Abstract
Halophytes are able to thrive in salt concentrations that would kill 99% of other plant species, and identifying their salt-adaptive mechanisms has great potential for improving the tolerance of crop plants to salinized soils. Much research has focused on the physiological basis of halophyte salt tolerance, whereas the elucidation of molecular mechanisms has traditionally lagged behind due to the absence of a model halophyte system. However, over the last decade and a half, two Arabidopsis (Arabidopsis thaliana) relatives, Eutrema salsugineum and Schrenkiella parvula, have been established as transformation-competent models with various genetic resources including high-quality genome assemblies. These models have facilitated powerful comparative analyses with salt-sensitive Arabidopsis to unravel the genetic adaptations that enable a halophytic lifestyle. The aim of this review is to explore what has been learned about halophytism using E. salsugineum and S. parvula. We consider evidence from physiological and molecular studies suggesting that differences in salt tolerance between related halophytes and salt-sensitive plants are associated with alterations in the regulation of basic physiological, biochemical, and molecular processes. Furthermore, we discuss how salt tolerance mechanisms of the halophytic models are reflected at the level of their genomes, where evolutionary processes such as subfunctionalization and/or neofunctionalization have altered the expression and/or functions of genes to facilitate adaptation to saline conditions. Lastly, we summarize the many areas of research still to be addressed with E. salsugineum and S. parvula as well as obstacles hindering further progress in understanding halophytism.
Securing food for a growing human population is a grand challenge to humanity, which will require solutions stemming from basic sciences (United Nations Framework Convention on Climate Change, 2016; Food and Agriculture Organization of the United Nations, 2017). Saline soils are one of the most serious environmental factors limiting crop productivity. Although assessments vary, it is estimated that up to 10% of the Earth’s surface is affected by salt and approximately 50% of irrigated land (Flowers and Yeo, 1995; Polle and Chen, 2015). In some areas, 67% of agricultural land can be potentially affected by transient salinity (Rengasamy, 2006). The economic cost of crop losses due to salinity, estimated at about US$27 billion annually, in addition to the serious impact on global food security due to increasing salinization of soils (Qadir et al., 2014), is driving the need to generate crops with improved tolerance to salt stress and to develop halophyte-based agriculture (Bressan et al., 2011; Ventura et al., 2015).
Much of our understanding of plant responses to salt stress has been gleaned from a limited number of model plants or economically valuable crop species. In particular, investigations using Arabidopsis (Arabidopsis thaliana) have provided a wealth of information on the physiological and molecular mechanisms of salt tolerance due to its many qualities as a model organism and unmatched genetic and genomic resources. However, Arabidopsis is by no means a stress-tolerant species and is unlikely to possess many of the salt tolerance determinants that are naturally selected and exist in halophytes. These plants grow and reproduce in salt concentrations that would kill 99% of other plant species (Flowers and Colmer, 2008), and studying how halophytes thrive in such saline environments could act as a guide to improving the performance of nonhalophytic crop species (Shabala, 2013).
Characterization of the physiological mechanisms of salt tolerance in halophytes has advanced over the decades, although much is still unknown (Flowers et al., 1977, 2015; Flowers and Colmer, 2008; Munns and Tester, 2008; Cheeseman, 2013). However, elucidation of the molecular mechanisms and adaptive evolutionary signatures of halophytism has lagged behind physiological characterization due to the absence of a model halophyte with all the attendant molecular and genetic tools. Thus, in 2001, the interest of the plant stress community was piqued by mention of an Arabidopsis relative, commonly referred to as salt cress, from the coast of the Chinese Shandong province that is tolerant to salt stress (Bressan et al., 2001; Zhu, 2001). Furthermore, reports showed that salt cress possessed many of the attributes of a model plant that are also associated with Arabidopsis: small size, short life cycle, self-pollination, copious seed production, relatively small genome (approximately twice that of Arabidopsis), and transformation feasibility (Bressan et al., 2001; Amtmann, 2009). The plant was referred to in the literature by the name Thellungiella halophila but later determined to be the species Thellungiella salsuginea, now named Eutrema salsugineum (Al-Shehbaz and Warwick, 2005; Koch and German, 2013).
Realizing the huge potential for comparative analysis of stress tolerance mechanisms between closely related salt-sensitive and halophytic species, a workshop was held in 2004 to promote E. salsugineum as an Arabidopsis relative model system (Amtmann et al., 2005). Since that pioneering meeting, over 125 articles have been published on E. salsugineum and the related (and even more salt-tolerant [Orsini et al., 2010]) Schrenkiella parvula (formally Thellungiella parvula and Eutrema parvulum). Furthermore, various genetic resources have been generated, including chromosome-level genome assemblies, natural accession collections, full-length cDNA and EST collections, transformation protocols, a plant transformation-competent large-insert DNA library, Arabidopsis lines expressing an E. salsugineum cDNA library, nuclear and organelle genome sequences, microRNA sequences, cDNA microarrays, RNA sequencing data sets (Wang et al., 2004, 2010, 2016, 2018a, 2018b; Wong et al., 2005; Du et al., 2008; Taji et al., 2008; Zhang et al., 2008, 2013; Amtmann, 2009; Dassanayake et al., 2011a; Oh et al., 2012; Wu et al., 2012; Bartels and Dinakar, 2013; Champigny et al., 2013; Lee et al., 2013; Yang et al., 2013; Batelli et al., 2014; Fukami-Kobayashi et al., 2014; Guo et al., 2016; He et al., 2016; Yin et al., 2018), and dedicated Web resources (http://extremeplants.org/). Given their increasing importance as model systems (Box 1), E. salsugineum and S. parvula were ranked by the journal Cell as one of the next top models (Zhu et al., 2015).
The aim of this review, therefore, is to survey what progress has been made in understanding halophytism using comparative physiological and molecular analyses of Arabidopsis and E. salsugineum and/or S. parvula. In general, work that has only analyzed the halophytes alone is not covered, as it is difficult to draw conclusions regarding adaptation to salt stress without reference to a salt-sensitive relative. Similarly, we do not cite reports on the salt tolerance of transgenic Arabidopsis overexpressing E. salsugineum genes, as these articles rarely, if ever, examine whether overexpressing these genes has a different effect from overexpressing the Arabidopsis orthologs.
ABIOTIC STRESS TOLERANCE OF E. SALSUGINEUM AND S. PARVULA
E. salsugineum or S. parvula can be synchronously grown with Arabidopsis for comparative analysis in controlled environments (Fig. 1). On the one hand, E. salsugineum is similar to Arabidopsis in that it possesses a rosette leaf ontology during vegetative growth and it bolts during reproductive growth. However, E. salsugineum morphology can differ greatly in its natural habitat, displaying a complete absence of rosette leaves and developing only cauline leaves on multiple stems (Guevara et al., 2012; Fig. 1D). E. salsugineum also has a different developmental program from Arabidopsis; for instance, one of the principal ecotypes under study (Shandong ecotype) requires vernalization to accelerate flowering, although the plant will flower without vernalization after approximately 10 to 12 months (Inan et al., 2004). It also exhibits seed set and germination over an extended period (Inan et al., 2004). S. parvula shows similar growth rates to Arabidopsis but has an elongated stem with alternate leaves and displays indeterminate growth during vegetative and reproductive phases.
Figure 1.
Growth of Arabidopsis, S. parvula, and E. salsugineum on increasing levels of soil NaCl. A, Seven-day-old Arabidopsis plants were treated with various concentrations of NaCl incrementally until final salt concentrations (0, 100, 250, and 500 mm) were reached 1 week later. Plants were imaged after another 1 week. B, E. salsugineum plants treated in the same manner as in A. C, Forty-five-day-old S. parvula plants were treated with various concentrations of NaCl (0, 50, 100, 400, and 600 mm) for 21 d and imaged on day 70. S. parvula plants were able to complete their life cycle and yield seeds even at extreme salt levels. D, E. salsugineum in the salt-crust soils of the Yukon, Canada. Note the presence of cauline leaves while rosette leaves are absent. (Photograph by Jeff Dedrick.)
At the tissue level, E. salsugineum possesses several features that could contribute to its salt tolerance, such as a second layer of palisade mesophyll cells, a higher stomatal density, and differences in leaf epicuticular wax and lipid concentration/composition, all of which could affect water loss under saline conditions (Teusink et al., 2002; Inan et al., 2004; Xu et al., 2014). S. parvula has a higher stomatal density as well as a higher epicuticular wax content compared with Arabidopsis (Teusink et al., 2002; Orsini et al., 2010). Similar to other halophytes, E. salsugineum roots develop a second layer of endodermis and an additional cortex layer that could restrict ion flow from roots to shoots (Flowers et al., 1986; Inan et al., 2004).
E. salsugineum is more tolerant than Arabidopsis to a number of abiotic stresses, including salt stress (Inan et al., 2004), low-nitrogen stress (Kant et al., 2008), phosphate deprivation (Velasco et al., 2016), high boron levels (Lamdan et al., 2012), and heat stress (Higashi et al., 2013). E. salsugineum also exhibits tolerance to chilling and freezing. However, in terms of comparison with Arabidopsis, the relative tolerance of E. salsugineum depends upon the accessions employed, whether plants are preacclimated, and whether the treatment is short or long term (Lee et al., 2012; Khanal et al., 2015). No firm evidence exists for the increased drought tolerance of E. salsugineum compared with Arabidopsis, although natural variation exists in drought tolerance between E. salsugineum accessions (MacLeod et al., 2015). However, E. salsugineum leaves do have constitutively lower water content than Arabidopsis and are able to lose more water in response to osmotic stress, suggesting that E. salsugineum can tolerate dehydration to a greater extent than Arabidopsis (Lugan et al., 2010). Accordingly, E. salsugineum also exhibits greater tolerance to polyethylene glycol-induced osmotic stress than Arabidopsis (Yu and Li, 2014). In addition to high-Na+ stress, S. parvula is adapted to multiple-ion stresses, including tolerance to K+, Li+, and Mg2+ at levels toxic to most plants, including E. salsugineum and Arabidopsis (Oh et al., 2014).
The great majority of studies have focused upon E. salsugineum responses to salt stress. Unlike Arabidopsis, this plant is able to survive and complete its life cycle in extreme salt concentrations up to 500 mm NaCl (Inan et al., 2004; Taji et al., 2004; Vera-Estrella et al., 2005; Kant et al., 2006; Ghars et al., 2008; Lugan et al., 2010; Guo et al., 2012). These reports have compared fresh and dry weight reductions as a marker of stress tolerance and have shown that, while salt stress-mediated growth reduction does take place in E. salsugineum, it occurs at higher salt concentrations than for Arabidopsis. However, a more detailed comparative phenomics study unmasked a reduction in E. salsugineum total rosette area by salt concentrations as mild as 50 mm NaCl (Kazachkova et al., 2013). This is likely an adaptive response, in agreement with findings that plants actively reduce their growth in response to stress independently of photosynthesis (Skirycz and Inzé, 2010; Skirycz et al., 2010). The more recently introduced halophytic model, S. parvula, shows a greater capacity to withstand higher NaCl levels than E. salsugineum without compromising growth (Orsini et al., 2010; Oh et al., 2014).
CONTROL OF Na+-K+ ION TRANSPORT
It has long been dogma that all plants must prevent the accumulation of toxic levels of salts in the cytosol, although what exactly constitutes toxic cytosolic levels of Na+ ions is still unclear (Cheeseman, 2013; Flowers et al., 2015). Nevertheless, halophytes have evolved more efficient mechanisms to control cytosolic Na+ levels by a combination of exclusion, sequestration, osmotic adjustment, and redistribution (Flowers and Colmer, 2015).
Several studies have shown that, under control conditions, E. salsugineum and S. parvula shoots possess higher levels of Na+ than Arabidopsis (Inan et al., 2004; Kant et al., 2006; Wang et al., 2006; Orsini et al., 2010; Oh et al., 2014). Consistent with higher Na+ accumulation, E. salsugineum exhibits a constitutively more negative leaf osmotic potential than Arabidopsis in the absence of external NaCl (Inan et al., 2004). Under increasing levels of external NaCl, both E. salsugineum and S. parvula accumulate Na+ to a far lower extent than Arabidopsis while maintaining shoot K+ levels, suggesting that the halophytes can more tightly control Na+ uptake (Inan et al., 2004; Kant et al., 2006; Orsini et al., 2010; Oh et al., 2014). This notion has been confirmed by radiotracer studies demonstrating that the steady-state unidirectional Na+ influx into the roots is approximately 50% lower in E. salsugineum than in Arabidopsis (Wang et al., 2006). Consistent with the radiotracer data, electrophysiology studies have revealed a significantly smaller Na+ inward current in E. salsugineum than in Arabidopsis, due to the higher selectivity for K+ over Na+ of a putative E. salsugineum voltage-independent channel (Volkov and Amtmann, 2006). In fact, a general feature of E. salsugineum stress tolerance appears to be its ability to tightly control the accumulation of various ions, including Na+, K+, B(OH)3, PO43−, and NO3− (Inan et al., 2004; Kant et al., 2006, 2008; Lamdan et al., 2012; Velasco et al., 2016).
