3 complex with bound actin‐WCA. Structure 16: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczkowska M, Rebowski G, Kast DJ, Dominguez R (2014) Structural analysis of the transitional state of Arp2/3 complex activation by two actin‐bound WCAs. Nat Commun 5: 3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Makhov AM, Schafer DA, Bear JE (2008) Coronin 1B antagonizes cortactin and remodels Arp2/3‐containing actin branches in lamellipodia. Cell 134: 828–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD (2010) A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol 11: 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhaimer P, Pollard TD (2010) Molecular dynamics simulations of Arp2/3 complex activation. Biophys J 99: 2568–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang I, Gorelik R, Sousa‐Blin C, Derivery E, Guérin C, Linkner J, Nemethova M, Dumortier JG, Giger FA, Chipysheva TA, Ermilova VD, Vacher S, Campanacci V, Herrada I, Planson A‐G, Fetics S, Henriot V, David V, Oguievetskaia K, Lakisic G et al (2013) Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature 503: 281–284 [DOI] [PubMed] [Google Scholar]
- Doublié S (2007) Production of selenomethionyl proteins in prokaryotic and eukaryotic expression systems. Methods Mol Biol 363: 91–108 [DOI] [PubMed] [Google Scholar]
- Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS (2007) Dia‐interacting protein modulates formin‐mediated actin assembly at the cell cortex. Curr Biol 17: 579–591 [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza Sanchez S, Metskas LA, Chou SZ, Rhoades E, Pollard TD (2018) Conformational changes in Arp2/3 complex induced by ATP, WASp‐VCA and actin filaments. Proc Natl Acad Sci USA 115: E8642–E8651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka M, Suetsugu S, Miki H, Fukami K, Endo T, Takenawa T (2001) A novel neural Wiskott‐Aldrich syndrome protein (N‐WASP) binding protein, WISH, induces Arp2/3 complex activation independent of Cdc42. J Cell Biol 152: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Vos MR, Schröder GF, Egelman EH (2015) Near‐atomic resolution for one state of F‐actin. Structure 23: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M, Smith BA, Bovellan M, Paavilainen V, Daugherty‐Clarke K, Gelles J, Lappalainen P, Goode BL (2010) GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr Biol 20: 861–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Welch MD (2006) The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol 7: 713–726 [DOI] [PubMed] [Google Scholar]
- Groves MR, Barford D (1999) Topological characteristics of helical repeat proteins. Curr Opin Struct Biol 9: 383–389 [DOI] [PubMed] [Google Scholar]
- Gul IS, Hulpiau P, Saeys Y, van Roy F (2017) Metazoan evolution of the armadillo repeat superfamily. Cell Mol Life Sci 74: 525–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeson LA, Nolen BJ (2013) Mechanism of synergistic activation of Arp2/3 complex by cortactin and N‐WASP. eLife 2: e00884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeson LA, Prendergast JG, Wagner AR, Rodnick‐Smith M, Nolen BJ (2014) Interactions with actin monomers, actin filaments, and Arp2/3 complex define the roles of WASP family proteins and cortactin in coordinately regulating branched actin networks. J Biol Chem 289: 28856–28869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick B, Han MS, Helgeson LA, Nolen BJ (2013) Small molecules CK‐666 and CK‐869 inhibit actin‐related protein 2/3 complex by blocking an activating conformational change. Chem Biol 20: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI (1997) Three‐dimensional structure of the armadillo repeat region of beta‐catenin. Cell 90: 871–882 [DOI] [PubMed] [Google Scholar]
- Humphries CL, Balcer HI, D'Agostino JL, Winsor B, Drubin DG, Barnes G, Andrews BJ, Goode BL (2002) Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J Cell Biol 159: 993–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgenson CT, Pollard TD (2015) Crystals of the Arp2/3 complex in two new space groups with structural information about actin‐related protein 2 and potential WASP binding sites. Acta Crystallogr Sect F Struct Biol Commun 71: 1161–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DJ, Kim SH, Lim CS, Choi KY, Park CS, Sung BH, Yeo MG, Chang S, Kim J‐K, Song WK (2006) Interaction of SPIN90 with the Arp2/3 complex mediates lamellipodia and actin comet tail formation. J Biol Chem 281: 617–625 [DOI] [PubMed] [Google Scholar]
