GmINS1, a candidate gene underlying a major QTL for nodulation, functions in nodule development and is important for nitrogen fixation in soybean.
Abstract
Nodulation is crucial for biological nitrogen fixation (BNF) in legumes, but the molecular mechanisms underlying BNF have remained elusive. Here, we cloned a candidate gene underlying a major nodulation quantitative trait locus in soybean (Glycine max), INCREASING NODULE SIZE1 (GmINS1). GmINS1 encodes a cell wall β-expansin and is expressed primarily in vascular bundles, along with cortical and parenchyma cells of nodules. Four single-nucleotide polymorphisms distinguishing the two parents were found in the GmINS1 promoter region. Among them, single-nucleotide polymorphism A/C has a significant effect on GmINS1 expression in the parental genotype P2, based on β-glucuronidase activity and promoter deletion analysis. The expression of GmINS1 and the P2 genotype promoter was strongly associated with nodule development, not only in the parents but also in 40 progeny lines and 40 genotypes selected from a soybean core collection. Overexpression of GmINS1 resulted in increases in the number, biomass, infection cell abundance, and nitrogenase activity of large nodules and subsequently changed the nitrogen content and biomass of soybean plants. GmINS1 suppression via RNA interference had the opposite effect. Double suppression of GmEXPB2 and GmINS1 dramatically inhibited soybean nodulation. Our results reveal that GmINS1 is a critical gene in nodule development and that GmEXPB2 and GmINS1 synergistically control nodulation in soybean. Our findings shed light on the genetic basis of soybean nodulation and provide a candidate gene for optimizing BNF capacity through molecular breeding in soybean.
Biological nitrogen fixation (BNF) proceeding from symbiotic associations between legumes and rhizobia is a prominent natural source of nitrogen (N) in agroecosystems (Herridge et al., 2008). In legumes, BNF occurs in nodules, highly specialized organs harboring bacterial symbionts, and provides enormous amounts of fixed nitrogen (N2) to both domesticated crops and wild plant species (Graham and Vance, 2003; Herridge et al., 2008; Köpke and Nemecek, 2010; Qin et al., 2012). Due to the extensive area of land cultivated with soybean (Glycine max) as well as its high BNF capacity, this crop not only plays important roles in providing proteins and oils for humans and animals but also serves as a central player in sustainable agriculture (Alves et al., 2003; Herridge et al., 2008). The capacity for BNF, which varies among soybean genotypes, is a complex trait controlled by multiple quantitative trait loci (QTLs; Okereke and Unaegbu, 1992; Nicolás et al., 2006; Santos et al., 2013; Yang et al., 2017). To date, the genetic and molecular mechanisms underlying BNF remain largely unknown.
Nodulation is a multistep process, starting with a molecular dialogue between host plants and bacterial partners followed by a cascade of responses, including root hair curling, bacterial colonization, infection thread (IT) differentiation, and nodule development (Oldroyd and Downie, 2008; Oldroyd et al., 2011). The process of nodulation is tightly regulated in host roots through coordinated gene networks (Kouchi et al., 2010). In past decades, legume nodulation-deficient mutants have yielded insight into the molecular bases underlying the recognition of rhizobial infection signals, early symbiosis signaling pathways, and IT formation (Limpens and Bisseling, 2003; Oldroyd and Downie, 2004; Kouchi et al., 2010; Murray, 2011). Nodule development from the initiation of nodule primordia through maturation and the formation of bacteroids is essential for N2 fixation (Oke and Long, 1999). Although several genes, small peptides, and noncoding RNAs involved in nodule development in different legume species have been characterized (Boualem et al., 2008; Imin et al., 2013; Djordjevic et al., 2015; Mohd-Radzman et al., 2016; Nanjareddy et al., 2016; Cai et al., 2017; Di Giacomo et al., 2017; Hobecker et al., 2017), the regulatory elements of nodule development still need to be explored further.
Legumes have two major types of nodules with distinctive morphology, namely indeterminate and determinate nodules (Lauridsen et al., 1993). For indeterminate nodules, such as in Medicago truncatula, pea (Pisum sativum), and alfalfa (Medicago sativa), the initiation of cell division occurs first in the root pericycle followed by inner cortical cells, which leads to the formation of nodule primordia. A persistent meristem in each primordium produces a meristem zone (I), infection zone (II), interzone (II-III), fixation zone (III), and senescence zone (IV). In contrast, determinate nodules, as in soybean and peanut (Arachis hypogaea), are derived from cell divisions in the outer root cortex, where meristematic activity is lost very early in nodule development (Crespi and Gálvez, 2000; Popp and Ott, 2011). Therefore, cell expansion and cell wall extension might play crucial roles in the organogenesis of determinate nodules.
A class of cell wall-loosening proteins known as expansins was identified previously in a variety of plant species (Taiz, 1994; Cosgrove, 1998; Cosgrove et al., 2002). Expansins are critical for the pH-dependent extension of cell walls known as acid growth, which acts through the loosening of cell wall matrix polymers or the weakening of noncovalent binding between cell wall polysaccharides (McQueen-Mason et al., 1992; McQueen-Mason and Cosgrove, 1994; Cosgrove, 2000). To date, several expansin genes have been associated with nodulation. For example, the expression of a sweet clover (Melilotus alba) α-expansin gene, MaEXP1, is enhanced within hours of rhizobial infection, and MaEXP1 also is expressed in the nodule cortex, meristem, invasion zone, and interzone II-III (Giordano and Hirsch, 2004). Western-blot analysis revealed that an expansin-like protein, PsEXP1, is localized in IT walls of pea nodules (Sujkowska et al., 2007), suggesting that PsEXP1 might function in IT growth. More recently, a β-expansin gene, GmEXPB2, was found to be critical for soybean nodulation (Li et al., 2015). Overexpression of GmEXPB2 enhances infection events and increases nodule numbers, nodule mass, and nitrogenase activity, thus elevating plant N and phosphorus content as well as biomass. Expression analysis has demonstrated that most of the soybean β-expansin gene family members are expressed in nodules (Li et al., 2014), suggesting that differentially expressed β-expansins might fulfill special functions during soybean nodule formation and development. However, no soybean β-expansin other than GmEXPB2 has been analyzed functionally in nodulation.
Quantitative trait locus (QTL) analysis is a powerful approach for understanding genetic variation and mechanisms of host participation in symbiotic interactions. QTLs related to BNF have been identified in several legumes, including pea (Bourion et al., 2010), Lotus japonicas (Tominaga et al., 2012), and common bean (Phaseolus vulgaris; Tsai et al., 1998; de Souza et al., 2000). At present, few QTLs controlling nodulation traits in soybean have been described. Santos et al. (2013) identified three QTLs for nodule number and one for individual nodule weight in a population of 157 F2:7-derived lines using multiple trait composite interval mapping. Hwang et al. (2014) detected five QTLs for total nodule weight and seven for nodule size from a dense and complete linkage map in KS4895×Jackson recombinant inbred lines (RILs) tested in field experiments. Despite these recent advances in QTL analysis of BNF-associated traits in soybean, the molecular mechanisms underlying significant QTLs for BNF are still poorly understood, because the genes responsible for the QTLs have not yet been identified via map-based cloning. To date, only the genetic mechanism regulating the symbiosis specificity between soybean and rhizobia has been documented, with a class of plant resistance (R) genes being identified as the key candidates for controlling the specificity of nodulation in soybean (Tang et al., 2014, 2016).
Previously, we identified several new QTLs for BNF using 175 F9:11 RILs derived from a P1×P2 cross in soybean (Yang et al., 2017). However, none of the related genes were cloned and characterized. In this study, a gene potentially accountable for variation at one QTL associated with both nodule number and nodule weight was identified as INCREASING NODULE SIZE1 (GmINS1), an ortholog of GmEXPB2. Five single-nucleotide polymorphisms (SNPs) were detected between the two parental genotypes, and the transcription of GmINS1 was strongly associated with the number of large nodules and individual nodule size in the field. Altering GmINS1 expression significantly changed the size and mass of nodules, the number of infection cells, and nitrogenase activity, subsequently affecting soybean N content and biomass. These results indicate that GmINS1 plays an important role in the development of soybean nodules. This information might be valuable for the marker-assisted selection of elite soybean varieties that optimize BNF capacity and yield.
RESULTS
GmINS1 Is the Candidate Gene in QTL qBNF-11 Affecting Soybean Nodulation in the Field
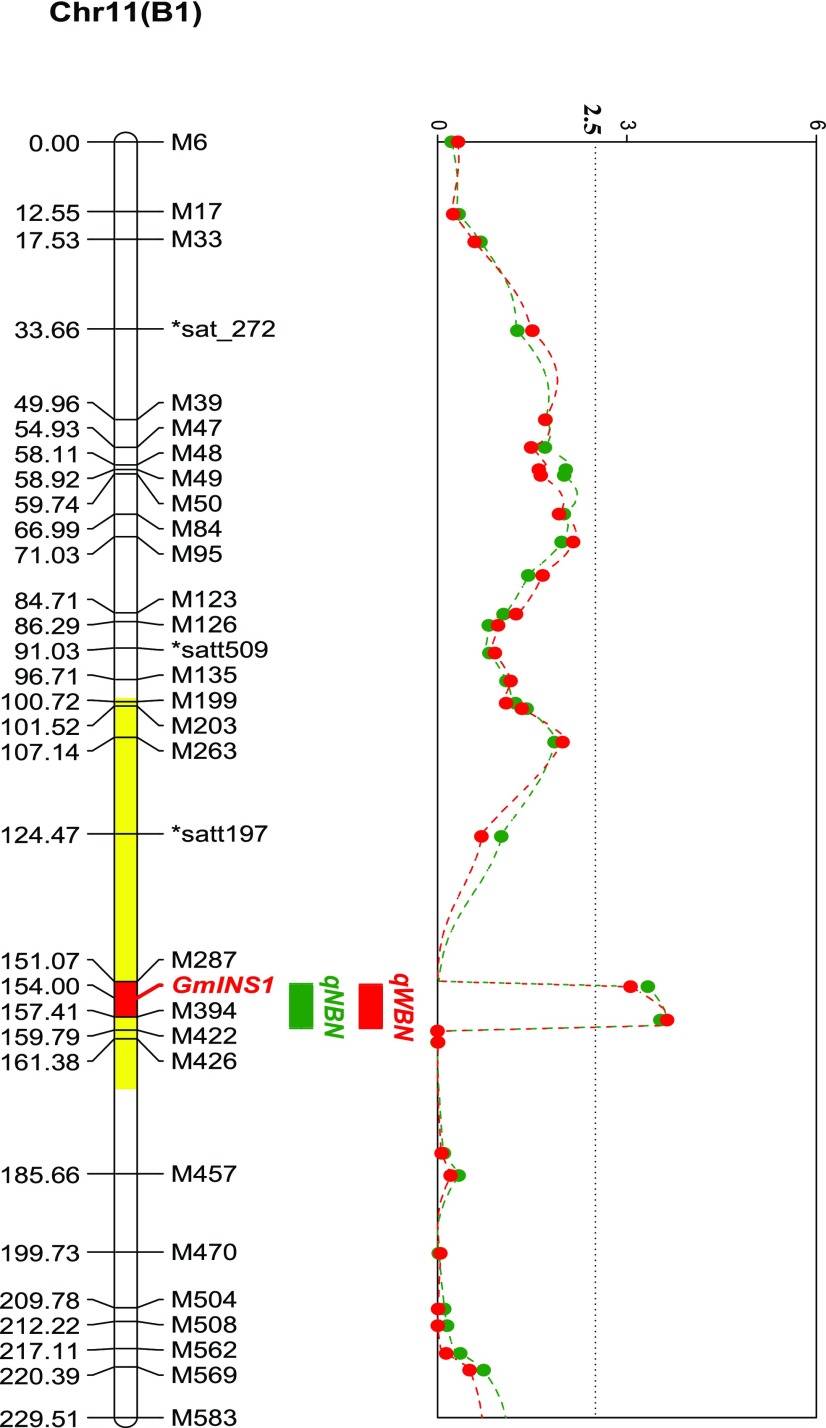
A major QTL, qBNF-11, for the number and weight of large nodules was identified previously (Yang et al., 2017). However, only three simple sequence repeat markers cover this linkage group. To narrow the target gene location, a high-resolution genetic map using 27 derived cleaved-amplified polymorphic sequence (dCAPs) markers for linkage group B1 was constructed (Supplemental Table S1). This genetic map covered 248.6 centimorgan (cM) with an average density of 7.31 cM per marker. As expected, qBNF-11 also was detected on the new map with a log of the odds value of 3.67, explaining 9.65% of the variation for the number of large nodules. Importantly, qBNF-11 was localized to a 6.3-cM genetic region, with the target gene possibly located between markers M287 and M394, in an approximately 3,000-kb physical region based on the Wm82.a1.v1 genome (http://www.phytozome.net ; Fig. 1).