Several transporters have been studied that could contribute to E. salsugineum salt tolerance. In Arabidopsis, the plasma membrane Na+/H+ antiporter, SALT OVERLY SENSITIVE1 (SOS1), functions in the exclusion of Na+ ions from root cells as well as controlling the loading and retrieval of Na+ into and from the xylem stream (Shi et al., 2002). An E. salsugineum ortholog of AtSOS1 is essential for E. salsugineum salt tolerance, and RNA interference-based interference of EsSOS1 expression essentially converts E. salsugineum from a halophyte into a salt-sensitive plant (Oh et al., 2007, 2009a, 2009b). Moreover, when expressed in a salt-sensitive yeast strain, EsSOS1 confers greater salt tolerance than SpSOS1 and AtSOS1 (Jarvis et al., 2014). SOS1 appears to have evolved under positive selection in E. salsugineum, with a single amino acid change in the putative autoinhibitory domain necessary but not sufficient for EsSOS1 to confer enhanced salt tolerance. The Na+/K+ HIGH-AFFINITY K+ TRANSPORTERs (EsHKT1 and SpHKT1) also are essential for E. salsugineum and S. parvula salt adaptation (Ali et al., 2012, 2018; Oh et al., 2014). For instance, RNA interference knockdown of EsHKT1;2 leads to increased hypersensitivity to NaCl, greater accumulation of Na+ in the shoots, and reduced K+ accumulation (Ali et al., 2012, 2013). Conversely, overexpression of SpHKT1;2 in Arabidopsis causes enhanced tolerance to NaCl, decreased Na+ accumulation, and increased K+ content (Ali et al., 2018). Thus, EsHKT1;2 and SpHKT1;2 have a vital role in Na+/K+ balance and an essential function in salt tolerance of the halophytes. Moreover, as detailed below (see “E. salsugineum and S. parvula Genomes: A Window into the Genetic Basis of Adaptation to Extreme Environments”), genomic sequence and expression analyses of the HKT gene family in E. salsugineum and S. parvula reveal specific features of adaptation to saline environments.
An increase in tonoplast Na+/H+ exchange has been observed in E. salsugineum plants under salt stress (Vera-Estrella et al., 2005), suggesting that compartmentalization of Na+is also an important feature of E. salsugineum salt tolerance. The Na+/K+ (NHX1) transporter was identified originally as an Arabidopsis tonoplast Na+/H+ antiporter (Apse et al., 1999), although it has been suggested that it functions mainly as a K+/H+ transporter (Jiang et al., 2010; Leidi et al., 2010). An ortholog of AtNHX1 was isolated from E. salsugineum, and loss-of-function transgenic plants exhibited greater sensitivity to salt and osmotic stress than wild-type plants (Wu et al., 2009). Consistent with these results, it also was demonstrated that Na+ ions are compartmentalized primarily within the vacuole in E. salsugineum (Wang et al., 2013). However, neither of these reports provided evidence regarding whether EsNHX1 improved salt tolerance compared with AtNHX1 or how much vacuolar Na+ was accumulated relative to Arabidopsis.
HALOPHYTIC PHOTOSYNTHETIC PERFORMANCE AND PROTECTIVE ANTIOXIDANT SYSTEMS
In general, salt stress leads to reduced photosynthetic performance due to various factors, including decreased CO2 diffusionbecause of diminished stomatal aperture, reduced mesophyll CO2 transport, altered leaf photochemistry and carbon metabolism, and down-regulation of photosynthetic gene expression (Chaves et al., 2009; Bartels and Dinakar, 2013). A detailed comparison of Arabidopsis and E. salsugineum (Shandong ecotype) photosynthetic responses to salt provided insight into the halophyte’s strategy for maintaining photosynthetic performance under saline conditions (Stepien and Johnson, 2009). This study showed that, in Arabidopsis, salt stress causes a reduction in stomatal conductance, net CO2 assimilation, PSII electron transport, greater cyclic electron flow around PSI, and increased nonphotochemical quenching (NPQ). Thus, Arabidopsis behaves in a similar manner to other species under stress; namely, a stress-mediated decrease in carbon fixation leads to the down-regulation of linear electron transport, reflecting a reduced need for electrons and the need to minimize the generation of damaging reactive oxygen species (ROS; Golding and Johnson, 2003). Simultaneously, there is a rise in NPQ to dissipate excess energy, thereby protecting PSII. This rise in NPQ is supported by a ∆pH generated by cyclic electron transport. On the other hand, E. salsugineum behaves quite differently from Arabidopsis. Even at extreme salt levels, gas exchange is inhibited only marginally, while no increase in cyclic electron transport or NPQ is observed. There is, however, a large rise in PSII electron flow, and at least 30% of this flow can be accounted for by increased levels of a plastid terminal oxidase (PTOX) compared with Arabidopsis under both control and salt stress conditions (Stepien and Johnson, 2009; Wiciarz et al., 2015). The PTOX could act as an alternative electron sink by transferring electrons from plastoquinone to molecular oxygen without generating ROS.
Limitation of Calvin cycle reactions and the consequent leakage of electrons from the photosynthetic electron transport chain are associated with ROS generation leading to cellular damage (Miller et al., 2010). Several reports suggest that E. salsugineum and S. parvula have highly active antioxidant systems. For instance, Uzilday et al. (2015) showed that a number of enzymes involved in ROS scavenging are active in S. parvula under nonsaline conditions, with only a slightly higher activity under a range of soil NaCl concentrations. When activities are measured specifically in chloroplasts, however, they increase in a NaCl dose-dependent manner. Nevertheless, since there was no comparison with enzyme activities in Arabidopsis, it is difficult to conclude anything about ROS-scavenging mechanisms that are unique to the halophyte. Where such comparative analyses have been performed, antioxidant metabolites such as ascorbate, dehydroascorbate, and caffeic acid are either constitutively higher or accumulate to a greater extent in soil-grown E. salsugineum compared with Arabidopsis (Kazachkova et al., 2013; Eshel et al., 2017). Indeed, the avoidance of singlet oxygen production at PSII has been attributed to induction of the ascorbate-glutathione cycle in E. salsugineum (Wiciarz et al., 2018). Similarly, higher levels of thioredoxin are maintained in salt-stressed E. salsugineum, and this finding is correlated with reduced lipid peroxidation (M’rah et al., 2007). Accordingly, E. salsugineum is more tolerant than Arabidopsis to ROS-inducing methyl viologen (Taji et al., 2004). Higher expression of E. salsugineum genes encoding components of the antioxidant machinery under both control and salt stress conditions might be responsible for a more active and efficient capacity for coping with ROS (Taji et al., 2004; Gong et al., 2005).
Induction of plant defenses against various stresses can be due to fast and transient production of ROS, such as H2O2, which act as a stress signal (Apel and Hirt, 2004). A similar amount of H2O2 production is induced in Arabidopsis and E. salsugineum leaves a few hours after a 400 mm NaCl shock treatment, whereas higher induction of H2O2 is observed in E. salsugineum roots (Ellouzi et al., 2014). However, a more detailed analysis using plants adapted to milder saline conditions (150 mm for Arabidopsis and 300 mm for E. salsugineum), employing a variety of complementary methods, demonstrated that both isolated thylakoid membranes and chloroplasts from intact E. salsugineum leaf tissue produce more H2O2 in both control and saline conditions compared with Arabidopsis (Pilarska et al., 2016). Furthermore, under control conditions, a lower amount of damaging superoxide is generated in E. salsugineum than in Arabidopsis, which is correlated with reduced lipid peroxidation in isolated thylakoids and intact leaves of the halophyte. These findings led the authors to speculate that enhanced leakage of H2O2 from E. salsugineum plastids preadapts the halophyte to salt stress by maintaining an up-regulated antioxidant system while preventing ROS damage. This hypothesis is further supported by the following observations: (1) the reduced activity of H2O2-detoxifying enzymes, such as ascorbate peroxidase (APX) in E. salsugineum and similar activities of catalase in E. salsugineum and Arabidopsis (Pilarska et al., 2016); and (2) the ability of E. salsugineum to sustain a highly reduced plastoquinone pool (Wiciarz et al., 2015, 2018). This could facilitate the persistence of the cellular H2O2 signal (Bose et al., 2014). On the other hand, the chloroplasts of E. salsugineum are better protected from ROS damage: thylakoid membranes from the halophyte display a higher abundance of tAPX and Fe superoxide dismutase, while the stroma contains greater levels of peroxiredoxins compared with Arabidopsis. Taken together with other reports suggesting the increased abundance/activity of chloroplast redox components and Calvin-Benson cycle proteins in E. salsugineum (Gong et al., 2005; M’rah et al., 2007; Stepien and Johnson, 2009; Chang et al., 2015; Wiciarz et al., 2015), it is suggested that specific ROS signaling signatures and fine-tuning of specific antioxidants could be a hallmark of stress anticipation in the halophyte.
SALT-MEDIATED TRANSCRIPTOME RESPONSES AND METABOLIC REPROGRAMMING
Metabolism
Several groups have shown that salt-mediated metabolic reprogramming of E. salsugineum is clearly distinct from that of Arabidopsis (Gong et al., 2005; Arbona et al., 2010; Lugan et al., 2010; Pedras and Zheng, 2010; Guevara et al., 2012; Kazachkova et al., 2013; Eshel et al., 2017). In general, the same metabolic pathways are regulated in response to salt stress, but most of the detected metabolites accumulate to higher levels in E. salsugineum under control and salt treatments compared with Arabidopsis. The much lower response to stress observed for E. salsugineum metabolites compared with that of Arabidopsis suggests that the halophyte is primed for stress, a theme that also is observed at the transcriptome level (see below). This idea is emphasized by the finding that over 70% of the detected metabolites that are more abundant in E. salsugineum under control conditions exhibit a salt-mediated increase in Arabidopsis while over 50% of metabolites that are less abundant in E. salsugineum display a salt-mediated decrease in Arabidopsis (Lugan et al., 2010). In other words, the Arabidopsis salt metabolic profile partially resembles that of E. salsugineum under control conditions. Comparing salt stress- and osmotic stress-mediated metabolic reprogramming in Arabidopsis and E. salsugineum also reveals that the expected accumulation of solutes to maintain a negative osmotic potential for water uptake is balanced by a decrease in other solutes such that, overall, no osmotic adjustment takes place in either species (Lugan et al., 2010). The more negative osmotic potential of E. salsugineum compared with Arabidopsis is due to the ability of the halophyte to lose more water. Indeed, inspection of global physiochemical properties suggests that the E. salsugineum metabolome is reconfigured to osmoprotection and tolerance of dehydration.
Several metabolic signatures are maintained independent of growth condition or type of stress in E. salsugineum. For instance, the levels of the tricarboxylic acid cycle intermediates, malate and citrate, are higher in E. salsugineum than in Arabidopsis, whereas fumarate and the osmoprotectants raffinose and galactinol display constitutively low amounts in E. salsugineum (Lugan et al., 2010; Kazachkova et al., 2013; Eshel et al., 2017). The levels of these compounds in E. salsugineum could represent E. salsugineum core stress tolerance mechanisms (Kant et al., 2008; Kazachkova et al., 2013). The functional significance of these signatures is unclear at present. It is tempting to speculate that high malate levels might facilitate the increased production of oxaloacetate to provide carbon skeletons for the increased levels of amino acids observed under control conditions in E. salsugineum. Moreover, the continuous use of oxaloacetate could eventually lead to a deficiency in tricarboxylic acid cycle intermediates, such as the observed reduced accumulation of fumarate. It is interesting that E. salsugineum does not accumulate raffinose or its precursor galactinol in response to salt stress, because raffinose and other members of the raffinose family of oligosaccharides accumulate to high levels under control and stress conditions in other extremophytes (Peters et al., 2007; Farrant et al., 2009; Gechev et al., 2012; Benina et al., 2013). It is unclear why galactinol and raffinose do not accumulate in E. salsugineum, but this might be a distinctive trait similar to that in the desiccation-tolerant spike moss Selaginella lepidophylla (Yobi et al., 2012).
