Abstract
Background.
Discharge of textile dyes into the environment poses a significant threat. They are poorly biodegradable and toxic due to their complex composition and aromatic structures. In the search for alternatives to physical and chemical treatments, biodegradation of synthetic dyes by various microbes is emerging as an effective and promising approach.
Objectives.
The decolorization of synthetic dyes by yeast co-cultures and consortia from leaves and fruit peels was assessed at a 50 μg/mL dye concentration.
Methods.
Yeasts isolates from leaves and fruit peels were screened for potential decolorization of synthetic dyes at 25–50 μg/mL. Decolorization parameters were optimized for synergistic properties and development of yeast co-cultures and consortium. Possible decolorization reactions were initially assessed by cell immobilization, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and Fourier transform infrared spectroscopy (FTIR) analysis.
Results.
A total of 16 organisms were isolated from rose, mango, and pineapple leaves and pineapple fruit peels. Only 4 organisms showed high decolorization of four synthetic dyes: Direct Pink B, Disperse Yellow 5G, Direct Fast Orange S, and Reactive Turquoise Blue G. The optimum condition for best decolorizers of selected dyes at 50 μg/mL were Candida guilliermondii (Y011) for Direct Pink B at pH 9, 37°C; C. dubliniensis (Y014) for Disperse Yellow 5G at pH 4, 25°C; C. guilliermondii (Y004) for Direct Fast Orange S at pH 7, 25°C, and C. famata (Y003) for Reactive Turquoise Blue G at pH 4, 35°C. None of the 4 yeast isolates showed any antagonistic activity when subjected to the lawn-spotting method for the formation of co-cultures and consortium. The best co-cultures obtained 61% decolorization of Direct Pink B, 65% decolorization of Disperse Yellow 5G, 41% decolorization of Direct Fast Orange S, and 50–51% decolorization of Reactive Turquoise Blue G. Immobilized yeast cells were active in decolorizing the dyes and SDS-PAGE analysis confirmed the presence of an extracellular protein. The results of FTIR also showed changes in the functional group of Direct Pink B, but minimal changes in the functional groups of Reactive Turquoise Blue G, indicating a different decolorization pathway.
Conclusions.
Yeasts in co-cultures and consortia can decolorize toxic synthetic dyes through different decolorization pathways such as enzyme degradation and bioaccumulation. This technique may have a use in the treatment of wastewater systems.
Keywords: dyes, biodegradation, decolorization, microbial consortia
Introduction
The textile industry has flourished along with the rapid increase in modernization and urbanization of industries in terms of yearly production. Azo and anthraquinone dyes are major organic (synthetic) textile dyes that are commonly used and often preferred in the textile industry because they are more stable, easier to produce, and have a wider variety of colors than natural dyes. However, discharge or waste disposal of these dyes into the environment poses a significant threat due to their poor degradability. These dyes are also reported to be toxic and carcinogenic due to their complex chemical composition and aromatic structures.1 When these colored effluents enter the human system, they may inhibit biological activity and can even cause illnesses such as nausea, skin ulceration, and dermatitis.2 Therefore, treatments for dye-containing effluents are needed. There are several chemical and physical methods that are used for the removal of dyes in waste water systems such as adsorption, oxidation, and electrochemical methods. However, these processes lead to high-energy costs, sludge production, and formation of toxic by-products.3
Microorganisms are now being tapped as potential agents for the decolorization and degradation of dyes because of their environmentally friendly process and cost-effective treatment capability. Studies have reported the capability of microorganisms to decolorize a wide range of dyes. In the course of the biological treatment of industrial effluents, a diverse microbial community is also often noted. Different microorganisms living in the same environment may have variations in their physiological attributes, wherein they may either compliment or inhibit one another. Microbial strains that are complimentary can lead to a more efficient biodegradation due to their synergistic interactions.4 In the Philippines and elsewhere, however, few studies have been conducted to date to elaborate on the effectiveness of microbial consortia to decolorize and detoxify hazardous synthetic dyes.
Thus, this study provides new insights on pollution management in textile plants. It focuses on the use of solitary and combinations of yeast isolates from leaves and fruit peels as potential decolorizers of the synthetic dyes Direct Pink B, Direct Fast Orange S, Disperse Yellow 5G, and Reactive Turquoise Blue G.
Methods
Culture Media and Dyes
Sabouraud dextrose agar (SDA) and Saboraud dextrose broth (SDB) used for the isolation and maintenance of yeasts was purchased from Belman Laboratories (Philippines).
Textile dyes Direct Pink B, Direct Fast Orange S, Disperse Yellow 5G, and Reactive Turquoise Blue G were obtained from L.G. Atkimson Import-Export, Inc., (Quezon City, Philippines). Unfortunately, no information was available on either the exact composition or purity of these specific dyes.
Dye Solution in Decolorization Studies
The decolorization medium was plain distilled water, to which the dyes were added to arrive at aqueous dye solutions. This solution and the media for yeasts were autoclaved at 121°C, 15 pound-force per square inch for 20 minutes.
Abbreviations
- ANOVA
Analysis of variance
- API
Analytical profile index
- FTIR
Fourier transform infrared
- OD
Optical density
- SDA
Sabouraud dextrose agar
- SDB
Sabouraud dextrose broth
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
Isolation and Identification of Top Decolorizer Yeasts
Source substrata of yeasts such as fresh leaves of rose, pineapple and mango, and pineapple fruit peels were collected from market waste. These substrates, even in the form of wastes, may be a good source of microorganisms. These samples were cut into appropriate sizes, washed three times in sterile distilled water, and then imprinted (upper surface of leaf pieces) onto plated SDA for 1 minute. Isolation plates were incubated for 24–48 hours at room temperature. Resulting yeast colonies were confirmed by microscopic examination in wet-mount preparations. Confirmed yeast cells were re-streaked onto new SDA for purification and eventual isolation into pure axenic culture in SDA slants maintained in a refrigerated condition.
Only the four yeast isolates (one for each of the four dyes) with the highest decolorization percentages were identified using the analytical profile index (API) system 20 C Aux (BioMérieux, Marcy l'Etoile, France). Each isolate was grown by streaking on SDA for 24 hours and one colony was inoculated in normal saline solution. Cell suspension (1 mL) from the normal saline solution was transferred to API C medium and 200 μL from the latter was then seeded to each cupule in the identification kit. All identification kits were read after 48 and 72 hours of incubation at room temperature. Binomial identifications of chosen isolates were provided in the API database. In addition, the general colonial characteristics and cell morphology of these isolates were studied based on growth in SDA plates.
Screening of Yeasts for Growth Tolerance to and Decolorization of Dye
Growth tolerance. All yeast isolates (16) were initially screened for growth tolerance by streaking in distilled water agar (1.5% agar) with 25 μg/mL of the selected dyes Direct Pink B, Direct Fast Orange S, Dispersed Yellow 5G, and Reactive Turquoise Blue G. Presence or absence of growth was recorded after 24-hour incubation of culture plates at room temperature. Isolates with positive growth in triplicate plates for each of the four dyes under study were screened below for decolorization and used in subsequent experiments; those with no growth were excluded.
