Mutations in two glycosyltransferase-encoding genes severely impair the elongation of pectic rhamnogalacturonan I, resulting in hydrophobic seeds that do not release mucilage polymers.
Abstract
Pectin is a vital component of the plant cell wall and provides the molecular glue that maintains cell-cell adhesion, among other functions. As the most complex wall polysaccharide, pectin is composed of several covalently linked domains, such as homogalacturonan (HG) and rhamnogalacturonan I (RG I). Pectin has widespread uses in the food industry and has emerging biomedical applications, but its synthesis remains poorly understood. For instance, the enzymes that catalyze RG I elongation remain unknown. Recently, a coexpression- and sequence-based MUCILAGE-RELATED (MUCI) reverse genetic screen uncovered hemicellulose biosynthetic enzymes in the Arabidopsis (Arabidopsis thaliana) seed coat. Here, we use an extension of this strategy to identify MUCI70 as the founding member of a glycosyltransferase family essential for the accumulation of seed mucilage, a gelatinous wall rich in unbranched RG I. Detailed biochemical and histological characterization of two muci70 mutants and two galacturonosyltransferase11 (gaut11) mutants identified MUCI70 and GAUT11 as required for two distinct RG I domains in seed mucilage. We demonstrate that, unlike MUCI70, GAUT11 catalyzes HG elongation in vitro and, thus, likely is required for the synthesis of an HG region important for RG I elongation. Analysis of a muci70 gaut11 double mutant confirmed that MUCI70 and GAUT11 are indispensable for the production and release of the bulk of mucilage RG I and for shaping the surface morphology of seeds. In addition, we uncover relationships between pectin and hemicelluloses and show that xylan is essential for the elongation of at least one RG I domain.
Plant cell walls are largely composed of three major classes of polysaccharides: cellulose, hemicellulose, and pectin. While cellulose and hemicelluloses are largely built of neutral sugars connected by β-1,4 linkages, pectin is defined by its high content of GalA residues connected by α-1,4 linkages. Cellulose-hemicellulose networks have been thought to provide the tensile strength of the wall, while pectin was implicated mainly in cell-cell adhesion and determining the porosity of the wall (Cosgrove, 2016). However, recent evidence indicates that pectin-cellulose junctions are more prevalent than expected previously and, thus, that pectin may play additional structural roles (Wang et al., 2015). Furthermore, since mutations in several pectin-related genes are lethal, it is evident that this matrix polysaccharide has vital functions in plants (Caffall et al., 2009). Pectin also has widespread uses in the food industry and has emerging applications in the biomedical field, including use as a gelling agent for targeted drug delivery and as a bioactive molecule for cancer treatment (Maxwell et al., 2012; Munarin et al., 2012).
Pectin is the most complex polysaccharide in the plant cell wall, consisting of multiple glycan domains that may exist in one or more polymers linked via their backbones (Nakamura et al., 2002; Atmodjo et al., 2013). The backbone of the most abundant extractable pectin consists exclusively of d-GalA subunits and can be unbranched (homogalacturonan [HG]), substituted with d-Xyl residues (xylogalacturonan), or decorated with a conserved set of side chains (rhamnogalacturonan II [RG II]). In contrast, the backbone of RG I consists of a repeating α-d-1,4-GalA-α-l-1,2-Rha disaccharide. The rhamnose (Rha) residues in the RG I backbone frequently can be substituted with a wide variety of oligosaccharide or polysaccharide side chains. Around 40 different RG I side chain structures have been reported so far (Atmodjo et al., 2013), including linear β-1,4-linked d-galactan and α-1,5-linked l-arabinan or arabinogalactans containing both Gal and arabinose (Ara) units. Despite the biochemical evidence that HG and RG I are covalently linked in soybean (Glycine max; Nakamura et al., 2002), the full in vivo structure of the pectin macromolecules has yet to be determined due to the difficulty of extracting them in an intact form (Atmodjo et al., 2013). In addition, a complex proteoglycan purified from Arabidopsis (Arabidopsis thaliana) suspension cultures has been shown to contain covalently linked HG and RG I domains, which are further branched with the hemicellulose xylan (Tan et al., 2013). This finding suggests that certain pectin domains such as RG I may have a more central role in cell wall organization than thought previously.
Based on the large number of pectin structures that have been detected in plants, their biosynthesis is hypothesized to require at least 67 distinct enzymes that transfer glycosyl, methyl, or acetyl groups (Atmodjo et al., 2013). However, only four types of pectin biosynthetic enzymes have been identified and biochemically characterized so far. These include glycosyltransferase (GT) proteins that belong to four different Carbohydrate-Active Enzyme (CAZy; http://www.cazy.org/; Lombard et al., 2014) families: GT8, GT47-C, GT77, and GT92. Two GT8 proteins, GALACTURONOSYLTRANSFERASE1 (GAUT1) and GAUT7, form the core of a GAUT1:GAUT7 complex that catalyzes the elongation of the HG backbone (Sterling et al., 2006; Atmodjo et al., 2011). Additional GAUT and GAUT-LIKE (GATL) proteins from the GT8 family encode proven and putative HG galacturonosyltransferases (α-GalA transferases). For example, GAUT4 was shown recently to be an HG α-GalA transferase whose down-regulation results in reduced HG and RG II production (Biswal et al., 2018). Although GAUT1 and GAUT7 are predicted to have similar protein topologies, they have surprisingly distinct functions. In vivo, the GAUT1 enzyme is cleaved into a soluble form that is retained at the site of pectin synthesis via interactions with GAUT7, a Golgi membrane-bound protein anchor with no demonstrated catalytic activity (Atmodjo et al., 2011). Unlike GAUT4 and the GAUT1:GAUT7 complex, which synthesize the HG backbone, the other GTs known to be involved in pectin synthesis catalyze the synthesis of three distinct pectin side chains: the β-1,3-xylosyl branches of xylogalacturonan (GT47-C; Jensen et al., 2008), the α-1,3-xylosyl residues in RG II (GT77; Egelund et al., 2006), and the β-1,4-galactan side chains of RG I (GT92; Liwanag et al., 2012). Overall, these GT activities account for only a small fraction of the pectin structures found in nature. In addition, there is increasing evidence that seemingly distinct wall polymers, such as pectin and the hemicellulose xylan, are structurally dependent on one another (Hao and Mohnen, 2014). For example, the loss of GAUT12 (a GT8 protein) in the irregular xylem8 (irx8) mutant leads to dwarf plants that have significant reductions in both xylan and HG (Peña et al., 2007; Persson et al., 2007). Therefore, the production of pectin remains poorly understood on a mechanistic level, and most of the molecular players involved in this process remain unknown.
Although coexpression analysis has been a successful approach to identify GTs involved in cellulose and hemicellulose biosynthesis (Brown et al., 2005; Persson et al., 2005), it failed previously to predict obvious candidates for pectin production. Two potential challenges are that pectin biosynthetic enzymes may lack distinctive expression profiles in most plant tissues and that the relevant GTs are not part of classified CAZy families (Harholt et al., 2010). These obstacles were surpassed by the identification of novel GT-like plant proteins through Golgi proteomic studies (Nikolovski et al., 2012, 2014) and the establishment of Arabidopsis seed mucilage as a model for dissecting pectin synthesis (Haughn and Western, 2012). Within a narrow developmental window, Arabidopsis seed coat epidermal (SCE) cells produce copious amounts of RG I, along with minor amounts of cellulose, hemicellulose, arabinogalactans, and HG (Voiniciuc et al., 2015c). Since at least 90% of the mucilage extracted from Arabidopsis seeds consists of Rha and GalA units derived from pectin, the SCE cells can be exploited to identify pectin-related GTs. In addition, structural changes in seed mucilage polysaccharides can be monitored conveniently in situ with a variety of imaging techniques and specific probes (Voiniciuc et al., 2018).
Despite the great potential of this model system, only two GTs have been implicated so far in the synthesis of the pectin domains in mucilage. A screen of 26 gaut mutant lines for altered staining of seed mucilage found only one mutant (gaut11-2) that showed smaller mucilage capsules and reduced uronic acid content compared with the wild type (Caffall et al., 2009). Although the results indicated that GAUT11 might affect HG biosynthesis in SCE cells, the gaut11-2 phenotype was not supported by an independent knockdown gaut11-1 allele (Caffall et al., 2009). GATL5, another protein from the GT8 family, is the only other pectin-related GT that has been implicated in mucilage biosynthesis. A knockout T-DNA insertion in GATL5 increased the Mr of mucilage polysaccharides without dramatically altering the glycosidic linkage composition or the content of pectin epitopes bound by antibodies (Kong et al., 2013). Since GATL5 was proposed to simply regulate the final size of pectin polymers in mucilage, additional players must be required for the elongation of RG I in Arabidopsis SCE cells.
Recently, a coexpression- and sequence-based MUCILAGE-RELATED (MUCI) reverse genetic screen identified three GTs required for the synthesis of two distinct hemicellulosic polymers (xylan and galactoglucomannan) in Arabidopsis SCE cells (Voiniciuc et al., 2015a, 2015b). Using an extension of this strategy, we now report that the biosynthesis of pectin requires MUCI70, a putative GT from an unclassified CAZy family that was not known to affect cell wall structure. Through a detailed biochemical and histological characterization of muci70 mutants and two novel gaut11 alleles, we show that these two genes are required for the production of two distinct RG I domains essential for seed mucilage architecture. Finally, the analysis of a muci70 gaut11 double mutant and the demonstration that GAUT11 is an HG α-GalA transferase confirms that MUCI70 and GAUT11 are indispensable for the production of two RG I domains that represent the bulk of seed mucilage and shape the surface morphology of seeds.
RESULTS
MUCI70 Is a Novel Pectin-Related GT Localized in the Golgi Apparatus
To identify novel players involved in pectin production, we systematically profiled the expression of all 1,128 Arabidopsis members of the CAZy database (Lombard et al., 2014) in the seed coat using ATH1 microarray data in Genevestigator (Hruz et al., 2008). This strategy revealed more than 50 CAZy genes that are transcribed in the seed coat when mucilage is produced. The majority of these genes were not identified in the initial MUCI screen (Voiniciuc et al., 2015b) because they are not coexpressed significantly with known mucilage genes in GeneCAT (Mutwil et al., 2008), GeneMANIA (Warde-Farley et al., 2010), and ATTED-II (Obayashi et al., 2014). Among this collection of genes were MUCI64/IRX14 (Voiniciuc et al., 2015a) and four members of the GAUT family (Supplemental Fig. S1), including GAUT11 and GATL5. Interestingly, we also found one gene encoding a putative GT (At1g28240), which we named MUCI70, as a promising candidate for pectin production in the Arabidopsis seed coat.
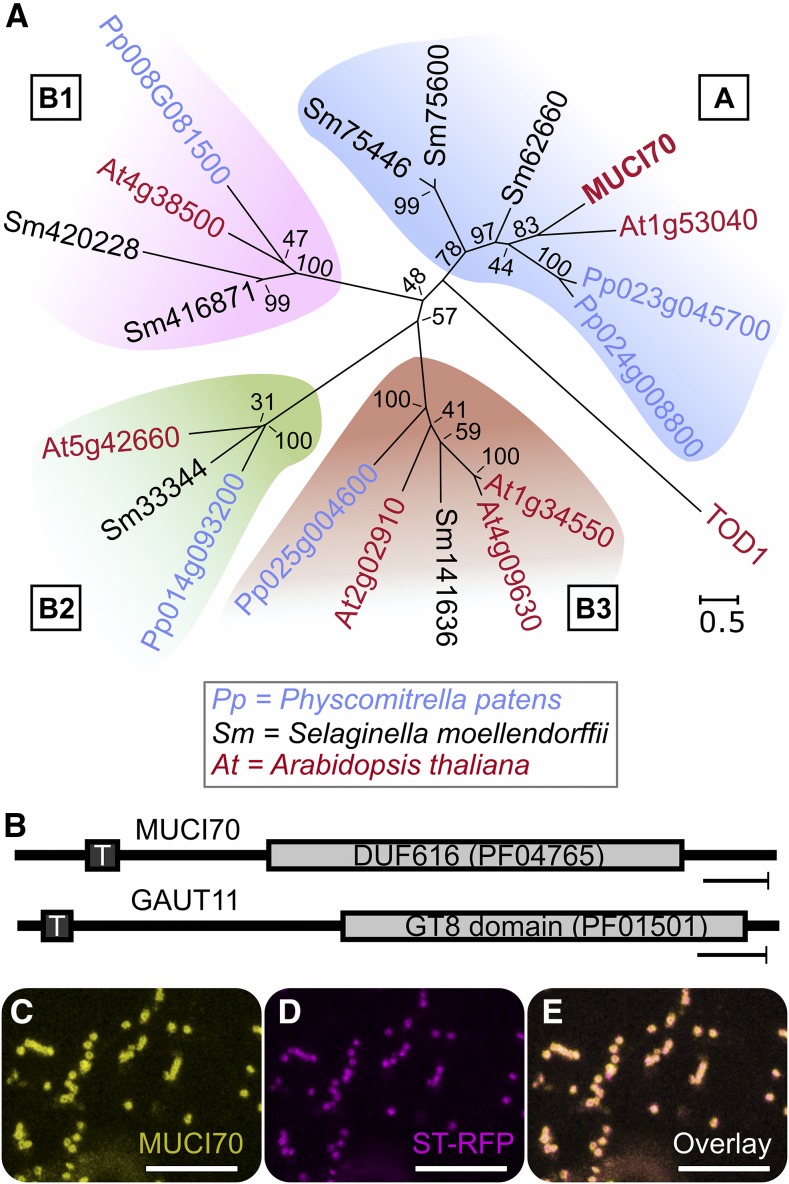
MUCI70 represents the founding member of a GT family whose roles in cell wall biology remain unclear (Fig. 1A). The MUCI70 protein contains a single transmembrane domain (AREMEMNON Consensus TM α-helix prediction, AramTmCon; Schwacke et al., 2003) near its N terminus and a DUF616 (PF04765) conserved domain of unknown function (Fig. 1B). Phylogenetic analysis of DUF616 proteins organized MUCI70 and its six Arabidopsis paralogs into four clades (Fig. 1A). Each of these groups contains at least one ortholog in both Physcomitrella patens and Selaginella moellendorffii, members of two early diverging lineages of land plants (Fig. 1A). In contrast, TURGOR REGULATION DEFECT1 (TOD1; AT5G46220), the only other Arabidopsis protein containing a DUF616 motif, did not cluster with any of these clades (Fig. 1A) and appeared to be functionally distinct. Indeed, TOD1 was demonstrated to have alkaline ceramidase activity in vitro (Chen et al., 2015), rather than a CAZy-related function. At4g38500, a close paralog of MUCI70 (Fig. 1A), was identified previously in a Golgi proteomics study and showed little similarity in primary sequence and predicted 3D structure to the GT8 family in Arabidopsis (Nikolovski et al., 2012). Based on the tight coexpression with GAUT genes, At4g38500 was hypothesized to be involved in pectin biosynthesis (Voxeur et al., 2012).