A number of metabolites behave differently according to growth conditions, exemplifying the high degree of metabolic plasticity in E. salsugineum (Kazachkova et al., 2013). The levels of many metabolites (including several known to be involved in stress tolerance) are repressed in E. salsugineum when plants are grown in vitro on nutrient agar plates compared with soil-grown plants. This finding is particularly interesting when considering Pro levels. Several groups have reported increased Pro content in control and salt-stressed E. salsugineum compared with Arabidopsis (Inan et al., 2004; Taji et al., 2004; Kant et al., 2006; Ghars et al., 2008), and it has been suggested that Pro contributes to E. salsugineum salt tolerance as an osmoprotectant and possible ROS scavenger. However, all these studies were performed with soil-grown plants in growth rooms/cabinets or plants grown using hydroponics. Yet, on the saline soil in its native habitat (Yukon ecotype), E. salsugineum exhibits a negligible Pro content (Guevara et al., 2012), and it was surmised that the low levels of nitrogen present in the Yukon soils might be responsible for the naturally low E. salsugineum Pro content in the field. This hypothesis was supported by demonstrating that, under nitrogen-limiting conditions in growth cabinets using soil-grown plants, Pro no longer accumulates in response to salt stress. Similarly, under in vitro conditions, E. salsugineum (Shandong ecotype) does not display increased Pro levels in response to salt stress (Kazachkova et al., 2013). Although the in vitro nutrient medium in that report did not contain reduced nitrogen levels, it did contain Suc, which may have increased the carbon-nitrogen ratio in a similar manner to reducing nitrogen levels. The important point here is that, regardless of growth condition-dependent metabolic profiles (and growth phenotypes), E. salsugineum still retains its salt tolerance, and Guevara et al. (2012) suggested that such phenotypic and metabolic adaptive plasticity would allow the flexibility required for an extremophyte lifestyle.
Transcriptome
The salt responsiveness of the E. salsugineum and S. parvula transcriptomes largely mirrors the metabolome (Taji et al., 2004; Gong et al., 2005; Oh et al., 2014). Both microarray and proteome studies show that fewer transcripts and proteins respond to salt stress in E. salsugineum than in Arabidopsis (Taji et al., 2004; Gong et al., 2005; Pang et al., 2010; Vera-Estrella et al., 2014). Exemplifying a stress-primed state at stress-neutral growth conditions, many genes known for their induction upon salt stress in stress-sensitive plants are found at high constitutively expressed levels in E. salsugineum. The stress-mediated induction of transcriptional responses in E. salsugineum does occur but at higher salinity levels than are required for Arabidopsis, indicating a difference in sensitivity to salt between the two species (Gong et al., 2005; Amtmann, 2009). However, it is important to note that preadaptation is not a universal halophytic trait. Comparison of metabolic profiles of the halophytic legume Lotus creticus with its salt-sensitive relatives Lotus corniculatus and Lotus tenuis has demonstrated that the salt response is conserved globally among the three species (Sanchez et al., 2011). Another interesting observation from global studies of E. salsugineum transcript responses is that there is very little overlap of gene expression between salinity, drought, and cold conditions, suggesting that E. salsugineum is more specific in its stress response than stress-sensitive species (Wong et al., 2005, 2006).
The comparative global analyses of Arabidopsis and E. salsugineum transcriptome/proteome/metabolome responses suggest that the differential regulation of a basic set of stress tolerance genes is a crucial component of E. salsugineum salt tolerance. This notion is supported by comparative RNA sequencing and targeted quantitative PCR analyses between Arabidopsis and the halophytes. Orthologous genes with higher basal expression in S. parvula compared with Arabidopsis are enriched in ion transport functions, whereas pathogen-related genes display higher expression in Arabidopsis (Oh et al., 2014). For example, SOS1 expression is constitutively higher in E. salsugineum roots under control conditions compared with Arabidopsis and is induced by salt to a greater extent in E. salsugineum shoots (Taji et al., 2004, 2010; Kant et al., 2006; Oh et al., 2010; Dassanayake et al., 2011b). This increased EsSOS1 expression is correlated with a reduced uptake of Na+ ions into the xylem in response to salt stress compared with Arabidopsis (Kant et al., 2006). Another example is the down-regulation of an E. salsugineum PROLINE DEHYDROGENASE (PDH) gene encoding a Pro catabolic enzyme. This down-regulation is correlated with increased levels of the osmoprotectant, Pro, under control and salt stress conditions in E. salsugineum shoots compared with Arabidopsis (Kant et al., 2006). No such down-regulation of PDH is observed in E. salsugineum roots. Accordingly, Pro levels are similar in E. salsugineum and Arabidopsis roots. It is also notable that the early phospholipase C and D lipid signaling pathways in E. salsugineum control Pro accumulation under non-salt stress or salt stress conditions in an opposite manner to Arabidopsis (Ghars et al., 2012). However, this control does not appear to be exerted via the transcriptional regulation of genes involved either in Pro biosynthesis or catabolism.
E. SALSUGINEUM AND S. PARVULA GENOMES: A WINDOW INTO THE GENETIC BASIS OF ADAPTATION TO EXTREME ENVIRONMENTS
The phenotypic, physiological, biochemical, molecular, and omics studies described above have provided evidence to account for halophyte stress tolerance. However, the missing tool to link genetic adaptation to stress-tolerant phenotypes has been provided by the genome sequences of E. salsugineum and S. parvula and the powerful comparative analysis of these genomes with each other and with that of Arabidopsis (Oh et al., 2010, 2012, 2014; Dassanayake et al., 2011a; Dittami and Tonon, 2012; Wu et al., 2012; Bressan et al., 2013; Yang et al., 2013). The two extremophyte genomes provide high-quality references (Table 1) because they span large intergenic regions beyond the fragmented short scaffolds and minimum curation released for many draft genomes. The S. parvula genome in particular provides one of the most contiguous gap-free assemblies available among plant genomes (Dassanayake et al., 2011a). Thus, the extremophyte genomes are powerful platforms for comparative genomic analyses with established models such as Arabidopsis, rice (Oryza sativa), and poplar (Populus trichocarpa). The two extremophyte genomic resources are being actively updated. For example, new transcripts were reported for E. salsugineum (Yin et al., 2018), and an updated genome assembly and annotation for S. parvula was released via http://thellungiella.org/data/. The genomic resources also are shared via publicly accessible databases, including NCBI, PLAZA, CoGe, Phytozome, and eXtremeplants (http://extremeplants.org).
Table 1. Comparison of Arabidopsis, S. parvula, and E. salsugineum reference genomes.
NA, Not applicable, when the reference publications did not include the relevant value.
| Parameter | Arabidopsis | S. parvula | E. salsugineum | |
|---|---|---|---|---|
| Arabidopsis Genome Initiative | Dassanayake et al. (2011a) | Wu et al. (2012) | Yang et al. (2013) | |
| Predicted genome size (Mb) | 150 | 160 | 260 | NA |
| Actual genome size (Mb) | 115.4 | 137.09 | 233.7 | 241 |
| Total number of genes | 27,655 | 26,847 | 28,457 | 26,531 |
| Haploid chromosome number | 5 | 7 | 7 | 7 |
| Total number of assembled scaffolds | NA | 7 | 515a | 639 |
| N50 (bp), contigs | NA | 5.29 Mb | NA | 272 kb |
| N50 (bp), scaffolds | NA | NA | 403,516 | 8 Mb |
| Number of scaffolds at least N50 | NA | 8 | 119 | NA |
| Transposons (% of total genome size) | 13.2 | 7.5 | 52 | NA |
| Number of orthologous genesb | 19,834 | 19,280 | 19,582 | NA |
| Number of orphan genesc | 5,019 | 5,844 | 6,810 | NA |
| Percentage of functionally unknown genes | 36.5 | 62.8 | 54.7 | NA |
| Average gene length (bp) | 2,374 | 2,110 | 2,041 | NA |
| Longest open reading frame (bp) | NA | 16,758 | NA | NA |
| Average exon length (bp) | 224 | 230 | 228 | NA |
| Average intron length (bp) | 157 | 191 | 200 | NA |
Scaffolds anchored to seven chromosomes.
Genes were considered orthologs if they shared more than 50% amino acid sequence similarity using BLASTP among the three genomes, e < 0.00001.
Genes lacking orthologs in the other two species.
Several features of the halophyte genomes provide hypotheses to explain the genetic basis for plant adaptations to extreme environments. Both the E. salsugineum and S. parvula genomes show extensive macrosynteny with the Arabidopsis genome (Dassanayake et al., 2011a; Wu et al., 2012). However, local microsynteny between the species is disrupted by tandem gene duplications, translocations, deletions, and insertions (Oh et al., 2014). Comparisons between the Arabidopsis and S. parvula genomes show that orthologous pairs that have undergone such genome structural variation in either species exhibit less similarity in the putative promoter regions (upstream of the translation start site) than promoters between collinear orthologs. Accordingly, noncollinear genes between the two species exhibit differential basal expression compared with collinear genes (Oh et al., 2014). Moreover, the genes that show genome structural variation in the S. parvula and E. salsugineum genomes via tandem duplications or translocation-duplications are enriched in abiotic stress-associated functions compared with the orthologs in Arabidopsis (Dassanayake et al., 2011a; Wu et al., 2012; Oh et al., 2014). Most of these genomic structural variations also affect gene copy number variation (CNV), which, for decades, has been proposed as a major driver of adaptation to new environments (Ohno, 1970; DeBolt, 2010). Table 2 lists genes associated with stress responses in Arabidopsis that show CNV unique to S. parvula and E. salsugineum based on a network analysis using synteny information from Brassicaceae genomes (Oh and Dassanayake, 2018).
Table 2. Genes that have experimentally verified functions associated with stress responses in Arabidopsis and that show increased copy number in S. parvula and E. salsugineum.
The selected duplication events were not detected in other Brassicaceae lineage I and II genomes in a multispecies comparison including Arabidopsis, Arabidopsis lyrata, Capsella rubella, and Sisymbrium irio genomes (selected subset from Oh and Dassanayake, 2018).
| Arabidopsis Locus | Gene Name | Copy Number (At, Es, Sp) | Experimentally Verified Functions | References |
|---|---|---|---|---|
| AT2G26430 | ARGININE-RICH CYCLIN1 (RCY1) | 1, 3, 2 | Confers tolerance to LiCl and NaCl | Forment et al. (2002) |
| AT4G30380 | BARWIN-RELATED ENDOGLUCANASE | 1, 2, 2 | Induced during hypoxia | Yang et al. (2011) |
| AT1G10570 | OVERLY TOLERANT TO SALT2 (OTS2) | 1, 2, 2 | ots1ots2 double mutants show extreme sensitivity to salt | Conti et al. (2008) |
| AT4G26080 | ABA INSENSITIVE1 (ABI1) | 1, 2, 2 | Reduced sensitivity to salt and osmotic stress during germination | Krzywińska et al. (2016) |
| AT5G11260 | ELONGATED HYPOCOTYL5 (HY5) | 1, 3, 3 | Role in cold acclimation | Catalá et al. (2011); An et al. (2017) |
| AT4G20370 | TWIN SISTER OF FT (TSF) | 2, 4, 5 | Role in drought-escape response | Riboni et al. (2013) |
| AT1G47510 | INOSITOL POLYPHOSPHATE 5-PHOSPHATASE11 (5PTASE11) | 1, 2, 2 | Role in salt-induced ROS production | Kaye et al. (2011) |
| AT5G59820 | RESPONSIVE TO HIGH LIGHT41 (RHL41) | 3, 4, 4 | Role in reactive oxygen and abiotic stress signaling | Rizhsky et al. (2004); Davletova et al. (2005) |
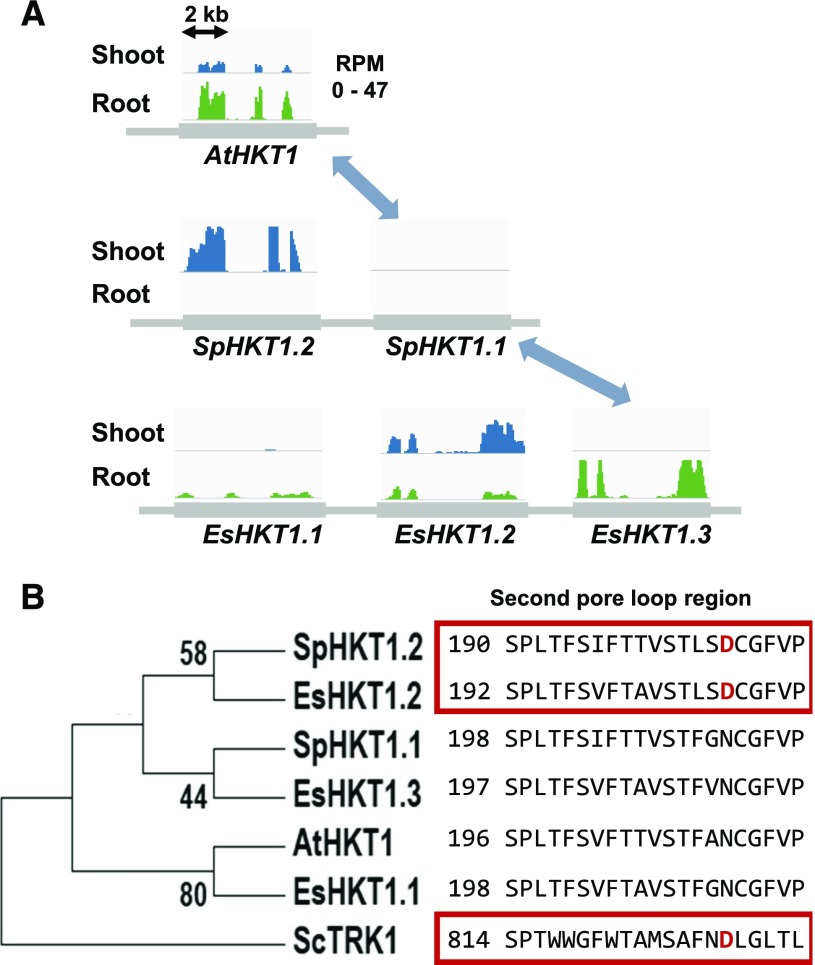
CNV can affect the transcriptome response by the acquisition of new regulatory elements or modification of existing elements in the duplicated genes, leading to gene subfunctionalization (Duarte et al., 2006; Galimba and Di Stilio, 2015). Notably, a number of ion transporters that show CNV in S. parvula suggest gene subfunctionalization, as manifested by organ-specific expression in roots and shoots of the paralogs compared with the single-copy orthologs in Arabidopsis, which are expressed in both organs (Oh et al., 2014; Fig. 2).