Decolorization
Included isolates were screened in 50 μg/mL aqueous solutions of the dyes. Cell suspensions enriched for 24 hours in SDB under agitation condition were centrifuged at 7000 rpm. Pellets of biomass were washed two times with distilled water and 0.1 g of each per yeast isolate was added in test vials with 30 mL aqueous dye solution. Control vials with dye solutions were not seeded. All vials were incubated with agitation (150 rpm, Gerhadt Laboshake) at room temperature and 1.5 mL aliquot samples were taken after 7 days. Cell biomass was removed from all samples by centrifugation at 7000 rpm for 30 minutes and the supernate in test vials and dye solution in control vials were pipetted to corresponding wells in a 96-well microtiter plate for ultraviolet-visible spectrophotometry at different wavelengths of 343, 376, 399, and 400 nm, respectively, for Disperse Yellow 5G, Direct Pink B, Reactive Turquoise Blue G, and Direct Fast Orange S. Decolorization percentages after 7 days were computed as follows:
 |
where, initial OD(0h) and Final OD(t) are absorbance readings at 0 and 48 hours of incubation, respectively.
Optimization of Assays for Decolorization by the Best Decolorizers
Only the top four decolorizer yeast isolates in the assay above were re-assayed for optimum decolorization, each tried with only the specific dye it decolorized most in the decolorization assay described above. Triplicate sets of test and control vials were prepared for each yeast. First-set vials were for optimization of pH (4, 7, and 9) at room temperature, the 2nd set for optimization of temperature (25, 37, and 45°C), and the 3rd set for optimization of dye concentration (50, 100, and 150 μg/mL), likewise in aqueous solution. Aliquot samples from test and control vials were processed as for the initial decolorization assay. Ultraviolet-visible spectrophotometry for optical density readings of processed samples in 96-well microtiter plates was performed on days 1, 3, 5, and 7. Decolorization percentages were calculated as described above.
Assay for Synergistic and Antagonistic Interactions
This assay was done prior to tests for decolorization and degradation of the dyes by the yeasts, singly or in combinations (dual and in consortia of all four top isolates). The “lawn-spotting” assay by Long and Azam was adapted with modification because not all the yeasts could be homogenized completely in cell suspension in normal saline solution.5 SDA plates were used for all possible dual combinations (“antagonist” and “antagonized”, and roles in reverse). The “antagonized” sample (0.1 g biomass in 10 mL normal saline solution) was swabbed on the agar medium, and the “antagonist” sample (1.0 g in 10 mL normal saline solution), spotted on the lawn of the “antagonized” sample. Presence or absence of zone of inhibition was noted after 24 hours incubation of assay plates at room temperature. Absence of zone of inhibition indicated possible synergistic interaction, and its presence, antagonism.
Decolorization of Dyes by Yeasts, Singly, and in Combinations
Isolates which did not exhibit any antagonistic interactions to one another in dual combinations were subjected again to decolorization assay. The top four individual isolates, co-cultures, and consortium were used. The co-cultures were composed of two decolorizers for each dye, while the yeast consortia contained all the four isolates together in one medium. The isolates were pre-enriched individually in flasks containing SDB. The flasks were placed in a rotary shaker (150 rpm) for 24 hours at room temperature. After incubation, the cells were harvested at 7000 rpm for 30 minutes and washed twice with distilled water. For single isolates, the organism was standardized to 0.1 g biomass and then inoculated in the distilled water containing 50 μg/mL of the selected dye. For co-cultures and consortia, the isolates were also standardized to 0.1 g biomass and then inoculated as well in dye solution. The vials containing the dye solutions were placed again in a rotary shaker (150 rpm) at room temperature for one week. Control vials with dye solutions were not seeded. Decolorizations were monitored on days 1, 3, 5, and 7.
Decolorization with Immobilized Yeasts
The method used in the present study was adapted from a study by Tan et al.6 Calcium alginate was used for the entrapment of yeast cells. Individual isolates were pre-enriched in separate flasks containing SDB under agitation condition (150 rpm) for 24 hours. Cell suspensions were centrifuged at 7000 rpm for 30 minutes. Pellets of biomass were washed twice with distilled water and 0.1 g of each per yeast isolate was used. The cell suspension was mixed with an equal volume of calcium alginate solution. The mixture was dropped by means of a syringe into a calcium chloride solution to form the beads. The beads were washed with sterile normal saline solution and then placed in vials containing 30 mL of aqueous dye solutions. All vials were incubated with agitation (150 rpm, Gerhadt Laboshake) at room temperature and 1.5-mL aliquot samples were monitored likewise with readings on the 1st, 3rd, 5th, and 7th day of incubation. For control tests, another batch of beads was prepared, but without the cell suspension.
Partial Characterization of Extracellular Enzymes
The extracellular enzymes of each top decolorizer were assessed through electrophoresis using the Novex® Bolt Mini Gel Tank. Individual isolates were placed again in separate vials containing 30 mL of aqueous dye solutions. The vials were incubated at agitated conditions (150 rpm) for 7 days. A 1.5-mL aliquot from the decolorization media used was collected and centrifuged at 7000 rpm for 30 minutes. A 1-μL sample from the supernatant was taken and mixed with 2.5 μL of 4X Bolt® LDS Sample Buffer, 1 μL of 10X Bolt® Reducing Agent, and 5.5 μL of deionized water. The provided cassette was removed from the pouch and placed in the well. The well was washed with 1X running buffer twice and was filled with 1X running buffer again. Meanwhile, the chamber containing the gel tank was filled with buffer just above the electrode. The sample was loaded on the gel. The machine was run at 80 V for 40 minutes. The gel was de-stained after being in buffer for 24 hours and the bands were analyzed by obtaining the approximate molecular weights of the protein.
Analysis of Changes in the Functional Groups of Dyes
Fourier Transform Infrared Spectroscopy (Shimadzu IR Prestige 21) was used to determine changes in the components of the selected dyes after being treated by individual yeast isolates. Aqueous dye solutions containing 50 μg/mL of dyes were treated with individual yeast isolates for 7 days. A 1.5-mL aliquot was sampled from the treated solutions and centrifuged at 7000 rpm for 30 minutes. The supernatant of the centrifuged solution was placed in a tube and freeze dried (GoldSim International). The freeze-dried samples were then placed in a desiccator overnight prior to Fourier Transform Infrared (FTIR) analysis. The samples were then mixed with dried potassium bromide powder and then processed to form a disc. The formed discs were placed inside the FTIR instrument (Shimadzu IR Prestige-21) and then analyzed. Untreated aqueous dye solutions were also processed and analyzed to determine if there were changes in the functional groups of the dyes before and after yeast treatment.
Statistical Analyses
The mean values in the screening of isolates, optimization of different parameters, and degradation of dyes using free cells and immobilized cells were subjected to analysis of variance (ANOVA) at a 0.5% significance level, as well as post hoc Tukey's range (HSD) test for decolorization using the software PAST 13.0 and Statistical Analysis System (SAS).
Results
Yeast Isolation and Identification
Yeasts are unicellular fungi that can thrive in different environments. In the present study, a total of 16 isolates (Y001-Y016) were obtained from the leaves and fruit peel samples as shown in Figure 1. Eleven (11) isolates came from rose, pineapple, and mango leaves, while 6 came from pineapple fruit peels. The number of isolates is low considering the fact that the leaf area is an essential plant part that determines light interception, transpiration, productivity, and photosynthesis. Photosynthesis is strongly associated with chlorophyll content and total sugar concentration.7 In addition, pineapple wastes such as peels and leaves are also rich in sugar and other carbohydrates and these may be factors in good microbial growth. Although yeasts may also be present in leaves, it has been reported that perennial trees such as mango trees have low orchard efficiency and the overall content may be influenced by the seasons. When the leaves were picked for processing, the samples were almost dried up and this may have contributed to the low number of isolated yeasts from mango leaf samples.
Figure 1.
