A trihelix family transcription factor regulates the expression of a mixed-linkage glucan biosynthesis gene and a gene encoding a MLG:xyloglucan endotransglucosylase.
Abstract
Mixed-linkage glucan (MLG) is a polysaccharide that is highly abundant in grass endosperm cell walls and present at lower amounts in other tissues. Cellulose synthase-like F (CSLF) and cellulose synthase-like H genes synthesize MLG, but it is unknown if other genes participate in the production and restructuring of MLG. Using Brachypodium distachyon transcriptional profiling data, we identified a B. distachyon trihelix family transcription factor (BdTHX1) that is highly coexpressed with the B. distachyon CSLF6 gene (BdCSLF6), which suggests that BdTHX1 is involved in the regulation of MLG biosynthesis. To determine the genes regulated by this transcription factor, we conducted chromatin immunoprecipitation sequencing (ChIP-seq) experiments using immature B. distachyon seeds and an anti-BdTHX1 polyclonal antibody. The ChIP-seq experiment identified the second intron of BdCSLF6 as one of the most enriched sequences. The binding of BdTHX1 to the BdCSLF6 intron sequence was confirmed using electrophoretic mobility shift assays (EMSA). ChIP-seq also showed that a gene encoding a grass-specific glycoside hydrolase family 16 endotransglucosylase/hydrolase (BdXTH8) is bound by BdTHX1, and the binding was confirmed by EMSA. Radiochemical transglucanase assays showed that BdXTH8 exhibits predominantly MLG:xyloglucan endotransglucosylase activity, a hetero-transglycosylation reaction, and can thus produce MLG-xyloglucan covalent bonds; it also has a lower xyloglucan:xyloglucan endotransglucosylase activity. B. distachyon shoots regenerated from transformed calli overexpressing BdTHX1 showed an abnormal arrangement of vascular tissue and seedling-lethal phenotypes. These results indicate that the transcription factor BdTHX1 likely plays an important role in MLG biosynthesis and restructuring by regulating the expression of BdCSLF6 and BdXTH8.
Mixed-linkage (1,3;1,4)-β-ᴅ-glucan (MLG) is widely distributed in grasses, especially in the cell walls of cereals. It is also found in the vascular plant Equisetum and some algae and fungi, seemingly as a result of convergent evolution (Popper and Fry, 2003; Fry et al., 2008b; Sørensen et al., 2008; Pettolino et al., 2009). MLG is most abundant in the endosperm of grains, where it serves as an energy source for seedling growth. The MLG content in grain varies considerably among various species with Brachypodium distachyon endosperm accumulating more than 40% (w/w) MLG by dry weight (Guillon et al., 2011). MLG is also found in vegetative tissues, especially in expanding cells of young tissues in cereals (Kim et al., 2000; Gibeaut et al., 2005; Fincher, 2009). The high levels of MLG in the walls of growing cells suggest that MLG may play an important role in cell expansion. MLG is also found in secondary cell walls of mature tissues at lower amounts compared to primary cell walls of young tissues (Vega-Sánchez et al., 2012, 2013)
MLG consists of several adjacent (1,4)-linked Glc residues separated by a single (1,3)-glucosyl linkage that occurs every third or fourth residue (Woodward et al., 1983). The digestion of MLG with lichenase, an enzyme that hydrolyzes the 1,4-glucosidic bond on the reducing side of a 1,3 glucosidic bond of MLG, produces trisaccharide and tetrasaccharide residues containing single (1,3)-β-linkages (Lazaridou and Biliaderis, 2004). The trisaccharide:tetrasaccharide ratios in MLG vary between species and affect its water solubility (Burton et al., 2010). This ratio is relatively high in B. distachyon endosperm compared with other cereal grains, which renders B. distachyon MLG insoluble in water (Guillon et al., 2011; Burton and Fincher, 2012).
Recently, several members of the cellulose synthase-like gene family have been shown to be involved in the synthesis of MLG. The accumulation of MLG in transgenic Arabidopsis (Arabidopsis thaliana) expressing rice (Oryza sativa) cellulose synthase-like F (CSLF) genes provided direct evidence that the enzymes encoded by these genes synthesize MLG (Burton et al., 2006). Using a similar experimental approach, cellulose synthase-like H (Doblin et al., 2009) and cellulose synthase-like J (Little et al., 2018) genes were also shown to be involved in MLG biosynthesis. In the cellulose synthase-like gene family, CSLF genes are only found in grasses and CSLF6 is the most highly expressed member of this gene family (Burton et al., 2008; Christensen et al., 2010; Kaur et al., 2017). The location of CSLF6 and MLG synthesis is controversial, as one study in barley (Hordeum vulgare) found that MLG is synthesized at the plasma membrane (Wilson et al., 2015), while another study using tobacco (Nicotiana tabacum) epidermal cells expressing B. distachyon CSLF6 and B. distachyon plants overexpressing CSLF6 demonstrated that MLG is synthesized in the Golgi apparatus (Kim et al., 2015, 2018), supporting the conclusion of Carpita and McCann (2010), who had shown by in vivo radiolabeling that MLG is synthesized in the Golgi apparatus in maize (Zea mays).