Figure 1.
Colocation analysis of the putative QTLs for nodulation traits and GmINS1. Genetic localization analysis of GmINS1 was performed by using 27 dCAPs and three simple sequence repeat markers. The area highlighted in yellow indicates a QTL for nodule size as identified by Hwang et al. (2014). Nodulation traits, including the number of large nodules (NBN; green line) and the weight of large nodules (WBN; red line), were used in QTL detection. The dotted line represents the threshold value of 2.5-log of the odds score.
Interestingly, in a separate field trial, Hwang et al. (2014) also identified a unique QTL for nodule size that colocated with qBNF-11 (Fig. 1). This region is located on the chromosome 11 scaffold sequence. To determine the gene conferring qBNF-11 regulation of nodulation, all polymorphic genes in this region were identified. Among them, Glyma11g17160 (update to Glyma.U014500 in Wm82.a2.v1), a homolog of GmEXPB2 encoding a cell wall β-expansin, was predicted to be possibly involved in soybean nodule organogenesis. Glyma.U014500 and GmEXPB2 exhibit 78% amino acid sequence similarity and were classified into the same subgroup in phylogenetic analysis (Supplemental Fig. S1). Given that determinate nodule growth largely relies on cell expansion, and considering that GmEXPB2 appears to function in nodule formation and development, Glyma.U014500 was selected as a candidate gene for qBNF-11 QTL regulation of nodulation. Since qBNF-11 controls the number and weight of large nodules, Glyma.U014500 might function in increasing nodule size in soybean and, thus, was named GmINS1.
GmINS1 Protein Is Located in the Cell Wall
The full-length open reading frame of GmINS1 is 834 bp long and encodes a protein of 277 amino acids, with a calculated mass of 29.37 kD and a pI of 4.95. Using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/), it was predicted that GmINS1 has a signal peptide for secretion into the cell wall. To verify the subcellular localization of GmINS1, tobacco (Nicotiana tabacum) leaves were infiltrated with Agrobacterium tumefaciens harboring a translational fusion of GmINS1 and GFP cDNA as well as a plasma membrane marker, all under the control of a cauliflower mosaic virus 35S (35S) promoter. When expressing GFP alone, the signal was distributed in the nucleus and cytosol (Supplemental Fig. S2). In contrast, the GmINS1-GFP fusion was localized exclusively at the periphery of the cell. To check whether GmINS1 was located on the cell wall or plasma membrane, transfected tobacco cells were plasmolyzed in 10% (w/v) NaCl for 5 min. After cell plasmolysis, GmINS1-GFP localized to the cell wall but not the plasma membrane, which was readily distinguishable from GFP control fluorescence (Supplemental Fig. S2, white arrow). These results indicate that GmINS1 is a cell wall protein.
GmINS1 Is Expressed Preferentially in the P2 Parental Genotype, Which Possesses Higher BNF Capacity in Hydroponics
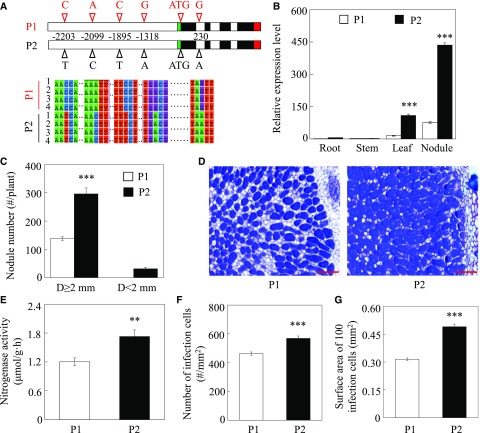
Qualitative PCR analysis revealed four SNPs within the promoter region of GmINS1: G/A, C/T, A/C, and C/T at −1,318, −1,895, −2,099, and −2,203 bp upstream of the start codon, respectively (Fig. 2A). According to PLACE and PlantCARE analysis (Higo et al., 1999; Lescot et al., 2002), the four SNPs are located within predicted cis-elements including Arabidopsis (Arabidopsis thaliana) response regulators (ARR1; NGATT), activation sequence-1 (ASF-1; TGACG), an ACGT sequence required for etiolation-induced expression of ERD1 (early responsive to dehydration) in Arabidopsis, a GA-responsive element (GARE; TAACAAR), and a WRKY domain defined by the conserved amino acid sequence WRKYGOK at its N-terminal end, (WRKY71OS; TGAC) in the P2 genotype (Supplemental Table S2). One additional SNP was located in the first intron, namely G/A for P1 and P2, respectively (Fig. 2A). These SNPs suggest that the variation of GmINS1 effects on nodule growth and development among the observed RILs might be related to differences in the regulation of its expression.
Figure 2.
GmINS1 expression and variation in nodule development between parental lines. A, SNPs of GmINS1 between parental lines. Four SNPs were found in the promoter region and one was located in the first intron of GmINS1. The green, red, and black boxes represent 3′ untranslated region, 5′ untranslated region, and exon sequences of GmINS1, respectively. B, GmINS1 expression in various plant tissues. C, Nodule number. Nodules were classified into two groups according to their diameter (D): large (D ≥ 2 mm) and small (D ˂ 2 mm) nodule groups. D, Toluidine Blue-stained nodule cross sections of P1 and P2. Bars = 200 µm. E, Nitrogenase activity. F, Number of infection cells. G, Surface area of 100 infection cells. Soybean seedlings inoculated with rhizobia were grown in low-N nutrient solution for 45 d. Each bar represents the mean of four biological replicates with se. Ten nodules were selected from each replicate for infection cell analysis (F and G). Asterisks represent significant differences between P1 and P2 for the same trait in Student’s t tests (**, 0.001 < P ≤ 0.01 and ***, P ≤ 0.001).
Since four SNPs were found in the promoter region of GmINS1 between parents P1 and P2, the expression of GmINS1 was examined in different organs using quantitative reverse transcription PCR (RT-qPCR). GmINS1 was expressed primarily in nodules and leaves, where transcription levels in P2 were 7.46 and 5.79 times higher than in P1, respectively (Fig. 2B). Meanwhile, almost no GmINS1 expression was detected in roots or stems of either parent.
To further characterize nodulation and the BNF capacities of the two parental genotypes, P1 and P2 were inoculated with rhizobia in hydroponics for 45 d. At this time, P2 had 115% more large nodules (diameter, D ≥ 2 mm; large nodule group) than P1 (Fig. 2C), along with a 65.6% higher nitrogenase activity (Fig. 2E). Furthermore, cross sections of large nodules revealed that those in P2 contained more and larger infection cells (Fig. 2D). The number of infection cells and the surface area of 100 infection cells were 22.8% and 55.7% higher in P2 than in P1 large nodules, respectively (Fig. 2, F and G). Taken together, these data demonstrate that the two parental genotypes differ significantly in nodulation and BNF capacity, with P2 outperforming P1 in both.
Deletion and Histochemical Analysis of the GmINS1 Promoter
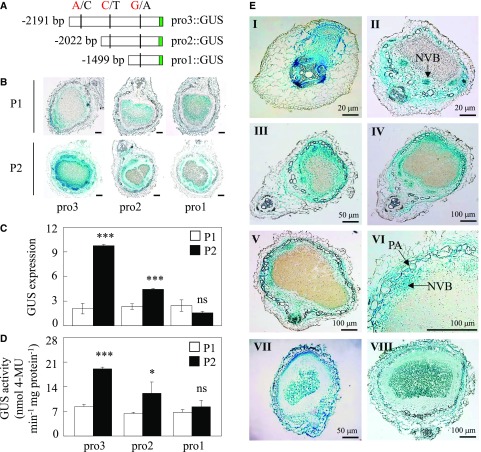
To investigate which SNP in the promoter region of GmINS1 is critical for its gene expression, a series of 5′ end promoter deletions were fused to the GUS reporter gene and then transferred to both tobacco leaves and hairy roots of transgenic composite soybean plants to analyze its transient expression. The deletion promoters were constructed for both parental genotypes and named pro3 (−2,191 to −1), pro2 (−2,022 to −1), and pro1 (−1,499 to −1), carrying three, two, and one SNPs, respectively (Fig. 3A). Although all three truncated GmINS1 promoters were able to drive GUS gene expression, transgenic nodules and tobacco leaves harboring pro3::GUS and pro2::GUS amplified from the parent P2 exhibited higher GUS activity than the P1 genotype (Fig. 3B; Supplemental Fig. S3). The expression level of GUS and its protein activity mediated by pro3 and pro2 from P2 were 4.69- and 2.28-fold and 1.89- and 1.9-fold higher in nodules and 1.49- and 2.19-fold and 1.46- and 1.78-fold higher in transient transgenic tobacco, compared with the same fragments from P1 (Fig. 3, C and D; Supplemental Fig. S3, B and C). Furthermore, the pro3 promoter from P2 displayed about 1.57- and 1.13-fold more GUS activity than pro2 in transgenic nodules and tobacco, respectively. These results suggest that the pro3 promoter region between −2,191 and −1 bp, which included the SNP A/C, was critical for the high transcript levels of GmINS1 observed in parent P2.