- Kim S‐M, Bae J, Cho IH, Choi KY, Park YJ, Ryu JH, Chun J‐S, Song WK (2011) Control of growth cone motility and neurite outgrowth by SPIN90. Exp Cell Res 317: 2276–2287 [DOI] [PubMed] [Google Scholar]
- Kobe B (1999) Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat Struct Biol 6: 388–397 [DOI] [PubMed] [Google Scholar]
- Lim CS, Park ES, Kim DJ, Song YH, Eom SH, Chun JS, Kim JH, Kim JK, Park D, Song WK (2001) SPIN90 (SH3 protein interacting with Nck, 90 kDa), an adaptor protein that is developmentally regulated during cardiac myocyte differentiation. J Biol Chem 276: 12871–12878 [DOI] [PubMed] [Google Scholar]
- Liu S‐L, Needham KM, May JR, Nolen BJ (2011) Mechanism of a concentration‐dependent switch between activation and inhibition of Arp2/3 complex by coronin. J Biol Chem 286: 17039–17046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S‐L, May JR, Helgeson LA, Nolen BJ (2013) Insertions within the actin core of actin‐related protein 3 (Arp3) modulate branching nucleation by Arp2/3 complex. J Biol Chem 288: 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Q, Nolen BJ (2013) Structural basis for regulation of Arp2/3 complex by GMF. Nat Struct Mol Biol 20: 1062–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Q, Zelter A, MacCoss MJ, Davis TN, Nolen BJ (2018) Identification of Wiskott‐Aldrich syndrome protein (WASP) binding sites on the branched actin filament nucleator Arp2/3 complex. Proc Natl Acad Sci USA 115: E1409–E1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD (1999) Scar, a WASp‐related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA 96: 3739–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand JB, Kaiser DA, Pollard TD, Higgs HN (2001) Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol 3: 76–82 [DOI] [PubMed] [Google Scholar]
- Maritzen T, Zech T, Schmidt MR, Krause E, Machesky LM, Haucke V (2012) Gadkin negatively regulates cell spreading and motility via sequestration of the actin‐nucleating ARP2/3 complex. Proc Natl Acad Sci USA 109: 10382–10387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul RS, Song Y, Amann KJ, Gerbin SC, Pollard TD, Chang DD (2003) EPLIN regulates actin dynamics by cross‐linking and stabilizing filaments. J Cell Biol 160: 399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Numazaki M, Takeuchi K, Uchibori Y, Ando‐Akatsuka Y, Tominaga M, Tominaga T (2004) DIP (mDia interacting protein) is a key molecule regulating Rho and Rac in a Src‐dependent manner. EMBO J 23: 760–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M (2006) HKL‐3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62: 859–866 [DOI] [PubMed] [Google Scholar]
- Moreau V, Madania A, Martin RP, Winson B (1996) The Saccharomyces cerevisiae actin‐related protein Arp2 is involved in the actin cytoskeleton. J Cell Biol 134: 117–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita EC, Murayama K, Kato‐Murayama M, Ishizuka‐Katsura Y, Tomabechi Y, Hayashi T, Terada T, Handa N, Shirouzu M, Akiyama T, Yokoyama S (2011) Crystal structures of the armadillo repeat domain of adenomatous polyposis coli and its complex with the tyrosine‐rich domain of Sam68. Structure 19: 1496–1508 [DOI] [PubMed] [Google Scholar]
- Murakami K, Yasunaga T, Noguchi TQP, Gomibuchi Y, Ngo KX, Uyeda TQP, Wakabayashi T (2010) Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell 143: 275–287 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum‐likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nakano K, Kuwayama H, Kawasaki M, Numata O, Takaine M (2010) GMF is an evolutionarily developed Adf/cofilin‐super family protein involved in the Arp2/3 complex‐mediated organization of the actin cytoskeleton. Cytoskeleton (Hoboken) 67: 373–382 [DOI] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maéda Y, Narita A (2009) The nature of the globular‐ to fibrous‐actin transition. Nature 457: 441–445 [DOI] [PubMed] [Google Scholar]