Figure 1.
MUCI70 is a DUF616 protein related to glycosyltransferases. A, Phylogenetic analysis of DUF616 proteins in Arabidopsis, P. patens, and S. moellendorffii. B, Schematic of conserved domains in MUCI70 and GAUT11 proteins. T, Transmembrane domain. C to E, Colocalization of MUCI70-sYFP with the Golgi marker ST-RFP (Teh and Moore, 2007) in stably transformed Arabidopsis rosette leaf epidermal cells. Bars = 50 amino acids (B) and 10 µm (C–E).
MUCI70 and GAUT11, a gene that was implicated in mucilage HG biosynthesis (Caffall et al., 2009), showed similar transcriptional profiles in developing seeds (Supplemental Fig. S1; Belmonte et al., 2013) and encode proteins with similar topologies (Fig. 1B). GAUT11 was found previously in the Golgi proteome (Parsons et al., 2012), but the subcellular localization of MUCI70 remained unknown. To address this, MUCI70 tagged with super yellow fluorescent protein (sYFP) was stably expressed in Arabidopsis using the constitutive 35S promoter. MUCI70-sYFP was observed in intracellular punctae (Fig. 1C) that colocalized with the Golgi marker sialyltransferase (ST) tagged with red fluorescent protein (ST-RFP; Fig. 1, D and E), which marks the site of pectin production in plants.
Mutations in MUCI70 and GAUT11 Cause Severe Mucilage Defects
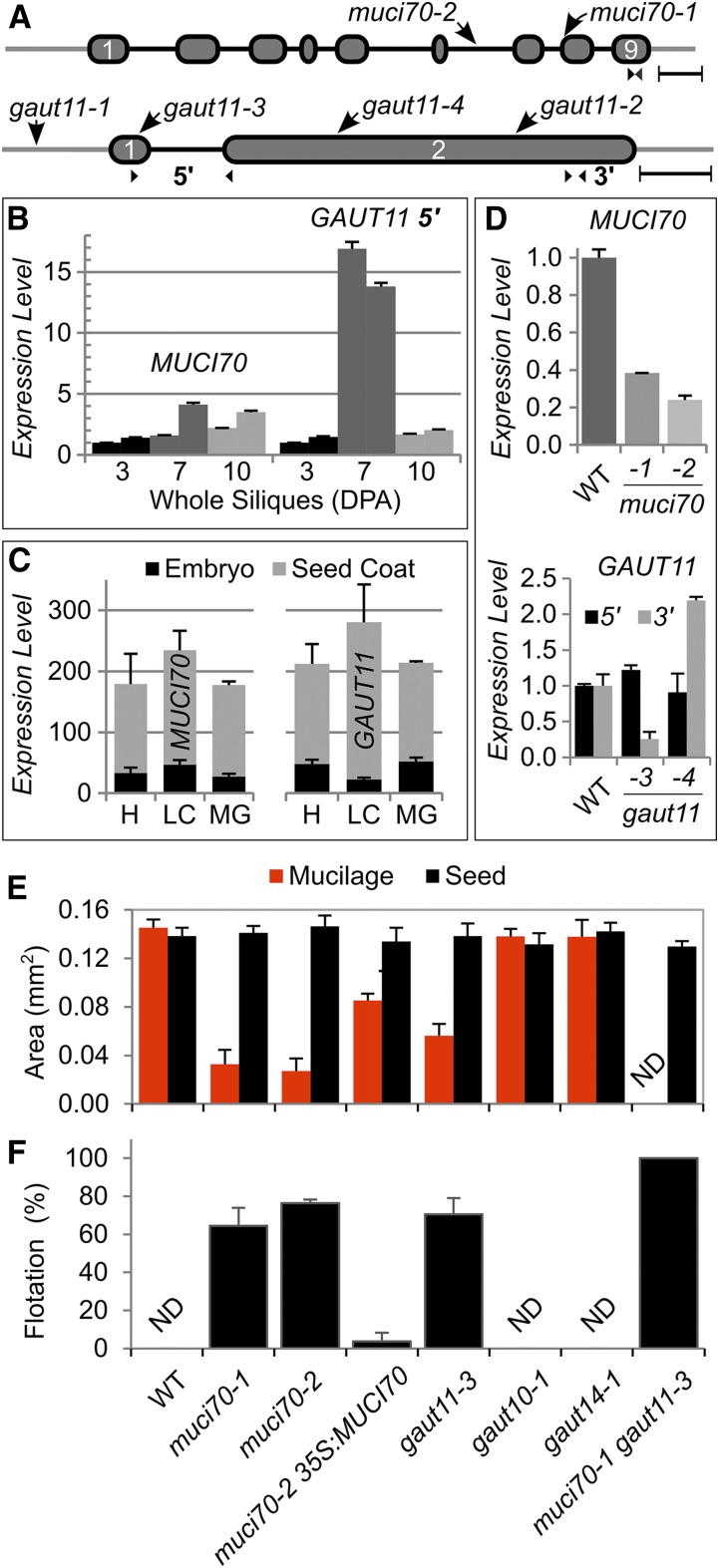
To investigate the biological role of MUCI70 in SCE cells, we obtained two independent T-DNA lines and identified homozygous mutants (Fig. 2A; Supplemental Table S1). While GATL5 was shown unambiguously to be required for mucilage pectin structure (Kong et al., 2013), only one of two transcriptional knockdown mutants (gaut11-2; Fig. 2A) previously indicated that GAUT11 influences mucilage structure (Caffall et al., 2009). Therefore, we analyzed two muci70 insertional mutants alongside two gaut11 mutants, gaut11-3 and gaut11-4, with insertions in GAUT11 exons (Fig. 2A). Using RT-qPCR, we validated that both MUCI70 and GAUT11 were transcribed in developing Arabidopsis siliques from 3 to 10 DPA. GAUT11 showed a dramatic increase (around 15-fold) in expression at 7 DPA, when pectin synthesis in SCE cells is at its peak (Fig. 2B). Based on the results of public microarray data sets (Winter et al., 2007; Belmonte et al., 2013), both genes were expressed preferentially in the seed coat relative to the embryo (Fig. 2C) and had similar transcript levels from the heart stage (∼3 DPA) to the mature green stage (∼10 DPA). Each insertion in the MUCI70 gene reduced its expression by at least 60% (Fig. 2D). Although gaut11-3 and gaut11-4 did not significantly alter GAUT11 transcription at either the 5′ or 3′ end (Fig. 2D), these alleles and the previously described gaut11-2 (Caffall et al., 2009) are exonic insertions (Fig. 2A) that likely disrupt the GAUT11 protein sequence.
Figure 2.
Analysis of T-DNA insertions in MUCI70 and GAUT genes. A, Positions of T-DNA insertions in MUCI70 and GAUT11 genes. Ovals represent exons, connecting lines show introns, and outer lines depict untranslated regions. Small arrowheads indicate the positions of reverse transcription quantitative PCR (RT-qPCR) primers. Bars = 250 bp. B, Gene expression in wild-type (WT) siliques at three different stages (DPA; two biological replicates per time point). C, ATH1 GeneChip expression levels (means + sd) in general seed coats and embryos at heart (H), linear cotyledon (LC), and maturation green (MG) stages. Data obtained by Belmonte et al. (2013) were extracted from the eFP Browser (Winter et al., 2007). D, Effects of T-DNA insertions on MUCI70 and GAUT11 transcript abundance in whole siliques at 7 DPA. In B and D, data show means + sd of two technical (B) or biological (D) replicates, normalized to the geometric mean of the UBQ5 and elF4A1 reference genes, and the relative expression of the first sample was set as 1 in each series. E, Dimensions of Ruthenium Red (RR)-stained mucilage capsules released from seeds in water. Data show means + sd of five biological replicates (more than 20 seeds each). The 35S:MUCI70-sYFP transgene partially rescued the mucilage defect of the muci70-2 mutant. ND denotes not detected. F, Percentage of seeds that float on water. Data show means + sd of three biological replicates (more than 35 seeds each).
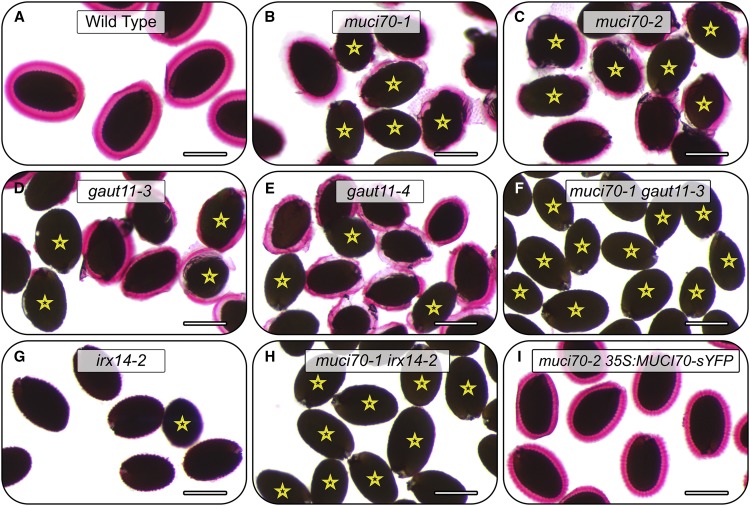
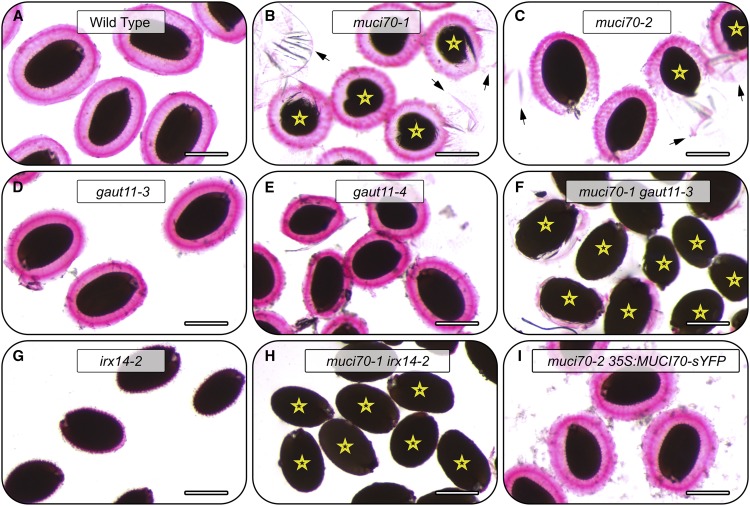
In contrast to wild-type seeds, which are surrounded by large mucilage capsules (Fig. 3A), two muci70 and two gaut11 homozygous mutants showed severe RR staining defects (Fig. 3, B–E), consisting of patchy or completely impaired mucilage release. Consequently, the muci70-1, muci70-2, and gaut11-3 seeds were surrounded by significantly smaller mucilage capsules (Fig. 2E), whose surface area was only 19% to 39% of the wild-type value. At least 65% of muci70 and gaut11 seeds floated on water (Figs. 2F and 3, marked by stars), whereas wild-type seeds did not float (Fig. 2F), despite having similar dimensions (Fig. 2E). Besides GAUT11, three other GAUT genes (GAUT8, GAUT10, and GAUT14) were expressed in the developing seed coat (Supplemental Fig. S1). The gaut8 mutant was found previously to be lethal, but the gaut10-1 and gaut14-1 transcriptional knockout mutants were viable (Caffall et al., 2009) and reexamined in this study. In contrast to muci70 and gaut11 mutants, gaut10-1 and gaut14-1 did not disrupt the dimensions of the seeds or the surrounding RR-stained mucilage capsules (Fig. 2E; Supplemental Fig. S2). Therefore, only one of the GAUT genes tested was essential on its own for maintaining mucilage architecture, consistent with the previous study of the whole GAUT family (Caffall et al., 2009).
Figure 3.
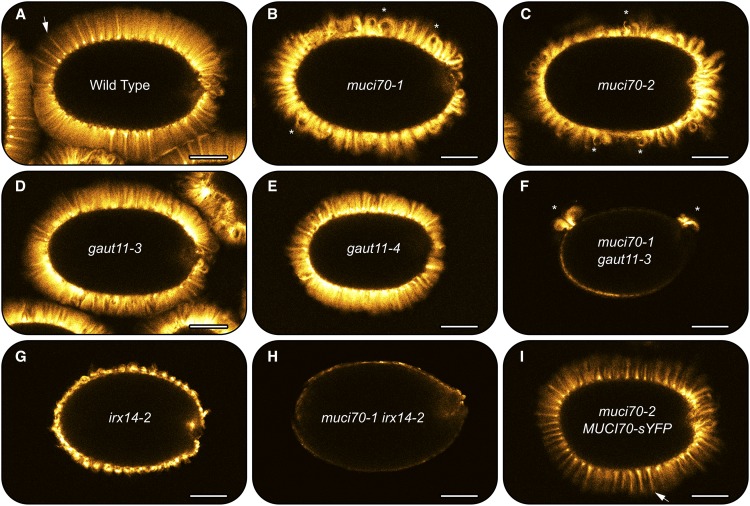
RR staining of mucilage polysaccharides around seeds hydrated in water. Images show RR staining of mucilage released from seeds. Stars mark seeds that float on water. Relative to wild-type seeds (A), muci70 and gaut11 single mutants release less mucilage (B–E). No mucilage is released from the muci70 gaut11 double mutant (F) or muci70 irx14 seeds (H). In the irx14 single mutant (G), mucilage is released but detaches from the seed surface. The 35S:MUCI70-sYFP transgene rescues the impaired mucilage release and the seed flotation defects of the muci70-2 mutant (I). Bars = 0.4 mm.