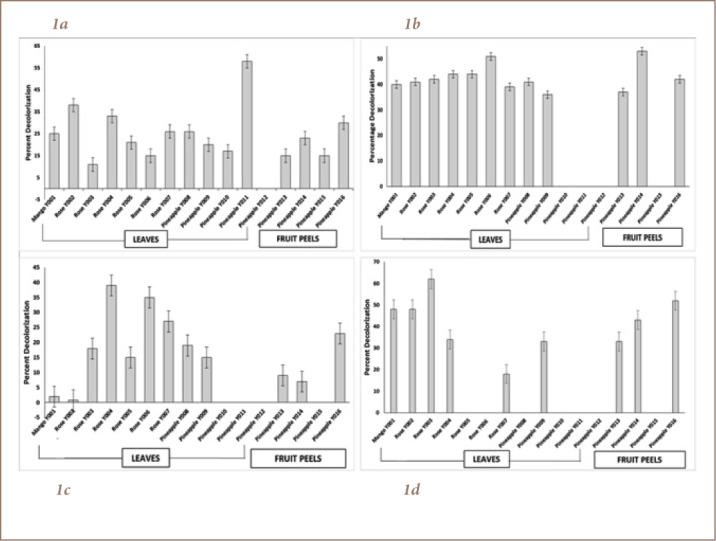
Screening of yeast isolates to different dyes. (A) Direct Pink B, (B) Disperse Yellow 5G, (C) Direct Fast Orange S, (D) Reactive Turquoise Blue G. Blank spaces indicate that the corresponding isolate did not grow on agar with 25 μg/mL of dye and was not included on the seconding screening at 50 μg/mL of dye. (Note: * signifies that data was statistically significant)
Yeasts that were actively decolorizing the selected dyes were identified by API C Aux 20. Based on the biochemical test results, all isolates used in the study belong to the genus Candida and the best decolorizer for Direct Pink B and Direct Fast Orange S was identified as C. guilliermondii (Y004 and Y011), the best decolorizer for Disperse Yellow 5G was C. dubliniensis (Y014), and the best decolorizer for Reactive Turquoise Blue G was C. famata (Y003). All four isolates showed similar morphological characteristics, which were spherical and unicellular in smooth, compact, and creamy white colonies.
Screening of Isolates for Dye Decolorization
Yeasts were screened for the ability to thrive in synthetic dyes and to eliminate those that cannot tolerate even the lowest concentration of dyes. Results showed that for Direct Pink B, C. guilliermondii (Y011) obtained the highest decolorization rate of 57.61% after 7 days (p=0.013) while C. guilliermondii (Y004) achieved a decolorization rate of 39.22% for Direct Fast Orange S (p=0.003). On the other hand, C. dubliniensis (Y014) had a 53.21% decolorization rate (p=0.0009) for Disperse Yellow 5G and C. famata (Y003) had a rate of 62.31% for Reactive Turquoise Blue G (p=0.002), as shown in Figure 1.
Yeast cultures possess a number of benefits in the application of dye decolorization as they grow faster than molds. This is an advantage in industrial settings where fast treatment of the wastewater system is environmentally and economically important. Moreover, yeasts have different mechanisms of decolorization as they can accumulate dyes or degrade them through biocatalytic activities, or a combination of both. A number of studies have been conducted to investigate the potential of yeast as a decolorizing agent.8–10
Some studies have examined whether microorganisms can use dye as their sole carbon source. It has also been reported that yeast cannot grow without the presence of a carbon or energy source. However, the results obtained from the screening assays in the present study proved that yeasts can thrive in the presence of dyes even without the addition of glucose or another easily metabolized carbon source since their metabolic activities may change and adapt to another source of carbon in the screening medium.
Optimization of Decolorization Activity
The culture conditions were optimized to determine the best parameters for the decolorization of dyes or degradation condition of the isolates, as summarized in Table 1. First, incubation temperature was an important parameter as it contributes to the growth of microorganisms, enzyme production, and rate of dye decolorization by the microorganisms. Direct Pink B dye with C. guilliermondii (Y011) had high decolorization at 37°C, Disperse Yellow 5G with C. dubliniensis showed the highest decolorization at 25°C, Direct Fast Orange S showed highest decolorization at 25°C and for Reactive Turquoise Blue G, the highest decolorization of dye was obtained at 45°C.
Table 1.
Optimum Conditions (Temperature, pH, Top Yeast Decolorizers
| Organisms | Dyes | Optimum Conditions for Each Parameter |
| C. guilliermondii Y011 | Direct Pink B | 50 μg/mL, 37°C, pH 9 |
| C. dublienensis Y014 | Disperse Yellow 5G | 50 μg/mL, 25°C, pH 4 |
| C. guilliermondii Y011 | Direct Fast Orange S | 50 μg/mL, 25°C, pH 7 |
| C. famata Y003 | Reactive Turquoise | 50 μg/mL, 45°C, pH 4 |
Second, varying dye concentrations have an effect on the activity of a microorganism to decolorize and degrade them. All isolates in the present study yielded better decolorization at 50 μg/mL compared to higher dye concentrations. This result was similar to the findings in a study by Mahmoud, which reported a reduction in the dye removal capability of isolates with an increase in dye concentration.11 This may be further explained by the fact that the sorption sites were already saturated and could not take any more dyes. In addition, increasing dye concentrations decreased specific growth rates.12 A study by Phugare et al. also indicated that higher dye concentrations decreased decolorization efficiency due to the toxicity of dye to the organism.13
Another essential parameter was pH, since dye accumulation and dye absorption was dependent on it. The experiments were carried out at 50 μg/mL since the optimization assay yielded better results at this concentration. Results indicated good decolorization activity for Direct Pink B at pH 9, Direct Fast Orange S at pH 7, Reactive Turquoise Blue G at pH 4, and Disperse Yellow 5G at pH 4. Most of the decolorization activities of yeast isolates were better at a lower pH, except for Direct Pink B. Yeast isolates tend to favor lower pH since the positive charges of the biomass create an attraction between its surface and the anionic nature of the dye.12 In the reaction between yeast isolates and the dyes, when pH increases, the electrostatic attraction between the biomass and the dye decreases. This is because at lower pH, decolorization becomes more favorable since the biomass becomes positively charged, giving way to the adsorption of the negatively-charged dye. Negative charges are observed on the functional groups attached on the heterocyclic group of the compound. Direct dyes are organic compounds that have functional groups of hydroxyl, sulfino, and amino, dispersed dyes are organic dyes that have functional groups of nitro and cyano, while reactive dyes are also organic compounds that have chloro and amino groups.
Antagonistic-Synergistic Interactions and Decolorization Assay
No clearing zone was observed among the four isolates, with greater activity on selected dyes. This may be a result of synergistic interactions with the yeast isolates. Most consortium studies focused only on bacterial consortium or fungal-bacterial consortium and this is the first study to subject yeast strains into a co-culture or a consortium for dye decolorization. Investigating the potential interactions within a co-culture or a consortium was important even if the organisms came from the same genus. The biocatalytic activity of a single organism varies in the presence of other organisms and the activity of a co-culture or consortium is different compared to an individual one.
Decolorization of Dyes by Yeasts, Singly, and in Combinations
The decolorization ability of the pure isolates was tested, as well as that of the developed co-culture and consortium. Although single strains are known to decolorize dyes, there is still a considerable advantage in using a combination of isolates. Single cultures may only use one or two processes to achieve an efficient decolorization result. However, the use of two or more organisms in the developed co-cultures and consortium may help to attack the dyes at different positions and may even produce different metabolites that could help achieve chemical decomposition of dyes, provided that they are not antagonistic to one another. The benefits of using different organisms in one system in decolorization studies have already been elucidated. However, most of these studies have focused on bacterial consortium or mixed bacterial consortium.14 This is the first study to attempt to combine different Candida species in a single system.