Figure 2.
CNV in the HKT1 gene family indicating subfunctionalization and neofunctionalization following tandem duplication in S. parvula and E. salsugineum. A, The single copy of HKT1 in Arabidopsis (At) is tandemly duplicated in S. parvula (Sp) and E. salsugineum (Es). Collinear genomic regions are shown in At, Sp, and Es, with blue and green histograms indicating normalized RNA coverage (RPM) for shoots and roots under stress-neutral conditions. Orthologous genes are connected with arrows. The tissue-specific expression among halophyte HKT1 paralogs suggests gene subfunctionalization. B, Amino acid alignment and tree structure of the HKT1 proteins. ScTRK1, a yeast K+ transporter, was used as an outgroup. The sequences selected are a subset of those published by Ali et al. (2012). The alignment includes Es- and Sp-specific HKT1 copies, with the amino acid substitution from Asn (N) to Asp (D) in the second pore-loop domain (depicted in the red boxes for EsHKT1;2, SpHKT1;2, and ScTRK1 copies, with red letters for the substituted D). This N→D substitution was shown to confer the unique K+ transport activity (Ali et al., 2012, 2018; Ali and Yun, 2016) indicating neofunctionalization in the HKT1;2 copies compared with the Arabidopsis ortholog. The maximum likelihood tree based on sequence alignment was generated using MEGA7 with a consensus tree based on 100 bootstrap values (Kumar et al., 2016).
A good example of subfunctionalization is Na+/H+ EXCHANGER8 (NHX8), encoding a putative Li+/H+ antiporter that is present as a single-copy gene in Arabidopsis (An et al., 2007). However, S. parvula possesses three tandem duplicates of SpNHX8. Genes encoding SpNHX8;1 and SpNHX8;3 exhibit higher basal expression than AtNHX8 in shoots and roots, while SpNHX8;2, which shows a comparable basal expression to AtNHX8 in roots, is induced by LiCl treatment. Accordingly, S. parvula displays much greater tolerance to high levels of LiCl (Oh et al., 2014), consistent with high Li+ levels present in its natural habitat and indicating local edaphic adaptation (Helvaci et al., 2004; Nilhan et al., 2008). Although not yet confirmed with targeted transgenic studies, expansion and subfunctionalization of the SpNHX8 genes may represent a CNV event that underlies the genetic basis for the high Li tolerance in S. parvula.
Certain genes that appear to have strict dosage-dependent functions generally are maintained as a single copy in plant genomes (De Smet et al., 2013). The limitations to their expansion are not always clear, and our knowledge of how these gene networks functionally apply restrictions to CNV is limited. However, genes present as single copies in both Arabidopsis and the two halophytes may have significantly different expression profiles at the transcriptome level (i.e. transcriptional CNV) enabled by the insertion or modifications of regulatory elements. For example, the acquisition of new regulatory elements is well illustrated by the AtSOS1 and VACUOLAR H+-PYROPHOSPHATASE (AtAVP1) orthologs in E. salsugineum and S. parvula. SOS1 basal and salt-induced expression is greater in the halophytes than in Arabidopsis. Interestingly, EsSOS1, SpSOS1, and indeed all halophytic SOS1 orthologs examined possess a pyrimidine-rich stretch in the 5′ untranslated region (UTR) as well as a predicted stem-loop structure close to the start codon (Dassanayake et al., 2011b). Although the significance of these observations has yet to be demonstrated, such sequences and structures in 5′ UTRs have been associated with enhanced basal gene expression, mRNA stability, and translation efficiency (An and Meagher, 2010; Grillo et al., 2010; Wever et al., 2010). Similarly, the promoter of EsAVP1 can drive the salt-meditated induction of gene expression, unlike the AtAVP1 promoter (Sun et al., 2010). Deletion analysis of the EsAVP1 promoter revealed a 130-bp region that is sufficient for the salt stress response. Further examination of conserved motifs in this region indicated the presence of novel regulatory elements that may provide a genetic basis for this salt-adapted response.
In contrast to the challenge of maintaining high K+ uptake while lowering Na+ intake that is faced by E. salsugineum and many other plants, S. parvula must limit the uptake of both ions because soils in its natural habitat contain toxic levels of K+ and Na+ (Ozfidan-Konakci et al., 2016). Gene duplication followed by neofunctionalization and subfunctionalization of the Na+/K+-transporter gene HKT1 in E. salsugineum and S. parvula illustrates how the two halophytes differentially express HKT1 copies to adapt to specific challenges unique to their niches (Fig. 2). HKT1, a single-copy gene in Arabidopsis, is tandemly duplicated in E. salsugineum and S. parvula (Dassanayake et al., 2011a; Wu et al., 2012). A single amino acid substitution in the duplicated E. salsugineum paralog shifts its function from being a more Na+-specific transporter to a more K+-specific transporter (Ali et al., 2012, 2016; Ali and Yun, 2016). Under high-Na+ stress, E. salsugineum down-regulates the expression of the Na+-transporting HKT1 while up-regulating the expression of the K+-transporting HKT1 copy, thereby enabling greater K+ uptake to facilitate growth and development. In contrast, the expression of the S. parvula Na+-transporting HKT1 ortholog is down-regulated in both shoots and roots, similar to E. salsugineum, but the K+-transporting S. parvula HKT1 paralog displays further gene subfunctionalization: its expression is down-regulated in the roots to limit K+ uptake in a high-K+ soil but up-regulated in the shoots to allow K+ uptake into developing tissue (Oh et al., 2014). Accordingly, S. parvula indeed maintains higher K+ levels in shoots compared with E. salsugineum and Arabidopsis (Orsini et al., 2010; Oh et al., 2014). The HKT1 gene duplication may reveal only one contributing factor in understanding the complex trait of maintaining a suitable K+-Na+ balance in different environments, but it illustrates how comparative genomic and transcriptomic analyses coupled with functional genetic studies using not just Arabidopsis but also the halophytes could lead to the discovery of naturally selected, stress-adaptive mechanisms.
SALT INHIBITION OF HALOPHYTE SEED GERMINATION
It has been known for decades that, counterintuitively, seeds of many halophytes show reduced germination on saline growth media (Gul et al., 2013, and refs. therein). However, research into halophyte seed germination has focused mainly on surveying germination rates in a large variety of halophytes. Nothing was understood regarding the physiological and biochemical basis of the salt inhibition of halophyte seed germination, let alone the molecular mechanisms regulating this process.
Freshly harvested E. salsugineum seeds (Hebei ecotype) that have not undergone cold treatment display primary physiological dormancy and require up to several months of after-ripening to achieve dormancy release (Li et al., 2015). However, cold stratification of seeds for a few days leads to rapid germination of the principal research ecotypes (e.g. Shandong; Kazachkova et al., 2016). Similar to other halophytes, E. salsugineum and S. parvula seed germination is reduced by NaCl and inhibited completely above a certain NaCl threshold concentration (Inan et al., 2004; Orsini et al., 2010; Guo et al., 2012; Li et al., 2015; Kazachkova et al., 2016). In contrast, at a NaCl concentration that fully inhibits E. salsugineum and S. parvula seed germination, Arabidopsis seeds are still able to germinate. Kazachkova et al. (2016) performed a detailed examination of the physiological, biochemical, and gene expression changes taking place during salt inhibition of E. salsugineum seed germination and showed that seeds respond to the osmotic component of the salt treatment. The seeds remain viable on the saline medium but seed protein and lipid reserves are not mobilized, and the progression of germination-associated metabolic changes is either reduced or halted between 24 and 48 h after stratification. Furthermore, salt-treated seeds exhibit molecular features associated with dormancy, including a reduced GA3-abscisic acid ratio, induction of stress tolerance-linked processes, and increased expression of dormancy-related genes such as DELAY OF GERMINATION1 (DOG1), compared with non-salt-treated control seeds. However, the observation that salt-inhibited seeds germinate rapidly on salt if the seed coat is removed or if intact seeds are transferred to nonsaline medium suggests that salt does not cause secondary dormancy. Rather, the seeds enter into a dormancy-like state. The physical constraint to seed germination provided by the seed coat is consistent with the higher expression of DOG1 in salt-treated seeds, because DOG1 is involved in repressing the germination-associated weakening of the micropylar endosperm cap (Graeber et al., 2014). Overall, the entry of germinating E. salsugineum seeds into a dormancy-like state in response to the osmotic component of salinity could allow seeds to remain viable in the soil seed bank until rain or melting snow causes a rise in soil water potential and conditions arise that are more favorable to seedling establishment and survival. E. salsugineum now provides an excellent model system to unravel the molecular mechanisms controlling the salt inhibition of halophytic seed germination, and screening of an ethyl methanesulfonate-derived mutant population to identify genes involved in this process is under way (G. Batelli, M. Dassanayake, and S. Barak, unpublished data).
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
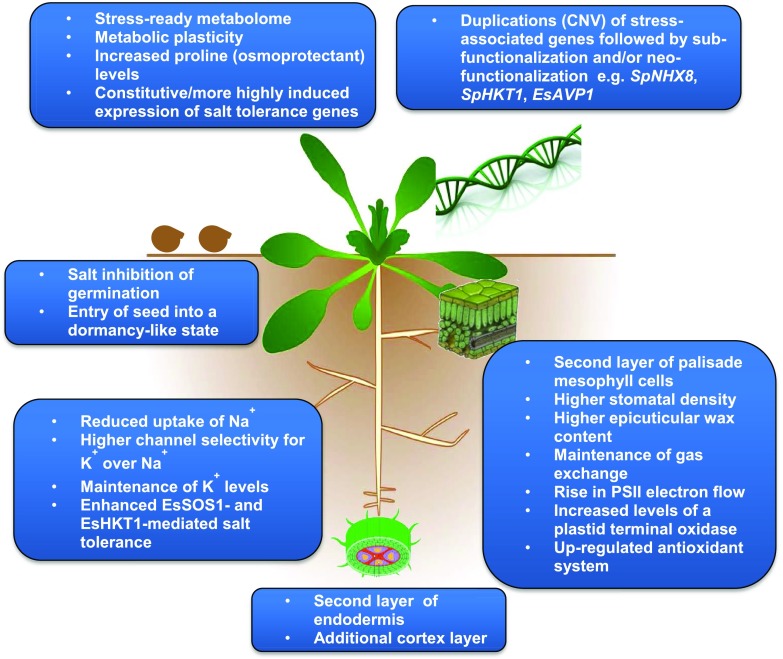
We started this review by asking what has been learned about halophytism by comparing glycophytic Arabidopsis with its halophytic relatives, E. salsugineum, and S. parvula. First, it should be noted that much of the work with these halophytes has confirmed previous findings with other halophytic species. These findings include tighter control of Na+ uptake, compartmentalization of Na+, the importance of plasma membrane and tonoplast Na+ transporters in maintaining ionic balance, synthesis of compatible osmolytes, and activation of ROS signaling and detoxification systems (Fig. 3). On the other hand, salt-sensitive plants also employ similar mechanisms to cope with salt stress, suggesting that differences in salt tolerance between halophytes and salt-sensitive species are related to alterations in the regulation of basic physiological and biochemical processes. This hypothesis has been borne out by the advances made studying E. salsugineum and S. parvula (Fig. 3). (1) The halophytic Arabidopsis relatives appear to exhibit global preadaptation of the transcriptome, proteome, and metabolome to stress and differential expression/accumulation of specific stress tolerance genes, proteins, and metabolites. At the same time, they exhibit phenotypic and metabolic flexibility in the face of changing environmental conditions while maintaining salt tolerance. (2) The maintenance of energy supply under saline conditions is clearly a crucial component of halophytism and is at least partly achieved by a combination of alternative pathways for the flow of excess electrons, such as increased levels of PTOX, and the protection of chloroplasts via an up-regulated plastid antioxidant system. (3) Many of the physiological and molecular salt tolerance mechanisms of E. salsugineum and S. parvula are reflected at the genome level. Here, the power of using Arabidopsis relatives in understanding halophytism, at least in Brassicaceae, really comes into its own. Thus, comparative analysis of E. salsugineum and S. parvula genomes with that of Arabidopsis has shown that tandem duplications, insertions, and translocations in the halophytes have led to the expansion of selected genes and alterations in the expression and/or function of these genes via subfuctionalization and/or neofuctionalization. Many of the altered genes can be identified readily as possessing functions that would be important for adaptation to a stressful environment in general and saline conditions specifically.