Since both gaut11-3 and gaut11-4 mutants showed similar mucilage staining defects to the previously described gaut11-2 allele (Caffall et al., 2009), we primarily used gaut11-3 for further experiments. To investigate if MUCI70 and GAUT11 function in the same pathway, we crossed the muci70-1 and gaut11-3 single mutants. While muci70 and gaut11 single mutants showed smaller RR-stained mucilage capsules than the wild type (Fig. 3, A–E), all muci70 gaut11 double mutant seeds failed to release mucilage (Fig. 3F) and, thus, floated on water (Fig. 2F). Despite the severe mucilage defects, the muci70 gaut11 seeds were only 6% smaller than the wild type (Fig. 2E). This suggested that both MUCI70 and GAUT11 might be required for the biosynthesis of pectin in SCE cells, which is ultimately released as a hydrophilic capsule from mature seeds.
MUCI70 and GAUT11 Are Important for Pectin Production in SCE Cells
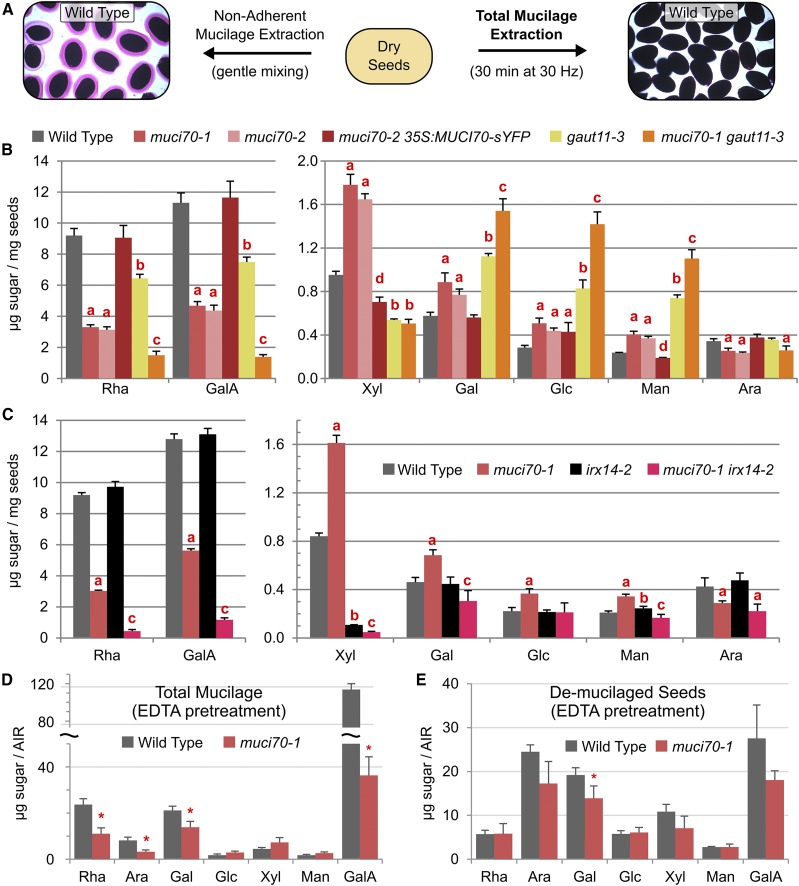
To identify the underlying biochemical defects that lead to impaired mucilage release, total mucilage was extracted from seeds vigorously mixed using a ball mill (Voiniciuc et al., 2015b; Voiniciuc and Günl, 2016). As described previously, this intensive mechanical agitation effectively removes all mucilage polysaccharides, resulting in seeds that are no longer stained by RR (Fig. 4A). The monosaccharide composition of the total mucilage extracted from hydrated seeds was quantified using high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD; Supplemental Table S2). Rha and GalA, the building blocks of the RG I backbone, represent around 90% of total mucilage extracted from wild-type Arabidopsis seeds (Fig. 4; Voiniciuc et al., 2015c). The muci70-1 and muci70-2 mutations reduced the absolute levels of Rha and GalA in total mucilage extracts by more than 50% compared with the wild-type control (Fig. 4B). The gaut11-3 single mutant reduced pectin content by around 30% compared with the wild type (Fig. 4B), similar to the gaut11-4 allele (Supplemental Table S2). Interestingly, the absolute abundance of the minor sugars in the total mucilage extracts increased by more than 40% in the muci70 and gaut11 mutants compared with the wild type (Supplemental Table S2). This suggests that both MUCI70 and GAUT11 are particularly important for the production and release of RG I but are not indispensable for the release of the minor mucilage components. For comparison, a knockout insertion in the MYB5 transcription factor, which promotes seed coat differentiation and mucilage production (Li et al., 2009; Voiniciuc et al., 2015c), significantly decreased the content of all sugars found in total mucilage extracts (Supplemental Table S2). Therefore, muci70 and gaut11 mutants are deficient in the production and release of pectic polysaccharides. In contrast to the gaut11-3 and gaut11-4 mutants, the gaut10-1 and gaut14-1 knockout mutants, identified by genotyping (Supplemental Table S3), reduced Rha and GalA levels by only 8% to 13% (Supplemental Table S2). Consistent with their normal RR staining phenotypes (Supplemental Fig. S2), gaut10-1 and gaut14-1 thus had a relatively minor influence on mucilage production. Indeed, analysis of the muci70-1 gaut11-3 double revealed that MUCI70 together with GAUT11 accounted for the biosynthesis and release of 88% of GalA-containing polymers in total seed mucilage extracts (Fig. 4B). Two-factor ANOVA of the HPAEC-PAD data (Supplemental Table S4) indicated that the muci70-1 and gaut11-3 mutations had purely additive effects on GalA abundance. Since the muci70-1 gaut11-3 total mucilage extracts also contained 84% less Rha than the wild type, the mutated genes controlled the content of mucilage pectin in a nonredundant manner (Fig. 4B). Compared with the single mutants, the muci70-1 gaut11-3 double mutant released even more minor sugars in total mucilage extracts (Supplemental Table S2). Since the minor sugars are derived primarily from hemicelluloses (Voiniciuc et al., 2015a, 2015b), the observed chemotype is consistent with the specific loss of pectin.
Figure 4.
Carbohydrate analysis of total mucilage extracted with water. A, Overview of the total mucilage extraction, which removes all polysaccharides from the seed surface that can be stained with RR. B and C, Monosaccharide composition of total mucilage extracted from seeds. Data show means + sd of four biological replicates per genotype. Significant changes from the wild type and between mutants are indicated by different red letters (Student’s t test, P < 0.05). The monosaccharide composition of the lines shown in B is provided in Supplemental Table S2, along with the data for gaut11-4, gaut10-1, and gaut14-1 mutants. D and E, Monosaccharide composition of the alcohol-insoluble residue (AIR) isolated from total mucilage extracts, following EDTA pretreatment, and the remaining seeds. Data show means + sd of three biological replicates. Asterisks indicate significant changes relative to the wild type (Student’s t test, P < 0.05).
Besides the drastic deficiency of RG I backbone sugars, mutations in MUCI70 and GAUT11 significantly increased the absolute amounts of Gal, Glc, and Man in total mucilage extracts (Fig. 4B; Supplemental Table S2) but had distinct effects on the content of Ara and Xyl. Based on ANOVA, the muci70-1 and gaut11-3 mutations had purely additive effects on the content of Gal, while the increases in Glc and Man content were higher than expected (Supplemental Table S4). The two muci70 alleles decreased Ara content significantly (26%–32%) relative to the wild type and gaut11 mutants. ANOVA confirmed that only MUCI70 influenced the presence of Ara (Supplemental Table S4). Surprisingly, muci70 and gaut11 single mutants had polarizing effects on Xyl content. Relative to the wild type, muci70 single mutants increased Xyl abundance by 73% to 87%, while the gaut11 single mutants and the muci70-1 gaut11-3 double mutant decreased Xyl content by 43% to 47% (Supplemental Table S2).
To further investigate the structure of pectin and other polysaccharides, glycosyl linkage analysis was performed on total mucilage extracts (Table 1). Relative to the wild-type control, the total mucilage extracts of both the muci70-1 and gaut11-3 mutants contained significant reductions in 4-linked GalA, the main building block of all pectin, and 2-Rha, characteristic of unbranched RG I (Pettolino et al., 2012; Voiniciuc et al., 2015c). The abundance of 2-Rha and 4-GalA linkages was decreased by around 75% in muci70-1 and 25% in the gaut11-3 mutant relative to the wild type (Table 1), consistent with the impaired production of RG I and HG, the two most abundant pectic domains in seed mucilage (Voiniciuc et al., 2015c). In contrast to their consistent reduction of pectin linkages, the muci70 and gaut11 mutants had distinct changes in the abundance of minor mucilage components. Only the muci70-1 mutant showed significant decreases in both 3-Ara and 5-Ara (Table 1), two linkages that could be derived from arabinan side chains on RG I (Atmodjo et al., 2013). Based on the ratio of 5-Ara to t-Ara linkages, arabinan chains in muci70-1 mucilage were estimated to be 30% shorter than in the wild type. While muci70-1 had a significant increase in the Xyl linkages associated previously with a highly branched xylan polymer (Voiniciuc et al., 2015a), gaut11-3 mucilage had significantly less xylan (Table 1), consistent with changes in Xyl detected with HPAEC-PAD (Fig. 4B). The reduced xylan content of the gaut11-3 mutant occurred with the presence of significantly more glycosyl linkages associated with galactoglucomannan (t-Gal, 4-Glc, 4-Man, and 4,6-Man) compared with the wild type (Table 1). To further investigate the distribution of polysaccharides, we immunolabeled whole seeds using the anti-mucilage CCRC-M30 and CCRC-M36 antibodies and the anti-xylan CCRC-M139 antibody. CCRC-M36 is specific for unbranched RG I (Ruprecht et al., 2017), while CCRC-M30 binds a yet-to-be-identified epitope unique to seed mucilage (Pattathil et al., 2010). All three antibodies labeled a uniform halo around wild-type seeds (Supplemental Fig. S3). In contrast, muci70-1 seeds typically displayed only faint, irregular patches of CCRC-M36 and CCRC-M30 epitopes but more intense and broader labeling of xylan (Supplemental Fig. S3). Both the immunolabeling and glycosyl linkage data indicated that mutations in MUCI70 resulted in a major decrease in RG I content accompanied by increased xylan content in seed mucilage.
Table 1. Glycosyl linkages in total mucilage extracted with water.
Linkage abundance was normalized to the absolute monosaccharide levels (µg mg−1 seed) of the same mucilage extracts. Data show means ± sd of three biological replicates per genotype. Boldface values are significantly different from the wild type (Student’s t test, P < 0.05).
| Linkage | Wild Type | muci70-1 | gaut11-3 |
|---|---|---|---|
| Rha | |||
| t-Rha | 0.113 ± 0.000 | 0.175 ± 0.069 | 0.204 ± 0.093 |
| 2-Rha | 9.082 ± 0.048 | 1.949 ± 0.141 | 6.277 ± 0.319 |
| 2,3-Rha | 0.220 ± 0.025 | 0.066 ± 0.013 | 0.156 ± 0.031 |
| 2,4-Rha | 0.093 ± 0.065 | 0.051 ± 0.010 | 0.095 ± 0.007 |
| Ara | |||
| t-Ara | 0.056 ± 0.007 | 0.030 ± 0.009 | 0.042 ± 0.014 |
| 5-Ara | 0.069 ± 0.000 | 0.021 ± 0.008 | 0.047 ± 0.009 |
| 3-Ara | 0.167 ± 0.012 | 0.114 ± 0.018 | 0.208 ± 0.021 |
| Gal | |||
| t-Gal | 0.153 ± 0.012 | 0.288 ± 0.071 | 0.313 ± 0.034 |
| 2-Gal | 0.072 ± 0.016 | 0.127 ± 0.010 | 0.270 ± 0.024 |
| 4-Gal | 0.097 ± 0.001 | 0.052 ± 0.007 | 0.108 ± 0.008 |
| 6-Gal | 0.024 ± 0.002 | 0.021 ± 0.007 | 0.035 ± 0.006 |
| 2,4-Gal | 0.036 ± 0.008 | 0.026 ± 0.003 | 0.045 ± 0.004 |
| 4,6-Gal | 0.092 ± 0.001 | 0.075 ± 0.026 | 0.168 ± 0.009 |
| Glc | |||
| t-Glc | 0.013 ± 0.009 | 0.014 ± 0.007 | 0.029 ± 0.009 |
| 4-Glc | 0.246 ± 0.037 | 0.373 ± 0.052 | 0.767 ± 0.089 |
| 3,4-Glc | 0.014 ± 0.003 | 0.014 ± 0.003 | 0.036 ± 0.010 |
| 4,6-Glc | 0.029 ± 0.003 | 0.037 ± 0.013 | 0.101 ± 0.025 |
| Xyl | |||
| t-Xyl | 0.172 ± 0.005 | 0.335 ± 0.062 | 0.108 ± 0.014 |
| 4-Xyl | 0.640 ± 0.034 | 1.070 ± 0.090 | 0.361 ± 0.009 |
| 2,4-Xyl | 0.258 ± 0.018 | 0.372 ± 0.026 | 0.125 ± 0.013 |
| Man | |||
| 4-Man | 0.061 ± 0.002 | 0.092 ± 0.012 | 0.157 ± 0.013 |
| 4,6-Man | 0.159 ± 0.018 | 0.228 ± 0.044 | 0.562 ± 0.041 |
| GalA | |||
| t-GalA | 0.075 ± 0.002 | 0.049 ± 0.007 | 0.096 ± 0.028 |
| 4-GalA | 12.175 ± 0.647 | 3.793 ± 0.394 | 9.252 ± 0.625 |
| 2,4-GalA | 0.128 ± 0.003 | 0.060 ± 0.018 | 0.085 ± 0.007 |
| 4,6-GalA | 0.165 ± 0.001 | 0.053 ± 0.007 | 0.146 ± 0.033 |
We further validated that the muci70 defects resulted from the loss of a Golgi-localized putative GT via the complementation of muci70 with a recombinant MUCI70 construct. The 35S:MUCI70-sYFP construct, which was used to confirm the Golgi localization of the MUCI70 protein (Fig. 1, C–E), at least partially rescued the mucilage defects of the muci70-2 mutant. Multiple independent muci70-2 35S:MUCI70-sYFP transformants produced seeds with uniform RR-stained mucilage capsules (Fig. 3I) and without the flotation defect that was observed frequently for the muci70 mutant seeds (Figs. 2F and 3C). The constitutive expression of MUCI70-sYFP proteins tripled the RR-stained mucilage area of muci70-2 seeds hydrated in water, although this still fell short of the wild-type level (Fig. 2E). In addition, the 35S:MUCI70-sYFP construct fully rescued the abundance of the Rha and GalA pectic sugars extracted from muci70-2 mutant seeds (Fig. 4B) but unexpectedly reduced the content of Xyl and Man relative to the wild type. Therefore, the constitutive expression MUCI70 appeared to negatively affect hemicellulose biosynthesis, consistent with the significant increases of Xyl and Man in muci70 mutants.