Overexpression of CSLF6 often results in growth defects in plants, which may be due to excessive amounts of MLG produced, its misregulation in specific cell types, or the lack of additional enzymes required to integrate MLG into the cell wall. Enzymes that serve the latter function for hemicelluloses include xyloglucan endotransglucosylases/hydrolases (XTHs; Hrmova et al., 2007; Fincher, 2009). XTHs are members of the glycoside hydrolase family 16 (GH16), most of which predominantly exert xyloglucan:xyloglucan endotransglucosylase (XET) activity, catalyzing the exchange of covalent linkages between xyloglucan chains, i.e. showing homo-transglucanase activity (Hrmova et al., 2007; Franková and Fry, 2013; Hara et al., 2014). In contrast, hetero-transβ-glucanase, an XTH preferentially catalyzing MLG:xyloglucan endotransglucosylase (MXE) activity, i.e. exchanging linkages between MLG and xyloglucan (MXE:XET ratio ≈ 5), exists in Equisetum but was not detected in barley shoots (Fry et al., 2008a; Mohler et al., 2013; Simmons and Fry, 2017). A purified barley XTH (HvXET5) showed transglucanase activity between xyloglucan and MLG but at a very low rate (MXE:XET ratio ≈ 0.002; Hrmova et al., 2007, 2009). Hara et al. (2014) found that a rice XTH gene has negligible xyloglucan:MLG hetero-transglucanase activity compared to XET activity. MLG homo-transglycosylation activity has not been found in any plant but may exist in Poaceae, as Poaceae cell walls accumulate large amounts of MLG (Carpita and Gibeaut, 1993; Carpita, 1996; Vogel, 2008). Discovering such transglucanase genes should lead to a better understanding of MLG biosynthesis and could prove useful in engineering plants with increased MLG content or modified integration of MLG within the cell wall.
Processes such as cell wall biosynthesis require the coordinated action of a large number of enzymes. The expression of genes encoding such enzymes is often coordinately regulated both spatially and temporally by specific transcription factors. Studies that examine the coexpression of genes have been successful in elucidating transcription factors involved in cell wall biosynthesis (Brown et al., 2005; Hirano et al., 2013; Ferreira et al., 2016). Using this strategy, we have identified a transcription factor of the trihelix family that is highly coexpressed with the MLG biosynthetic enzyme CSLF6 in B. distachyon. Our intention was to use such a transcription factor to discover additional genes involved in MLG biosynthesis or restructuring.
Trihelix transcription factors are plant specific, with 34 members in Arabidopsis, 40 members in rice, and 54 members in B. distachyon (Jin et al., 2017). They are also known as GT factors defined according to a highly conserved DNA-binding domain that specifically binds to GT elements, which are highly variable elements with the consensus sequence GGT(A/T) (A/T) (A/T) (Gilmartin et al., 1990; Dehesh et al., 1992; Zhou, 1999). These conserved DNA binding domains are found at the N terminus, while the C terminus is less conserved but enriched in hydrophilic amino acids. This hydrophilic region is likely the activation domain of trihelix factors (Ma, 2011; Kaplan-Levy et al., 2014). Additionally, trihelix factors can form heterodimers and homodimers as well as participate in interactions with other classes of transcription factors (Xie et al., 2009; Li et al., 2015). The trihelix factors have been found to control the expression of light-regulated genes as well as genes involved in plant development and stress responses (Kaplan-Levy et al., 2012).
Here we report that the B. distachyon trihelix factor, BdTHX1, is highly coexpressed with BdCSLF6, a gene encoding MLG synthase. Further biochemical experiments show that BdTHX1 directly binds to an intronic region of BdCSLF6 and also to the 3′ proximal region of BdXTH8, a gene that encodes a grass-specific endotransglucosylase that preferentially uses MLG as a substrate.