Figure 3.
Deletion analysis of the GmINS1 promoter and histochemical detection of GUS expression in soybean transgenic nodules. A, Schematic outlines showing the truncated GmINS1 promoters (pro1::GUS to pro3::GUS) harboring different SNPs from P1 (A/C/G) and P2 (C/T/A) parents. B, GUS staining of the transgenic nodules. Bars = 100 µm. C, Relative expression of the GUS gene. D, Quantitative GUS activity analysis of the transgenic nodules by fluorimetric assay. Each bar represents the mean of six biological replicates with se. Asterisks represent significant differences between promoters from P1 and P2 for the same trait in Student’s t tests (*, 0.01 < P ≤ 0.05 and ***, P ≤ 0.001). ns, Not significant at the 0.05 value. 4-MU, 4-Methylumbelliferyl-β-d-glucuronide. E, Histochemical localization analysis of GmINS1 in cross sections of transgenic soybean nodules at different developmental stages. Soybean transgenic composite plants harboring a 2,323-bp fragment of the GmINS1 promoter (proGmINS1::GUS) PCR amplified from P2 inoculated with rhizobia were grown in sand culture irrigated with low-N nutrient solution for 4 d (I), 7 d (II), 14 d (III), 21 d (IV), and 30 d (V). VI, Magnified image from IV. NVB, Nodule vascular bundle; PA, parenchymatous cell. VII and VIII, Transgenic nodules expressing 35S::GUS were analyzed at 7 d (VII) and 30 d (VIII) after rhizobia inoculation as a control. Bars = 20 µm (I and II), 50 µm (III and VII), and 100 µm (all other images).
To more precisely determine the tissue localization of GmINS1 transcripts, transgenic composite plants carrying a 2,323-bp fragment of the GmINS1 promoter amplified from P2 (proGmINS1::GUS) or 35S::GUS as the control were inoculated with rhizobia, and nodules were harvested at various growth stages for GUS staining. Unlike 35S::GUS expression, which was detected in all nodule tissues (Fig. 3E, VII and VIII), proGmINS1::GUS expression started upon the initiation of nodule primordia (Fig. 3E, I), which was followed by expression in NVB tissues during early nodule development (Fig. 3E, II) and then extended to the nodule cortex and PA in elder nodules (Fig. 3E, III–V). The fact that GmINS1 expression was restricted to the mature NVB and, to a lesser extent, PA (Fig. 3E, VI) suggests that GmINS1 is closely involved in soybean nodule development.
GmINS1 Expression Is Positively Associated with Soybean Nodule Development in the Field
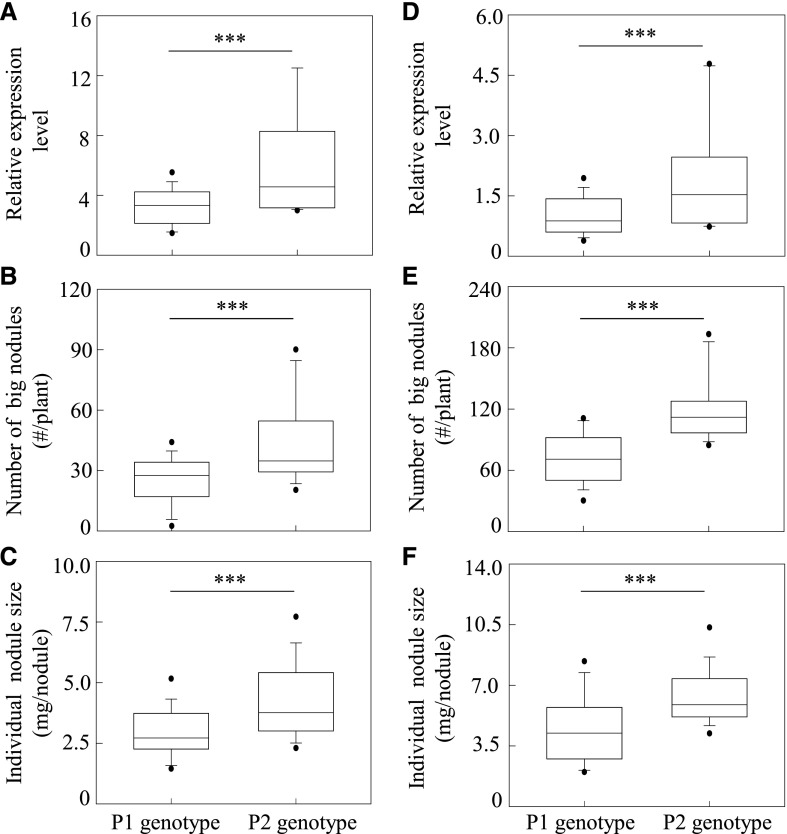
Since the GmINS1 promoter carrying SNP A/C from the parental genotype P2 mediated higher GUS activity than the other fragments, a dCAPs marker was developed based on the different base pair A/C in the promoter region of GmINS1 to examine the relationship between GmINS1 expression and nodule growth (Supplemental Table S1). Forty F9:11 RILs were screened and divided into two groups based on the dCAPs marker. Among these RILs, the expression of GmINS1 was significantly associated with the number of large nodules as well as individual nodule size (Fig. 4, A–C). The 20 RILs with the parent P2 genotype had higher expression of GmINS1, accompanied by a higher number of large nodules than the 20 RILs with the parent P1 genotype.
Figure 4.
Relationship between GmINS1 expression and nodule development in RILs and in genotypes from a soybean core collection. A and D, Expression levels of GmINS1. B and E, Number of large nodules. C and F, Individual nodule size. Forty individual progeny from an F9:11 RIL population (A–C) and a soybean core collection (D–F) were selected and divided into two groups based on the SNP A/C in the promoter region of GmINS1. Black horizontal lines within the boxes represent median values. The lower and upper edges, and bars above or below the boxes represented 25th, 75th, 5th and 95th percentiles of all data, respectively. Asterisks indicate the significance of differences between two groups in Student’s t tests (***, P ≤ 0.001).
To investigate whether the SNP A/C in the promoter region of GmINS1 also displayed a strong association with the expression of GmINS1 and nodulation in other soybean varieties, we further screened 40 genotypes from a soybean core collection (Zhao et al., 2004) and grouped them into the P1 and P2 genotypes based on the dCAPs marker. The 20 genotypes with P2 had higher transcript levels of GmINS1 and also exhibited more and larger nodules than the 20 genotypes with P1 (Fig. 4, D and E), suggesting that GmINS1 transcription at least partially explains the phenotypic differences in soybean nodule development observed in the field.
Alteration of GmINS1 Expression Influences Nodulation in Transgenic Soybean Roots
To investigate the physiological roles of GmINS1 in soybean nodulation, the effects of GmINS1 overexpression and RNA interference (OX and Ri lines) on soybean growth and nodulation were evaluated using transgenic composite plants at 30 d after inoculation (dai). After checking the quality of gene transformation in transgenic hairy roots through qualitative PCR (Supplemental Fig. S4A), single hairy root plants expressing the target gene were selected. Further RT-qPCR analysis showed that the transcription of GmINS1 was 2.88 times higher in OX lines and 8.24 times lower in Ri lines than in nodules transformed with the empty vector (Ev), but the expression of GmEXPB2 did not show significant differences among the GmINS1 transgenic nodules (Supplemental Fig. S4, B and C). Subsequent investigations were conducted to determine whether changes in GmINS1 expression could affect IT formation upon inoculation with GFP-labeled rhizobium strain USDA110. It was found that ITs formed in root hairs at 3 dai (Supplemental Fig. S5A). Suppression of GmINS1 decreased IT numbers by 17.4%, while no significant differences were found in OX lines compared with Ev plants (Supplemental Fig. S5B). These observations imply that GmINS1 might be more involved in nodule growth and development than in rhizobium infection.
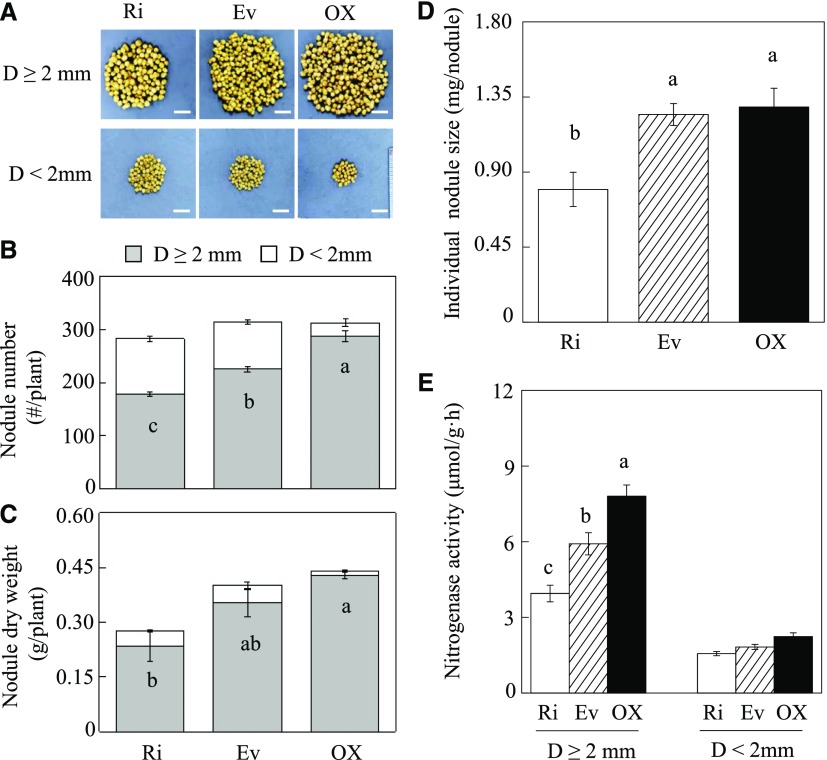
The effect of GmINS1 expression on nodule organogenesis was assessed in soybean transgenic composite plants at 30 dai. To more clearly characterize how GmINS1 affects nodule growth, nodules diameters were separated into large (D ≥ 2 mm) and small (D < 2 mm). Overexpression of GmINS1 facilitated nodule expansion, as reflected by an increase in the number of large nodules, while suppression of GmINS1 led to more numerous and smaller nodules when compared with Ev plants (Fig. 5A). In OX lines, 91.9% of the nodules belonged to the large nodule group, while only 63.2% and 71.9% of the nodules on Ri and Ev roots, respectively, were large (Fig. 5B). The number and dry weight of large nodules were 27.6% and 20.9% higher on OX roots and 20.7% and 33.4% lower on Ri roots, respectively, than on Ev roots (Fig. 5, B and C). Furthermore, in looking at individual nodules, the suppression of GmINS1 significantly inhibited nodule development. The individual nodule size of transgenic Ri lines was 36.1% lower than that of control nodules, while nodule size was not affected in OX lines relative to those on control roots (Fig. 5D). Moreover, altering the expression of GmINS1 significantly influenced nitrogenase activity in large nodules, as indicated by a 31.9% increase in OX lines and a 33.3% decrease in Ri lines relative to control lines. On the other hand, no significant differences in nodule nitrogenase activity were observed in the small nodule group among transgenic lines (Fig. 5E).