Figure 3.
Summary of physiological and molecular adaptations that facilitate a halophytic lifestyle. Only those mechanisms demonstrated so far to exhibit differences in E. salsugineum and S. parvula compared with Arabidopsis are depicted. CNV, copy number variation; EsSOS1, E. salsugineum ortholog of the AtSOS1 Na+/H+ antiporter; EsHKT1, E. salsugineum ortholog of the AtHKT1 Na+/K+ co-transporter. The genes encoding these transporters have undergone subfunctionalization, neofunctionalization, or both.
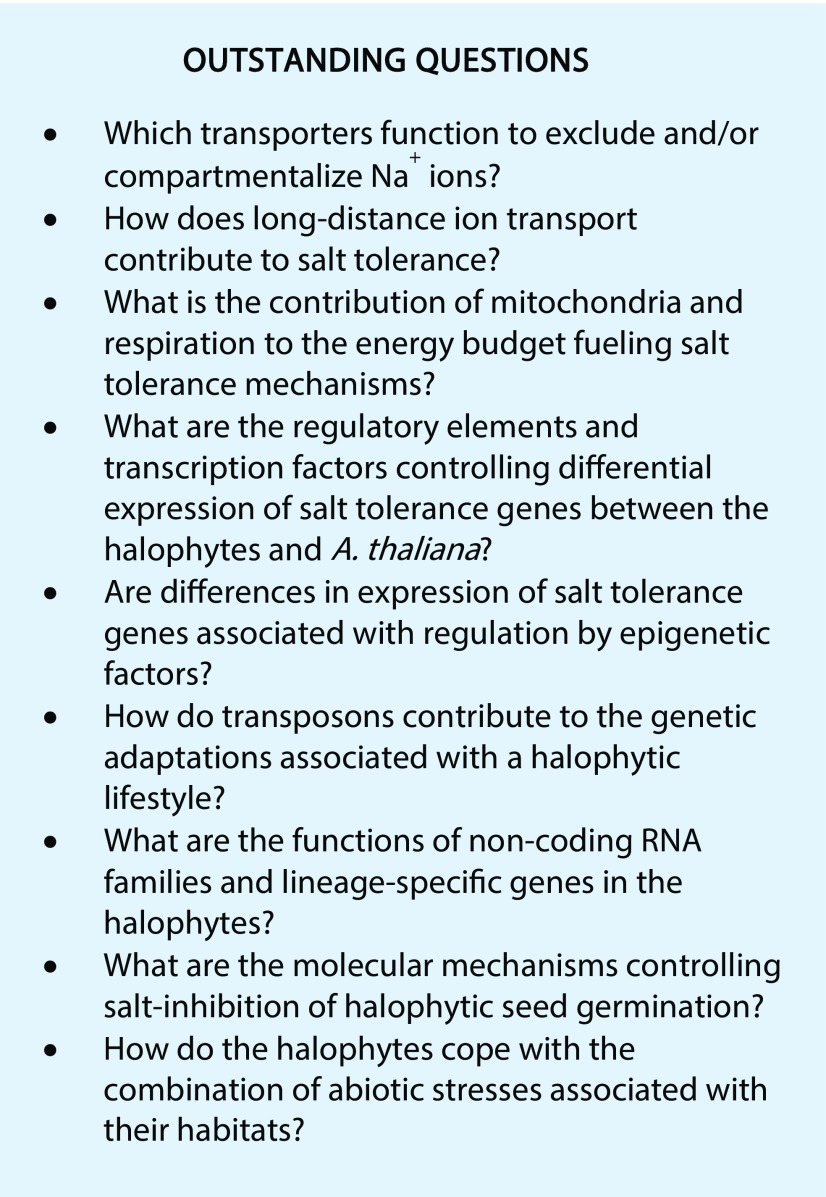
Yet, for all that has been revealed about halophytism by studying E. salsugineum and S. parvula, much regarding salt tolerance that is still unexplained in other halophytes (Cheeseman, 2013; Flowers et al., 2015) remains to be elucidated in the halophytic Arabidopsis relatives (see “Outstanding Questions”). Furthermore, detailed physiological investigations that have been carried out in other halophytes need to be performed with the Arabidopsis relatives. This is particularly true for the more salt-tolerant S. parvula, which lags behind the physiological and molecular studies performed with E. salsugineum. For example, rigorous studies of Na+ and Cl− ion concentrations in various cell compartments are required to understand the cellular basis of salt tolerance in E. salsugineum and S. parvula (Flowers et al., 2015). Short-distance ion transport and the various transporters that perform this function (Cheeseman, 2013) are poorly understood, while long-distance ion transport (via xylem loading and its regulation) that could be essential for osmotic adjustment in the shoots (Shabala, 2013) has not been investigated at all in the Arabidopsis relatives. We have already seen how E. salsugineum is able to maintain photosynthetic energy supply, but the subject of energy budgets to fuel salt tolerance processes (e.g. Na+ vacuolar sequestration, compatible osmolyte production, metabolome and transcriptome reprogramming, etc.) needs to be addressed. Indeed, the contribution of respiration and mitochondria in general has not yet been researched at all in the halophytic Arabidopsis relatives.
One of the general themes from studies with both E. salsugineum and S. parvula is the differential expression of genes between the halophytes and Arabidopsis, whether it is constitutive or more highly salt-induced/-repressed expression of selected genes or at the global transcriptome level. However, the promoter elements, associated transcription factors, and UTR structures involved in this differential halophytic gene expression are unknown, although we have hints of their importance from work on halophytic SOS1 genes and the EsAVP1 gene.
In addition to the acquisition of new regulatory elements, epigenetic factors (DNA methylation and histone modifications) might also be critical for the expression of stress-related genes (Grafi and Barak, 2015; Kim et al., 2015). There are very few reports that have examined epigenetic modifications in extremophytes, but of those that have, some interesting features have emerged. For instance, white mangrove (Laguncularia racemose) located near a salt marsh exhibits global hypomethylation of DNA compared with the same species living next to a freshwater river (Lira-Medeiros et al., 2010). Zygophyllum spp. inhabiting dry and semidry regions lack dimethylation and trimethylation of histone H3 at Lys-9 and Lys-27 (Granot et al., 2009; Granot and Grafi, 2014). Furthermore, the summer dormancy of this plant (leaflets are shed, leaving thick, wax-covered petioles) is associated with the disappearance of permissive H3K4 methylation, leading to a compact chromatin conformation (Khadka et al., 2018). A recent report showed that E. salsugineum has lost gene body DNA methylation (gbM) from its genome and is missing the CHROMOMETHYLASE3 gene that is required for gbM (Bewick et al., 2016). However, no correlation between the loss of gbM and an effect on gene transcription was found. Yet, gbM is only one type of epigenetic feature. Methylation of the promoter regions, histone modifications both globally and locally, and nucleosome occupancy can affect the expression of genes including those involved in stress responses (Grafi and Barak, 2015). The methylation status of transposon remnants also can affect the expression of stress-related genes via transcriptional read-through (Popova et al., 2013). This could be particularly pertinent because of the importance of transposons in generating S. parvula orphan genes (Oh et al., 2012). Thus, epigenetic factors are a topic ripe for investigation in the halophytic Arabidopsis relatives.
Three other features of the halophyte genomes also merit further investigation. (1) Comparison of the E. salsugineum and S. parvula genomes reveals significant differences in repeat-rich transposable elements, where they contribute up to ∼50% of the sequenced genome in E. salsugineum whereas S. parvula has a more compact genome, with less than 10% of the sequenced genome comprising transposable elements (Dassanayake et al., 2011a; Wu et al., 2012). Indeed, several specific transposable element families are underrepresented in the S. parvula genome (Ito et al., 2013; Gao et al., 2016). A preliminary analysis of the two genomes suggested a higher likelihood of differential expression in genes adjacent to transposable elements (Oh et al., 2014). Transposable elements make up a larger fraction of plant genomes than protein-coding genes, thereby providing much of the genomic diversity even in closely related species, with mounting evidence for their role in genome evolution and adaptations (Hollister et al., 2011; Fedoroff, 2012; Oliver et al., 2013; Bennetzen and Wang, 2014; Joly-Lopez et al., 2016, 2017). Yet, the direct influence of transposable elements in contributing to an extremophyte lifestyle is, at present, unexplored. (2) Nearly one-fourth of the genomic sequences under selection in the Arabidopsis genome are made up of noncoding sequences, and a significant fraction of these noncoding sequences are conserved across lineages in Brassicaceae, including in the E. salsugineum and S. parvula genomes (Haudry et al., 2013). Moreover, a large proportion of the conserved noncoding sequences are adjacent to coding regions, yet their functional significance is unknown. In contrast, there is increasing evidence for the functional and evolutionary significance of noncoding RNA families in the E. salsugineum and S. parvula genomes, but this evidence remains in the realm of in silico predictions (Zhang et al., 2013; Rathore et al., 2016; Wu et al., 2016). (3) While there can be many novel or modified functions in the orthologous genes found in the extremophytes that are not yet described in Arabidopsis, there also is a significant fraction of lineage-specific genes with unknown functions in the extremophyte genomes (Dassanayake et al., 2011a; Wu et al., 2012; Yang et al., 2013). Eleven percent of the annotated, nontransposon putative protein-coding genes from S. parvula show no sequence similarity with Arabidopsis genes, while about 7% of these orphan genes show no similarity with any known plant sequence (Dassanayake et al., 2011a; Oh et al., 2012). These genes could represent a pool of unique stress tolerance determinants.
The research areas discussed above are only a few of the many topics that must be addressed both individually and, ultimately, in a holistic manner so that we can understand how the various physiological, biochemical, and molecular mechanisms are integrated to allow halophytes to flourish in saline environments (Cheeseman, 2013). However, at least two obstacles currently prevent greater progress in researching halophytic Arabidopsis relatives. The first problem relates to the variety of nonecologically relevant growth regimes and salt treatments, ranging from hydroponics to soil and from salt shock at extreme salt levels to milder treatments allowing plants to adapt to the applied treatment. E. salsugineum displays very different phenotypic and metabolic characteristics when grown in the laboratory versus plants found in the field, where concentrations of other ions in the soil are of major importance (Guevara et al., 2012; Kazachkova et al., 2013). It is vital, then, that we grow the halophytic Arabidopsis relatives under growth and treatment regimes that reflect at least a modicum of their natural environment. As pointed out by Cheeseman (2013), salt shock treatments should stop. The level of salt given also needs careful consideration, because the E. salsugineum transcriptome/proteome/metabolome respond to concentrations of salt that are considerably higher than those at which Arabidopsis responds. We also must recognize that the halophytes do not experience just salinity in their natural habitats but rather a combination of abiotic stresses. A few groups have begun such combined stress experiments (Khanal et al., 2015; Shamustakimova et al., 2017), but much more needs to be done in this area.