RESULTS
BdTHX1 Is Coexpressed with BdCSLF6, and This Expression Pattern Correlates with MLG Accumulation in B. distachyon
To discover additional components involved in the process of MLG biosynthesis and deposition, we obtained B. distachyon transcriptional profiling data from a series of tissues at different development stages and analyzed these data to identify genes coexpressed with CSLF6 (Supplemental Fig. S1). An examination of these data shows that BdCSLF6 (Bradi3g16307.1) is highly expressed in elongating leaves, elongating stem internodes (internodes 1.1–1.4 and internode 3), and endosperm but is expressed at low levels in older stem tissues (internodes 2, 4, 5, and 6) and in the mature leaf (Supplemental Fig. S1B). Coexpression analyses revealed that BdTHX1 (Bradi5g17150.1), a transcription factor of the trihelix family, showed the highest coexpression with BdCSLF6 of any gene (R2 = 0.9688). Trihelix factors are divided into five clades based on the trihelix domain (Kaplan-Levy et al., 2012). BdTHX1 is a member of the GT-2 subfamily, a clade whose proteins have two trihelix DNA-binding domains (Kaplan-Levy et al., 2012; Jin et al., 2014). Using expression databases for rice and maize (Jain et al., 2007; Sekhon et al., 2011; Wang et al., 2014; Stelpflug et al., 2016), we found that THX1 is coexpressed with CSLF6 in these plants as well as in B. distachyon (Supplemental Fig. S2). This high level of coexpression with BdCSLF6 identified BdTHX1 as a candidate transcription factor for the regulation of BdCSLF6.
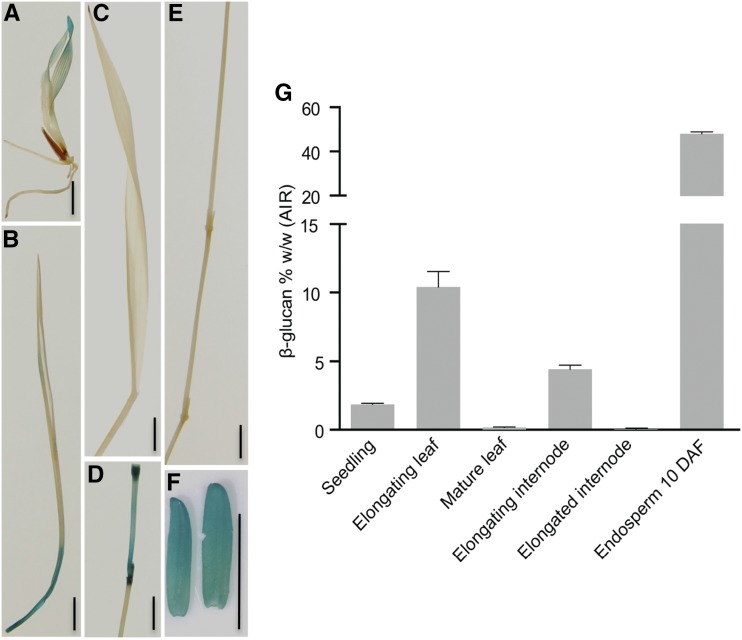
To explore the correlation of the expression pattern of BdTHX1 with MLG levels in different B. distachyon tissues, we generated transgenic plants with 3.5 kb of the BdTHX1 upstream sequence driving the expression of GUS. A variety of tissues from these plants were assayed by GUS staining and 11 of 15 independent lines showed a similar staining pattern as shown in Figure 1. This analysis indicated that pBdTHX1::GUS was expressed in seedlings, young leaves, young internodes, and endosperm but was absent or expressed at very low levels in mature leaves or internodes (Fig. 1, A–F). The expression pattern driven by the BdTHX1 promoter is similar to the expression profile of BdTHX1 from the transcriptome profiling data. In order to investigate the MLG content in the same tissues, alcohol-insoluble residue (AIR) was prepared from those tissues and analyzed (Supplemental Fig. S3). Consistent with previous findings in other grasses (Kim et al., 2000; Gibeaut et al., 2005; Fincher, 2009), young, elongating vegetative tissues and endosperm of B. distachyon accumulate large amounts of MLG (Fig. 1G). The AIR from elongating leaves and young internodes contained 10% (w/w) and 5% (w/w) MLG, respectively. Whole seedlings contained approximately 1% (w/w) MLG, while mature leaves and senesced internodes have very low levels (Fig. 1G). The endosperm showed the strongest GUS staining signal and accumulates up to 45% (w/w) MLG (Fig. 1G). Both the expression profile data and the promoter activity of BdTHX1 driving GUS expression indicate that the expression of BdTHX1 is correlated with MLG accumulation.