Unlike MUCI70, GAUT11 Functions as an HG α-1,4 GalA Transferase in Vitro
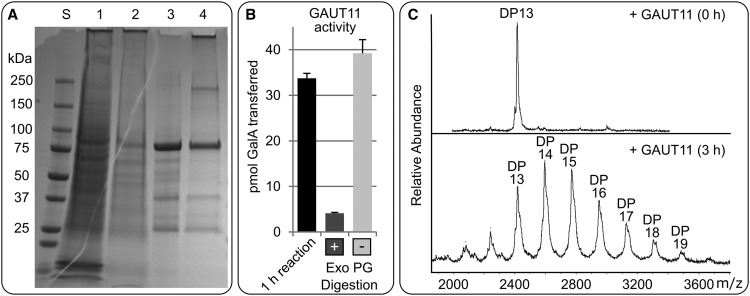
As mentioned above, GAUT11 belongs to the GAUT family of proven and putative HG α-1,4 GalA transferases (Sterling et al., 2006; Atmodjo et al., 2011; Biswal et al., 2018). Since, in addition to RG I, Arabidopsis seed mucilage is known to contain HG (Macquet et al., 2007a; Voiniciuc et al., 2013), particularly in the inner layer, we tested whether GAUT11 had HG α-1,4 GalA transferase activity that could account for the mucilage defects observed in the gaut11 mutants. A recombinant GAUT11∆39 protein, containing N-terminal His8X and GFP tags instead of the predicted transmembrane domain (Fig. 1B), was expressed in the Human Embryonic Kidney (HEK293) cell system (Moremen et al., 2018). Purification of the expressed His8X-GFP-GAUT11∆39 from the medium of the HEK293 cells, followed by SDS-PAGE of the protein under both reducing and nonreducing conditions (Fig. 5A), revealed that GAUT11 does not form a disulfide-linked dimeric or larger protein complex but rather exists primarily as a monomer in vitro. To determine if GAUT11 catalyzed HG elongation, we tested whether the recombinant protein incorporated radiolabeled GalA from UDP-[14C]GalA onto HG acceptors with degrees of polymerization (DP) 7 to 23 (Fig. 5B). Under these conditions, measurable amounts of GalA[14C] were detected in the product, suggesting that GAUT11 is an HG:GalA transferase. Treatment of the products with exopolygalacturonase (Exo PG), which specifically cleaves α-1,4 GalA linkages, confirmed that the products synthesized by GAUT11 were HG (Fig. 5B). The incorporation of GalA into HG by GAUT11 was linear over 45 min, with a specific activity of 1,473 ± 349 pmol GalA transferred min−1 mg−1 GAUT11 (Supplemental Fig. S4A). To confirm that HG was elongated and to identify the size of the products formed, GAUT11 was incubated with a fluorescently labeled HG acceptor of DP 13 (GalA13x-2AB) and UDP-GalA for 3 h and the products were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). The resulting peak masses showed that GAUT11 catalyzed the addition of up to six GalA residues or more onto the HG acceptor (Fig. 5C), confirming that GAUT11 is an HG α-1,4 GalA transferase. Since putative GTs containing a DUF616 domain have unknown biochemical functions (Fig. 1A), we also tested whether MUCI70 had HG:GalAT activity. A recombinant MUCI70∆77 protein, without its transmembrane domain (Fig. 1B), was expressed using the HEK293 cell system, purified, and assayed for HG:GalA transferase activity by MALDI-TOF MS. No elongation of the GalA13X-2AB acceptor by MUCI70 was observed (Supplemental Fig. S4B), while, under the same conditions, GAUT11 exhibited significant GalA13X-2AB acceptor elongation (Fig. 5C). These results strongly suggest that reduced synthesis of HG is the defect underlying the gaut11 mucilage phenotype. In contrast, MUCI70 lacks HG:GalAT activity and exerts its effects via a different mechanism.
Figure 5.
Purification and enzymatic characterization of His8X-GFP-GAUT11∆39. A, Coomassie Blue-stained SDS-PAGE of protein standard (lane S), 40 µL of combined medium and HEK293 cells expressing His8X-GFP-GAUT11∆39 (lane 1), 40 µL of medium only from HEK293 cells expressing His8X-GFP-GAUT11∆39 (lane 2), 5 µg of purified recombinant protein under reducing (lane 3), and nonreducing conditions (lane 4). The expected molecular mass of His8X-GFP-GAUT11∆39 is 91.1 kD. B, Incorporation of [14C]GalA by His8X-GFP-GAUT11∆39 into products sensitive to Exo PG. The purified protein, HG oligosaccharides (DP 6–23), and UDP-[14C]GalA were incubated for 1 h. An aliquot of the products was treated with (+) or without (−) Exo PG for 18 h. Data show means + se of two independent assays, each with duplicate samples. Exo PG treatment significantly degraded the product (P < 0.001), based on ANOVA followed by Tukey’s honestly significant difference test. C, MALDI-TOF MS of the products resulting from the incubation of His8X-GFP-GAUT11∆39, GalA13x-2AB acceptor, and UDP-GalA for 0 h (top) and 3 h (bottom). The mass differences between each peak are consistent with the sequential addition of one GalA residue (176 D) for each catalytic transfer. Spectra are representative of two independent assays.
Residual Mucilage Pectins in the muci70 Mutant Require Xylan Produced by IRX14
GAUT11 and MUCI70 both were required for pectin synthesis in Arabidopsis seed mucilage, but they had contrasting effects on xylan abundance. The constitutive expression of MUCI70-sYFP restored the mucilage RG I content to wild-type levels but reduced Xyl content, while mutations in MUCI70 elevated xylan production based on mucilage biochemical analysis and immunolabeling (Fig. 4; Supplemental Fig. S3). These results prompted us to further investigate the relationship between pectin and xylan production in SCE cells. The irx14-1 mutant, shown previously to be essentially devoid of xylan (Voiniciuc et al., 2015a), produced a normal amount of pectin that detached from the seed surface following hydration in water (Figs. 3G and 4C). We crossed the irx14-1 mutant to the muci70-1 mutant and isolated homozygous double mutant plants by genotyping. Relative to the single mutants, the muci70-1 irx14-1 double mutant showed more severe reductions than expected in both xylan- and pectin-related sugars in total mucilage extracts (Fig. 4C). Data evaluation using ANOVA revealed that MUCI70 and IRX14 interact to control the abundance of most mucilage sugars (Supplemental Table S5). As a notable exception, only the muci70-1 mutation significantly altered the Ara content (Fig. 4C), which could be derived from arabinan.
Cellulose Staining Reveals the Extent of Impaired Mucilage Release
To further investigate the underlying causes for the observed RR staining defects (Fig. 3), seeds were stained with Pontamine Fast Scarlet S4B (abbreviated S4B), a cellulose-specific fluorescent dye (Anderson et al., 2010), and examined with confocal microscopy (Fig. 6). The distribution of cellulose stained with S4B around seeds hydrated in water provides a clear overview of the primary cell wall and mucilage architecture. Wild-type mucilage capsules stained with S4B were characterized by long and regularly spaced cellulosic rays (Fig. 6A). Although some muci70 and gaut11 seeds released mucilage after prolonged shaking in water, they showed altered distribution of cellulose compared with the wild type. The muci70-1 and muci70-2 seeds were surrounded by shorter rays, which were curled rather than straight (Fig. 6, B and C). The gaut11-3 and gaut11-4 mutants showed an intermediate defect, with short but relatively straight rays (Fig. 6, D and E). The curly ray phenotype of the muci70-2 mutant was complemented by the constitutive expression of MUCI70s-YFP (Fig. 6I), although the overall intensity of S4B staining remained lower than in the wild type. Unlike either single mutant, the muci70-1 gaut11-3 double mutant displayed no S4B staining or only small patches around the seed (Fig. 6F), suggesting that most SCE cells did not release or produce mucilage. While the irx14-2 single mutant displayed clear S4B-labeled cellulosic regions (Fig. 6G), despite the loss of pectin adherence to the seed surface (Fig. 3G), the muci70-1 irx14-2 double mutant was essentially devoid of any S4B staining beyond the seed surface (Fig. 6H).
Figure 6.
S4B staining of cellulose in mucilage capsules of seeds hydrated in water. Single optical sections of fluorescent signals were detected with a confocal microscope. Arrows show well-defined cellulosic rays (A and I). Asterisks indicate short, curly rays observed in mutants with muci70 insertions. No straight rays are observed in F to H. Bars = 150 µm.
MUCI70 and GAUT11 Are Essential for Mucilage Accumulation in Seeds
To further investigate if the observed RR staining defects (Fig. 3) resulted from reduced pectin biosynthesis rather than only poor extrusion in water, dry seeds were pretreated with EDTA prior to water washes and RR staining. Cation chelators such as EDTA disrupt Ca2+-mediated pectic cross-links to promote mucilage release from mutants that synthesize normal amounts of pectin, but with a lower degree of methylesterification (Rautengarten et al., 2008; Voiniciuc et al., 2013). Although the impaired mucilage-release defects of muci70 and gaut11 single mutants were partially suppressed by the EDTA pretreatment (Fig. 7, A–E), many muci70 seeds still floated on water (Fig. 7, B and C) and displayed the detachment of outer tangential primary cell walls as large sheets. To confirm that MUCI70 is indispensable for RG I biosynthesis, we analyzed the composition of total mucilage extracts (Fig. 4A), following the EDTA pretreatment, and of the remaining (demucilaged) seeds. For the wild-type seeds, the use of EDTA increased the relative proportion of GalA and the absolute content of carbohydrates in total mucilage extracts (Fig. 4D; compare with Fig. 4, B and C). Nevertheless, the muci70-1 total mucilage extracts contained at least 53% less Rha and GalA than the wild type with the EDTA pretreatment (Fig. 4D) or without it (Fig. 4, B and C). In contrast to the pectin-deficient total mucilage extracts, the Rha and GalA content of muci70-1 demucilaged seeds was similar to that of the wild type (Fig. 4E). In addition, the reduced Ara content of muci70-1 total mucilage extracts was detected consistently with or without the EDTA pretreatment (Fig. 4, B–D). Except for reduced Gal in the mucilage and remaining seeds of muci70-1 following EDTA pretreatment, the abundances of the other minor sugars were not significantly different from those of the wild type (Fig. 4, D and E). Therefore, the EDTA pretreatment partially enhanced the extraction of pectic polysaccharides from seeds (Fig. 7) but could not rescue the Rha and GalA deficiency of the muci70-1 mutant. In addition, the 35S:MUCI70-sYFP transgene complemented the defects of muci70-2 seeds pretreated with EDTA (Fig. 7I), including the aberrant primary cell wall detachment, small RR-stained mucilage capsules, and seed flotation phenotypes. Unlike the muci70 alleles, the EDTA pretreatment rescued the flotation phenotype (Figs. 2F and 3, D and E) of gaut11-3 and gaut11-4 seeds (Fig. 7, D and E). Nevertheless, both gaut11 mutants released mucilage capsules that were still smaller than those of the wild type (Fig. 7A) and surrounded by debris that may originate from the primary cell wall (Fig. 7, D and E).
Figure 7.
RR staining of mucilage polysaccharides around seeds hydrated in EDTA. RR staining of seeds after EDTA pretreatment is shown. Arrows indicate detached sheets from the seed surface. Stars mark floating seeds. Bars = 0.4 mm.