Figure 5.
Effects of RNA interference or overexpression of GmINS1 on soybean nodulation. A, Phenotypes of nodules. Bars = 1 cm. B, Nodule number. C, Nodule dry weight. D, Individual nodule size. E, Nitrogenase activity. Soybean transgenic composite plants inoculated with rhizobia were grown in sand culture under low-N conditions for 30 d. Nodules were classified into two groups according to their diameter (D): large (D ≥ 2 mm) and small (D ˂ 2 mm). Each bar represents the mean of four biological replicates with se. Different letters indicate significant differences between Ri or OX lines and Ev control plants for the same trait in a two-way ANOVA test (P < 0.05).
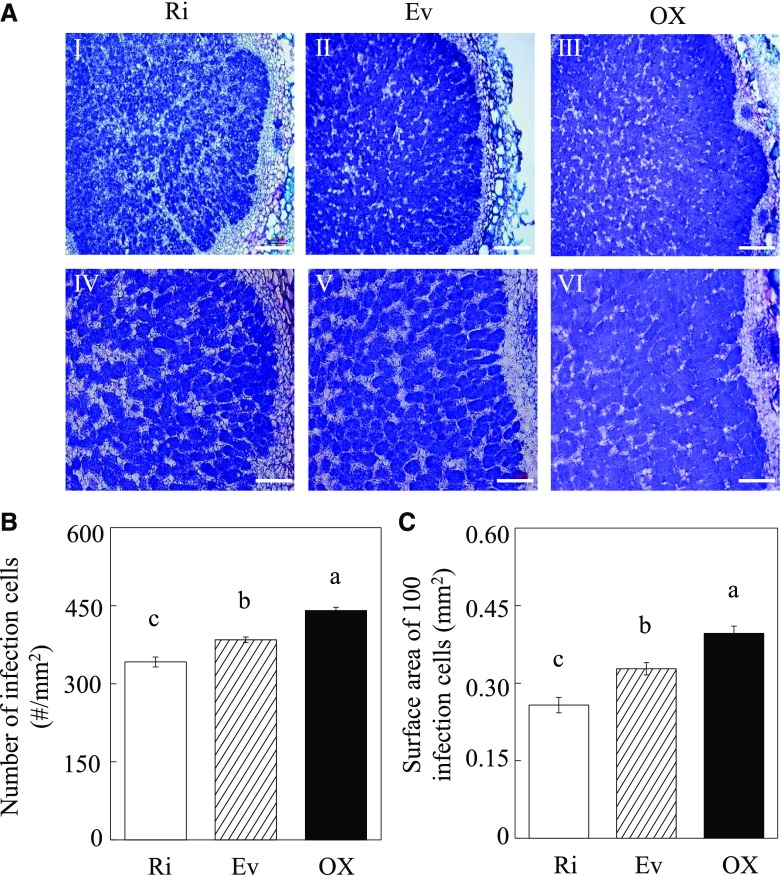
Imaging of sectioned nodules revealed that GmINS1 overexpression significantly affected nodule morphological development (Fig. 6A). In comparison with control plants, the number of infection cells increased by 14.7% in OX lines and decreased by 12.4% in Ri lines (Fig. 6B), and the surface area of 100 infection cells increased by 20.9% in OX lines and decreased by 21.5% in Ri lines (Fig. 6C).
Figure 6.
Effects of RNA interference and overexpression of GmINS1 on infection cell development in nodules. A, Toluidine Blue-stained nodule cross sections of Ri and OX lines of GmINS1. IV to VI are magnified images of the respective images I to III showing the infection zone. Bars = 200 µm (I–III) and 100 µm (IV–VI). B, Number of infection cells. C, Surface area of 100 infection cells. Soybean transgenic composite plants inoculated with rhizobia were grown in sand culture irrigated with low-N nutrient solution for 30 d. Each bar represents the mean of five biological replicates with se. Five nodules were selected and measured from each replicate. Different letters indicate significant differences between Ri or OX lines and Ev control plants for the same trait in a two-way ANOVA test (P < 0.05).
Soybean growth was enhanced significantly in OX lines and inhibited in Ri lines compared with Ev plants at 30 dai. Overexpression of GmINS1 led to 26.5% and 29.9% increases in fresh weight and N content, respectively, relative to controls (Fig. 7). In contrast, suppression of GmINS1 resulted in 23.6% and 30.1% decreases in plant fresh weight and N content, respectively (Supplemental Fig. S6, A and B). Taken together, these results suggest that the expression of GmINS1 in soybean nodules directly affects nodule growth and development and, subsequently, influences plant growth and N nutrition.
Figure 7.
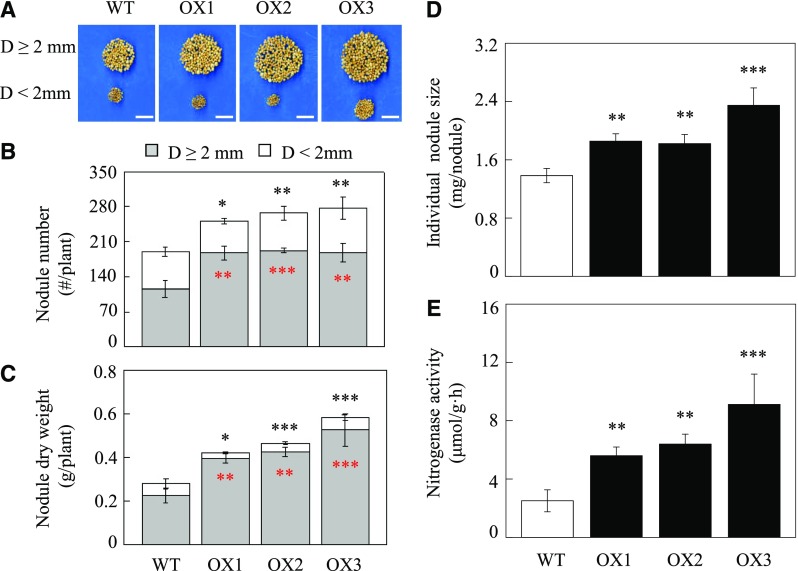
Effects of overexpressing GmINS1 on the nodulation of soybean stable transgenic lines. A, Phenotypes of nodules. Bars = 2 cm. B, Nodule number. Nodules were classified into two groups according to their diameter (D): large (D ≥ 2 mm) and small (D ˂ 2 mm) nodule groups. C, Nodule dry weight. D, Individual nodule size. E, Nitrogenase activity. OX1 to OX3, Independent soybean stable transgenic lines overexpressing GmINS1; WT, wild-type plants. Soybean plants inoculated with rhizobia were grown in low-N nutrient solution for 40 d. Each bar represents the mean of five replicates with se. Asterisks represent significant differences between OX lines and wild-type plants for the same trait in Student’s t tests (*, 0.01 < P ≤ 0.05; **, 0.001 < P ≤ 0.01; and ***, P ≤ 0.001).
Promotion of Nodulation and Growth in GmINS1-Overexpressing Stable Transgenic Soybean Plants
To further evaluate the effects of overexpressing GmINS1 on nodulation, plant growth, and N content at the whole plant level, three T2 OX lines of GmINS1 and wild-type plants were inoculated with rhizobia, BXYD3, under low-N solution conditions for 40 d. The transgenic plants were checked for the presence of the bar gene through qualitative PCR, then the mRNA accumulation of GmINS1 in both leaves and nodules was monitored using RT-qPCR (Supplemental Fig. S7). The transcript levels of GmINS1 in leaves and nodules of three OE transgenic lines were 48.6, 565.2, and 997.9 times, and 145.4, 75.6, and 96.1 times, higher than in wild-type plants, respectively (Supplemental Fig. S7, B and C). Overexpression of GmINS1 significantly promoted soybean nodulation and produced more numerous and large nodules (D ≥ 2 mm) when compared with wild-type plants (Fig. 7A). Relative to the wild type, the three GmINS1 OX lines showed increases of 62.5%, 62.6%, and 66.9% in the number of large nodules and increases of 74.7%, 87.7%, and 131.4% in nodule dry weight, respectively (Fig. 7, B and C). The percentage of large nodules was calculated as the ratio of the large nodule group to the total number of nodules. Overexpression of GmINS1 resulted in 67.7%, 72%, and 74.7% large nodules, while wild-type plants showed 60.7% large nodules. Consistently, the individual nodule size increased by 34.1%, 31.7%, and 69.6% in OX1 to OX3 lines; therefore, the three OX lines significantly promoted BNF capacity, as indicated by 123.7%, 155.4%, and 163.4% increases in nitrogenase activity compared with wild-type nodules, respectively (Fig. 7, D and E). These results further illustrate that overexpression of GmINS1 facilitates nodule development as well as N2 fixation in soybean.
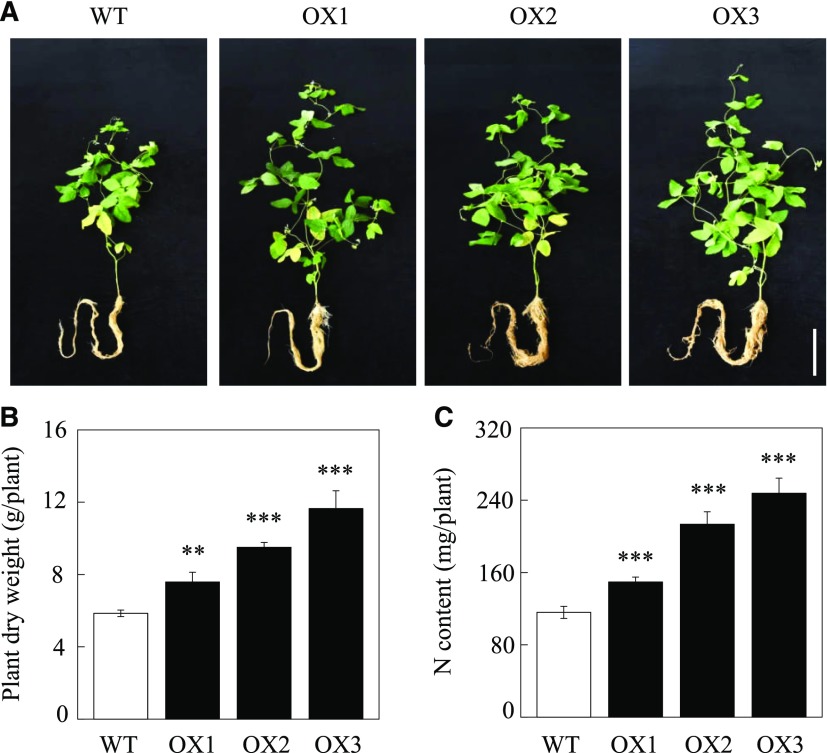
Moreover, overexpression of GmINS1 in soybean significantly promoted plant growth and N content after rhizobia inoculation for 40 d. Photographs show that the three OX lines grew much better than wild-type plants (Fig. 8A), and compared with wild-type lines, overexpressing GmINS1 led to 29.5%, 62.3%, and 98.8% increases in plant dry weight and 29%, 83.9%, and 113.4% increases in N content, respectively (Fig. 8, B and C). Taken together, these results suggest that overexpression of GmINS1 in soybean can boost soybean growth and N efficiency through the promotion of nodule growth and development.