Figure 1.
Gus expression driven by the 3.5-kb upstream region of BdTHX1 and MLG content in different B. distachyon tissues. A, GUS staining of 5-d-old pBdTHX1::GUS T1 seedling. B to F, GUS staining of elongating leaf (B), mature leaf (C), elongating internode (D), elongated internode (E), and 10 d-after-fertilization (DAF) endosperm (F) of pBdTHX1::GUS T1 plant. Scale bars, 5 mm. G, MLG amounts in various B. distachyon tissues are shown as means ± sd from three biological replicates. Alcohol-insoluble residue (AIR) extracted from seedlings and different tissues of Bd21-3 plant were subjected to MLG assay.
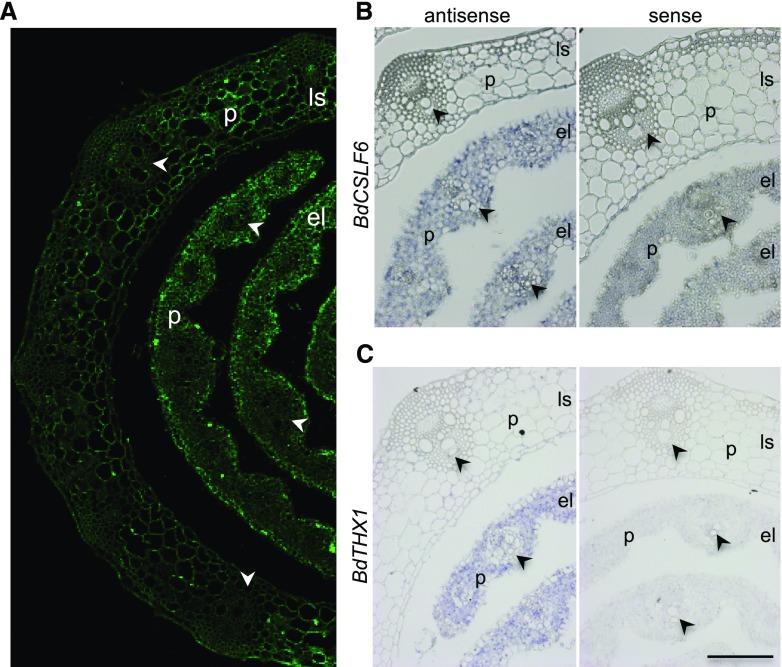
To determine the correlation between MLG accumulation and the expression pattern of BdCSLF6 and BdTHX1 at the cellular level, we examined MLG deposition in young leaves by immunohistochemistry and the expression of BdCSLF6 and BdTHX1 by in situ hybridization. Use of an anti-MLG monoclonal antibody indicated that the highest amounts of MLG were in parenchyma and epidermal cells of elongating leaves. There was weak labeling in mature leaf cells (Fig. 2A). RNA in situ hybridization showed that BdCSLF6 is most highly expressed in parenchyma cells in elongating leaves. Low levels of BdCSLF6 were found in leaf sheath tissues (Fig. 2B). BdTHX1 showed a similar pattern of expression as BdCSLF6 by RNA in situ hybridization (Fig. 2C). GUS staining of sections of pBdTHX1::GUS plants showed that BdTHX1 was expressed in parenchyma cells and immature vascular bundles of the elongating leaf, but not in the leaf sheath or mature vascular bundles in the elongating leaf (Supplemental Fig. S4A). In sections of young internodes, GUS staining was observed in all cells except vascular bundles (Supplemental Fig. S4B). There was strong GUS staining throughout endosperm tissue (Supplemental Fig. S4C). These results indicate that both BdTHX1 and BdCSLF6 are predominantly expressed in the parenchyma cells of young vegetative tissues and that these cells accumulate large amounts of MLG. The close correlation of BdTHX1 and BdCSLF6 expression and MLG accumulation at the cellular level is consistent with the hypothesis that BdTHX1 is involved in MLG biosynthesis by regulating BdCSLF6 expression.
Figure 2.
Immunolabeling and expression of BdCSLF6 and BdTHX1 in leaf. A, Immunolabeling of elongating leaf and leaf sheath with MLG monoclonal antibody. B and C, RNA in situ hybridization of BdCSLF6 (B) and BdTHX1 (C) in elongating leaf and leaf sheath. Sections hybridized with sense probes are shown as controls. The arrowheads indicate the vascular bundle. P, Parenchyma cells; ls, leaf sheath; el, elongating leaf. Scale bars, 100 μm.