For Direct Pink B, the highest decolorizer was the co-culture of C. guilliermondii (Y011)–C. famata (Y003) and C. guilliermondii (Y011)–C. guillermondii (Y004), with a total of 61% color reduction after 7 days and 60% decolorization with the combination of C. guilliermondii (Y011) and C. dubliniensis (Y014). Meanwhile, the consortium achieved a total of 47.3% decolorization, but only 39% with the individual isolate of C. guilliermondii (Y011) (Figure 2A and Figure 3A–B).
Figure 2.
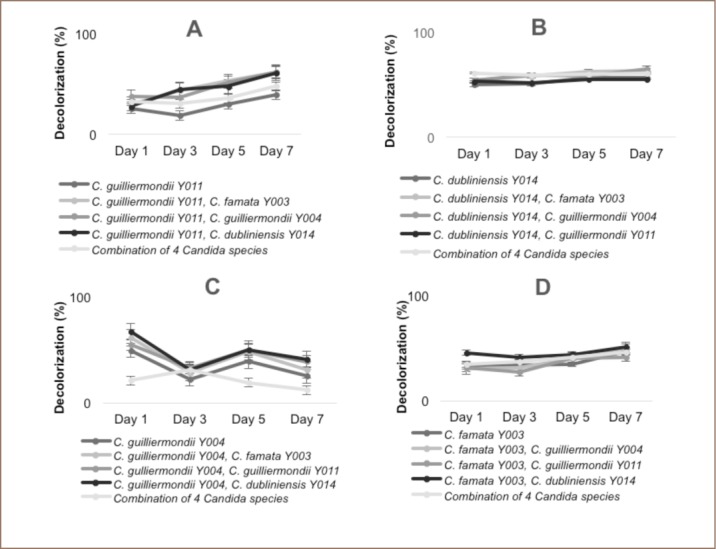
Decolorization of dyes by individual, co-cultures, and consortium for (A) Direct Pink B, (B) Disperse Yellow 5G, (C) Direct Fast Orange S, (D) Reactive Turquoise Blue G.
Figure 3.
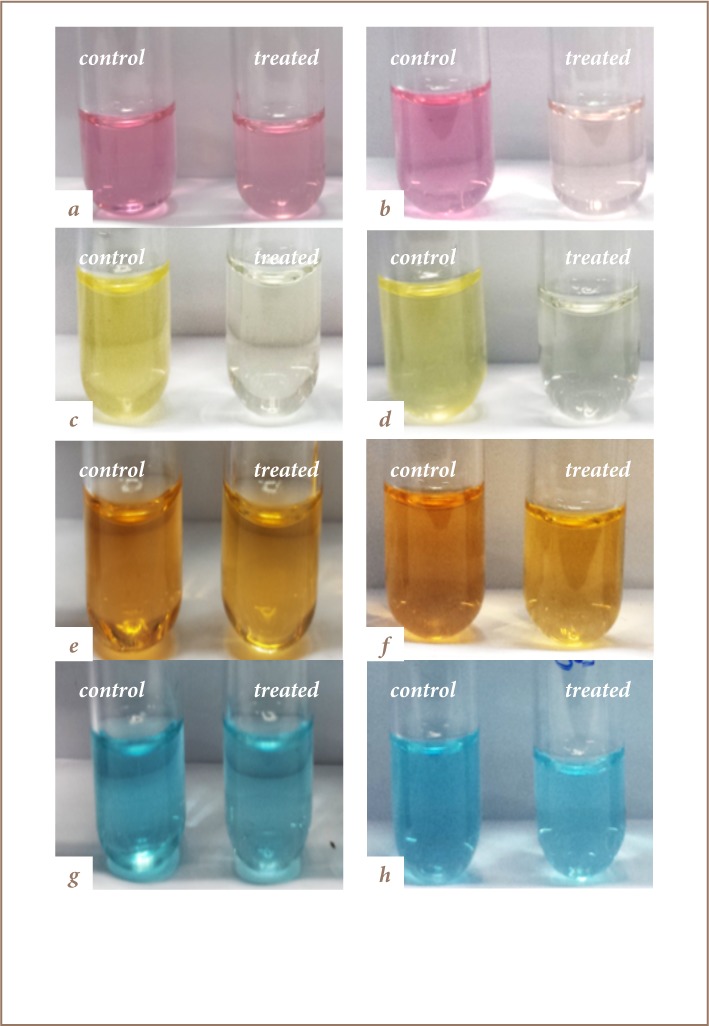
Decolorization of dyes by yeast co-cultures Direct Pink B Treated with C. guilliermondii Y011 and C. famata Y003 at Day 1 (A) and at Day 7 (B); Disperse Yellow 5G treated with C. dubliniensis Y014 and C. guilliermondii Y004 at Day 1 (C) and at Day 7 (D); Direct Fast Orange S treated with C. guilliermondii Y004 and C. dubliniensis Y014 at Day 1 (E) and at Day 7 (F); Reactive Turquoise Blue G treated with C. famata Y003 and C. guilliermondii Y004 at Day 1 (G) and at Day 7 (H).
For Disperse Yellow 5G, regardless of the combination of isolates, all samples achieved more than 50% decolorization after 7 days. The combination of C. dubliniensis (Y014)–C. guilliermondii (Y004) achieved 65% decolorization of this dye, while C. dubliniensis (Y014)–C. famata (Y003) achieved 63% decolorization. The consortium exhibited 61% decolorization compared to 57% with the individual C. dubliniensis (Y014). The lowest decolorization achieved from this dye was the co-culture C. dubliniensis (Y014)–C. guilliermondii (Y011) with 55% decolorization after one week (Figure 2B and Figure 3C–D).
As for Direct Fast Orange, the highest decolorization was achieved with C. guilliermondiii (Y004)–C. guilliermondii (Y014), with a total of 41% decolorization after one week compared to 39% from C. guilliermondii (Y004)–C. guilliermondiii (Y011). Meanwhile, C. guilliermondii (Y004)–C. famata (Y003) showed 31% decolorization on this dye compared to 25% from C. guilliermondii (Y004) alone. The lowest decolorization rate was from the consortium, with only 12% decolorization after the 7-day incubation time (Figure 2C and Figure 3E–F).
For Reactive Turquoise Blue G, the best decolorization rate was achieved from the co-culture of C. famata (Y003)–C. dubliniensis (Y014) and C. famata (Y003)–C. guilliermondii (Y004), with a total of 51% and 50% decolorization after 7 days of incubation, respectively. The individual isolate and the consortium showed a total of 47% and 46% decolorization, respectively, while the lowest rate was 41% from the co-culture C. famata (Y003)–C. guilliermondii (Y011) (Figure 2D and Figure 3G–H).
The best decolorization rate came from the co-culture setup compared to the consortium or the individual isolates. According to Hesselman et al., some cells do not yield good physiological behaviors in experimental setups, but the incorporation of another culture may lead to more successful results in vivo.15 In addition, some cell cultures may exhibit cooperative relationships, as seen in the present results. While it may seem intuitive that a consortium would yield better results compared to fewer cell populations, cell interactions involving more than three populations may lead to unpredictable and unstable levels. It has been suggested by Goers et al. that it is best to avoid using a large number of populations, as most of the time this is not advantageous.16 The mixture of different Candida species in one system may have led to the production of various metabolites inhibiting the other species present in the consortium setup.