The second obstacle to progress with halophytic Arabidopsis relatives is the poor availability of functional genomics tools. There are no widely available mutant populations for detailed study of individual genes, transgenic lines for tissue/cell specific expression analysis, or the plethora of other tools and databases available for Arabidopsis (Box 1). The published genome sequences of E. salsugineum and S. parvula are a large step in the right direction, but the development of a rich research platform is vital if we are to truly tap the huge potential of the halophytic Arabidopsis relatives.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We express our appreciation to Elizabeth Weretilnyk for providing the photograph by Jeff Dedrick of E. salsugineum in its wild habitat, which is shown in Figure 1. We also thank the reviewers for aiding us in improving the article. Our thanks to Guannan Wang for help with curation of genes mentioned in this article.
Footnotes
This work was supported by the Goldinger Trust Jewish Fund for the Future to S.B., the I-CORE Program of the Planning and Budgeting Committee to S.B., and the National Science Foundation award MCB 1616827 to M.D.
Articles can be viewed without a subscription.
References
- Ali A, Yun DJ (2016) Differential selection of sodium and potassium ions by TsHKT1;2. Plant Signal Behav 11: e1206169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Park HC, Aman R, Ali Z, Yun DJ (2013) Role of HKT1 in Thellungiella salsuginea, a model extremophile plant. Plant Signal Behav 8: e25196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Raddatz N, Aman R, Kim S, Park HC, Jan M, Baek D, Khan IU, Oh DH, Lee SY, et al. (2016) A single amino-acid substitution in the sodium transporter HKT1 associated with plant salt tolerance. Plant Physiol 171: 2112–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A, Khan IU, Jan M, Khan HA, Hussain S, Nisar M, Chung WS, Yun DJ (2018) The high-affinity potassium transporter EpHKT1;2 from the extremophile Eutrema parvula mediates salt tolerance. Front Plant Sci 9: 1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z, Park HC, Ali A, Oh DH, Aman R, Kropornicka A, Hong H, Choi W, Chung WS, Kim WY, et al. (2012) TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiol 158: 1463–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shehbaz IA, Warwick SI (2005) A Synopsis of Eutrema (Brassicaceae). Harv Pap Bot 10: 129–135 [Google Scholar]
- Amtmann A. (2009) Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol Plant 2: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A, Bohnert HJ, Bressan RA (2005) Abiotic stress and plant genome evolution: search for new models. Plant Physiol 138: 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An YQC, Meagher R (2010) Strong expression and conserved regulation of ACT2 in Arabidopsis thaliana and Physcomitrella patens. Plant Mol Biol Rep 28: 481–490 [Google Scholar]
- An JP, Yao JF, Wang XN, You CX, Wang XF, Hao YJ (2017) MdHY5 positively regulates cold tolerance via CBF-dependent and CBF-independent pathways in apple. J Plant Physiol 218: 275–281 [DOI] [PubMed] [Google Scholar]
- An R, Chen QJ, Chai MF, Lu PL, Su Z, Qin ZX, Chen J, Wang XC (2007) AtNHX8, a member of the monovalent cation:proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li/H antiporter. Plant J 49: 718–728 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Arbona V, Argamasilla R, Gómez-Cadenas A (2010) Common and divergent physiological, hormonal and metabolic responses of Arabidopsis thaliana and Thellungiella halophila to water and salt stress. J Plant Physiol 167: 1342–1350 [DOI] [PubMed] [Google Scholar]
- Bartels D, Dinakar C (2013) Balancing salinity stress responses in halophytes and non-halophytes: a comparison between Thellungiella and Arabidopsis thaliana. Funct Plant Biol 40: 819–831 [DOI] [PubMed] [Google Scholar]
- Batelli G, Oh DH, D’Urzo MP, Orsini F, Dassanayake M, Zhu JK, Bohnert HJ, Bressan RA, Maggio A (2014) Using Arabidopsis-related model species (ARMS): growth, genetic transformation, and comparative genomics. Methods Mol Biol 1062: 27–51 [DOI] [PubMed] [Google Scholar]
- Benina M, Obata T, Mehterov N, Ivanov I, Petrov V, Toneva V, Fernie AR, Gechev TS (2013) Comparative metabolic profiling of Haberlea rhodopensis, Thellungiella halophyla, and Arabidopsis thaliana exposed to low temperature. Front Plant Sci 4: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Wang H (2014) The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol 65: 505–530 [DOI] [PubMed] [Google Scholar]
- Bewick AJ, Ji L, Niederhuth CE, Willing EM, Hofmeister BT, Shi X, Wang L, Lu Z, Rohr NA, Hartwig B, et al. (2016) On the origin and evolutionary consequences of gene body DNA methylation. Proc Natl Acad Sci USA 113: 9111–9116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose J, Rodrigo-Moreno A, Shabala S (2014) ROS homeostasis in halophytes in the context of salinity stress tolerance. J Exp Bot 65: 1241–1257 [DOI] [PubMed] [Google Scholar]
- Bressan RA, Zhang C, Zhang H, Hasegawa PM, Bohnert HJ, Zhu JK (2001) Learning from the Arabidopsis experience: the next gene search paradigm. Plant Physiol 127: 1354–1360 [PMC free article] [PubMed] [Google Scholar]
- Bressan RA, Reddy MP, Chung SH, Yun DJ, Hardin LS, Bohnert HJ (2011) Stress-adapted extremophiles provide energy without interference with food production. Food Secur 3: 93–105 [Google Scholar]
- Bressan RA, Park HC, Orsini F, Oh DH, Dassanayake M, Inan G, Yun DJ, Bohnert HJ, Maggio A (2013) Biotechnology for mechanisms that counteract salt stress in extremophile species: a genome-based view. Plant Biotechnol Rep 7: 27–37 [Google Scholar]
- Catalá R, Medina J, Salinas J (2011) Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci USA 108: 16475–16480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champigny MJ, Sung WWL, Catana V, Salwan R, Summers PS, Dudley SA, Provart NJ, Cameron RK, Golding GB, Weretilnyk EA (2013) RNA-Seq effectively monitors gene expression in Eutrema salsugineum plants growing in an extreme natural habitat and in controlled growth cabinet conditions. BMC Genomics 14: 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Guo A, Jin X, Yang Q, Wang D, Sun Y, Huang Q, Wang L, Peng C, Wang X (2015) The beta subunit of glyceraldehyde 3-phosphate dehydrogenase is an important factor for maintaining photosynthesis and plant development under salt stress: based on an integrative analysis of the structural, physiological and proteomic changes in chloroplasts in Thellungiella halophila. Plant Sci 236: 223–238 [DOI] [PubMed] [Google Scholar]
- Chaves MM, Flexas J, Pinheiro C (2009) Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot 103: 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman JM. (2013) The integration of activity in saline environments: problems and perspectives. Funct Plant Biol 40: 759–774 [DOI] [PubMed] [Google Scholar]
- Conti L, Price G, O’Donnell E, Schwessinger B, Dominy P, Sadanandom A (2008) Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20: 2894–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake M, Oh DH, Haas JS, Hernandez A, Hong H, Ali S, Yun DJ, Bressan RA, Zhu JK, Bohnert HJ, et al. (2011a) The genome of the extremophile crucifer Thellungiella parvula. Nat Genet 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake M, Oh DH, Hong H, Bohnert HJ, Cheeseman JM (2011b) Transcription strength and halophytic lifestyle. Trends Plant Sci 16: 1–3 [DOI] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBolt S. (2010) Copy number variation shapes genome diversity in Arabidopsis over immediate family generational scales. Genome Biol Evol 2: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R, Adams KL, Vandepoele K, Van Montagu MCE, Maere S, Van de Peer Y (2013) Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc Natl Acad Sci USA 110: 2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittami SM, Tonon T (2012) Genomes of extremophile crucifers: new platforms for comparative genomics and beyond. Genome Biol 13: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Huang YP, Xi J, Cao MJ, Ni WS, Chen X, Zhu JK, Oliver DJ, Xiang CB (2008) Functional gene-mining for salt-tolerance genes with the power of Arabidopsis. Plant J 56: 653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte JM, Cui L, Wall PK, Zhang Q, Zhang X, Leebens-Mack J, Ma H, Altman N, dePamphilis CW (2006) Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol 23: 469–478 [DOI] [PubMed] [Google Scholar]
- Ellouzi H, Ben Hamed K, Hernández I, Cela J, Müller M, Magné C, Abdelly C, Munné-Bosch S (2014) A comparative study of the early osmotic, ionic, redox and hormonal signaling response in leaves and roots of two halophytes and a glycophyte to salinity. Planta 240: 1299–1317 [DOI] [PubMed] [Google Scholar]
- Eshel G, Shaked R, Kazachkova Y, Khan A, Eppel A, Cisneros A, Acuna T, Gutterman Y, Tel-Zur N, Rachmilevitch S, et al. (2017) Anastatica hierochuntica, an Arabidopsis desert relative, is tolerant to multiple abiotic stresses and exhibits species-specific and common stress tolerance strategies with its halophytic relative, Eutrema (Thellungiella) salsugineum. Front Plant Sci 7: 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrant JM, Lehner A, Cooper K, Wiswedel S (2009) Desiccation tolerance in the vegetative tissues of the fern Mohria caffrorum is seasonally regulated. Plant J 57: 65–79 [DOI] [PubMed] [Google Scholar]
- Fedoroff NV. (2012) Presidential address. Transposable elements, epigenetics, and genome evolution. Science 338: 758–767 [DOI] [PubMed] [Google Scholar]
- Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. New Phytol 179: 945–963 [DOI] [PubMed] [Google Scholar]
- Flowers TJ, Colmer TD (2015) Plant salt tolerance: adaptations in halophytes. Ann Bot 115: 327–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers TJ, Yeo AR (1995) Breeding for salinity resistance in crop plants: where next? Aust J Plant Physiol 22: 875–884 [Google Scholar]
- Flowers TJ, Troke PF, Yeo AR (1977) The mechanism of salt tolerance in halophytes. Annu Rev Plant Physiol 28: 89–121 [Google Scholar]
- Flowers TJ, Hajibagheri MA, Clipson NJW (1986) Halophytes. Q Rev Biol 61: 313–337 [Google Scholar]
- Flowers TJ, Munns R, Colmer TD (2015) Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann Bot 15: 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations (2017) SOFI 2017: The State of Food Security and Nutrition in the World. http://www.fao.org/state-of-food-security-nutrition/en/
- Forment J, Naranjo MA, Roldán M, Serrano R, Vicente O (2002) Expression of Arabidopsis SR-like splicing proteins confers salt tolerance to yeast and transgenic plants. Plant J 30: 511–519 [DOI] [PubMed] [Google Scholar]
- Fukami-Kobayashi K, Nakamura Y, Tamura T, Kobayashi M (2014) SABRE2: a database connecting plant EST/full-length cDNA clones with Arabidopsis information. Plant Cell Physiol 55: e5. [DOI] [PubMed] [Google Scholar]
- Galimba KD, Di Stilio VS (2015) Sub-functionalization to ovule development following duplication of a floral organ identity gene. Dev Biol 405: 158–172 [DOI] [PubMed] [Google Scholar]
- Gao D, Li Y, Kim KD, Abernathy B, Jackson SA (2016) Landscape and evolutionary dynamics of terminal repeat retrotransposons in miniature in plant genomes. Genome Biol 17: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Dinakar C, Benina M, Toneva V, Bartels D (2012) Molecular mechanisms of desiccation tolerance in resurrection plants. Cell Mol Life Sci 69: 3175–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghars MA, Parre E, Debez A, Bordenave M, Richard L, Leport L, Bouchereau A, Savouré A, Abdelly C (2008) Comparative salt tolerance analysis between Arabidopsis thaliana and Thellungiella halophila, with special emphasis on K+/Na+ selectivity and proline accumulation. J Plant Physiol 165: 588–599 [DOI] [PubMed] [Google Scholar]
- Ghars MA, Richard L, Lefebvre-De Vos D, Leprince AS, Parre E, Bordenave M, Abdelly C, Savouré A (2012) Phospholipases C and D modulate proline accumulation in Thellungiella halophila/salsuginea differently according to the severity of salt or hyperosmotic stress. Plant Cell Physiol 53: 183–192 [DOI] [PubMed] [Google Scholar]