Identification of BdTHX1 Binding Sites by Chromatin Immunoprecipitation-Sequencing
To identify the direct targets of BdTHX1 transcriptional regulation, we conducted chromatin immunoprecipitation-sequencing (ChIP-seq) experiments with chromatin extracted from B. distachyon immature seeds (12 to 20 d after fertilization) as this tissue has a high level of THX1 expression and accumulates large amounts of MLG. We optimized the nuclei isolation method used for DNase-rich tissues as described previously (Peterson et al., 2000; Lutz et al., 2011) and successfully obtained preparations of high Mr nuclear DNA from B. distachyon using a buffer containing EDTA, spermine, and spermidine.
To obtain an antibody specific to BdTHX1, the C-terminal 572 to 758 amino acid region of BdTHX1 was chosen because this region is specific to BdTHX1. The sequence of this region was compared with all protein sequences from B. distachyon using the BLAST program, and only short sequences with very low scores were found (Supplemental Fig. S5A). The resulting polyclonal antibody revealed a single band of the correct size by immunoblot analysis of total protein from immature seeds, which have high levels of the BdTHX1 transcript, while no visible band was detected with protein extracted from 20-d-old whole plant tissues that have very low levels of the BdTHX1 transcript (Supplemental Fig. S5B). To further test the specificity of the BdTHX1 antibody, we examined the binding of this antibody to BdTHX2 (Bradi1g77610.1), a trihelix member that has the highest protein sequence identity and a similar expression pattern as BdTHX1 (Supplemental Fig. S6). Both proteins fall into the GT-2 clade and have two trihelix DNA binding domains, one central domain, and a C terminus that may act as an activation domain (Jin et al., 2014). The recombinant proteins 6×HIS-BdTHX1 and 6×HIS-BdTHX2 were synthesized using a cell-free wheat germ expression system (Takai et al., 2010). Immunoblot analyses of the cell-free synthesized proteins showed that BdTHX1 antibody detected 6×HIS-BdTHX1, but not 6×HIS-BdTHX2 (Supplemental Fig. S5C). These results indicate that the antibody produced is specific to BdTHX1. The affinity purified polyclonal anti-BdTHX1 and preimmune serum were used in the ChIP assays as the immunoprecipitated (IP) sample and negative control, respectively. Isolated ChIP-DNA was subjected to DNA sequencing. The top 100 DNA-binding sites identified from two independent ChIP-seq experiments using program MACS are shown in Supplemental Table S1.
BdTHX1 Binds to GT Elements in the Second Intron of BdCSLF6
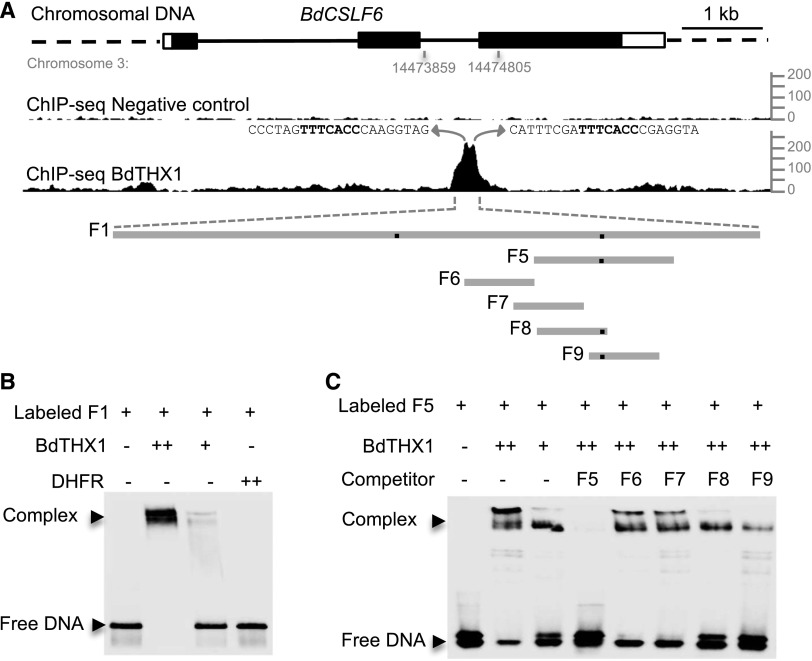
One of the highest scoring candidate genes in the ChIP-seq data was BdCSLF6 (Supplemental Table S1). None of the genes with a higher score showed coexpression with BdTHX1 nor did their expression correlate with MLG accumulation in different B. distachyon tissues, while BdTHX1 and BdCSLF6 have very similar expression patterns (Fig. 1G; Supplemental Figs. S1 and S7). It is often observed that transcription factor binding does not lead to gene regulation (MacQuarrie et al., 2011), so the combination of binding and coexpression was used as a filter to select candidate genes regulated by BdTHX1. BdTHX1 binds to the second intron of BdCSLF6 and therefore likely functions as an enhancer (Fig. 3A). The binding of BdTHX1 to the BdCSLF6 intron sequence was confirmed using electrophoretic mobility shift assays (EMSA). A 276-bp fragment of the second intron F1 (Supplemental Fig. S8A) was labeled with biotin and used for EMSA along with BdTHX1 protein produced from a cell-free wheat germ expression system. The shift of the labeled DNA was dependent on the amount of BdTHX1 protein added, while no change in mobility of the probe was observed when the control protein dihydrofolate reductase (DHFR), also synthesized in the wheat germ system, was assayed (Fig. 3B).