Decolorization Assay using Immobilized Cells
Microorganisms used in bioremediation may not persist for a long time in the area being treated as they can be washed out in the process. Immobilization of microorganisms has been proposed to solve this problem as this technique makes the decolorizer more efficient and increases its potential enzymatic activities.17 As shown in Figure 4, individual isolates are still active in decolorizing dyes even in an immobilized state. The distribution of decolorization among the dyes was statistically different (p=0.0001). However, the best results were achieved from Reactive Turquoise Blue G with a total of 25% decolorization, with significantly different results from Direct Pink B, Direct Fast Orange, and Disperse Yellow 5G (p-value >0.05). The results achieved were low compared to those of free yeast cells, but decolorization was still recorded, indicating that there was something that the yeast cells were capable of producing or excreting to degrade the dyes present in their environment, even if the yeast cells were immobilized, such as extracellular enzyme secretion.17,18 In addition, there could be another pathway that the yeast cells performed when they were not immobilized. On the other hand, no carbon source was present in the solution, which plays an important role in the growth of yeasts and decolorization activity. This may indicate that yeasts may remain active despite the absence of a carbon source and perform decolorization activities in different pathways apart from extracellular enzyme secretion.
Figure 4.
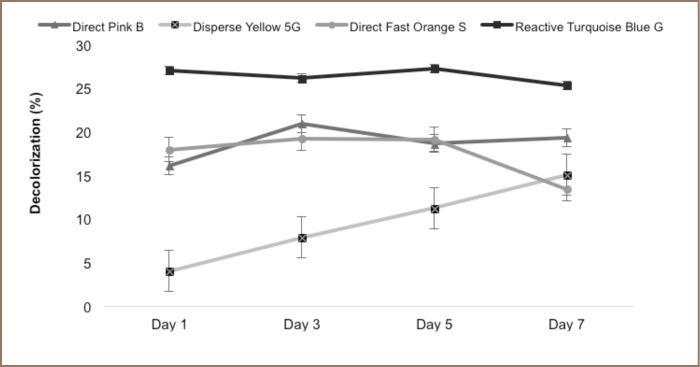
Immobilized individual yeast decolorizers on their respective dyes
Proposed Mechanism of Dye Decolorization
Decolorization of dyes can be achieved through adsorption, absorption, or both. Adsorption is defined as the accumulation of toxicants through the cells of living or dead cells. The capacity of yeasts to remove dyes can be in the manner of biosorption, in which dyes adhere in a non-specific manner to cell peripheries and then eventually into the cell.19 It is generally based on the interactions made by the functional groups present on a cell surface through electrostatic interactions, ion exchange or ion chelation. Different mechanisms of dye decolorization have been reported depending on the organisms used in different experiments. In bacteria, mineralization is the main mechanism of dye decolorization, but aerobic and anaerobic decolorization, followed by utilization of carbon and nitrogen in the degraded products as nutrient sources has also been reported.18 Fungi have also been reported to decolorize dyes by biosorption as the major mechanism, while biodegradation is also a possibility because of the extracellular enzymes it releases. Some studies have reported that both biosorption and biodegradation are performed by some fungi as part of the mechanism of action for dye degradation; utilization of degraded products of carbon and nitrogen as nutrient sources has also been reported, but mineralization remains unexplored as a fungal mechanism of dye degradation.18 There have been few reports of dye degradation by yeasts and only the mechanisms of bioaccumulation, biosorption and biodegradation using enzymes in dye degradation have been reported by Rana et al.18 Enzymatic system degradation of dye using Issatchenkia occidentals, Saccharomyces cerevisiae, Galactomyces geotrichum, Candida krusei, and bioaccumulation using Candida tropicalis have been reported for yeast dye degradation.20,21,11
The dye components of Reactive Turquoise Blue G were found to be attached to the yeast cell wall and some were adsorbed into the cytoplasm, resulting in colored cells of C. famata Y003 under the microscope (Figure 5). Results from decolorization using immobilized cells showed no intracellular adsorption, but the observed decolorization of dye indicated that the mechanism of action of C. famata Y003 may be both biosorption and release of extracellular enzymes in dye decolorization. The behavior of yeasts in the present study may be further explained as a process through which yeast cells adsorb metallic ions and other pollutants, such as textile dyes, as the chemical makeup of the microbial cells consists of different molecular structures or biomolecules that play a role in their metabolic activity and dictate their ability to perform functions in order to reduce the sudden change in their environment and enable them to adapt to it. Yeasts typically have chitin, β-glucan, and mannoproteins on their cell walls. There are several chemical groups in the biomass that could attract and seize charged pollutants such as acetamide groups of chitin, amine, amide, sulfohydryl, as well as carboxyl groups in proteins, and hydroxyls in polysaccharide. These functional groups are signaling molecules that eventually attach on the differently charged polysaccharides and proteins present on the extracellular side of the cell wall or signal the opening of an ion-gated channel in which dyes may be sequestered by cell surface sorption followed by intracellular accumulation and possibly by degradation using intracellular enzymes.22 FTIR analysis (Figures 6 and 7) also showed that there was no change in the structural components of Reactive Turquoise Blue G and Direct Fast Orange S before and after treatment of certain yeast organisms corresponding to the optimum decolorization of dye. Comparing the results of the FTIR analysis and the image of the yeast cells under microscope (Figure 5) showed that there was no extracellular degradation of dye, but instead the dye was intracellularly accumulated and degraded.
Figure 5.
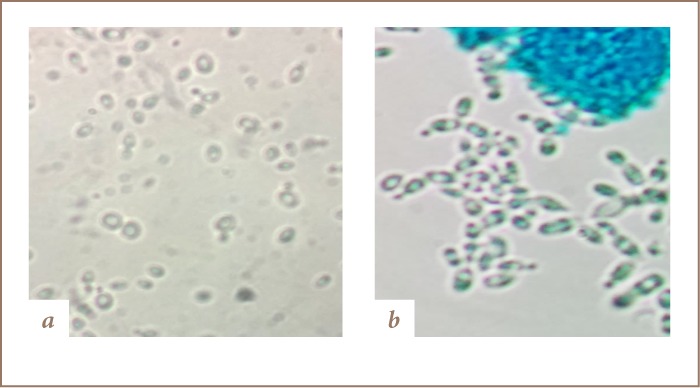
Cells of (A) Candida guilliermondii Y011 and (B) Candida famata Y003 after being treated with synthetic dyes
Figure 6.