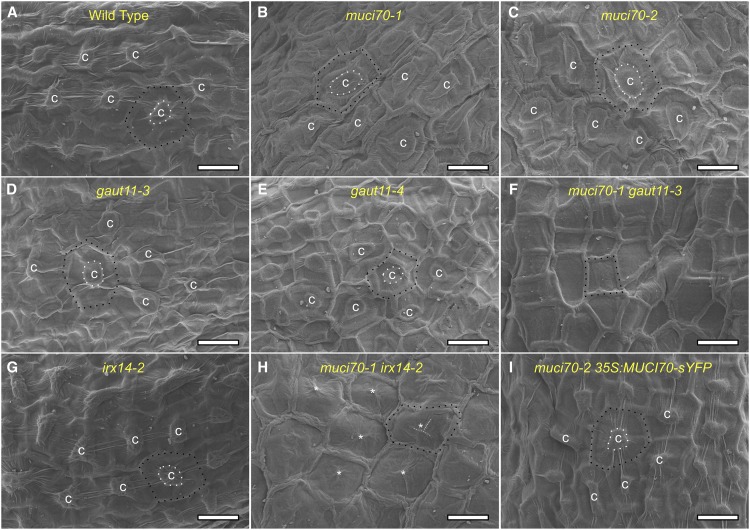
To investigate how the severe defects in pectin structure (Figs. 3, 6, and 7) affected the surface morphology of SCE cells, dry seeds were examined using scanning electron microscopy (SEM) and wet seeds were examined with the transmitted light detector of a confocal microscope. The mutant seeds isolated in this study displayed wild-type surface area (Fig. 2E) and overall seed shape (Supplemental Fig. S5). However, close examination of SCE cells with SEM revealed defective architecture of the primary and secondary cell walls in the RG I-deficient single and double mutants examined (Fig. 8). In the wild type, cellulose-rich columellae are observed in the center of every SCE cell (Fig. 8A) and protrude like volcanoes from the surface of hydrated seeds (Supplemental Fig. S6A). The characteristic shape of the columellae is established by the polar secretion of copious amounts of pectin early in seed coat development, when mucilage is produced (Young et al., 2008). Mutations in RHM2/MUM4, which supplies UDP-Rha for RG I synthesis, were shown previously to have flattened columellae as a result of reduced pectin accumulation and smaller mucilage pockets (Usadel et al., 2004; Western et al., 2004). Similarly, the muci70 and, to a lesser extent, gaut11 mutants showed flatter columellae compared with the wild type in transmitted light images of hydrated seeds (Supplemental Fig. S6) as well as in SEM micrographs of dry seeds (Fig. 8). The impaired SCE cell surface morphology of the muci70-2 mutant (Fig. 8C) was fully rescued by the 35S:MUCI70s-YFP transgene (Fig. 8I). Consistent with their severe reductions in mucilage production (Fig. 4), seeds of the muci70-1 gaut11-3 double mutant and the muci70-1 irx14-2 double mutant lacked detectable columellae structures in both SEM (Fig. 8, F and H) and transmitted light images (Supplemental Fig. S6, F and H). The SCE cells of the muci70-1 gaut11-3 double mutant, in particular, lacked the hexagonal appearance of the wild type and instead were surrounded by radial primary walls with highly irregular shapes (Fig. 8F). Therefore, the loss of both MUCI70 and GAUT11 completely flattened the landscape characteristic of the mucilage-secreting Arabidopsis seed coat.
Figure 8.
Surface morphology of Arabidopsis SCE cells. Scanning electron micrographs of mature, dry seeds are shown. The letter c marks the center of volcano-shaped columellae, which are not detected in F. Asterisks mark small remnants of columellae in H. White dashed lines highlight the size of columellae, while black dashed lines highlight primary walls surrounding epidermal cells. Bars = 20 µm.
DISCUSSION
GTs Indispensable for Mucilage RG I Elongation Are Uncovered
Even though Arabidopsis seed mucilage consists primarily of unbranched RG I, little to no insight into its production has been gained in recent years. While pectin production in SCE cells remains enigmatic, several studies in the last 4 years have characterized Arabidopsis seed mucilage mutants that shed new light on the production of cellulose (Ben-Tov et al., 2015; Griffiths et al., 2015), xylan (Voiniciuc et al., 2015a; Hu et al., 2016a, 2016b; Ralet et al., 2016b), and galactoglucomannan (Yu et al., 2014; Voiniciuc et al., 2015b, 2016). Since cellulose and hemicellulose represent relatively minor components of mucilage (Voiniciuc et al., 2015c, 2016), we hypothesized that screens for mucilage mutants have not been saturated and that novel pectin-deficient mutants remained to be identified. Therefore, we expanded the previously described MUCI reverse genetic screen to systematically profile the expression of all Arabidopsis CAZy genes during seed coat development. This strategy identified MUCI70, a member of a previously uncharacterized GT family, as a promising candidate for mucilage biosynthesis (Fig. 1). Compared with the wild type, two independent mutations in MUCI70 resulted in seeds that released smaller mucilage capsules (Fig. 2E), floated on water (Fig. 2F), and contained at least 60% less pectin in total mucilage extracts (Fig. 4, B and C). The reverse genetic screen also yielded several GT8 family members (Supplemental Fig. S1), including the GATL5 and GAUT11 genes that were already linked to mucilage structure. Although a gatl5 knockout mutant and a transgene complemented line have been analyzed in detail (Kong et al., 2013), two gaut11 knockdown lines previously showed inconsistent mucilage phenotypes (Caffall et al., 2009). Therefore, we examined muci70 mutants alongside two novel gaut11-3 and gaut11-4 alleles, which showed similar defects in mucilage staining with RR (Fig. 3).
Out of all the candidate genes screened, MUCI70 and GAUT11 were found to be the most important players for the biosynthesis and release of mucilage from seeds (Fig. 2; Supplemental Fig. S2). The SCE cells of muci70 and gaut11 single mutants produced significantly less RG I compared with the wild type based on their impaired mucilage staining phenotypes (Fig. 3), their Rha and GalA monosaccharide deficiency in total mucilage extracts (Fig. 4B), and their glycosyl linkage composition (Table 1). Previously, gaut11-2 nonadherent mucilage appeared only to have decreased HG content, but the content of Rha and uronic acids was determined via separate techniques (Caffall et al., 2009). By extracting the total mucilage polysaccharides (Fig. 4A) and quantifying neutral and uronic sugars with a single HPAEC-PAD method (Voiniciuc and Günl, 2016), we found that two independent mutations in GAUT11 showed significant reductions in GalA as well as Rha monosaccharides, which corresponded to lower amounts of glycosyl linkages found in RG I and HG backbones (Table 1). To rule out that mucilage accumulated normally but was not released effectively upon hydration, we pretreated seeds with EDTA, a cation chelator capable of rescuing mucilage defects dependent on HG-calcium cross-links (Rautengarten et al., 2008; Voiniciuc et al., 2013). While EDTA pretreatment extracted more mucilage from muci70 and gaut11 seeds (Fig. 7) than water alone (Fig. 3), all of the single mutants still displayed RR staining defects relative to the wild type. Indeed, muci70-1 total mucilage extracts contained less than half of the Rha and GalA found in the wild type, with (Fig. 4D) or without (Fig. 4, B and C) the EDTA pretreatment. In contrast, after EDTA pretreatment and total mucilage extraction, wild-type and muci70-1 seeds contained similar amounts of Rha and GalA (Fig. 4E). Therefore, MUCI70 was indispensable for the production of RG I in SCE cells. Both muci70 and gaut11 single mutants showed noticeably flatter columellae in confocal images of hydrated seeds (Supplemental Fig. S6) as well as SEM micrographs of dry seeds (Fig. 8), consistent with the accumulation of significantly less mucilage than in the wild type. In contrast to the major defects that resulted from the loss of either MUCI70 or GAUT11, a gatl5 knockout mutant was reported previously to have wild-type mucilage monosaccharide and glycosyl linkage composition (Kong et al., 2013). Therefore, we propose that MUCI70 and GAUT11 are indispensable for the production of the majority of pectin in Arabidopsis seed mucilage, while GATL5 might influence only the final organization or macromolecular size of these polymers.
MUCI70 and GAUT11 Are Required for the Production of Distinct RG I Domains
Despite containing putative GT domains with distinct primary structures, MUCI70 and GAUT11 have similar protein topologies (Fig. 1B) and transcriptional profiles in developing seeds and embryos (Fig. 2C). Insertions in either MUCI70 or GAUT11 significantly reduced the content of RG I- and HG-derived monosaccharides by around 60% and 30%, respectively (Fig. 4; Supplemental Table S2). The muci70-1 gaut11-3 double mutant nearly eliminated the production of RG I in SCE cells, as only 12% to 16% of the wild-type Rha and GalA sugars remained (Fig. 4B; Supplemental Table S2), and seeds hydrated in EDTA or water released little to no mucilage (Figs. 3, 5, and 6). ANOVA of the mucilage monosaccharide composition indicated that the muci70-1 and gaut11-3 mutations had purely additive effects on GalA abundance but partially overlapping effects on Rha content (Supplemental Table S4). Furthermore, while muci70 and gaut11 single mutants still displayed columellae, albeit flatter and wider than in the wild type, the muci70-1 gaut11-3 double mutant completely flattened the surface of SCE cells (Supplemental Fig. S6) and impaired the shape of their radial walls (Fig. 8). The defects in seed surface morphology are consistent with severely impaired mucilage accumulation in the SCE cells, as reported previously for the pectin-deficient mum4 mutant (Western et al., 2004) and the myb5-1 transcription factor mutant (Li et al., 2009). The pattern of cellulose deposition in wild-type SCE cells is determined by the polarized secretion of copious amounts of pectin into donut-shaped mucilage pockets (Voiniciuc et al., 2015c). The resulting volcano-shaped cytoplasmic columns are circled by cellulose synthases (Griffiths et al., 2015), leading to the deposition of cellulose-rich columellae (Mendu et al., 2011). Therefore, the absence of cellulosic rays (Fig. 6) and volcano-shaped collumellae (Fig. 8; Supplemental Fig. S6) around muci70-1 gaut11-3 double mutant seeds likely resulted from reduced pectin accumulation rather than from direct changes in cellulose synthesis. Overall, the results suggest that MUCI70 and GAUT11 are essential for the production of RG I domains, whose structures or biosynthesis are at least partially distinct but make up the bulk of Arabidopsis seed mucilage.
In addition to their significant decreases in the glycosyl residues of the RG I backbone, muci70 and gaut11 mutants had distinct effects on Ara and Xyl, two minor mucilage components. Besides Rha and GalA, total mucilage extracts from both muci70 alleles also were significantly deficient in Ara, which corresponded to decreases in the arabinan side chain of RG I (Table 1). The 5-linked Ara content was reduced by 70% in the muci70-1 mutant compared with the wild type (Table 1). In contrast, the gaut11 mutants had normal Ara content but a significant decrease in Xyl (Fig. 4B), derived from a highly branched xylan polymer found in wild-type total mucilage extracts (Table 1; Voiniciuc et al., 2015a). Although most of the RG I found in mucilage released from mature seeds is unbranched (Voiniciuc et al., 2015c), its backbone is likely synthesized in a branched form in the Golgi apparatus and is modified subsequently in the extracellular space. Mutant seeds deficient in β-galactosidase (Dean et al., 2007; Macquet et al., 2007b) or α-arabinofuranosidase activity (Arsovski et al., 2009) contain more galactan or arabinan RG I branches and display severely impaired mucilage release. Therefore, we hypothesize that MUCI70 and GAUT11 participate in the production of two distinct RG I domains that contain arabinan and xylan side chains, respectively. Mucilage was demonstrated recently to contain xylan branches on RG I that mediate the adherence of pectin to seeds (Ralet et al., 2016b).
Novel Links between Pectin and Hemicellulose Biosynthesis
While the biological function of mucilage in Arabidopsis seeds remains unclear, the architecture of this gelatinous wall is determined primarily by the structure of RG I, its major component. With the exception of upstream transcriptional regulators (Voiniciuc et al., 2015c), the mutants that display the most severe defects in mucilage release are involved directly in the production of nucleotide sugars for RG I biosynthesis or its metabolism in the wall (Usadel et al., 2004; Dean et al., 2007; Macquet et al., 2007b; Arsovski et al., 2009). As discussed in the preceding paragraph, we found compelling evidence that MUCI70 and GAUT11 are required for the synthesis and release of mucilage pectin. By demonstrating that GAUT11 catalyzes HG elongation in vitro (Fig. 5; Supplemental Fig. S4A), we propose that the synthesis of HG or of an HG-glycan region is essential for mucilage RG I production. However, we cannot exclude the alternative hypothesis that GAUT11 could utilize additional donor and acceptor substrates and, therefore, might play a more direct role in RG I backbone elongation. In contrast to GAUT11, MUCI70 purified from HEK293 cells did not appear to be involved in the elongation of HG domains (Supplemental Fig. S4B). The severe deficiency of RG I in muci70 total mucilage extracts suggests that MUCI70 may be involved more directly in its synthesis. So far, the other GTs known to be involved in the production of mucilage were found to only affect the structure of a single class of polysaccharides: pectin, hemicellulose, or cellulose. For instance, the irx14 mutant SCE cells had a nearly complete loss of xylan but did not significantly alter the content of other mucilage polymers (Fig. 4C; Voiniciuc et al., 2015a). In contrast, mutations in MUCI70 and/or GAUT11 reduced Rha and GalA content and significantly increased the absolute amounts of Gal, Glc, and Man in mucilage extracts (Fig. 4B), the building blocks of galactoglucomannan (Table 1). The greater abundance of minor sugars in total mucilage extracts indicates that muci70 and gaut11, unlike the myb5-1 transcription factor mutant (Supplemental Table S2), are not deficient in the release of all mucilage polymers but are involved specifically in pectin production. Relative to the wild type, the gaut11-3 single mutant contained a 3-fold increase in the content of galactoglucomannan, while the muci70-1 gaut11-3 double mutant had a 4-fold increase (Supplemental Table S2). Since highly branched galactoglucomannans have gelling properties akin to those of pectin and are known to control the architecture of wild-type mucilage (Voiniciuc et al., 2015b), a potential explanation for the observed changes is that SCE cells may attempt to compensate for the reduced synthesis of pectic domains by producing more hemicellulosic polymers with mucilaginous properties.