Figure 8.
Plant growth and N content as affected by overexpressing GmINS1 in soybean stable transgenic lines. A, Photographs showing soybean growth performance. Bar = 20 cm. B, Plant dry weight. C, N content. OX1 to OX3, Independent soybean stable transgenic lines overexpressing GmINS1; WT, wild-type plants. Soybean plants inoculated with rhizobia were grown in low-N nutrient solution for 40 d. Each bar represents the mean of five replicates with se. Asterisks represent significant differences between OX lines and wild-type plants for the same trait in Student’s t tests (**, 0.001 < P ≤ 0.01 and ***, P ≤ 0.001).
Double Suppression of GmINS1 and GmEXPB2 Severely Restricts Soybean Nodulation
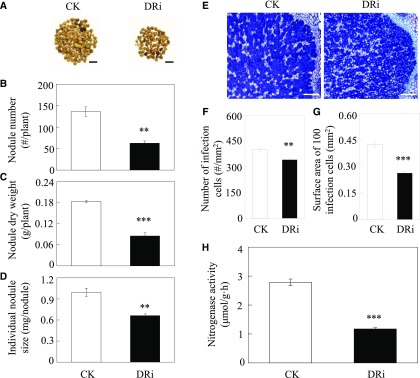
A homolog of GmINS1, GmEXPB2, was reported previously to be involved in both nodule formation and development, and they share 78% identity in the coding sequence (Li et al., 2015). To dissect the role of cell wall expansins, including GmINS1 and GmEXPB2, in the organogenesis of determinate nodules, we generated soybean transgenic composite plants with double RNA interference of both GmINS1 and GmEXPB2 (DRi). The quality of transformation in transgenic hairy roots was assessed through qualitative PCR (Supplemental Fig. S8A). RT-qRCR analysis showed that the expression of GmINS1 and GmEXPB2 in the double suppression lines was reduced by 97.4% and 77.9%, respectively, in comparison with the expression in control lines (Supplemental Fig. S8B). Cosuppression of GmINS1 and GmEXPB2 significantly inhibited nodule development (Fig. 9A). The number of large nodules, nodule dry weight, and individual nodule size decreased by 54%, 53.5%, and 33.4%, respectively, compared with control values (Fig. 9, B–D). Furthermore, similar effects were observed in the formation of infection cells in nodules (Fig. 9E), with 14.7% fewer infection cells and a 38.2% reduction in surface area of 100 infection cells observed in DRi nodules relative to control values (Fig. 9, F and G). As a result, double suppression of GmINS1 and GmEXPB2 significantly restricted N2 fixation efficiency as well as soybean growth and N content (Supplemental Fig. S9). The nitrogenase activity, soybean biomass, and N content decreased by 57.9%, 46.4%, and 48.1%, respectively, in DRi lines compared with control plants (Fig. 9H; Supplemental Fig. S9, B and C). These results indicate that the expression of GmINS1 and GmEXPB2 is associated with determinate nodule formation and expansion and, therefore, is crucial for soybean growth and N nutrition.
Figure 9.
Effects of DRi on the nodulation of soybean transgenic composite plants. A, Phenotypes of control and DRi nodules. Bars = 1 cm. B, Nodule number. C, Nodule dry weight. D, Individual nodule size. E, Growth performance of infection cells. Bars = 200 µm. F, Number of infection cells. G, Surface area of 100 infection cells. H, Nitrogenase activity. CK, Soybean transgenic plants harboring empty vector as the control. Soybean transgenic composite plants inoculated with rhizobia were grown in sand culture irrigated with low-N nutrient solution for 30 d. Each bar represents the mean of four biological replicates with se. Ten nodules were selected from each replicate for infection cell analysis (F and G). Asterisks represent significant differences between DRi lines and control plants for the same trait in Student’s t tests (**, 0.001 < P ≤ 0.01 and ***, P ≤ 0.001).
DISCUSSION
Legumes play crucial roles in agricultural sustainability, in large part due to symbiotic BNF in root nodules. Nodule organogenesis incorporates a number of processes that are critical for nodule formation, differentiation, and maturation, in which rhizobia are partitioned into intracellular N2-fixing bacteroids (Oke and Long, 1999; Oldroyd et al., 2011). Therefore, nodule development is vital and tightly associated with BNF capacity. Although a series of QTLs for BNF-associated traits have been reported for legumes (Bourion et al., 2010; Santos et al., 2013; Yang et al., 2017), the genes responsible for major BNF QTLs and the related molecular mechanisms underlying nodule development are not well understood.
Previous work has identified three new QTLs for nodulation traits using 175 F9:11 RILs derived from a P1×P2 cross in soybean (Yang et al., 2017). Plus, one of these QTLs is colocated on chromosome 11 with an independently reported QTL for individual nodule weight (Fig. 1; Hwang et al., 2014). In this study, we constructed a high-resolution genetic map consisting of 27 dCAPs markers, and the gene GmINS1 was selected as a candidate gene for explaining qBNF-11 QTL effects on nodulation (Fig. 1). Interestingly, RT-qPCR analysis indicated that GmINS1 expression is associated significantly with the number of large nodules and individual nodule weight, not only in parental genotypes but also in progeny RILs as well as in genotypes from a soybean core collection (Figs. 2 and 4). This indicates that the expression level of GmINS1 is indeed an important contributor to the nodulation QTL identified in two field studies.
Gene expression is regulated by cis-regulatory elements in upstream promoter regions and cognate transcription factors (Sun et al., 2013; Liu et al., 2014; Bilas et al., 2016). The cis-motifs are involved in a variety of regulatory networks and ultimately determine the phenotypic traits exhibited in response to different environmental conditions (Kohli, 2005; Priest et al., 2009). Here, we found that GmINS1 harbors four SNPs between the two parental genotypes in a 2,323-bp region upstream of the ATG start codon but not in exons (Fig. 3A). The four SNPs, included in the motifs ARR1 (NGATT), ASF-1 (TGACG), ACGT, GARE (TAACAAR), and WRKY71OS (TGAC; Supplemental Table S1), are located within important cis-elements involved in diverse biological processes, such as phytohormone signaling, UV light regulation, and responses to biotic and abiotic stresses (Sakai et al., 2000; Ibraheem et al., 2010; Kumar et al., 2012; Yuan et al., 2012; Mehrotra et al., 2013; Bhalothia et al., 2016). Promoter deletion analysis revealed that the sequence between −2,191 and −1 (pro3::GUS) of GmINS1 from the parent P2, carrying an A/C SNP, had the highest GUS activity among all the deletion promoters (Fig. 3, A–D; Supplemental Fig. S3). This result suggests that the SNP A/C in the promoter region might play an important role in the expression of GmINS1. Subsequently, the expression of GmINS1 was strongly associated with large nodule number and individual nodule weight in a group of 40 progeny lines as well as germplasms from a soybean core collection carrying the P2 genotype (Fig. 4). These results suggest that the observed effects of GmINS1 on nodule development might be the result of variation in transcriptional regulation, due to sequence polymorphism in one or more of the five motifs described above. Another possible explanation is that linked genes in this QTL region might regulate the transcription of GmINS1 during nodule organogenesis. Due to the complex regulation of promoter in vivo, understanding of the molecular mechanisms underlying the modulation of GmINS1 expression still requires further study.
Soybeans produce determinate nodules, with the activity of nodule meristems apparently ceasing early in nodule development (Crespi and Gálvez, 2000; Popp and Ott, 2011). Thus, determinate nodule growth depends largely on cell expansion and cell wall extension rather than cell division. Previous studies have revealed that GmEXPB2, a cell wall β-expansin, is expressed preferentially in the nodule vascular trace and NVB in early stages of nodule development (Li et al., 2015). Altering GmEXPB2 expression significantly affects both rhizobial infection events and nodule numbers through modifications in root architecture and nodule development. Here, GmINS1, the candidate gene for explaining the qBNF-11 QTL-associated effects on BNF (Fig. 1), is identified as a homolog of GmEXPB2, which belongs to the cell wall β-expansin family (Supplemental Fig. S1). In contrast to GmEXPB2 expression, GUS staining showed that GmINS1 is expressed primarily during the rapid developing stage of nodules and is localized mainly within the nodule cortex and parenchymatous cells, as well as in the NVB, but not in the nodule vascular trace (Fig. 3E). Overexpression of GmINS1 did not affect IT and nodule formation; however, it did significantly facilitate nodule development and led to a higher number of large nodules (D ≥ 2 mm) and larger infection cells, which eventually promoted BNF activity and plant biomass (Figs. 5 and 6; Supplemental Figs. S5 and S6). Further studies using stably transformed soybean plants showed that GmINS1 overexpression also led to significant increases in the number of large nodules, individual nodule size, and nitrogenase activity, resulting in significantly enhanced biomass and N content (Figs. 7 and 8). In contrast, the suppression of GmINS1 limited nodule enlargement and reduced individual nodule size and the number of infection cells, which, subsequently, inhibited BNF capacity and soybean growth (Figs. 5 and 6; Supplemental Fig. S6).
BNF capacity is determined largely by processes including (1) rhizobial infection to produce more nodules and (2) nodule organogenesis to form more expanded nodules. Our previous studies showed that overexpression of GmEXPB2 resulted in increased nodule number through enhancing infection events (Li et al., 2015). In addition, GmINS1 expression significantly promoted nodule development and expansion. We also generated transgenic composite soybean lines with double suppression of GmEXPB2 and GmINS1 in order to further explore the role of expansins in BNF. As expected, DRi compounded severely inhibited soybean nodulation, as reflected by decreases in the number and weight of large nodules, as well as the size of infection cells, nitrogenase activity, biomass, and N content of plants (Fig. 9; Supplemental Fig. S9). These results demonstrate that the two β-expansin proteins might coordinately regulate soybean nodulation and, thereby, influence plant growth and N nutrition.