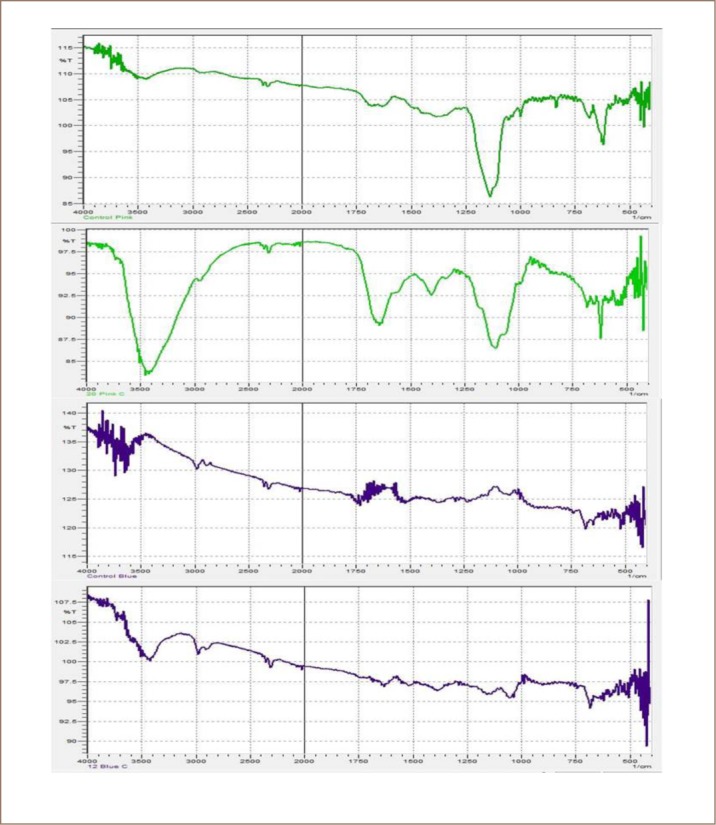
FTIR Analysis of (A) Direct Pink B before treatment and (B) Direct Pink B after treatment with an observable change in the functional groups; (C) Reactive Turquoise Blue G before treatment and (D) Reactive Turquoise Blue G after treatment showing no change in the functional group.
Figure 7.
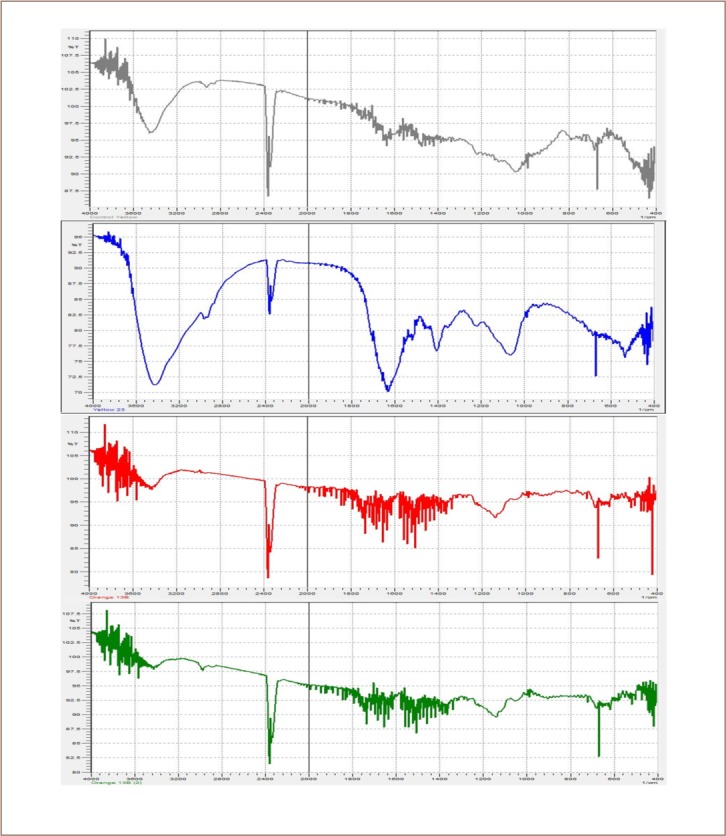
FTIR Analysis of (A) Disperse Yellow 5G before treatment and (B) Disperse Yellow 5G after treatment with an observable change in the functional group; (C) Direct Fast Orange S before treatment and (D) Direct Fast Orange S after treatment showing no change.
In addition, for Direct Pink B, the cells of C. guilliermondii (Y011) appeared colorless (Figure 5A) under the microscope, suggesting that there are other ways the yeast cells decolorize dyes aside from biosorption. The microscopic view showed that no adsorption happened between the yeast cells and Direct Pink B dye, and no adsorption was shown on the decolorization results using immobilized cells, but the decolorization of the dye still proceeded. As FTIR analyses have shown (Figures 6 and 7), there are some changes in the functional groups of the dye structure. This may indicate that aside from the capacity of yeasts to act as an adsorbent, they may also have other factors such as extracellular release of enzymes that could be responsible for the decolorization of dyes. Most fungal strains used for decolorization studies have been known to produce and secrete different extracellular enzymes like laccases, manganese peroxidases, lignin peroxidases, tyrosinase, azoreductases and dichlorophenolindophenol reductases.23 Comparative analysis of the FTIR results and the yeast cell image under microscope showed that the functional group of Direct Pink B was changed after treatment with C. guilliermondii (Y011), while the image under the microscope showed that there was no visible intracellular accumulation. These results indicate that the C. guilliermondii (Y011) mechanism of action in treating toxic dyes was not intracellular biosorption or intracellular degradation, but degradation with the help of an enzyme that was released extracellularly by the yeast cells in its environment, since there was no observed biosorption in the tests using immobilized and free yeast cells in dye decolorization.
Partial Characterization of Enzymes Involved in Decolorization
Enzymes may be involved in the decolorization of synthetic dyes because of their highly specific and reactive characteristics to the target molecules.24 There have been reports of the specific enzymes produced by microorganisms for synthetic dye decolorization or toxicity reduction like laccases produced by Schyzohyllum commune, Pseudomonas putida; lignin peroxidases and manganese peroxidases produced by Schyzohyllum commune and azoreductases produced by Bacillus latrosporus and Xenophilus azovorans.25–29 Most of the studies of synthetic dye decolorization or detoxification have focused on bacteria and fungi because of their high diversity and ease of producing extracellular enzymes; yeast are rarely used in bioremediation studies and extracellular enzyme production for decolorization. Partial characterization of enzymes was done through the assessment of molecular weight based on the results obtained on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The molecular weight was compared with other available published studies (Table 2).
Isolate C. guilliermondii (Y011) exhibited 4 different enzymes that are possibly involved in the decolorization of Direct Pink B. These enzymes are peroxiredoxin-like, cytochrome c peroxidase-like, and two unknown proteins. Meanwhile, the other three top decolorizers, C. guilliermondii (Y004), C. dubliniensis (Y014), and C. famata (Y003) showed only 2 bands each. C. guilliermondii (Y004) and C. dubliniensis (Y014) may have involved peroxiredoxin-like and another unknown enzyme in the decolorization activity of Direct Fast Orange S and Disperse Yellow 5G. Isolate C. famata (Y003) may have used the activities of a peroxiredoxin-like and cytochrome-c peroxidase-like enzyme in the decolorization process of Reactive Turquoise Blue G.
Peroxidases have been widely involved in the treatment of synthetic dyes and other xenobiotic compounds. They can participate in oxygenated reactions in a large number of different substrates. They can catalyze oxidative reactions through the presence of hydrogen peroxides.30 Lignin peroxidases and manganese peroxidases have been widely regarded in their capacity to decolorize synthetic dyes. However, little is known of the involvement of cytochrome-c peroxidase.
Peroxiredoxins are known to be present in yeasts, in which they can both act as a peroxidase and as a molecular chaperone.29,31 The former is present in low molecular weight complexes, while the latter forms in high molecular weight complexes. As reactive oxygen species can be formed through aerobic and stressful conditions, these proteins may help to protect cells from oxidative stress and other damage.29 With all yeast isolates being exposed to aerobic conditions and producing a 126.45 kDa complex, it could be concluded that these enzymes help yeast cells survive when subjected to synthetic dyes. However, there have been no further studies of the capacity of peroxiredoxins to decolorize dyes.
Meanwhile, two unknown possible enzymes have been produced by C. guilliermondii (Y011), C. guilliermondii (Y004), and C. dubliniensis (Y014). It is difficult to identify the possible enzyme involved since there are no published studies of these two proteins and no study focusing on the possible enzyme decolorization of dyes using yeasts, especially in the genus Candida.