- Oh H, Kim H, Chung K‐H, Hong NH, Shin B, Park WJ, Jun Y, Rhee S, Song WK (2013) SPIN90 knockdown attenuates the formation and movement of endosomal vesicles in the early stages of epidermal growth factor receptor endocytosis. PLoS One 8: e82610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK (2011) Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci USA 108: E472–E479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD (1984) Polymerization of ADP‐actin. J Cell Biol 99: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD (1986) Rate constants for the reactions of ATP‐ and ADP‐actin with the ends of actin filaments. J Cell Biol 103: 2747–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD (2001) Crystal structure of Arp2/3 complex. Science 294: 1679–1684 [DOI] [PubMed] [Google Scholar]
- Rodnick‐Smith M, Liu S‐L, Balzer CJ, Luan Q, Nolen BJ (2016a) Identification of an ATP‐controlled allosteric switch that controls actin filament nucleation by Arp2/3 complex. Nat Commun 7: 12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnick‐Smith M, Luan Q, Liu S‐L, Nolen BJ (2016b) Role and structural mechanism of WASP‐triggered conformational changes in branched actin filament nucleation by Arp2/3 complex. Proc Natl Acad Sci USA 113: E3834–E3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW (1999) The interaction between N‐WASP and the Arp2/3 complex links Cdc42‐dependent signals to actin assembly. Cell 97: 221–231 [DOI] [PubMed] [Google Scholar]
- Rouiller I, Xu X‐P, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D (2008) The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol 180: 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Padrick SB, Doolittle LK, Daugherty‐Clarke K, Corrêa IR, Xu M‐Q, Goode BL, Rosen MK, Gelles J (2013) Three‐color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. eLife 2: e01008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova OS, Chemeris A, Guo S, Alioto SL, Gandhi M, Padrick S, Pechnikova E, David V, Gautreau A, Goode BL (2017) Structural basis of Arp2/3 complex inhibition by GMF, coronin, and arpin. J Mol Biol 429: 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R (2012) The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J Cell Biol 197: 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti S‐C, Jurgenson CT, Nolen BJ, Pollard TD (2011) Structural and biochemical characterization of two binding sites for nucleation‐promoting factor WASp‐VCA on Arp2/3 complex. Proc Natl Acad Sci USA 108: E463–E471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AR, Luan Q, Liu S‐L, Nolen BJ (2013) Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr Biol 23: 1990–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, Cooper JA (2001) Cortactin promotes and stabilizes Arp2/3‐induced actin filament network formation. Curr Biol 11: 370–374 [DOI] [PubMed] [Google Scholar]
- Weaver AM, Heuser JE, Karginov AV, Lee W, Parsons JT, Cooper JA (2002) Interaction of cortactin and N‐WASp with Arp2/3 complex. Curr Biol 12: 1270–1278 [DOI] [PubMed] [Google Scholar]
- Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE (2012) Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell 148: 973–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X‐P, Rouiller I, Slaughter BD, Egile C, Kim E, Unruh JR, Fan X, Pollard TD, Li R, Hanein D, Volkmann N (2012) Three‐dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J 31: 236–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Unruh JR, Deng M, Slaughter BD, Rubinstein B, Li R (2011) Dynamic maintenance of asymmetric meiotic spindle position through Arp2/3‐complex‐driven cytoplasmic streaming in mouse oocytes. Nat Cell Biol 13: 1252–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lin K, Gao L, Chen L, Shi X, Wu G (2011) Crystal structure of the armadillo repeat domain of adenomatous polyposis coli which reveals its inherent flexibility. Biochem Biophys Res Commun 412: 732–736 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Review Process File
Data Availability Statement
The three x‐ray crystal structures reported here have been deposited in the Protein Data Bank under accession codes 6DEC,6DEE, and 6DED (http://www.rcsb.org/pdb/explore/explore.do?structureId=6DEC, http://www.rcsb.org/pdb/explore/explore.do?structureId=6DEE, and http://www.rcsb.org/pdb/explore/explore.do?structureId=6DED, respectively).