For other nodulation traits, such as individual nodule size, RNA interference of GmINS1 alone had similar effects to double suppression of GmEXPB2 and GmINS1 (Figs. 5D and 9D). This result suggests that GmINS1 might play a dominant role in nodule enlargement, which also explains why the QTL qBNF-11 was identified in two independent field trials for individual nodule weight (Hwang et al., 2014) as well as the number and weight of large nodules (Yang et al., 2017). Nodulation in legumes is a very complicated process that also is affected by many biotic and abiotic factors, such as host specificity, indigenous rhizobia, and soil physical and chemical conditions (Almendras and Bottomley, 1987; Cevallos et al., 1989; Chmelíková and Hejcman, 2014). To date, only two studies on nodulation in legumes have been conducted in the field, and few QTLs for nodulation have been identified (Hwang et al., 2014; Yang et al., 2017). In addition, no corresponding genes have been cloned and functionally characterized. Given that soybean nodules are determinate and that nodule size largely controls BNF capacity (Crespi and Gálvez, 2000; Popp and Ott, 2011; Li et al., 2015), the results herein strongly suggest that GmINS1 might be a promising target in efforts to optimize BNF capacity through molecular breeding in soybean.
CONCLUSION
In summary, we identified and characterized a gene encoding a cell wall β-expansin, GmINS1, that is putatively responsible for the qBNF-11 QTL for soybean nodulation traits in the field. Altering the expression of GmINS1 significantly modified nodule expansion and infection cell development and subsequently affected BNF capacity and soybean growth. GmEXPB2 and GmINS1 coordinately control soybean nodulation with distinctive divisions of functions in the initiation and development of nodules. We conclude that GmINS1 functions primarily in nodule development, especially during the enlargement of nodules and infection cells.
MATERIALS AND METHODS
Construction of a High-Resolution Genetic Map and Prediction of Candidate Genes
A QTL associated with the number and weight of large nodules (D ≥ 2 mm), qBNF-11, was identified previously on soybean (Glycine max Wm82.a1.v1) chromosome 11 between the M287 and M394 markers using a population of 175 F9:11 RILs (Yang et al., 2017). To identify and isolate the candidate genes responsible for this QTL, we developed 27 dCAPs markers (Supplemental Table S1) to construct a high-resolution genetic map.
All PCRs were carried out in 25-μL volumes containing 1 μL of 1:50 diluted genomic DNA, 0.5 μL of specific primers (Supplemental Table S1), 13 μL of distilled, deionized water, and 10 μL of PCR Mix (TransStart KD Plus DNA Polymerase). The reaction conditions were as follows: denaturation at 94°C for 2 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 51°C to 57°C for 30 s, and extension at 72°C for 20 s; and a final extension at 72°C for 10 min. PCR products were visualized after electrophoresis on a 3% polyacrylamide gel.
Plant Growth Conditions
Experiments in this study included plants grown in the field, sand culture, and hydroponics. For the field experiment, 175 F9:11 RILs derived from P1 and P2 parents contrasting in nodulation were grown at the Dishang experimental farm (E114.48°, N38.03°) of the Institute of Cereal and Oil Crops, Hebei Academy of Agricultural and Forestry Sciences, Shijiazhuang City, Hebei Province, China (Yang et al., 2017). The genotypes from a soybean core collection were grown at the Boluo experimental farm (E114.28°, N23.18°) of South China Agricultural University, Huizhou City, Guangdong Province, China (Zhao et al., 2004). At the seed-filling stage, nodules were harvested to analyze nodule number, weight, and individual size. Then, 40 progeny RILs as well as 40 genotypes from the soybean core collection were selected and divided into two groups based on the dCAPs marker developed from SNP A/C in the promoter of GmINS1. The primers GmINS1-dCAPS-F and GmINS1-dCAPS-R are listed in Supplemental Table S1.
For the sand culture experiment, seeds of 40 selected F9:11 soybean RILs were germinated in sand culture inoculated with the rhizobium strain Bradyrhizobium sp. BXYD3. Seedlings were irrigated daily with 530 µm low-N nutrient solution for 30 d. The low-N nutrient solution included KNO3, Ca(NO3)2·4H2O, NH4NO3, and (NH4)2SO4 (15:12:4:3) as well as 1,200 µm CaCl2, 1,000 µm K2SO4, 500 µm MgSO4·7H2O, 25 µm MgCl2, 2.5 µm NaB4O7·10H2O, 1.5 µm MnSO4·H2O, 1.5 µm ZnSO4·7H2O, 0.5 µm CuSO4·5H2O, 0.15 µm (NH4)6Mo7O24·4H2O, 40 µm Fe-Na-EDTA, and 250 µm KH2PO4. Nodules were harvested for RT-qPCR assays to test the relationship of GmINS1 expression and nodule development.
For the hydroponic experiment, seeds of the parental genotypes, P1 and P2, were surface sterilized in 3% H2O2 for 1 min, rinsed with distilled water, and germinated in sand irrigated with one-half-strength nutrient solution for 7 d. Uniform seedlings were inoculated with BXYD3 by immersing roots in a rhizobial suspension for 1.5 h, prior to transplanting into a low-N nutrient solution containing 530 µm N as described above. Plants were grown in growth chambers (day/night: 13 h/11 h, 26°C/24°C) for 45 d. Nutrient solution was refreshed weekly, and the pH was maintained at 5.8 to 6 with diluted H2SO4 or KOH. Upon harvest, nodules were separated into two groups based on nodule diameter, specifically large (D ≥ 2 mm) and small (D < 2 mm) nodules, and the number and weight of nodules were then measured for each group. Large nodules also were used for nitrogenase activity analysis and infection cell survey in nodules.
For expression analysis of GmINS1 in P1 and P2 parent line plants, seedlings were grown in hydroponics for 3 weeks after transplanting as described above, and then roots, nodules, stems, and leaves were harvested separately. All plant tissues were rapidly frozen in liquid nitrogen and stored at −80°C for RNA extraction and RT-qPCR analysis.
Observation of Infection Cells in Nodules and Nitrogenase Activity Analysis
For the observation of infection cells, the nodules were embedded in paraffin and sectioned transversely to a thickness of 9 µm with a microtome (Leica RM2235). After dewaxing, the transverse nodule sections were stained with 0.1% Toluidine Blue and examined with a light microscope (AXIO Imager A2m; Carl Zeiss). Ten nodules were selected randomly and measured. Four cross sections were made on each nodule, so that a total of 40 cross sections were used to calculate the number of infection cells per 1 mm2 and surface area of 100 infection cells with ImageJ software (http://rbs.info.nih.gov/ij/).
An acetylene reduction assay was used for nitrogenase activity analysis (David et al., 1980). Briefly, nodules were collected separately into 8-mL air-tight glass bottles and injected with 1 mL of acetylene for 2 h, then the reaction was terminated with 0.5 m NaOH. Subsequently, 0.3 mL of gas was measured using a gas chromatograph (GC-2014AF) with a flame ionization detector to calculate the amount of acetylene catalyzed by nitrogenase in nodules per hour and per nodule fresh weight.
RNA Extraction and RT-qPCR Analysis
High-quality total RNA was isolated from soybean roots, nodules, stems, and leaves using RNAiso Plus reagent (Takara Bio) according to the manufacturer’s instructions. After removing genomic DNA with DNase, 1 μg of RNA was used for first-strand cDNA synthesis using oligo(dT), deoxyribonucleotide triphosphates, and Moloney murine leukemia virus reverse transcriptase (Promega) based on protocols from the supplier. RT-qPCR was performed in 20-μL volumes containing 2 μL of 1:50 diluted cDNA, 0.6 μL of specific primers, 6.8 μL of distilled, deionized water, and 10 μL of Trans Start Top Green qPCR SuperMix (Trans) using a LightCycler96 (Roche Diagnostics) PCR system. The reaction conditions for thermal cycling were 95°C for 1 min followed by 40 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 30 s. The elongation factor EF-1α gene from soybean (TefS1; accession no. X56856) or from tobacco (Nicotiana tabacum; NtEF1a; accession no. AF120093; Schmidt and Delaney, 2010) was used as a reference gene to evaluate relative transcript abundance. Relative transcript abundance was calculated as the ratio of the expression value of the target gene to that of TefS1 or NtEF1a using the 2−△△CT method. Primers used for detecting target genes are listed in Supplemental Table S3.
Subcellular Localization of GmINS1 in Tobacco Cells
For subcellular localization analysis, the coding region of GmINS1 was amplified using the specific primers GmINS1-GFP-F and GmINS1-GFP-R as listed in Supplemental Table S3. The PCR product was digested with XbaI and inserted into the modified pCAMBIA1300 vector with a 35S promoter. After checking by sequencing, 35S::GmINS1-GFP, the plasma membrane marker, pm-rb CD3-1008 (Nelson et al., 2007), and the 35S::GFP control vector were transformed separately into Agrobacterium tumefaciens strain GV3101, then transiently transformed into leaf epidermal cells of tobacco as described previously (Fraley et al., 1983). Epidermal cells were plasmolyzed by adding 10% NaCl solution for 5 min prior to observation by confocal microscopy. The GFP/RFP florescence was observed using a confocal scanning microscope (LSM880; Carl Zeiss) with 488 nm excitation and 500- to 525-nm emission filter wavelengths for GFP observation and with 543 nm excitation and 560- to 615-nm emission filter wavelengths for RFP observation.
Vector Construction and Generation of Soybean Transgenic Composite Plants
To characterize the function of the GmINS1 promoter, a series of 5′ end deletions from parents P1 and P2 were generated separately by PCR amplification. Three deletion fragments were named pro3 (−2,191 to −1), pro2 (−2,022 to −1), and pro1 (−1,499 to −1) carrying three, two, and one SNPs from P1 and P2 parental genotypes, respectively, and these fragments as well as the 2,323-bp promoter region of GmINS1 from P2 (−2,323 to −1) were amplified using the common reverse primer proGmINS1-R and the forward primers pro3-F, pro2-F, pro1-F, and proGmINS1-F (Supplemental Table S4). After digestion with BamHI and EcoRI, the different fragments upstream of the GmINS1 ATG start codon were fused separately into the pTF102 vector with a GUS reporter gene. Transgenic plants with the GUS reporter gene driven by a 35S promoter in the pTF102 vector were used as control plants.
To generate soybean transgenic composite plants overexpressing or suppressing GmINS1, the open reading frame of GmINS1 was amplified using the GmINS1OX-F and GmINS1OX-R primers. The PCR fragment was digested with HindIII and MluI and cloned subsequently into the binary vector pYLRNAi with a 35S promoter. For the RNA interference construct, 442 bp of the GmGINS1 coding region was amplified with the primers GmINS1Ri-F1 and GmG3PT3Ri-R1 (sense) and GmINS1Ri-F2 and GmG3PT3Ri-R2 (antisense). PCR products were digested separately and ligated into the BamHI and HindIII as well as the PstI and MluI sites of the pYLRNAi vector in the sense and antisense orientations.