Synthetic organic dyes such as azo and anthraquinone dyes have been known to be generally toxic, carcinogenic, and mutagenic. The results of the present study showed that dyes were transformed to a lesser toxic form, except for Disperse Yellow 5G, after being treated aerobically by different yeast co-cultures and consortium.
Conclusions
The top four yeast isolates obtained from rose, mango and pineapple leaves and pineapple fruit peels have the ability to decolorize synthetic textile dyes such as Direct Pink B, Direct Fast Orange S, Disperse Yellow 5G, and Reactive Turquoise Blue G. Optimized conditions of dye decolorization were assessed at a 50 μg/mL dye concentration at pH 4, 7, and 9, with varying incubation temperatures of 25 to 45°C. The four individual isolates did not exhibit any antagonistic interactions and the best decolorization using co-cultures and consortium were determined to be 41 to 65% decolorization. FTIR results also revealed that there were changes in the functional groups for Direct Pink B and minimal changes for Reactive Turquoise Blue G, which might have been decolorized through extracellular activities. Immobilization of yeast cells confirmed that decolorization of Direct Pink B was due to the help of extracellular enzymes and accompanied by the result of SDS-PAGE analysis, which showed the possible molecular weight of the secreted enzyme. Immobilized cells that were not able to decolorize the dyes were assumed to degrade the dyes through biosorption and internal degradation, which was also supported by the microscopic analysis of the yeast cells. This suggests that yeasts can be used as potential agents of bioremediation, especially in the treatment of effluents with synthetic dyes. Due to the threat of physical and chemical treatments to human and animal health, the use of a biological approach is a promising safer alternative.
Acknowledgments
We thank the Research Center for the Natural and Applied Sciences, University of Santo Tomas for providing the equipment and facilities for this research, the Department of Science and Technology-Science Education Institute and the Accelerated Science and Technology Human Resource Development Program (ASTHRDP) for the funds needed for this research.
References
- 1.Banat IM, Nigam P, Singh D, Marchant R. Microbial decolorization of textile-dyecontaining effluents: a review. Bioresour Technol [Internet] 1996 Dec [cited 2014 Oct 27];58(3):217–27. Available from: http://www.sciencedirect.com/science/article/pii/S0960852496001137 Subscription required to view. [Google Scholar]
- 2.Shah V, Garg N, Madamwar D. Exopolysaccharide production by marine cyanobacterium cyanothece sp.: application in dye removal by its gelation phenomenon. Appl Biochem Biotechnol [Internet] 1999 [cited 2015 Jan 4];82(2):81–90. Available from: http://eurekamag.com/research/013/122/013122952.php Subscription required to view. [Google Scholar]
- 3.Wang H, Su JQ, Zheng XW, Tian Y, Xiong XJ, Zheng TL. Bacterial decolorization and degradation of the reactive dye Reactive Red 180 by Citrobacter sp. CK3. Int Biodeterior Biodegrad [Internet] 2009 Jun [cited 2014 Oct 27];63(4):395–99. Available from: http://www.sciencedirect.com/science/article/pii/S0964830508001996 Subscription required to view. [Google Scholar]
- 4.Alkhatib MF, Alam MZ, Muyibi SA, Husain IA. An isolated bacterial consortium for crude oil biodegradation. Afr J Biotechnol [Internet] 2011 Dec 16 [cited 2014 Oct 27];10(81):18763–7. Available from: http://www.academicjournals.org/article/article1380891575_Alkhatib%20et%20al.pdf. [Google Scholar]
- 5.Long RA, Azam F. Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol [Internet] 2001 Nov [cited 2015 Jan 4];67(11):4975–83. doi: 10.1128/AEM.67.11.4975-4983.2001. Available from: http://aem.asm.org/content/67/11/4975.full.pdf+html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan L, Li H, Ning S, Xu B. Aerobic decolorization and degradation of azo dyes by suspended growing cells and immobilized cells of a newly isolated yeast Magnusiomyces ingens LH-F1. Bioresour Technol [Internet] 2014 Apr [cited 2014 Oct 27];158:321–8. doi: 10.1016/j.biortech.2014.02.063. Available from: http://www.sciencedirect.com/science/article/pii/S096085241400234X Subscription required to view. [DOI] [PubMed] [Google Scholar]
- 7.Rymbai H, Laxman RH, Dinesh MR, Sunoj VS, Ravishankar KV, Jha AK. Diversity in leaf morphology and physiological characteristics among mango (Mangifera indica) cultivars popular in different agro-climatic regions of India. Sci Hortic [Internet] 2014 Sep 11 [cited 2015 Jan 4];176:189–93. Available from: http://www.sciencedirect.com/science/article/pii/S0304423814003550 Subscription required to view. [Google Scholar]
- 8.Vitor V, Corso CR. Decolorization of textile dye by Candida albicans isolated from industrial effluents. J Ind Microbiol Biotechnol [Internet] 2008 Nov [cited 2014 Oct 27];35(11):1353–7. doi: 10.1007/s10295-008-0435-5. Available from: http://link.springer.com/article/10.1007%2Fs10295-008-0435-5 Subscription required to view. [DOI] [PubMed] [Google Scholar]
- 9.Gonen F, Aksu Z. Predictive expressions of growth and Remazol Turquoise Blue-G reactive dye bioaccumulation properties of Candida utilis. Enzym Microb Technol [Internet] 2009 Jul 8 [cited 2015 Jan 4];45(1):15–21. Available from: http://www.sciencedirect.com/science/article/pii/S0141022909000672 Subscription required to view. [Google Scholar]
- 10.Charumathi D, Das N. Removal of synthetic dye basic Violet 3 by immobilised Candida tropicalis grown on sugarcane bagasse extract medium. Int J Eng Sci Technol [Internet] 2010 Jan [cited 2014 Nov 11];2(9):4325–35. Available from: http://www.researchgate.net/publication/50346357_removal_of_synthetic_dye_basic_violet_3_by_immobilised_Candida_tropicalis_grown_on_sugarcane_bagasse_extract_medium. [Google Scholar]
- 11.Mahmoud MS. Decolorization of certain reactive dye from aqueous solution using Baker's Yeast (Saccharomyces cerevisiae) strain. HBRC J [Internet] 2014 [cited 2014 Oct 27] Forthcoming. Available from: http://www.sciencedirect.com/science/article/pii/S1687404814000649. [Google Scholar]
- 12.Zulfikar MA, Setiyanto H. Adsorption of Congo Red from aqueous solution using powdered eggshell. Int J Chem Tech Res [Internet] 2013 Apr–Jun [cited 2014 Nov 11];5(4):1532–40. Available from: http://www.researchgate.net/profile/Henry_Setiyanto/publication/236263530_Study_of_the_adsorption_kinetics_and_thermodynamic_for_the_removal_of_Congo_red_from_aqueous_solution_using_powdered_eggshell/links/0deec526875c001962000000.pdf. [Google Scholar]
- 13.Phugare S, Patil P, Govindwar S, Jadhav J. Exploitation of yeast biomass generated as a waste product of distillery industry for remediation of textile industry effluent. Int Biodeterior Biodegrad [Internet] 2010 Dec [cited 2014 Oct 28];64(8):716–26. Available from: http://www.sciencedirect.com/science/article/pii/S0964830510001472 Subscription required to view. [Google Scholar]