- Golding AJ, Johnson GN (2003) Down-regulation of linear and activation of cyclic electron transport during drought. Planta 218: 107–114 [DOI] [PubMed] [Google Scholar]
- Gong Q, Li P, Ma S, Indu Rupassara S, Bohnert HJ (2005) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J 44: 826–839 [DOI] [PubMed] [Google Scholar]
- Graeber K, Linkies A, Steinbrecher T, Mummenhoff K, Tarkowská D, Turečková V, Ignatz M, Sperber K, Voegele A, de Jong H, et al. (2014) DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc Natl Acad Sci USA 111: E3571–E3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafi G, Barak S (2015) Stress induces cell dedifferentiation in plants. Biochim Biophys Acta 1849: 378–384 [DOI] [PubMed] [Google Scholar]
- Granot G, Grafi G (2014) Epigenetic information can reveal phylogenetic relationships within Zygophyllales. Plant Syst Evol 300: 1819–1824 [Google Scholar]
- Granot G, Sikron-Persi N, Gaspan O, Florentin A, Talwara S, Paul LK, Morgenstern Y, Granot Y, Grafi G (2009) Histone modifications associated with drought tolerance in the desert plant Zygophyllum dumosum Boiss. Planta 231: 27–34 [DOI] [PubMed] [Google Scholar]
- Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, et al. (2010) UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res 38: D75–D80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara DR, Champigny MJ, Tattersall A, Dedrick J, Wong CE, Li Y, Labbe A, Ping CL, Wang Y, Nuin P, et al. (2012) Transcriptomic and metabolomic analysis of Yukon Thellungiella plants grown in cabinets and their natural habitat show phenotypic plasticity. BMC Plant Biol 12: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul B, Ansari R, Flowers TJ, Khan MA (2013) Germination strategies of halophyte seeds under salinity. Environ Exp Bot 92: 4–18 [Google Scholar]
- Guo X, Hao G, Ma T (2016) The complete chloroplast genome of salt cress (Eutrema salsugineum). Mitochondrial DNA A DNA Mapp Seq Anal 27: 2862–2863 [DOI] [PubMed] [Google Scholar]
- Guo Y, Jia W, Song J, Wang D, Chen M, Wang B (2012) Thellungilla halophila is more adaptive to salinity than Arabidopsis thaliana at stages of seed germination and seedling establishment. Acta Physiol Plant 34: 1287–1294 [Google Scholar]
- Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, Forczek E, Joly-Lopez Z, Steffen JG, Hazzouri KM, et al. (2013) An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet 45: 891–898 [DOI] [PubMed] [Google Scholar]
- He Q, Hao G, Wang X, Bi H, Li Y, Guo X, Ma T (2016) The complete chloroplast genome of Schrenkiella parvula (Brassicaceae). Mitochondrial DNA A DNA Mapp Seq Anal 27: 3527–3528 [DOI] [PubMed] [Google Scholar]
- Helvaci C, Mordogan H, Colak M, Gundogan I, Colak M (2004) Presence and distribution of lithium in borate deposits and some recent lake waters of west-central Turkey. Int Geol Rev 46: 177–190 [Google Scholar]
- Higashi Y, Ohama N, Ishikawa T, Katori T, Shimura A, Kusakabe K, Yamaguchi-Shinozaki K, Ishida J, Tanaka M, Seki M, et al. (2013) HsfA1d, a protein identified via FOX hunting using Thellungiella salsuginea cDNAs improves heat tolerance by regulating heat-stress-responsive gene expression. Mol Plant 6: 411–422 [DOI] [PubMed] [Google Scholar]
- Hollister JD, Smith LM, Guo YL, Ott F, Weigel D, Gaut BS (2011) Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci USA 108: 2322–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inan G, Zhang Q, Li P, Wang Z, Cao Z, Zhang H, Zhang C, Quist TM, Goodwin SM, Zhu J, et al. (2004) Salt cress: a halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol 135: 1718–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Yoshida T, Tsukahara S, Kawabe A (2013) Evolution of the ONSEN retrotransposon family activated upon heat stress in Brassicaceae. Gene 518: 256–261 [DOI] [PubMed] [Google Scholar]
- Jarvis DE, Ryu CH, Beilstein MA, Schumaker KS (2014) Distinct roles for SOS1 in the convergent evolution of salt tolerance in Eutrema salsugineum and Schrenkiella parvula. Mol Biol Evol 31: 2094–2107 [DOI] [PubMed] [Google Scholar]
- Jiang X, Leidi EO, Pardo JM (2010) How do vacuolar NHX exchangers function in plant salt tolerance? Plant Signal Behav 5: 792–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly-Lopez Z, Hoen DR, Blanchette M, Bureau TE (2016) Phylogenetic and genomic analyses resolve the origin of important plant genes derived from transposable elements. Mol Biol Evol 33: 1937–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly-Lopez Z, Forczek E, Vello E, Hoen DR, Tomita A, Bureau TE (2017) Abiotic stress phenotypes are associated with conserved genes derived from transposable elements. Front Plant Sci 8: 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Kant P, Raveh E, Barak S (2006) Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ 29: 1220–1234 [DOI] [PubMed] [Google Scholar]
- Kant S, Bi YM, Weretilnyk E, Barak S, Rothstein SJ (2008) The Arabidopsis halophytic relative Thellungiella halophila tolerates nitrogen-limiting conditions by maintaining growth, nitrogen uptake, and assimilation. Plant Physiol 147: 1168–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye Y, Golani Y, Singer Y, Leshem Y, Cohen G, Ercetin M, Gillaspy G, Levine A (2011) Inositol polyphosphate 5-phosphatase7 regulates the production of reactive oxygen species and salt tolerance in Arabidopsis. Plant Physiol 157: 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazachkova Y, Batushansky A, Cisneros A, Tel-Zur N, Fait A, Barak S (2013) Growth platform-dependent and -independent phenotypic and metabolic responses of Arabidopsis and its halophytic relative, Eutrema salsugineum, to salt stress. Plant Physiol 162: 1583–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazachkova Y, Khan A, Acuña T, López-Díaz I, Carrera E, Khozin-Goldberg I, Fait A, Barak S (2016) Salt induces features of a dormancy-like state in seeds of Eutrema (Thellungiella) salsugineum, a halophytic relative of Arabidopsis. Front Plant Sci 7: 1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadka J, Yadav NS, Granot G, Grafi G (2018) Seasonal growth of Zygophyllum dumosum Boiss.: summer dormancy is associated with loss of the permissive epigenetic marker dimethyl H3K4 and extensive reduction in proteins involved in basic cell functions. Plants (Basel) 7: E59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanal N, Moffatt BA, Gray GR (2015) Acquisition of freezing tolerance in Arabidopsis and two contrasting ecotypes of the extremophile Eutrema salsugineum (Thellungiella salsuginea). J Plant Physiol 180: 35–44 [DOI] [PubMed] [Google Scholar]
- Kim JM, Sasaki T, Ueda M, Sako K, Seki M (2015) Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front Plant Sci 6: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, German DA (2013) Taxonomy and systematics are key to biological information: Arabidopsis, Eutrema (Thellungiella), Noccaea and Schrenkiella (Brassicaceae) as examples. Front Plant Sci 4: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywińska E, Bucholc M, Kulik A, Ciesielski A, Lichocka M, Dębski J, Ludwików A, Dadlez M, Rodriguez PL, Dobrowolska G (2016) Phosphatase ABI1 and okadaic acid-sensitive phosphoprotein phosphatases inhibit salt stress-activated SnRK2.4 kinase. BMC Plant Biol 16: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamdan NL, Attia Z, Moran N, Moshelion M (2012) The Arabidopsis-related halophyte Thellungiella halophila: boron tolerance via boron complexation with metabolites? Plant Cell Environ 35: 735–746 [DOI] [PubMed] [Google Scholar]
- Lee YP, Babakov A, de Boer B, Zuther E, Hincha DK (2012) Comparison of freezing tolerance, compatible solutes and polyamines in geographically diverse collections of Thellungiella sp. and Arabidopsis thaliana accessions. BMC Plant Biol 12: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YP, Giorgi FM, Lohse M, Kvederaviciute K, Klages S, Usadel B, Meskiene I, Reinhardt R, Hincha DK (2013) Transcriptome sequencing and microarray design for functional genomics in the extremophile Arabidopsis relative Thellungiella salsuginea (Eutrema salsugineum). BMC Genomics 14: 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidi EO, Barragán V, Rubio L, El-Hamdaoui A, Ruiz MT, Cubero B, Fernández JA, Bressan RA, Hasegawa PM, Quintero FJ, et al. (2010) The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J 61: 495–506 [DOI] [PubMed] [Google Scholar]
- Li W, Khan MA, Yamaguchi S, Liu X (2015) Hormonal and environmental regulation of seed germination in salt cress (Thellungiella halophila). Plant Growth Regul 76: 41–49 [Google Scholar]
- Lira-Medeiros CF, Parisod C, Fernandes RA, Mata CS, Cardoso MA, Ferreira PC (2010) Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS ONE 5: e10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugan R, Niogret MF, Leport L, Guégan JP, Larher FR, Savouré A, Kopka J, Bouchereau A (2010) Metabolome and water homeostasis analysis of Thellungiella salsuginea suggests that dehydration tolerance is a key response to osmotic stress in this halophyte. Plant J 64: 215–229 [DOI] [PubMed] [Google Scholar]
- M’rah S, Ouerghi Z, Eymery F, Rey P, Hajji M, Grignon C, Lachaâl M (2007) Efficiency of biochemical protection against toxic effects of accumulated salt differentiates Thellungiella halophila from Arabidopsis thaliana. J Plant Physiol 164: 375–384 [DOI] [PubMed] [Google Scholar]
- MacLeod MJR, Dedrick J, Ashton C, Sung WWL, Champigny MJ, Weretilnyk EA (2015) Exposure of two Eutrema salsugineum (Thellungiella salsuginea) accessions to water deficits reveals different coping strategies in response to drought. Physiol Plant 155: 267–280 [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Nilhan TG, Emre YA, Osman K (2008) Soil determinants for distribution of Halocnemum strobilaceum Bieb. (Chenopodiaceae) around Lake Tuz, Turkey. Pak J Biol Sci 11: 565–570 [DOI] [PubMed] [Google Scholar]
- Oh DH, Dassanayake M (2018) Landscape of gene transposition-duplication within the Brassicaceae family. DNA Res 10.1093/dnares/dsy035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Gong Q, Ulanov A, Zhang Q, Li Y, Ma W, Yun DJ, Bressan RA, Bohnert HJ (2007) Sodium stress in the halophyte Thellungiella halophila and transcriptional changes in a thsos1-RNA interference line. J Integr Plant Biol 49: 1484–1496 [Google Scholar]
- Oh DH, Leidi E, Zhang Q, Hwang SM, Li Y, Quintero FJ, Jiang X, D’Urzo MP, Lee SY, Zhao Y, et al. (2009a) Loss of halophytism by interference with SOS1 expression. Plant Physiol 151: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Zahir A, Yun DJ, Bressan RA, Bohnert HJ (2009b) SOS1 and halophytism. Plant Signal Behav 4: 1081–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Dassanayake M, Haas JS, Kropornika A, Wright C, d’Urzo MP, Hong H, Ali S, Hernandez A, Lambert GM, et al. (2010) Genome structures and halophyte-specific gene expression of the extremophile Thellungiella parvula in comparison with Thellungiella salsuginea (Thellungiella halophila) and Arabidopsis. Plant Physiol 154: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Dassanayake M, Bohnert HJ, Cheeseman JM (2012) Life at the extreme: lessons from the genome. Genome Biol 13: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DH, Hong H, Lee SY, Yun DJ, Bohnert HJ, Dassanayake M (2014) Genome structures and transcriptomes signify niche adaptation for the multiple-ion-tolerant extremophyte Schrenkiella parvula. Plant Physiol 164: 2123–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. (1970) Evolution by Gene Duplication. Springer-Verlag, New York [Google Scholar]
- Oliver KR, McComb JA, Greene WK (2013) Transposable elements: powerful contributors to angiosperm evolution and diversity. Genome Biol Evol 5: 1886–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsini F, D’Urzo MP, Inan G, Serra S, Oh DH, Mickelbart MV, Consiglio F, Li X, Jeong JC, Yun DJ, et al. (2010) A comparative study of salt tolerance parameters in 11 wild relatives of Arabidopsis thaliana. J Exp Bot 61: 3787–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozfidan-Konakci C, Uzilday B, Ozgur R, Yildiztugay E, Sekmen AH, Turkan I (2016) Halophytes as a source of salt tolerance genes and mechanisms: a case study for the Salt Lake area, Turkey. Funct Plant Biol 43: 575–589 [DOI] [PubMed] [Google Scholar]