Figure 3.
Binding of BdTHX1 to BdCSLF6 intron sequences. A, ChIP-seq profile for BdCSLF6 using BdTHX1 antibody and schematic diagram of the probes used in electrophoretic mobility-shift assays. The ChIP-seq reads were mapped to B. distachyon Bd21 v1.0 chromosome 3: 14473859 to 14474805, corresponding to the second intron of BdCSLF6. y axis shows normalized read counts. The exons and introns of BdCSLF6 were shown as rectangles (filled, translated regions; open, untranslated regions) and black lines, respectively. Preimmune serum was used as a negative control. The sequences that correspond to the two peaks are shown, and GT motifs are highlighted in bold. F1 is a 272-bp-long probe located in the second intron of BdCSLF6. Probe F5 is 60 bp long. Probes F6, F7, F8, and F9 are 30 bp long. The black dots on the gray lines represent the binding site. B and C, EMSA was performed with biotin-labeled probes and wheat germ cell-free synthesized BdTHX1. Two different amounts of BdTHX1 were used to test if the binding is BdTHX1 amount dependent. ++ represents that 4-fold excess amount of protein was added in the reaction. Free DNA and protein-DNA complex were separated by 5% TBE polyacrylamide gels. B, The binding of BdTHX1 to probe F1. Wheat germ cell-free synthesized protein DHFR was used as negative control. C, The specific binding of BdTHX1 to probe F5 was tested by using a competition assay. Excess amounts of unlabeled DNA fragments (200-fold molar excess over labeled DNA) were included in the assay as competitors.
To identify the exact binding sequence, smaller oligonucleotide probes were synthesized and labeled with biotin and used for EMSA. BdTHX1 binds to the 60-bp probe F5 (Fig. 3C). The 30-bp unlabeled probes F6, F7, F8, and F9 containing overlapping regions of F5 were used to compete for the binding of BdTHX1 to biotin-labeled F5. Probes F8 and F9 showed competition with biotin-labeled F5, with F9 showing stronger competition indicating that the binding site is located in the overlap region of F8 and F9 (Fig. 3C). Unlabeled mutant versions of F9 (M1 and M2) were used as competitors to refine the sequence of the binding site. The motif TTTCACC was found using this approach (Fig. 4, A and B). This sequence occurs twice in the second intron of BdCSLF6 and likely corresponds to the two peaks shown on the map of ChIP-seq reads (Fig. 3A). The binding sequence TTTCACC matches the GT motif previously identified for other trihelix family members having the consensus sequence GGT(A/T) (A/T) (A/T) (Zhou, 1999; Kaplan-Levy et al., 2012) with two exceptions: (1) the binding motif found in the second intron of BdCSLF6 occurs as the reverse complement of the GT motif, (2) the fourth position of this motif is a cytosine (C) instead of thymine (T). Enhancer binding sites are found in both forward- and reverse-complementary orientations and the binding orientation of transcription factors is not relevant to the transcription start site of the downstream gene (Cox et al., 1997; Lis and Walther, 2016). To examine the binding sequence of BdTHX1, we searched for frequently occurring sequence motifs in DNA regions defined by ChIP-seq using the BdTHX1-specific antibody. The top scoring motif generated by MEME using motif analysis of the top 100 candidate regions for BdTHX1 binding is shown in Figure 4C. This motif was subsequently found in 270 peak regions of the top 500 peak regions and matches the GT motif previously identified. To confirm specificity, each nucleotide of this motif was mutated and used as a competitor in an EMSA. None of the mutated probes competed for the binding of BdTHX1 to biotin-F5 with the exception of M9 (Fig. 4, A and B). The motif created by statistical analysis of ChIP-seq data are consistent with the fact that competitor M9 can compete for the binding of BdTHX1 to biotin-F5 because the mutated position is predicted to be either a cytosine (C) or thymine (T) and M9 has a thymine (T) instead of cytosine (C) (Fig. 4C). Competitor M6t with the fourth nucleotide of the GT motif mutated from cytosine (C) to thymine (T) can compete for the binding, further indicating that BdTHX1 binds to the GT motif (Supplemental Fig. S9). It also supports the idea that GT elements are highly degenerate (Zhou, 1999). The binding sequence we identified by EMSA and statistical analysis of ChIP-bound DNA therefore matches the core sequence of GT elements.