In addition to the elevated content of galactoglucomannan-related sugars when RG I content was reduced, we discovered that xylan biosynthesis is indispensable for at least one RG I domain. Mutations in several GAUT genes were found previously to impair the production of pectin as well as xylan (Orfila et al., 2005; Peña et al., 2007; Persson et al., 2007; Caffall et al., 2009). Although no requirement for xylan in pectin elongation was described previously, there is evidence that these two classes of polysaccharides can be covalently linked. Proteoglycans that contain both the pectins RG I and HG as well as xylan have been identified (Tan et al., 2013), providing an example of a polymer that could require an RG I domain as a possible primer for the synthesis of a xylan glycan. Consistent with previous reports, we found that gaut11 total mucilage was deficient in both pectin and xylan. To our initial surprise, two independent muci70 mutants contained significantly more xylan than the wild type in the total mucilage extracts, despite a more severe reduction of RG I compared with gaut11 alleles (Fig. 4; Supplemental Table S2). These findings were supported by the more intense labeling of mucilage xylan by CCRC-M139 and the reduced detection of RG I with CCRC-M36 (Supplemental Fig. S3). Although irx14 mutants alone had no effect on pectin content in total mucilage extracts (Fig. 4C; Voiniciuc et al., 2015a; Hu et al., 2016a), muci70 irx14 double mutant seeds were more deficient in RG I than the muci70 single mutants (Fig. 4C). ANOVA of monosaccharide composition indicated that the muci70 and irx14 mutations have synergistic effects on RG I production (Supplemental Table S5). Since the muci70 irx14 seeds did not release any mucilage and showed only traces of columellae (Figs. 6–8; Supplemental Fig. S6), the xylan-pectin connections were found to be especially important for mucilage production in the muci70 background.
Gaining Insight into the Biological and Biochemical Roles of DUF616 Proteins
An impasse in the biosynthesis of HG was solved 12 years ago by the first enzymatic characterization of a GT involved in its elongation (Bacic, 2006; Sterling et al., 2006). However, the production of the RG I backbone, the only polysaccharide in plants with a repeating disaccharide backbone, has remained a mystery since then. In this study, we identified MUCI70 as a putative GT from a novel CAZy family and demonstrated that it is indispensable for RG I elongation in the Golgi apparatus of SCE cells and its release upon seed hydration. We also showed that GAUT11 has HG α-1,4 GalA transferase activity (Fig. 5; Supplemental Fig. S4A), suggesting that the synthesis of HG also may be required for RG I elongation in mucilage. The enzymatic characterization of MUCI70 and the functional analysis of other DUF616 proteins should shed additional light on pectin biosynthesis. Only one plant protein containing a DUF616 domain, TOD1, has a known biochemical activity and functions as an alkaline ceramidase involved in regulating turgor in guard cells and pollen tubes (Chen et al., 2015). TOD1 appears to be an anomaly among DUF616-containing proteins in Arabidopsis because it was an outlier in our MUCI70 phylogenetic tree and lacks orthologs in early diverging land plants (Fig. 1A). A tod1 suppressor screen surprisingly identified that a mutation in GAUT13, which encodes a putative pectin GT, rescued the low seed set of the tod1 mutant (Chen et al., 2015). Since a gaut mutant was identified as a suppressor, tod1 mutant pollen tubes were hypothesized to contain more pectin, which may reduce their growth potential. Nevertheless, the cell wall composition of tod1 mutants was not tested, so the link between pectin biosynthesis and alkaline ceramidase activity is indirect and requires further investigation. Based on the results presented here, MUCI70 is involved directly in pectin biosynthesis and, thus, likely has an activity distinct from TOD1.
Our characterization of muci70 and gaut11 single and double mutants indicates that MUCI70 and GAUT11 are required for the synthesis of two distinct pectic regions associated with RG I, a view consistent with the latest model of pectin biosynthesis (Atmodjo et al., 2013). The additive effects of the muci70-1 and gaut11-3 mutations on GalA levels suggests that MUCI70 and GAUT11 do not function in consecutive steps of pectin elongation. Rather, with the demonstrated HG α-1,4 GalA transferase activity of GAUT11, the results suggest that GAUT11 synthesizes an HG region required for, or associated with, RG I. Meanwhile, MUCI70 potentially could facilitate the transfer of Rha and/or GalA, or possibly arabinan or RG I oligosaccharides, into or onto RG I. Although RG I is found in the walls of all growing plant cells, rhamnosyltransferases or galacturonosyltransferases involved RG I elongation have not yet been identified. Since MUCI70 is indispensable for the production of Arabidopsis seed mucilage, its biochemical activity should be comprehensively tested in future studies, as should the role of the GAUT11-synthesized HG glycan in mucilage RG I synthesis. To accomplish this will require technical advances in the purification of donor and acceptor substrates as well as the establishment of robust in vitro assays for RG I biosynthesis. Advancements in this area have emerged only recently (Uehara et al., 2017), and further developments should make it feasible to determine if the promising candidates identified in this study can incorporate Rha, GalA, or other carbohydrates into RG I.
MATERIALS AND METHODS
Plant Material
The Arabidopsis (Arabidopsis thaliana) T-DNA insertion mutants analyzed in this study are listed in Supplemental Table S1 and were selected from the SALK (Alonso et al., 2003) and SAIL (Sessions et al., 2002) collections, using the T-DNA Express tool (http://signal.salk.edu/cgi-bin/tdnaexpress). Mutant seeds and the ST-RFP (N799376) marker were obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info). Plants were grown in constant light as described previously (Voiniciuc et al., 2015b, 2015c), and seeds were harvested into separate bags for each plant. Mutants were genotyped by Touch-and-Go PCR (Berendzen et al., 2005) according to the SALK primer design tool (http://signal.salk.edu/tdnaprimers.2.html). The primers are listed in Supplemental Table S3.
In Silico Analysis of Proteins
MUCI70-related protein sequences from three species and Arabidopsis GAUT sequences were obtained from Phytozome (Goodstein et al., 2012). Phylogenetic analysis was conducted using the MEGA6.0 software (Tamura et al., 2013), as described previously (Hall, 2013). Alignments were performed using the MUSCLE method, and the evolutionary history was inferred using the maximum likelihood method. Trees were built using the best model found, including all sites (LG+G for MUCI70-related proteins and LG+G+I for the GAUT family). Tree reliability was evaluated by the bootstrap method (500 replicates). The topology of MUCI70 and GAUT11 proteins was assessed using the AramTmCon tool in ARAMEMNON (Schwacke et al., 2003).
RNA Isolation and RT-qPCR Analysis
Silique development was staged using nontoxic paint (Dean et al., 2011), and three 7-DPA siliques were harvested per plant (biological replicate). Silique RNA was isolated with the RNeasy Plant Mini Kit (Qiagen) and was treated with DNase I as recommended by the manufacturer. For each biological replicate, 200 ng of RNA was used as a template for the iScript cDNA Synthesis Kit (Bio-Rad), and the expression of each gene was quantified at least twice using iQ SYBR Green Supermix (Bio-Rad) and the Bio-Rad MyiQ system. Primers for transcript quantification (Supplemental Table S3) were designed with Primer-BLAST (Ye et al., 2012) or QuantPrime (Arvidsson et al., 2008). UBQ5 and elF4A1 served as reference genes (Gutierrez et al., 2008), and fold changes in target gene expression, normalized to the geometric mean of the two reference genes, were calculated in Microsoft Excel according to a published method (Fraga et al., 2008).
Seed Mucilage Staining
RR (VWR International; catalog no. A3488.0001) staining of pectin was performed as described recently (Voiniciuc et al., 2015a, 2015b) using cell culture plates with 24 wells (VWR International; catalog no. 734-2325). The effect of cation removal on mucilage release was tested by mixing seeds with water or 50 mm EDTA, pH 9.5, for 60 min at 125 rpm before rinsing with water twice and staining with 0.01% (w/v) RR. All RR images were acquired with a Leica DFC 295 camera equipped on a Leica MZ12 stereomicroscope and processed uniformly in Fiji (Schindelin et al., 2012). RR-stained mucilage and seed areas were quantified in Fiji using a semiautomated protocol (Voiniciuc et al., 2015b).
Mucilage cellulose staining was performed similarly to a published method (Voiniciuc et al., 2015a). Seeds were first mixed with water on a 24-well plate on a horizontal shaker (15 min, 100 rpm). After the water was removed, cellulose was stained with 0.025% (w/v) S4B (now sold as Direct Red 23; Sigma-Aldrich; 212490-50G) in 50 mm NaCl solution (60 min, 100 rpm). The dye then was removed and the seeds were mixed with 500 µL of water and transferred to glass slides. Optical sections were acquired with a Leica SP8 confocal system (552 nm excitation, 600–650 nm emission) equipped with photomultipliers for fluorescence as well as transmitted light.
Statistical Analyses
As described previously (Voiniciuc et al., 2015a), significant changes relative to the wild type were detected using Student’s t test (two-tailed distribution, assuming equal variance of two samples). The effects of two independent mutations on mucilage monosaccharide composition were evaluated using two-factor ANOVA, performed with the Real Statistics Resource Pack (http://www.real-statistics.com) for Microsoft Excel 2010.
Monosaccharide Composition of Total Mucilage Extracts
Total mucilage polysaccharides were extracted from 5 mg of seeds and analyzed as described in a recent method (Voiniciuc and Günl, 2016), except that polymers were hydrolyzed for 90 min at 120°C. For each genotype, the seeds of at least three different plants were examined as independent biological replicates. Monosaccharides were separated and quantified via HPAEC-PAD using a Dionex DX-600 system equipped with CarboPac PA20 guard and analytical columns (Voiniciuc et al., 2015b). For each data set, all genotypes were grown, harvested, processed, and analyzed simultaneously. For the EDTA pretreatment, 5 mg of dry seeds was hydrated in 500 µL of 50 mm EDTA (pH 9.5) and then used for the total mucilage extraction (Voiniciuc and Günl, 2016), Afterward, 300 µL of the supernatant was transferred to a 2-mL screw-cap tube. Polymers were precipitated by adding 1,500 µL of absolute ethanol and vortex mixing. Following centrifugation (2 min at 20,000g), the supernatant was discarded. The precipitated mucilage polymers were washed with 500 µL of 70% (v/v) ethanol and then resuspended in 300 µL of acetone before drying for 5 min at 60°C. The seeds remaining from the EDTA pretreatment and total mucilage extraction were washed twice with 1 mL of water and ground using steel balls at 30 Hz for 1.5 min using a ball mill (Retsch MM400). Demucilaged seed polysaccharides were washed twice with 70% (v/v) ethanol, once with chloroform:methanol (1:1, v/v), and once with acetone. The insoluble polymers then were resuspended in 300 µL of acetone and dried for 5 min at 60°C. The monosaccharide composition of total mucilage and demucilaged seeds after EDTA pretreatment was analyzed as described above, using ribose as an internal standard.
Whole Seed Immunolabeling
Monoclonal antibodies directed against xylan were obtained from CarboSource. Immunolabeling of seeds hydrated in water was performed as described previously in detail (Voiniciuc et al., 2015a; Voiniciuc, 2017) using Alexa Fluor 488 goat anti-mouse secondary antibodies (Molecular Probes, ThermoFisher Scientific). Images of immunolabeled seeds (with or without counterstaining with S4B) were acquired on a Leica SP8 confocal microscope using the following settings: Alexa Fluor signal (488 nm excitation, 500–530 nm emission) and S4B/intrinsic seed fluorescence (552 nm excitation, 590–700 nm emission). The muci70-1 mutant was analyzed alongside previously described wild-type seeds (Voiniciuc et al., 2015a; Voiniciuc, 2017).
Glycosyl Linkage Analysis of Seed Mucilage
Glycosyl linkage analysis of total mucilage extracted with water from 60 mg of seeds was performed as described previously (Voiniciuc et al., 2015a, 2015b). For genotype, three plants (biological replicates) were analyzed in parallel. After uronic acid reduction (Gibeaut and Carpita, 1991), extensive dialysis, dimethyl sulfoxide solubilization, and methylation (Gille et al., 2009), the polysaccharides were hydrolyzed, derivatized to the corresponding alditol acetates, and analyzed by gas chromatography-MS (Foster et al., 2010) using sodium borodeuteride for the reduction. The glycosyl linkage composition was normalized to the absolute abundance of each sugar residue, as quantified using HPAEC-PAD analysis of an aliquot of the extracted mucilage. Polysaccharide composition was calculated as described in a detailed protocol (Pettolino et al., 2012) with a minor modification (t-Xyl was assigned to xylan).
sem
Mature, dry Arabidopsis seeds were sputter coated with a gold layer (approximately 5 nm thickness, 60 mA current) using a Cressington Sputter Coater 208 HR integrated with thickness controller MTM-20 (Cressington Scientific Instruments). Afterward, several seeds for each genotype were mounted on a typical electron microscopy stub using a carbon adhesive tape. The SEM images were acquired using a LEO (Zeiss) 1550 field emission SEM device (Zeiss/LEO), with an in-lens or secondary electron detector at 5 to 15 kV acceleration voltage, at the Helmholtz Nano Facility at Forschungszentrum Jülich (Albrecht et al., 2017).
Expression and Analysis of MUCI70-sYFP Proteins
The 35S:MUCI70-sYFP construct was assembled using ligation-independent cloning (LIC) and the pCV01 vector (Voiniciuc et al., 2015b). Primers containing LIC adapters (Supplemental Table S3) and Phusion High-Fidelity DNA Polymerase (New England Biolabs) were used to amplify a 3,275-bp MUCI70 fragment (from ATG up to, but excluding, the stop codon) from Arabidopsis genomic DNA. The gel-purified MUCI70 PCR product was then used for LIC as described (De Rybel et al., 2011). The 35S:MUCI70-sYFP plasmid was verified by Sanger sequencing and introduced in Agrobacterium tumefaciens GV3101::pMP90::pSOUP cells. Arabidopsis plants were transformed using a modified floral spray method (Weigel and Glazebrook, 2006) with an infiltration medium containing 5% (w/v) Suc and 0.02% (v/v) Silwet L-77. T1 seedlings were selected with a 10 mg L−1 glufosinate-ammonium spray (Sigma-Aldrich; catalog no. 45520-100MG).