- Pang Q, Chen S, Dai S, Chen Y, Wang Y, Yan X (2010) Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J Proteome Res 9: 2584–2599 [DOI] [PubMed] [Google Scholar]
- Pedras MS, Zheng QA (2010) Metabolic responses of Thellungiella halophila/salsuginea to biotic and abiotic stresses: metabolite profiles and quantitative analyses. Phytochemistry 71: 581–589 [DOI] [PubMed] [Google Scholar]
- Peters S, Mundree SG, Thomson JA, Farrant JM, Keller F (2007) Protection mechanisms in the resurrection plant Xerophyta viscosa (Baker): both sucrose and raffinose family oligosaccharides (RFOs) accumulate in leaves in response to water deficit. J Exp Bot 58: 1947–1956 [DOI] [PubMed] [Google Scholar]
- Pilarska M, Wiciarz M, Jajić I, Kozieradzka-Kiszkurno M, Dobrev P, Vanková R, Niewiadomska E (2016) A different pattern of production and scavenging of reactive oxygen species in halophytic Eutrema salsugineum (Thellungiella salsuginea) plants in comparison to Arabidopsis thaliana and its relation to salt stress signaling. Front Plant Sci 7: 1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polle A, Chen S (2015) On the salty side of life: molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant Cell Environ 38: 1794–1816 [DOI] [PubMed] [Google Scholar]
- Popova OV, Dinh HQ, Aufsatz W, Jonak C (2013) The RdDM pathway is required for basal heat tolerance in Arabidopsis. Mol Plant 6: 396–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadir M, Quillerou E, Nangia V, Murtaza G, Singh M, Thomas RJ, Drechsel P, Noble AD (2014) Economics of salt-induced land degradation and restoration. Nat Resour Forum 38: 282–295 [Google Scholar]
- Rathore P, Geeta R, Das S (2016) Microsynteny and phylogenetic analysis of tandemly organised miRNA families across five members of Brassicaceae reveals complex retention and loss history. Plant Sci 247: 35–48 [DOI] [PubMed] [Google Scholar]
- Rengasamy P. (2006) World salinization with emphasis on Australia. J Exp Bot 57: 1017–1023 [DOI] [PubMed] [Google Scholar]
- Riboni M, Galbiati M, Tonelli C, Conti L (2013) GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol 162: 1706–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R (2004) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279: 11736–11743 [DOI] [PubMed] [Google Scholar]
- Sanchez DH, Pieckenstain FL, Escaray F, Erban A, Kraemer U, Udvardi MK, Kopka J (2011) Comparative ionomics and metabolomics in extremophile and glycophytic Lotus species under salt stress challenge the metabolic pre-adaptation hypothesis. Plant Cell Environ 34: 605–617 [DOI] [PubMed] [Google Scholar]
- Shabala S. (2013) Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Ann Bot 112: 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamustakimova AO, Leonova ТG, Taranov VV, de Boer AH, Babakov AV (2017) Cold stress increases salt tolerance of the extremophytes Eutrema salsugineum (Thellungiella salsuginea) and Eutrema (Thellungiella) botschantzevii. J Plant Physiol 208: 128–138 [DOI] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Inzé D (2010) More from less: plant growth under limited water. Curr Opin Biotechnol 21: 197–203 [DOI] [PubMed] [Google Scholar]
- Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R, Andriankaja M, Van Aken O, Van Breusegem F, Fernie AR, et al. (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepien P, Johnson GN (2009) Contrasting responses of photosynthesis to salt stress in the glycophyte Arabidopsis and the halophyte Thellungiella: role of the plastid terminal oxidase as an alternative electron sink. Plant Physiol 149: 1154–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Gao F, Zhao L, Li K, Zhang J (2010) Identification of a new 130 bp cis-acting element in the TsVP1 promoter involved in the salt stress response from Thellungiella halophila. BMC Plant Biol 10: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu JK, Shinozaki K (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135: 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Sakurai T, Mochida K, Ishiwata A, Kurotani A, Totoki Y, Toyoda A, Sakaki Y, Seki M, Ono H, et al. (2008) Large-scale collection and annotation of full-length enriched cDNAs from a model halophyte, Thellungiella halophila. BMC Plant Biol 8: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Komatsu K, Katori T, Kawasaki Y, Sakata Y, Tanaka S, Kobayashi M, Toyoda A, Seki M, Shinozaki K (2010) Comparative genomic analysis of 1047 completely sequenced cDNAs from an Arabidopsis-related model halophyte, Thellungiella halophila. BMC Plant Biol 10: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teusink RS, Rahman M, Bressan RA, Jenks MA (2002) Cuticular waxes on Arabidopsis thaliana close relatives Thellungiella halophila and Thellungiella parvula. Int J Plant Sci 163: 309–315 [Google Scholar]
- United Nations Framework Convention on Climate Change (2016) Views on Issues Relating to Agriculture. http://unfccc.int/resource/docs/2016/sbsta/eng/misc01.pdf
- Uzilday B, Ozgur R, Sekmen AH, Yildiztugay E, Turkan I (2015) Changes in the alternative electron sinks and antioxidant defence in chloroplasts of the extreme halophyte Eutrema parvulum (Thellungiella parvula) under salinity. Ann Bot 115: 449–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco VME, Mansbridge J, Bremner S, Carruthers K, Summers PS, Sung WWL, Champigny MJ, Weretilnyk EA (2016) Acclimation of the crucifer Eutrema salsugineum to phosphate limitation is associated with constitutively high expression of phosphate-starvation genes. Plant Cell Environ 39: 1818–1834 [DOI] [PubMed] [Google Scholar]
- Ventura Y, Eshel A, Pasternak D, Sagi M (2015) The development of halophyte-based agriculture: past and present. Ann Bot 115: 529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Estrella R, Barkla BJ, García-Ramírez L, Pantoja O (2005) Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiol 139: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Estrella R, Barkla BJ, Pantoja O (2014) Comparative 2D-DIGE analysis of salinity responsive microsomal proteins from leaves of salt-sensitive Arabidopsis thaliana and salt-tolerant Thellungiella salsuginea. J Proteomics 111: 113–127 [DOI] [PubMed] [Google Scholar]
- Volkov V, Amtmann A (2006) Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, has specific root ion-channel features supporting K+/Na+ homeostasis under salinity stress. Plant J 48: 342–353 [DOI] [PubMed] [Google Scholar]
- Wang B, Davenport RJ, Volkov V, Amtmann A (2006) Low unidirectional sodium influx into root cells restricts net sodium accumulation in Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana. J Exp Bot 57: 1161–1170 [DOI] [PubMed] [Google Scholar]
- Wang G, Pantha P, Tran KN, Oh DH, Dassanayake M (2018a) Plant growth and Agrobacterium-mediated floral-dip transformation of the extremophyte, Schrenkiella parvula. J Vis Exp (in press) [DOI] [PubMed] [Google Scholar]
- Wang W, Wu Y, Li Y, Xie J, Zhang Z, Deng Z, Zhang Y, Yang C, Lai J, Zhang H, et al. (2010) A large insert Thellungiella halophila BIBAC library for genomics and identification of stress tolerance genes. Plant Mol Biol 72: 91–99 [DOI] [PubMed] [Google Scholar]
- Wang X, Chang L, Wang B, Wang D, Li P, Wang L, Yi X, Huang Q, Peng M, Guo A (2013) Comparative proteomics of Thellungiella halophila leaves from plants subjected to salinity reveals the importance of chloroplastic starch and soluble sugars in halophyte salt tolerance. Mol Cell Proteomics 12: 2174–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bi C, Xu Y, Wei S, Dai X, Yin T, Ye N (2016) The whole genome assembly and comparative genomic research of Thellungiella parvula (extremophile crucifer) mitochondrion. Int J Genomics 2016: 5283628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Hu QJ, Guo XY, Wang K, Ru DF, German DA, Weretilnyk EA, Abbott RJ, Lascoux M, Liu JQ (2018b) Demographic expansion and genetic load of the halophyte model plant Eutrema salsugineum. Mol Ecol 27: 2943–2955 [DOI] [PubMed] [Google Scholar]
- Wang ZL, Li PH, Fredricksen M, Gong Z, Kim CS, Zhang C, Bohnert HJ, Zhu JK, Bressan RA, Hasegawa PM, et al. (2004) Expressed sequence tags from Thellungiella halophila, a new model to study plant salt-tolerance. Plant Sci 166: 609–616 [Google Scholar]
- Wever W, McCallum EJ, Chakravorty D, Cazzonelli CI, Botella JR (2010) The 5′ untranslated region of the VR-ACS1 mRNA acts as a strong translational enhancer in plants. Transgenic Res 19: 667–674 [DOI] [PubMed] [Google Scholar]
- Wiciarz M, Gubernator B, Kruk J, Niewiadomska E (2015) Enhanced chloroplastic generation of H2O2 in stress-resistant Thellungiella salsuginea in comparison to Arabidopsis thaliana. Physiol Plant 153: 467–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiciarz M, Niewiadomska E, Kruk J (2018) Effects of salt stress on low molecular antioxidants and redox state of plastoquinone and P700 in Arabidopsis thaliana (glycophyte) and Eutrema salsugineum (halophyte). Photosynthetica 56: 811–819 [Google Scholar]
- Wong CE, Li Y, Whitty BR, Díaz-Camino C, Akhter SR, Brandle JE, Golding GB, Weretilnyk EA, Moffatt BA, Griffith M (2005) Expressed sequence tags from the Yukon ecotype of Thellungiella reveal that gene expression in response to cold, drought and salinity shows little overlap. Plant Mol Biol 58: 561–574 [DOI] [PubMed] [Google Scholar]
- Wong CE, Li Y, Labbe A, Guevara D, Nuin P, Whitty B, Diaz C, Golding GB, Gray GR, Weretilnyk EA, et al. (2006) Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol 140: 1437–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Gao X, Kong X, Zhao Y, Zhang H (2009) Molecular cloning and functional analysis of a Na+/H+ antiporter gene ThNHX1 from a halophytic plant Thellungiella halophila. Plant Mol Biol Rep 27: 1–12 [Google Scholar]
- Wu HJ, Zhang Z, Wang JY, Oh DH, Dassanayake M, Liu B, Huang Q, Sun HX, Xia R, Wu Y, et al. (2012) Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc Natl Acad Sci USA 109: 12219–12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Guo J, Cai Y, Gong X, Xiong X, Qi W, Pang Q, Wang X, Wang Y (2016) Genome-wide identification and characterization of Eutrema salsugineum microRNAs for salt tolerance. Physiol Plant 157: 453–468 [DOI] [PubMed] [Google Scholar]
- Xu X, Feng J, Lü S, Lohrey GT, An H, Zhou Y, Jenks MA (2014) Leaf cuticular lipids on the Shandong and Yukon ecotypes of saltwater cress, Eutrema salsugineum, and their response to water deficiency and impact on cuticle permeability. Physiol Plant 151: 446–458 [DOI] [PubMed] [Google Scholar]
- Yang CY, Hsu FC, Li JP, Wang NN, Shih MC (2011) The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol 156: 202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Jarvis DE, Chen H, Beilstein MA, Grimwood J, Jenkins J, Shu S, Prochnik S, Xin M, Ma C, et al. (2013) The reference genome of the halophytic plant Eutrema salsugineum. Front Plant Sci 4: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Gosney MJ, Dilkes BP, Mikelbart MV (2018) Dark period transcriptomic and metabolic profiling of two diverse Eutrema salsugineum accessions. Plant Direct 2: e00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yobi A, Wone BWM, Xu W, Alexander DC, Guo L, Ryals JA, Oliver MJ, Cushman JC (2012) Comparative metabolic profiling between desiccation-sensitive and desiccation-tolerant species of Selaginella reveals insights into the resurrection trait. Plant J 72: 983–999 [DOI] [PubMed] [Google Scholar]
- Yu B, Li W (2014) Comparative profiling of membrane lipids during water stress in Thellungiella salsuginea and its relative Arabidopsis thaliana. Phytochemistry 108: 77–86 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhao C, Li M, Sun W, Liu Y, Xia H, Sun M, Li A, Li C, Zhao S, et al. (2013) Genome-wide identification of Thellungiella salsuginea microRNAs with putative roles in the salt stress response. BMC Plant Biol 13: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lai J, Sun S, Li Y, Liu Y, Liang L, Chen M, Xie Q (2008) Comparison analysis of transcripts from the halophyte Thellungiella halophila. J Integr Plant Biol 50: 1327–1335 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2001) Plant salt tolerance. Trends Plant Sci 6: 66–71 [DOI] [PubMed] [Google Scholar]
- Zhu JK, Whited J, Seluanov A, Gorbunova V, Kasahara M, Amdam GV, Ulanovsky N, Feng G, Brunet A, Margoliash D (2015) The next top models. Cell 163: 18–20 [Google Scholar]