Figure 4.
Analysis of BdTHX1 binding site. A and B, BdTHX1 binding site was analyzed by using mutant competitors in EMSA. The sequences of mutant competitors used in EMSA (B) are shown in A. A, The binding motif is highlighted in bold, and the mutated nucleotide of each competitor is lowercased. B, EMSA was performed with biotin-labeled probe F5 and wheat germ cell-free synthesized BdTHX1. Free DNA and protein-DNA complex were separated by 5% TBE polyacrylamide gels. Excess amounts of unlabeled probes F5, M1, M2, M3, M4, M5, M6, M7, M8, and M9 (200-fold molar excess over labeled DNA) were included as competitors. C, The motif created using program MEME motif analysis of the top 100 peaks by IDR score of ChIP-seq data. The height the label of each residues represents its frequency at that position. The GT-motif nucleotides are underlined.
To evaluate the conservation of the GT motif in CSLF6, the prevalence of the motif in CSLF6 introns of several grass species was analyzed. Our analysis showed that second introns of all except sorghum (Sorghum bicolor) CSLF6 contain GGTAAAA/T, T/ATTCACC, or T/ATTTACC. The first intron of all examined sequences contained the motif GGTGAAA/T (Supplemental Table S2).
BdTHX1 Binds to the GT Element of BdXTH8, a Gene Encoding a Grass-Specific Endotransglucosylase Acting on MLG and Xyloglucan
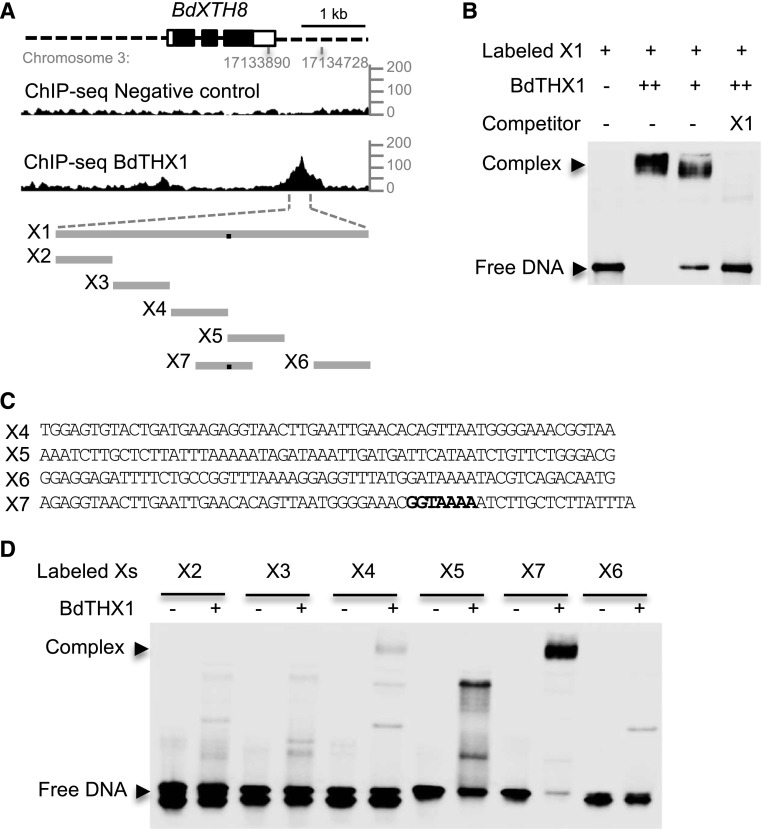
An analysis of the ChIP-seq data also identified a gene encoding a member of the GH16, here designated BdXTH8 (Bradi3g18600.1; Supplemental Table S1). The expression pattern of this gene is similar to that of both BdTHX1 and BdCSLF6, and BdXTH8 has a BdTHX1 binding site in the 3′ proximal region as determined from the ChIP-seq data (Supplemental Fig. S10B; Fig. 5, A and B). A 328-bp fragment of the BdXTH8 3′ region X1 (Supplemental Fig. S8B) was labeled with biotin and incubated with BdTHX1 synthesized from wheat germ extract. This probe showed an electrophoretic shift that was dependent on the amount of BdTHX1, and this binding was competed with unlabeled probe (Fig. 5B). To further identify the binding sequence, five 60-bp biotin-labeled probes (X2–X6) from the different regions of X1 were used separately with cell-free wheat germ synthesized BdTHX1 in EMSA. No probes showed electrophoretic shifts (Fig. 5, A and D). Further analysis showed that there is a GT element split between X4 and X5. The probe biotin-X7, which contains the intact GT element was assayed with BdTHX1 and produced an electrophoretic shift (Fig. 5, A, C, and D). This result shows that BdTHX1 binds to the 3′ proximal region of BdXTH8 and provides supporting evidence that BdTHX1 binds to the identified GT element in vivo (Fig. 4C).
Figure 5.