For soybean transgenic composite plants with double suppression of GmINS1 and GmEXPB2, a 442-bp segment of the GmINS1 coding sequence was amplified using the sense orientation primers GmGINS1DRi-F1 and GmINS1DRi-R1 and the antisense orientation primers GmGINS1DRi-F2 and GmINS1DRi-R2. The PCR products were inserted separately into the pSAT6-supP-RNAi vector, and then a long fragment containing supP and the 442-bp sense and antisense orientations of GmINS1 were cloned into PI-PspI sites of the pRCS2-ocs-nptII vector. Accordingly, a 400-bp fragment of the GmEXPB2 coding region was cloned using both sense (GmEXPB2DRi-F1 and GmEXPB2Ri-R1) and antisense (GmEXPB2DRi-F2 and GmEXPB2Ri-R2) orientation primers and inserted into the pSAT4-35S-RNAi vector prior to clone a long fragment containing 35S and the 400-bp sense and antisense orientations of GmEXPB2 into I-SceI sites of the pRCS2-ocs-nptII vector containing sense and antisense GmINS1 fragments. The primers used for the vector constructs are listed in Supplemental Table S4, and the restriction enzyme cutting sites are underlined in the corresponding primer sequences.
All of the above constructs were transformed separately into Agrobacterium rhizogenes strain K599 via the heat shock method. Then, composite plants with transgenic hairy roots were generated using the hypocotyl injection method as described previously (Guo et al., 2011). In tobacco leaves, the recombinant plasmids were introduced into the A. rhizogenes strain GV3101 and then transiently transferred into the tobacco leaves by infiltration. After 2 d, the transgenic leaves were harvested for GUS staining and activity analysis.
Histochemical GUS Staining of Tissue Sections and Fluorometric GUS Activity Assay
For histochemical analysis of GUS expression, soybean transgenic composite plants harboring pro3::GUS, pro2::GUS, pro1::GUS, proGmINS1::GUS, or 35S::GUS were inoculated with BXYD3 under low-N conditions. Given that soybean nodules belong to the determinate type, the size of the nodules represents nodule age. Therefore, nodule primordia were harvested 4 dai, and the largest nodules were harvested at 7, 14, 21, and 30 dai. All samples including nodules and tobacco leaves were incubated in GUS staining solution containing 50 mm inorganic phosphate-buffered saline (Na2HPO4-NaH2PO4 buffer, pH 7.2), 0.1% (v/v) Triton X-100, 2 mm K3Fe(CN)6, 2 mm K4[Fe(CN)6]·3H2O, 10 mm EDTA-2Na, and 2 mm 5-bromo-4-chloro-3-indolyl-β-d-GlcA at 37°C for 16 h and then washed three times with 70% ethanol. After GUS staining, roots and nodules carrying a 2,323-bp fragment of the GmINS1 promoter (proGmINS1::GUS) cloned from P2 were embedded in paraffin and sectioned transversely to a thickness of 9 µm with a microtome (Leica RM2235) as described previously (Qin et al., 2012). Cross sections were observed with a light microscope (Axio Imager A2m; Zeiss).
For fluorometric GUS assay, nodules and leaves of transgenic tobacco were used to determine GUS enzyme activity by measuring the fluorescence of 4-methylumbelliferone produced by GUS cleavage of 4-methylumbelliferyl-β-d-glucuronide according to the published procedure as described previously (Jefferson et al., 1987; Jefferson, 1988). GUS activity was calculated as nanomoles of methylumbelliferone per minute per milligram of protein. Protein amount was extracted and determined based on a published method using bovine serum albumin as a standard (Bradford, 1976).
Characterization of Nodule Organogenesis Using Soybean Transgenic Composite Plants
The soybean genotype HN66 was used as the plant material to generate transgenic composite plants including GmINS1 OX, Ri, and Ev control lines, as well as DRi lines, according to Guo et al. (2011). The main root was removed when the hairy roots emerging from the hypocotyl were approximately 10 cm long. Each hairy root was then checked for the quality of transformation using qualitative PCR to amplify hygromycin (for OX and Ri lines with primers Hyg-F and Hyg-R) or kanamycin (for DRi lines with primers Kan-F and Kan-R) resistance genes, which were on the vector harboring the target gene sequence (Supplemental Fig. S4). A single transgenic hairy root was kept and inoculated with rhizobium BXYD3 for 1.5 h and then transplanted into sand culture irrigated with 530 µm low-N nutrient solution for 30 d. After GmINS1 and GmEXPB2 expression assays by RT-qPCR, large nodules were collected to analyze the number and weight of nodules, nitrogenase activity, along with observations of bacteroids. The specific primers used in RT-qPCR are listed in Supplemental Table S3. Plants also were harvested separately to determine dry weight and N content.
Observation of ITs
For IT observations, OX and Ri lines of GmINS1, as well as Ev control soybean transgenic composite plants, were inoculated with the bacterial strain USDA110-GFP as constructed previously (Li et al., 2015) by immersing roots in a rhizobial suspension for 1.5 h and then transplanting into a sand culture system irrigated with N-free nutrient solution. The IT formation was observed at 3 dai with a confocal laser scanning microscope (LSM880; Carl Zeiss). Furthermore, 1-cm lengths of root segments from OX, RI, and Ev control transgenic soybean composite plants sampled at 3 dai were used to count IT numbers. Mean IT counts from 10 root segments from each plant were considered as one biological replicate. With four biological replicates, a total of 40 root segments were measured for each transgenic treatment. GFP fluorescence was viewed as described above.
Generation of Soybean Stable Transgenic Plants and Their Growth Conditions
For soybean stable plant transformation, the open reading frame region of GmINS1 was amplified using the GmINS1OX-F1 and GmINS1OX-R1 primers and then cloned into SacI and XbaI sites of the pTF101s vector with a 35S promoter to generate the overexpression construct. The expression clones then were transformed into the A. tumefaciens strain EHA105. The soybean genotype HN66 was used as the explant for the stable transformation as described previously (Wang et al., 2009). Primary transformants were established and screened by PCR amplification of bar DNA in the vectors using bar-F and bar-R primers (Supplemental Table S4).
To evaluate nodulation, seeds of transgenic and wild-type plants were germinated in sand for 7 d. Uniform seedlings were inoculated with BXYD3 by immersing roots in a rhizobial suspension for 1.5 h, then grown in nutrient solution with 530 µm low N for 40 d. Plants were harvested to determine nodule number, dry weight, and nitrogenase activity as well as plant dry weight and N content.
Measurement of Plant N Content
Following the manufacturer’s protocol, about 0.2 g of dried sample was digested and total N content was measured using a continuous flow analyzer (SAN++). Signals were output to a computer, and the results were analyzed in FlowAccess software (SAN++ FlowAccess V3 data acquisition Windows software package).
Data Analysis
Data from RT-qPCR results were normalized in each experiment. All data were analyzed statistically using Sigma Plot and Microsoft Excel 2010 to calculate means and se. Comparisons between groups were performed using Student’s t tests in Microsoft Excel 2010 or a two-way ANOVA test in SPSS (version 17.0).
Accession Number
Sequence data of GmINS1 used in this article can be found in GenBank with the accession number JQ303252.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of β-expansins in soybean and Arabidopsis.
Supplemental Figure S2. Subcellular localization of GmINS1 fused to GFP in tobacco cells.
Supplemental Figure S3. Deletion analysis of the GmINS1 promoter in tobacco leaves.
Supplemental Figure S4. Molecular identification of GmINS1 in soybean transgenic composite plants.
Supplemental Figure S5. Confocal microscopy of root hairs of soybean transgenic composite plants inoculated with rhizobium strain USDA110 carrying GFP.
Supplemental Figure S6. Effects of RNA interference and overexpression of GmINS1 on the growth of soybean transgenic composite plants.
Supplemental Figure S7. Molecular identification of GmINS1 in soybean stable transgenic lines.
Supplemental Figure S8. Molecular identification of DRi in soybean transgenic composite plants.
Supplemental Figure S9. Effects of DRi in growth and N content of soybean transgenic composite plants.
Supplemental Table S1. dCAPs markers developed for a higher resolution genetic map construction.
Supplemental Table S2. Putative cis-elements in the promoter of GmINS1 according to the four SNPs in the parents P1 and P2.
Supplemental Table S3. Gene-specific primers used for RT-qPCR analysis.
Supplemental Table S4. Gene-specific primers used for vector constructs and transgenic plant screening.
Acknowledgments
We thank all the staff and students for helping with field experiments, Dr. Jinxiang Wang at South China Agricultural University for providing the plasma membrane marker, and Dr. Thomas Walk at North Dakota State University for critical reviewing.
Footnotes
This work was financially supported by the China National Key Program for Research and Development (2016YFD0100700), the National Natural Science Foundation of China (31601814), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB15030202).
Articles can be viewed without a subscription.