- 14.Tony BD, Goyal D, Khanna S. Decolorization of textile azo dyes by aerobic bacterial consortium. Int Biodeterior Biodegrad [Internet] 2009 Feb [cited 2014 Nov 11];63(4):462–9. Available from: http://www.sciencedirect.com/science/article/pii/S096483050900016X Subscription required to view. [Google Scholar]
- 15.Hesselman MC, Odoni DI, Ryback BM, Groot S, Heck RG, Keijsers J, Kolkman P, Nieuwenhuijse D, Nuland YM, Sebus E, Spee R, Vries H, Wapenaar MT, Ingham CJ, Schroen K, Santos VA, Spaans SK, Hugenholtz F, Passel MW. A multi-platform flow device for microbial (Co-) cultivation and microscopic analysis. PLOS ONE [Internet] 2012 May 14 [cited 2014 Nov 11];7(5):e36982. doi: 10.1371/journal.pone.0036982. Available from: http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0036982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goers L, Freemont P, Polizzi KM. Co-culture systems and technologies: taking synthetic biology to the next level. J Royal Soc Interface [Internet] 2014 Jul [cited 2014 Nov 11];11(96):20140065. doi: 10.1098/rsif.2014.0065. Available from: http://rsif.royalsocietypublishing.org/content/11/96/20140065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milovanovic A, Bozic N, Vujcic Z. Cell wall invertase immobilization within calcium alginate beads. Food Chem [Internet] 2007 [cited 2014 Nov 11];104(1):81–6. Available from: http://www.sciencedirect.com/science/article/pii/S0308814606008636 Subscription required to view. [Google Scholar]
- 18.Rana S, Sharma R, Chandra S. Microbial degradation of synthetic textile dyes: a cost-effective and eco-friendly approach. Afr J Microbiol Res [Internet] 2013 Jun 11 [cited 2014 Oct 27];7(24):2983–9. Available from: http://www.academicjournals.org/article/article1380291977_Rana%20et%20al.pdf. [Google Scholar]
- 19.Donmez G. Bioaccumulation of the reactive textile dyes by Candida tropicalis growing in molasses medium. Enzym Microb Technol [Internet] 2002 Mar 13 [cited 2014 Oct 27];30(3):363–6. Available from: http://www.sciencedirect.com/science/article/pii/S0141022901005117 Subscription required to view. [Google Scholar]
- 20.Ramalho PA, Cardoso MH, Cavaco-Paulo A, Ramalho MT. Characterization of azo reduction activity in a novel ascomycete yeast strain. Appl Environ Microbiol [Internet] 2004 Apr [cited 2014 Nov 11];70(4):2279–88. doi: 10.1128/AEM.70.4.2279-2288.2004. Available from: http://aem.asm.org/content/70/4/2279.full.pdf+html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das D, Charumathi D, Das N. Combined effects of sugarcane bagasse extract and synthetic dyes on the growth and bioaccumulation properties of Pichia fermentans MTCC 189. J Hazard Mater [Internet] 2010 Nov 15 [cited 2014 Nov 11];183(1–3):497–505. doi: 10.1016/j.jhazmat.2010.07.051. Available from: http://www.sciencedirect.com/science/article/pii/S0304389410009362 Subscription required to view. [DOI] [PubMed] [Google Scholar]
- 22.Dhankhar R, Hooda A. Fungal biosorption – an alternative to meet the challenges of heavy metal pollution in aqueous solutions. Environ Technol [Internet] 2011 Apr [cited 2014 Oct 27];32(5):467–91. doi: 10.1080/09593330.2011.572922. Available from: http://www.tandfonline.com/doi/pdf/10.1080/09593330.2011.572922. [DOI] [PubMed] [Google Scholar]
- 23.Kariminiaae-Hamedaani H, Sakurai A, Sakakibara M. Decolorization of synthetic dyes by a new manganese peroxidase-producing white rot fungus. Dyes Pigments [Internet] 2007 [cited 2014 Dec 18];72(2):157–62. Available from: http://www.sciencedirect.com/science/article/pii/S0143720805002676 Subscription required to view. [Google Scholar]
- 24.Raghukumar C, Chandramohan D, Michel FC, Redd CA. Degradation of lignin and decolorization of paper mill bleach plant effluent (BPE) by marine fungi. Biotechnol Lett [Internet] 1996 Jan [cited 2014 Dec 18];18(1):105–6. Available from: http://link.springer.com/article/10.1007%2FBF00137820 Subscription required to view. [Google Scholar]
- 25.Asgher M, Yasmeen Q, Iqbal HM. Enhanced decolorization of solar brilliant red 80 textile dye by an indigenous white rot fungus Schizophyllum communa IBL-06. Saudi J Biol Sci [Internet] 2013 Oct [cited 2014 Dec 18];20(4):347–52. doi: 10.1016/j.sjbs.2013.03.004. Available from: http://www.sciencedirect.com/science/article/pii/S1319562X13000375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Xue M, Huang K, Liu Z. Textile dyeing wastewater treatment. In: Hauser PJ, editor. Advances in treating textile effluent [Internet] Rijeka, Croatia: InTech; 2011 Oct 26 [cited 2015 Jan 3]. In. editor. Chapter 5. Available from: http://www.intechopen.com/books/advances-in-treatingtextile-effluent/textile-dyeing-wastewater-treatment. [Google Scholar]
- 27.Sandhya S, Sarayu K, Uma B, Swaminathan K. Decolorizing kinetics of a recombinant Escherichia coli SS125 strain harboring azoreductase gene from Bacillus latrosporus RRK1. Bioresour Technol [Internet] 2008 May [cited 2015 Jan 3];99(7):2187–91. doi: 10.1016/j.biortech.2007.05.027. Available from: http://www.sciencedirect.com/science/article/pii/S0960852407004488 Subscription required to view. [DOI] [PubMed] [Google Scholar]
- 28.Blumel S, Knackmuss HJ, Stolz A. Molecular cloning and characterization of the gene coding for the aerobic azoreductase from Xenophilus azovorans KF46FF. Appl Environ Microbiol [Internet] 2002 Aug [cited 2015 Jan 3];68(8):3948–55. doi: 10.1128/AEM.68.8.3948-3955.2002. Available from: http://aem.asm.org/content/68/8/3948.full.pdf+html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, Choi YO, Kim WY, Kang JS, Cheong GW, Yun DJ, Rhee SG, Cho MJ, Lee SY. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell [Internet] 2004 May 28 [cited 2015 Jan 3];117(5):625–35. doi: 10.1016/j.cell.2004.05.002. Available from: http://www.sciencedirect.com/science/article/pii/S0092867404004878. [DOI] [PubMed] [Google Scholar]
- 30.Conesa A, Punt PJ, Hondel CA. Fungal peroxidases: molecular aspects and applications. J Biotechnol [Internet] 2002 Feb 14 [cited 2015 Jan 4];93(2):143–58. doi: 10.1016/s0168-1656(01)00394-7. Available from: http://www.sciencedirect.com/science/article/pii/S0168165601003947 Subscription required to view. [DOI] [PubMed] [Google Scholar]
- 31.Munhoz DC, Netto LE. Cytosolic thioredoxin peroxidase I and II are important defenses of yeast against organic hydroperoxide insult: catalases and peroxiredoxins cooperate in the decomposition of H2O2 by yeast. J Biol Chem [Internet] 2004 Aug 20 [cited 2015 Jan 4];279(34):35219–27. doi: 10.1074/jbc.M313773200. Available from: http://www.jbc.org/content/279/34/35219.full.pdf+html. [DOI] [PubMed] [Google Scholar]