The subcellular localization of fluorescently tagged proteins in stably transformed rosette leaf epidermal cells was examined using a Leica SP8 microscope as described previously (Voiniciuc et al., 2015b). Plants expressing both MUCI70-sYFP and ST-RFP were obtained through genetic crosses, and fluorescent signals were acquired sequentially for each line scan: sYFP (488 nm excitation, 505–550 emission) and RFP (552 nm excitation, 590–635 nm emission).
Expression and Purification of GAUT11∆39 and MUCI70Δ77 in HEK293 Cells
Gateway expression vectors for transient expression in HEK293 cells, and cloning and expression methods, were adapted from other publications (Moremen et al., 2018). The truncated coding sequences of GAUT11 and MUCI70 were PCR amplified, respectively, from TAIR clone U87017 (www.arabidopsis.org) and from 7-d-old Arabidopsis whole seedling cDNA (a gift from Dr. Melani Atmodjo, University of Georgia). Specifically, GAUT11 and MUCI70 were truncated to 3′ beyond their predicted transmembrane domains, ∆39 and ∆77, respectively, based on the AramTmCon tool from the ARAMEMNON database (Schwacke et al., 2003; http://aramemnon.botanik.uni-koeln.de/). For the first PCR amplification, the GAUT11∆39 F and R primers and the MUCI70∆77 F and R primers were used to amplify the respective genes (Supplemental Table S3). A second round of PCR amplification was performed using the attB F and R universal primers (Supplemental Table S3).
The attB PCR products were cloned into the Gateway pDONR221 entry vector using the Gateway BP Clonase II Enzyme (ThermoFisher) according to the manufacturer’s instructions. JM109 competent cells were transformed and plated on Luria-Bertani (LB) agar selection plates containing 50 µg mL−1 kanamycin. Colonies were selected and grown overnight at 37°C at 250 rpm in 3 mL of LB medium containing 50 µg mL−1 kanamycin. Plasmids were isolated using the GeneJet Plasmid Miniprep Kit (ThermoFisher) and sequence confirmed (Macrogen). The following primers were used for sequencing: M13F, M13R-pUC (Macrogen), and GAUT11 seq or MUCI70 seq primers (Supplemental Table S3). The sequence-confirmed GAUT11 and MUCI70 entry plasmids were cloned into the Gateway pGEn2-DEST vector using the Gateway LR Clonase II Enzyme (ThermoFisher) according to the manufacturer’s instructions. All steps were the same as in the BP Clonase II reaction, except that 100 µg mL−1 carbenicillin was used for selection. The following primers were used for sequencing: pG2F, pG2R, and the GAUT11 seq or MUCI70 seq primers (Supplemental Table S3). Glycerol stocks of sequence-confirmed colonies were stored at −80°C for future use.
Sequence-confirmed GAUT11∆39-pGEn2-DEST and MUCI70∆77-pGEn2-DEST cultures were grown in 3 mL of LB liquid medium containing 100 μg mL−1 carbenicillin at 250 rpm for 8 h. Two milliliters of the culture was added to 500 mL of LB liquid medium with carbenicillin, the culture was incubated at 37°C and 250 rpm for 18 h and centrifuged at 4,000g for 10 min at room temperature, and the supernatant was discarded. Plasmid isolation was performed using the Invitrogen PureLink HiPure Plasmid Filter Maxiprep Kit (ThermoFisher), and the final DNA concentration was measured using a NanoDrop spectrophotometer.
The transfection of sterile GAUT11∆39 pGEn2-DEST or MUCI70∆77-pGEn2-DEST DNA into HEK293 cells (Freestyle 293-F cells; ThermoFisher) was done at a total concentration of 3 µg mL−1 total culture volume (250 mL for GAUT11 and 20 mL for MUCI70) with 9 µg mL−1 polyethyleneimine (linear 25-kD polyethyleneimine; Polysciences) essentially as described previously (Moremen et al., 2018). A larger culture was required for GAUT11 due to lower purification yields. Following batch-mode production for 6 d, the cells were separated from the medium by centrifugation, and the resulting clarified medium was filtered through a 0.45-μm nylon filter. GAUT11∆39 was purified using the ÄKTA FPLC system equipped with a 1-mL His-Trap HP column (GE Healthcare). The column was equilibrated and washed with 50 mm HEPES, pH 8, 300 mm NaCl, and 10 mm imidazole, and GAUT11∆39 was eluted at 1 mL min−1 with 50 mm HEPES, pH 8, and 300 mm NaCl using a gradient of 0 to 500 mm imidazole (20 column volumes). MUCI70∆77 was purified using batch TALON metal affinity resin (Clontech) according to the manufacturer’s instructions using the same wash and elution buffers as for GAUT11∆39. Fractions containing the protein were exchanged into 50 mm HEPES, pH 7.2, 100 mm NaCl, and 15% (v/v) glycerol storage buffer using a PD-10 column (GE Healthcare). The eluted proteins were concentrated using a 30-kD cutoff Ultra Centrifugal Filter Unit (EMD Millipore), and their concentrations were measured by UV-Vis spectroscopy (Nanodrop). The resulting purified GAUT11∆39 (1.7 mg) and MUCI70∆77 (1.3 mg) were distributed into 50-μL aliquots, flash frozen in liquid nitrogen, and stored at −80°C until use.
Crude and purified protein preparations were separated by SDS-PAGE in the presence or absence of reducing agent (25 mm DTT), and the proteins were visualized by staining of the gels with Coomassie Brilliant Blue.
Biochemical Analyses of His8X-GFP-GAUT11Δ39 and His8X-GFP-MUCI70Δ77
The radioactive GAUT11 α-1,4 GalA transferase (HG:GalAT) activity assays (30 μL) contained GAUT11 (200 nm, 0.55 μg), 50 mm HEPES (pH 7.2), 0.25% (w/v) BSA, 0.25 mm MnCl2, 10 µm of a mixture of HG acceptors with DP of 7 to 23, and 5 μm UDP-[14C]GalA. For time-course assays, an additional 95 μm nonradiolabeled UDP-GalA was added (total of 100 µm UDP-GalA). Reactions were incubated at 30°C and terminated by the addition of 5 μL of 400 mm NaOH. For the GAUT11 HG:GalAT time course, the reactions were carried out from 0 to 240 min and terminated at the designated time points. Product formation was measured using the radioactive filter assay (Sterling et al., 2005).
The sensitivity of HG:GalAT reaction products to Exo PG was measured as follows. HG:GalAT reaction products produced in 1-h, 30-μL reactions were mixed with 3 µL of 1 m sodium acetate buffer, pH 4.2, and 15 µL of 2 m acetic acid. To half of the reaction tubes, 4 units of purified Exo PG was added. The reactions were incubated overnight at 30°C, and 30 µL of 1 m NaOH was added to stop the reaction. The final mixtures were assayed using the radioactive filter assay. Exo PG (EC 3.2.1.67) was purified from Aspergillus tubengensis using previously described methods (Kester et al., 1996), except that a 5-mL HiTrap DEAE FF column was used on the ÄKTA FPLC system (GE Healthcare).
UDP-d-[14C]GalpA was synthesized enzymatically from UDP-d-[14C]GlcpA (PerkinElmer) as described (Liljebjelke et al., 1995; Atmodjo et al., 2011). The HG acceptor mix, enriched for HG oligosaccharides of DP 7 to 23, and the homogenous 13-mer GalA acceptor (GalA13X) were generated by partial digestion of polygalacturonic acid with endopolygalacturonase and purified by HPAEC-PAD as described (Doong and Mohnen, 1998).
The analysis of HG-GalAT reaction products by MALDI-TOF MS was carried out as follows. HG:GalAT reactions (20 μL) containing GAUT11 (1 μg) or MUCI70 (5 μg), 50 mm HEPES (pH 7.2), 0.05% (w/v) BSA, 0.25 mm MnCl2, 100 μm GalA13X-2AB, and 1 mm UDP-GalA were analyzed using a Bruker LT mass spectrometer as described previously (Urbanowicz et al., 2014). Aliquots (1 μL) of the reaction mixture were diluted with 10 μL of water, and 1 μL was spotted on the target plate containing air-dried Nafion 117 solution (Sigma; Jacobs and Dahlman, 2001). The samples were overlaid with 1 μL of matrix solution (20 mg mL−1 2,5-dihydroxbenzoic acid in aqueous 50% [v/v] methanol), and the spot was crystallized under heat. The negative-ion spectra were recorded, and at least 300 laser shots were summed to generate each spectrum.
The GalA13X-2AB acceptor was generated by labeling GalA13X with the fluorescent probe 2-aminobenzamide on the reducing end as described (Ishii, 2002; Urbanowicz et al., 2014). The sample was dialyzed four times against water in 3,500 Mr cutoff tubing (VWR Scientific) and recovered by lyophilization.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Table S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Multiple GAUT genes are expressed in the seed coat.
Supplemental Figure S2. RR staining of mucilage capsules around gaut mutant seeds.
Supplemental Figure S3. Polysaccharide immunolabeling in seed mucilage capsules.
Supplemental Figure S4. Biochemical analyses of GAUT11 and MUCI70.
Supplemental Figure S5. Whole-seed morphology visualized with SEM.
Supplemental Figure S6. Protrusion of columellae from hydrated seeds.
Supplemental Table S1. Mutants examined for mucilage defects.
Supplemental Table S2. Monosaccharide composition of total mucilage extracted with water.
Supplemental Table S3. Primer sequences used for genotyping, RT-qPCR, and cloning.
Supplemental Table S4. ANOVA tables to test if MUCI70 and GAUT11 interact.
Supplemental Table S5. ANOVA tables to test if MUCI70 and IRX14 interact.
Acknowledgments
We thank Dr. Rainer Schwacke (Forschungszentrum Jülich) for helpful advice about the MUCI70 and GAUT11 protein topology and the evolutionary history of DUF616 domains. We also thank Robert Amos for advice on GAUT11 cloning, purification, and optimization of the MALDI and HG:GalAT activity assays and Melani Atmodjo for preparation of the UDP-[14C]GalA substrate. Charles “Graf” Exum is thanked for assistance with the initial cloning of GAUT11. Gerardo Gutierrez-Sanchez is thanked for providing the Exo PG A. tubengensis fungal stock.
Footnotes
The research was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC PGS-D3 to C.V.), Deutsche Forschungsgemeinschaft (US98/13-1) and by the Ministry of Innovation, Science, and Research of North-Rhine Westphalia within the framework of the NRW Strategieprojekt BioSC (No. 313/323‐400‐00213 to M.H.-W.S. and B.U.). The research was also supported by BioEnergy Science Center Grant (DE-PS02-06ER64304) and the Center for Bioenergy Innovation. The BioEnergy Science Center and the Center for Bioenergy Innovation are U.S. Department of Energy Bioenergy Research Centers supported by the Office of Biological and Environmental Research in the Department of Energy’s Office of Science. The research was also partially funded by the Department of Energy Center Grant DE-SC0015662 and U.S. National Institutes of Health grants P41GM103390 and P01GM107012. Generation of the CCRC series of monoclonal antibodies used in this work was supported by a grant from the NSF Plant Genome Program (DBI-0421683).
Articles can be viewed without a subscription.