References
- Almendras AS, Bottomley PJ (1987) Influence of lime and phosphate on nodulation of soil-grown Trifolium subterraneum L. by indigenous Rhizobium trifolii. Appl Environ Microbiol 53: 2090–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves BJR, Boddey RM, Urquiaga S (2003) The success of BNF in soybean in Brazil. Plant Soil 252: 1–9 [Google Scholar]
- Bhalothia P, Sangwan C, Alok A, Mehrotra S, Mehrotra R (2016) PP2C-like promoter and its deletion variants are induced by ABA but not by MeJA and SA in Arabidopsis thaliana. Front Plant Sci 7: 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilas R, Szafran K, Hnatuszko-Konka K, Kononowicz AK (2016) Cis-regulatory elements used to control gene expression in plants. Plant Cell Tiss Org 127: 269–287 [Google Scholar]
- Boualem A, Laporte P, Jovanovic M, Laffont C, Plet J, Combier JP, Niebel A, Crespi M, Frugier F (2008) MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J 54: 876–887 [DOI] [PubMed] [Google Scholar]
- Bourion V, Rizvi SMH, Fournier S, de Larambergue H, Galmiche F, Marget P, Duc G, Burstin J (2010) Genetic dissection of nitrogen nutrition in pea through a QTL approach of root, nodule, and shoot variability. Theor Appl Genet 121: 71–86 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cai Z, Wang Y, Zhu L, Tian Y, Chen L, Sun Z, Ullah I, Li X (2017) GmTIR1/GmAFB3-based auxin perception regulated by miR393 modulates soybean nodulation. New Phytol 215: 672–686 [DOI] [PubMed] [Google Scholar]
- Cevallos MA, Vázquez M, Dávalos A, Espín G, Sepúlveda J, Quinto C (1989) Characterization of Rhizobium phaseoli Sym plasmid regions involved in nodule morphogenesis and host-range specificity. Mol Microbiol 3: 879–889 [DOI] [PubMed] [Google Scholar]
- Chmelíková L, Hejcman M (2014) Effect of nitrogen, phosphorus and potassium availability on emergence, nodulation and growth of Trifolium medium L. in alkaline soil. Plant Biol (Stuttg) 16: 717–725 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. (1998) Cell wall loosening by expansins. Plant Physiol 118: 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2000) Loosening of plant cell walls by expansins. Nature 407: 321–326 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D (2002) The growing world of expansins. Plant Cell Physiol 43: 1436–1444 [DOI] [PubMed] [Google Scholar]
- Crespi M, Gálvez S (2000) Molecular mechanisms in root nodule development. J Plant Growth Regul 19: 155–166 [DOI] [PubMed] [Google Scholar]
- David KAV, Apte SK, Banerji A, Thomas J (1980) Acetylene reduction assay for nitrogenase activity: gas chromatographic determination of ethylene per sample in less than one minute. Appl Environ Microbiol 39: 1078–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza AA, Boscariol-Camargo RL, Moon DH, Camargo LEA, Tsai SM (2000) Effects of Phaseolus vulgaris QTL in controlling host-bacteria interactions under two levels of nitrogen fertilization. Genet Mol Biol 23: 155–161 [Google Scholar]
- Di Giacomo E, Laffont C, Sciarra F, Iannelli MA, Frugier F, Frugis G (2017) KNAT3/4/5-like class 2 KNOX transcription factors are involved in Medicago truncatula symbiotic nodule organ development. New Phytol 213: 822–837 [DOI] [PubMed] [Google Scholar]
- Djordjevic MA, Mohd-Radzman NA, Imin N (2015) Small-peptide signals that control root nodule number, development, and symbiosis. J Exp Bot 66: 5171–5181 [DOI] [PubMed] [Google Scholar]
- Fraley RT, Rogers SG, Horsch RB, Sanders PR, Flick JS, Adams SP, Bittner ML, Brand LA, Fink CL, Fry JS, et al. (1983) Expression of bacterial genes in plant cells. Proc Natl Acad Sci USA 80: 4803–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano W, Hirsch AM (2004) The expression of MaEXP1, a Melilotus alba expansin gene, is upregulated during the sweetclover-Sinorhizobium meliloti interaction. Mol Plant Microbe Interact 17: 613–622 [DOI] [PubMed] [Google Scholar]
- Graham PH, Vance CP (2003) Legumes: importance and constraints to greater use. Plant Physiol 131: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhao J, Li X, Qin L, Yan X, Liao H (2011) A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J 66: 541–552 [DOI] [PubMed] [Google Scholar]
- Herridge DF, Peoples MB, Boddey RM (2008) Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311: 1–18 [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobecker KV, Reynoso MA, Bustos-Sanmamed P, Wen J, Mysore KS, Crespi M, Blanco FA, Zanetti ME (2017) The microRNA390/TAS3 pathway mediates symbiotic nodulation and lateral root growth. Plant Physiol 174: 2469–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Ray JD, Cregan PB, King CA, Davies MK, Purcell LC (2014) Genetics and mapping of quantitative traits for nodule number, weight, and size in soybean (Glycine max L[Merr]). Euphytica 195: 419–434 [Google Scholar]
- Ibraheem O, Botha CEJ, Bradley G (2010) In silico analysis of cis-acting regulatory elements in 5′ regulatory regions of sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comput Biol Chem 34: 268–283 [DOI] [PubMed] [Google Scholar]
- Imin N, Mohd-Radzman NA, Ogilvie HA, Djordjevic MA (2013) The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J Exp Bot 64: 5395–5409 [DOI] [PubMed] [Google Scholar]
- Jefferson RA. (1988) Plant reporter genes: the GUS gene fusion system. Genet Eng 10: 247–263 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli A. (2005) Understanding plant gene expression through cis-regulatory elements for stress response and sustainable agriculture. Acta Physiol Plant 27: 13 [Google Scholar]
- Köpke U, Nemecek T (2010) Ecological services of faba bean. Field Crops Res 115: 217–233 [Google Scholar]
- Kouchi H, Imaizumi-Anraku H, Hayashi M, Hakoyama T, Nakagawa T, Umehara Y, Suganuma N, Kawaguchi M (2010) How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol 51: 1381–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Patro S, Ghosh J, Das A, Maiti IB, Dey N (2012) Development of a salicylic acid inducible minimal sub-genomic transcript promoter from Figwort mosaic virus with enhanced root- and leaf-activity using TGACG motif rearrangement. Gene 503: 36–47 [DOI] [PubMed] [Google Scholar]
- Lauridsen P, Franssen H, Stougaard J, Bisseling T, Marcker KA (1993) Conserved regulation of the soybean early nodulin ENOD2 gene promoter in determine and indeterminate transgenic root nodules. Plant J 3: 483–492 [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhao J, Walk TC, Liao H (2014) Characterization of soybean β-expansin genes and their expression responses to symbiosis, nutrient deficiency, and hormone treatment. Appl Microbiol Biotechnol 98: 2805–2817 [DOI] [PubMed] [Google Scholar]
- Li X, Zhao J, Tan Z, Zeng R, Liao H (2015) GmEXPB2, a cell wall β-expansin, affects soybean nodulation through modifying root architecture and promoting nodule formation and development. Plant Physiol 169: 2640–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Bisseling T (2003) Signaling in symbiosis. Curr Opin Plant Biol 6: 343–350 [DOI] [PubMed] [Google Scholar]
- Liu JH, Peng T, Dai WS (2014) Critical cis-acting elements and interacting transcription factors: key players associated with abiotic stress responses in plants. Plant Mol Biol Rep 32: 303–317 [Google Scholar]
- McQueen-Mason S, Cosgrove DJ (1994) Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA 91: 6574–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4: 1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra R, Sethi S, Zutshi I, Bhalothia P, Mehrotra S (2013) Patterns and evolution of ACGT repeat cis-element landscape across four plant genomes. BMC Genomics 14: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Radzman NA, Laffont C, Ivanovici A, Patel N, Reid D, Stougaard J, Frugier F, Imin N, Djordjevic MA (2016) Different pathways act downstream of the CEP peptide receptor CRA2 to regulate lateral root and nodule development. Plant Physiol 171: 2536–2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD. (2011) Invasion by invitation: rhizobial infection in legumes. Mol Plant Microbe Interact 24: 631–639 [DOI] [PubMed] [Google Scholar]
- Nanjareddy K, Blanco L, Arthikala MK, Alvarado-Affantranger X, Quinto C, Sánchez F, Lara M (2016) A legume TOR protein kinase regulates Rhizobium symbiosis and is essential for infection and nodule development. Plant Physiol 172: 2002–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nicolás MF, Hungria M, Arias CAA (2006) Identification of quantitative trait loci controlling nodulation and shoot mass in progenies from two Brazilian soybean cultivars. Field Crops Res 95: 355–366 [Google Scholar]
- Oke V, Long SR (1999) Bacteroid formation in the Rhizobium-legume symbiosis. Curr Opin Microbiol 2: 641–646 [DOI] [PubMed] [Google Scholar]
- Okereke GU, Unaegbu D (1992) Nodulation and biological nitrogen fixation of 80 soybean cultivars in symbiosis with indigenous rhizobia. World J Microbiol Biotechnol 8: 171–174 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA (2004) Calcium, kinases and nodulation signalling in legumes. Nat Rev Mol Cell Biol 5: 566–576 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Oldroyd GED, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144 [DOI] [PubMed] [Google Scholar]
- Popp C, Ott T (2011) Regulation of signal transduction and bacterial infection during root nodule symbiosis. Curr Opin Plant Biol 14: 458–467 [DOI] [PubMed] [Google Scholar]
- Priest HD, Filichkin SA, Mockler TC (2009) Cis-regulatory elements in plant cell signaling. Curr Opin Plant Biol 12: 643–649 [DOI] [PubMed] [Google Scholar]
- Qin L, Zhao J, Tian J, Chen L, Sun Z, Guo Y, Lu X, Gu M, Xu G, Liao H (2012) The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol 159: 1634–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24: 703–711 [DOI] [PubMed] [Google Scholar]
- Santos MA, Geraldi IO, Garcia AAF, Bortolatto N, Schiavon A, Hungria M (2013) Mapping of QTLs associated with biological nitrogen fixation traits in soybean. Hereditas 150: 17–25 [DOI] [PubMed] [Google Scholar]
- Schmidt GW, Delaney SK (2010) Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol Genet Genomics 283: 233–241 [DOI] [PubMed] [Google Scholar]
- Sujkowska M, Borucki W, Golinowski W (2007) Localization of expansin-like protein in apoplast of pea (Pisum sativum L) root nodules during interaction with Rhizobium leguminosarum bv Viciae 248. Acta Soc Bot Pol 76: 17–26 [Google Scholar]
- Sun L, Yang ZT, Song ZT, Wang MJ, Sun L, Lu SJ, Liu JX (2013) The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J 76: 274–286 [DOI] [PubMed] [Google Scholar]
- Taiz L. (1994) Expansins: proteins that promote cell wall loosening in plants. Proc Natl Acad Sci USA 91: 7387–7389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Yang S, Liu J, Gao M, Zhu H (2014) Fine mapping of the Rj4 locus, a gene controlling nodulation specificity in soybean. Mol Breed 33: 691–700 [Google Scholar]
- Tang F, Yang S, Liu J, Zhu H (2016) Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-like protein but not the one previously reported. Plant Physiol 170: 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga A, Gondo T, Akashi R, Zheng SH, Arima S, Suzuki A (2012) Quantitative trait locus analysis of symbiotic nitrogen fixation activity in the model legume Lotus japonicus. J Plant Res 125: 395–406 [DOI] [PubMed] [Google Scholar]
- Tsai SM, Nodari RO, Moon DH, Camargo LEA, Vencovsky R, Gepts P (1998) QTL mapping for nodule number and common bacterial blight in Phaseolus vulgaris L. Plant Soil 204: 135–145 [Google Scholar]
- Wang X, Wang Y, Tian J, Lim BL, Yan X, Liao H (2009) Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiol 151: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhao Q, Li X, Ai W, Liu D, Qi W, Zhang M, Yang C, Liao H (2017) Characterization of genetic basis on synergistic interactions between root architecture and biological nitrogen fixation in soybean. Front Plant Sci 8: 1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan SW, Wu XL, Liu ZH, Luo HB, Huang RZ (2012) Abiotic stresses and phytohormones regulate expression of FAD2 gene in Arabidopsis thaliana. J Integr Agric 11: 62–72 [Google Scholar]
- Zhao J, Fu JB, Liao H, He Y, Nian H, Hu YM, Qiu LJ, Dong YS, Yan XL (2004) Characterization of root architecture in an applied core collection for phosphorus efficiency of soybean germplasm. Chin Sci Bull 49: 1611–1620 [Google Scholar]