References
- Albrecht W, Moers J, Hermanns B (2017) HNF: Helmholtz Nano Facility. J Large-Scale Res Facil 3: A112 [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson CT, Carroll A, Akhmetova L, Somerville C (2010) Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152: 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsovski AA, Popma TM, Haughn GW, Carpita NC, McCann MC, Western TL (2009) AtBXL1 encodes a bifunctional beta-D-xylosidase/alpha-L-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiol 150: 1219–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B (2008) QuantPrime: a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA III, Orlando R, Scheller HV, Mohnen D (2011) Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci USA 108: 20225–20230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo MA, Hao Z, Mohnen D (2013) Evolving views of pectin biosynthesis. Annu Rev Plant Biol 64: 747–779 [DOI] [PubMed] [Google Scholar]
- Bacic A. (2006) Breaking an impasse in pectin biosynthesis. Proc Natl Acad Sci USA 103: 5639–5640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110: E435–E444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tov D, Abraham Y, Stav S, Thompson K, Loraine A, Elbaum R, De Souza A, Pauly M, Kieber JJ, Harpaz-Saad S (2015) COBRA-LIKE 2, a member of the glycosylphosphatidylinositol-anchored COBRA-LIKE family, plays a role in cellulose deposition in Arabidopsis seed coat mucilage secretory cells. Plant Physiol 167: 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendzen K, Searle I, Ravenscroft D, Koncz C, Batschauer A, Coupland G, Somssich IE, Ülker B (2005) A rapid and versatile combined DNA/RNA extraction protocol and its application to the analysis of a novel DNA marker set polymorphic between Arabidopsis thaliana ecotypes Col-0 and Landsberg erecta. Plant Methods 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal AK, Atmodjo MA, Li M, Baxter HL, Yoo CG, Pu Y, Lee YC, Mazarei M, Black IM, Zhang JY, et al. (2018) Sugar release and growth of biofuel crops are improved by downregulation of pectin biosynthesis. Nat Biotechnol 36: 249–257 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffall KH, Pattathil S, Phillips SE, Hahn MG, Mohnen D (2009) Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol Plant 2: 1000–1014 [DOI] [PubMed] [Google Scholar]
- Chen LY, Shi DQ, Zhang WJ, Tang ZS, Liu J, Yang WC (2015) The Arabidopsis alkaline ceramidase TOD1 is a key turgor pressure regulator in plant cells. Nat Commun 6: 6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2016) Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J Exp Bot 67: 463–476 [DOI] [PubMed] [Google Scholar]
- Dean GH, Zheng H, Tewari J, Huang J, Young DS, Hwang YT, Western TL, Carpita NC, McCann MC, Mansfield SD, et al. (2007) The Arabidopsis MUM2 gene encodes a beta-galactosidase required for the production of seed coat mucilage with correct hydration properties. Plant Cell 19: 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean G, Cao Y, Xiang D, Provart NJ, Ramsay L, Ahad A, White R, Selvaraj G, Datla R, Haughn G (2011) Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol Plant 4: 1074–1091 [DOI] [PubMed] [Google Scholar]
- De Rybel B, van den Berg W, Lokerse A, Liao CY, van Mourik H, Möller B, Peris CL, Weijers D (2011) A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol 156: 1292–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doong RL, Mohnen D (1998) Solubilization and characterization of a galacturonosyltransferase that synthesizes the pectic polysaccharide homogalacturonan. Plant J 13: 363–374 [Google Scholar]
- Egelund J, Petersen BL, Motawia MS, Damager I, Faik A, Olsen CE, Ishii T, Clausen H, Ulvskov P, Geshi N (2006) Arabidopsis thaliana RGXT1 and RGXT2 encode Golgi-localized (1,3)-α-D-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan-II. Plant Cell 18: 2593–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: carbohydrates. J Vis Exp 37: e1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga D, Meulia T, Fenster S (2008) Real-time PCR. In Gallagher SR, Wiley EA, eds, Current Protocols in Essential Laboratory Techniques. John Wiley & Sons, Hoboken, NJ, pp 1–33 [Google Scholar]
- Gibeaut DM, Carpita NC (1991) Tracing cell wall biogenesis in intact cells and plants: selective turnover and alteration of soluble and cell wall polysaccharides in grasses. Plant Physiol 97: 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Hänsel U, Ziemann M, Pauly M (2009) Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc Natl Acad Sci USA 106: 14699–14704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths JSJ, Šola K, Kushwaha R, Lam P, Tateno M, Young R, Voiniciuc C, Dean G, Mansfield SD, DeBolt S, et al. (2015) Unidirectional movement of cellulose synthase complexes in Arabidopsis seed coat epidermal cells deposit cellulose involved in mucilage extrusion, adherence, and ray formation. Plant Physiol 168: 502–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, et al. (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6: 609–618 [DOI] [PubMed] [Google Scholar]
- Hall BG. (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30: 1229–1235 [DOI] [PubMed] [Google Scholar]
- Hao Z, Mohnen D (2014) A review of xylan and lignin biosynthesis: foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes. Crit Rev Biochem Mol Biol 49: 212–241 [DOI] [PubMed] [Google Scholar]
- Harholt J, Suttangkakul A, Scheller HV (2010) Biosynthesis of pectin. Plant Physiol 153: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn GW, Western TL (2012) Arabidopsis seed coat mucilage is a specialized cell wall that can be used as a model for genetic analysis of plant cell wall structure and function. Front Plant Sci 3: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Li J, Wang X, Zhao X, Yang X, Tang Q, He G, Zhou G, Kong Y (2016a) Xylan synthesized by Irregular Xylem 14 (IRX14) maintains the structure of seed coat mucilage in Arabidopsis. J Exp Bot 67: 1243–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Li J, Yang X, Zhao X, Wang X, Tang Q, He G, Zhou G, Kong Y (2016b) Irregular Xylem 7 (IRX7) is required for anchoring seed coat mucilage in Arabidopsis. Plant Mol Biol 92: 25–38 [DOI] [PubMed] [Google Scholar]
- Ishii T. (2002) A sensitive and rapid bioassay of homogalacturonan synthase using 2-aminobenzamide-labeled oligogalacturonides. Plant Cell Physiol 43: 1386–1389 [DOI] [PubMed] [Google Scholar]
- Jacobs A, Dahlman O (2001) Enhancement of the quality of MALDI mass spectra of highly acidic oligosaccharides by using a nafion-coated probe. Anal Chem 73: 405–410 [DOI] [PubMed] [Google Scholar]
- Jensen JK, Sørensen SO, Harholt J, Geshi N, Sakuragi Y, Møller I, Zandleven J, Bernal AJ, Jensen NB, Sørensen C, et al. (2008) Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell 20: 1289–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kester HC, Kusters-van Someren MA, Müller Y, Visser J (1996) Primary structure and characterization of an exopolygalacturonase from Aspergillus tubingensis. Eur J Biochem 240: 738–746 [DOI] [PubMed] [Google Scholar]
- Kong Y, Zhou G, Abdeen AA, Schafhauser J, Richardson B, Atmodjo MA, Jung J, Wicker L, Mohnen D, Western T, et al. (2013) GALACTURONOSYLTRANSFERASE-LIKE5 is involved in the production of Arabidopsis seed coat mucilage. Plant Physiol 163: 1203–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, Koltunow AM, Parish RW (2009) The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21: 72–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljebjelke K, Adolphson R, Baker K, Doong RL, Mohnen D (1995) Enzymatic synthesis and purification of uridine diphosphate [14C]galacturonic acid: a substrate for pectin biosynthesis. Anal Biochem 225: 296–304 [DOI] [PubMed] [Google Scholar]
- Liwanag AJM, Ebert B, Verhertbruggen Y, Rennie EA, Rautengarten C, Oikawa A, Andersen MCF, Clausen MH, Scheller HV (2012) Pectin biosynthesis: GALS1 in Arabidopsis thaliana is a β-1,4-galactan β-1,4-galactosyltransferase. Plant Cell 24: 5024–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42: D490–D495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Kronenberger J, Marion-Poll A, North HM (2007a) In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol 48: 984–999 [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet MC, Loudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM (2007b) A naturally occurring mutation in an Arabidopsis accession affects a β-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 19: 3990–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell EG, Belshaw NJ, Waldron KW, Morris VJ (2012) Pectin: an emerging new bioactive food polysaccharide. Trends Food Sci Technol 24: 64–73 [Google Scholar]
- Mendu V, Griffiths JS, Persson S, Stork J, Downie AB, Voiniciuc C, Haughn GW, DeBolt S (2011) Subfunctionalization of cellulose synthases in seed coat epidermal cells mediates secondary radial wall synthesis and mucilage attachment. Plant Physiol 157: 441–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen KW, Ramiah A, Stuart M, Steel J, Meng L, Forouhar F, Moniz HA, Gahlay G, Gao Z, Chapla D, et al. (2018) Expression system for structural and functional studies of human glycosylation enzymes. Nat Chem Biol 14: 156–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munarin F, Tanzi MC, Petrini P (2012) Advances in biomedical applications of pectin gels. Int J Biol Macromol 51: 681–689 [DOI] [PubMed] [Google Scholar]
- Mutwil M, Obro J, Willats WGT, Persson S (2008) GeneCAT: novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res 36: W320–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Furuta H, Maeda H, Takao T, Nagamatsu Y (2002) Structural studies by stepwise enzymatic degradation of the main backbone of soybean soluble polysaccharides consisting of galacturonan and rhamnogalacturonan. Biosci Biotechnol Biochem 66: 1301–1313 [DOI] [PubMed] [Google Scholar]
- Nikolovski N, Rubtsov D, Segura MP, Miles GP, Stevens TJ, Dunkley TPJ, Munro S, Lilley KS, Dupree P (2012) Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol 160: 1037–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolovski N, Shliaha PV, Gatto L, Dupree P, Lilley KS (2014) Label-free protein quantification for plant Golgi protein localization and abundance. Plant Physiol 166: 1033–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Okamura Y, Ito S, Tadaka S, Aoki Y, Shirota M, Kinoshita K (2014) ATTED-II in 2014: evaluation of gene coexpression in agriculturally important plants. Plant Cell Physiol 55: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orfila C, Sørensen SO, Harholt J, Geshi N, Crombie H, Truong HN, Reid JSG, Knox JP, Scheller HV (2005) QUASIMODO1 is expressed in vascular tissue of Arabidopsis thaliana inflorescence stems, and affects homogalacturonan and xylan biosynthesis. Planta 222: 613–622 [DOI] [PubMed] [Google Scholar]
- Parsons HT, Christiansen K, Knierim B, Carroll A, Ito J, Batth TS, Smith-Moritz AM, Morrison S, McInerney P, Hadi MZ, et al. (2012) Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol 159: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, et al. (2010) A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol 153: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña MJ, Zhong R, Zhou GK, Richardson EA, O’Neill MA, Darvill AG, York WS, Ye ZH (2007) Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19: 549–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR (2005) Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA 102: 8633–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Caffall KH, Freshour G, Hilley MT, Bauer S, Poindexter P, Hahn MG, Mohnen D, Somerville C (2007) The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19: 237–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettolino FA, Walsh C, Fincher GB, Bacic A (2012) Determining the polysaccharide composition of plant cell walls. Nat Protoc 7: 1590–1607 [DOI] [PubMed] [Google Scholar]
- Ralet MCC Crépeau MJ, Vigouroux J, Tran J, Berger A, Sallé C, Granier F, Botran L, North HM (2016b) Xylans provide the structural driving force for mucilage adhesion to the Arabidopsis seed coat. Plant Physiol 171: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T (2008) A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J 54: 466–480 [DOI] [PubMed] [Google Scholar]
- Ruprecht C, Bartetzko MP, Senf D, Dallabernadina P, Boos I, Andersen MCF, Kotake T, Knox JP, Hahn MG, Clausen MH, et al. (2017) A synthetic glycan microarray enables epitope mapping of plant cell wall glycan-directed antibodies. Plant Physiol 175: 1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling JD, Lemons JA, Forkner IF, Mohnen D (2005) Development of a filter assay for measuring homogalacturonan: α-(1,4)-galacturonosyltransferase activity. Anal Biochem 343: 231–236 [DOI] [PubMed] [Google Scholar]
- Sterling JD, Atmodjo MA, Inwood SE, Kumar Kolli VS, Quigley HF, Hahn MG, Mohnen D (2006) Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci USA 103: 5236–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan C, Hao Z, Zhu X, Avci U, Miller JS, et al. (2013) An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25: 270–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh OK, Moore I (2007) An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448: 493–496 [DOI] [PubMed] [Google Scholar]
- Uehara Y, Tamura S, Maki Y, Yagyu K, Mizoguchi T, Tamiaki H, Imai T, Ishii T, Ohashi T, Fujiyama K, et al. (2017) Biochemical characterization of rhamnosyltransferase involved in biosynthesis of pectic rhamnogalacturonan I in plant cell wall. Biochem Biophys Res Commun 486: 130–136 [DOI] [PubMed] [Google Scholar]
- Urbanowicz BR, Peña MJ, Moniz HA, Moremen KW, York WS (2014) Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J 80: 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Kuschinsky AM, Rosso MG, Eckermann N, Pauly M (2004) RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol 134: 286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C. (2017) Whole-seed immunolabeling of Arabidopsis mucilage polysaccharides. Bio Protoc 7: e2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Günl M (2016) Analysis of monosaccharides in total mucilage extractable from Arabidopsis seeds. Bio Protoc 6: e1801 [Google Scholar]
- Voiniciuc C, Dean GH, Griffiths JS, Kirchsteiger K, Hwang YT, Gillett A, Dow G, Western TL, Estelle M, Haughn GW (2013) Flying saucer1 is a transmembrane RING E3 ubiquitin ligase that regulates the degree of pectin methylesterification in Arabidopsis seed mucilage. Plant Cell 25: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Günl M, Schmidt MHW, Usadel B (2015a) Highly branched xylan made by IRREGULAR XYLEM14 and MUCILAGE-RELATED21 links mucilage to Arabidopsis seeds. Plant Physiol 169: 2481–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Schmidt MHW, Berger A, Yang B, Ebert B, Scheller HV, North HM, Usadel B, Günl M (2015b) MUCILAGE-RELATED10 produces galactoglucomannan that maintains pectin and cellulose architecture in Arabidopsis seed mucilage. Plant Physiol 169: 403–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Yang B, Schmidt MHW, Günl M, Usadel B (2015c) Starting to gel: how Arabidopsis seed coat epidermal cells produce specialized secondary cell walls. Int J Mol Sci 16: 3452–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Zimmermann E, Schmidt MHW, Günl M, Fu L, North HM, Usadel B (2016) Extensive natural variation in Arabidopsis seed mucilage structure. Front Plant Sci 7: 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C, Pauly M, Usadel B (2018) Monitoring polysaccharide dynamics in the plant cell wall. Plant Physiol 176: 2590–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voxeur A, André A, Breton C, Lerouge P (2012) Identification of putative rhamnogalacturonan-II specific glycosyltransferases in Arabidopsis using a combination of bioinformatics approaches. PLoS ONE 7: e51129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Park YB, Cosgrove DJ, Hong M (2015) Cellulose-pectin spatial contacts are inherent to never-dried Arabidopsis primary cell walls: evidence from solid-state nuclear magnetic resonance. Plant Physiol 168: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38: W214–W220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2006) In planta transformation of Arabidopsis. CSH Protoc 2006: pdb.prot4668 [DOI] [PubMed]
- Western TL, Young DS, Dean GH, Tan WL, Samuels AL, Haughn GW (2004) MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol 134: 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RE, McFarlane HE, Hahn MG, Western TL, Haughn GW, Samuels AL (2008) Analysis of the Golgi apparatus in Arabidopsis seed coat cells during polarized secretion of pectin-rich mucilage. Plant Cell 20: 1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Shi D, Li J, Kong Y, Yu Y, Chai G, Hu R, Wang J, Hahn MG, Zhou G (2014) CELLULOSE SYNTHASE-LIKE A2, a glucomannan synthase, is involved in maintaining adherent mucilage structure in Arabidopsis seed. Plant Physiol 164: 1842–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]