Binding of BdTHX1 to the 3′ proximal region of BdXTH8. A, ChIP-seq profile for BdXTH8 using BdTHX1 antibody and schematic diagram of the probes used in EMSA. The ChIP-seq reads were mapped to B. distachyon Bd21 v1.0 chromosome 3: 17133890 to 17134728, corresponding to the 3′ proximal region of BdXTH8. y axis shows normalized read counts. The exons and introns of BdXTH8 are shown as rectangles (filled, translated regions; open, untranslated regions) and black lines, respectively. Dashed lines represents upstream and downstream sequences. X1 is a 328-bp sequence located downstream of BdXTH8. X2 to X7 vary between 57 and 60 bp long. The black dots on the gray lines represent the binding site. B, EMSA was performed with biotin-labeled probe X1 and wheat germ cell-free synthesized BdTHX1. Free DNA and protein-DNA complex were separated by 5% TBE polyacrylamide gels. Excess amounts of unlabeled X1 (200-fold molar excess over labeled X1) were included as specific competitor. Two different amounts of BdTHX1 were used. ++ represents that 4-fold excess amount of protein was added in the reaction. C, Probe sequence for X4, X5, X6, and X7. Both X4 and X5 contain part of GT motif. The binding motif in X7 is highlight in bold. D, Verification of binding motif using labeled probes from different regions of X1. BdTHX1 binds to X7, which probe contains a typical GT motif.
The GH16 family includes several endotransglucosylases likely required to integrate newly synthesized xyloglucan into the existing wall (Thompson et al., 1997) and to restructure existing wall-bound xyloglucan (Thompson and Fry, 2001). Phylogenetic analysis of B. distachyon GH16 members showed that four members have no close homologs in Arabidopsis and are unique to the grasses suggesting that they may be required for the synthesis or removal of a grass-specific cell wall polymer, such as MLG (Hayashi, 1989; Gibeaut and Carpita, 1993; Burton and Fincher, 2012). Intriguingly, BdXTH8 is a member of this grass-specific clade of GH16 genes in B. distachyon (Supplemental Fig. S10A). All of the four genes show the greatest expression in tissues with high MLG (Guillon et al., 2011, 2012; Supplemental Fig. S10B), suggesting that these proteins likely act on MLG, either as GH16 endohydrolases or endotransglucosylases.
To test this, BdXTH8 (25 to 301 amino acids, without the N-terminal signal peptide) was expressed in the yeast Pichia pastoris as a secreted protein using a P. pastoris signal sequence. A protein with a mass of approximately 60 kD that was specific to this transformed strain was detected (Supplemental Fig. S11A). Tryptic peptides of this protein isolated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis were analyzed by liquid chromatography with tandem mass spectrometry and confirmed the identity of this protein as BdXTH8. The increase in the Mr of this protein compared to that predicted from the sequence is likely due to glycosylation as has been observed for other XTHs expressed in P. pastoris (Henriksson et al., 2003; Simmons et al., 2015). In radiometric assays, BdXTH8 showed transglucosylase activity toward MLG and xyloglucan, when a xyloglucan-derived heptasaccharide ([3H]XXXGol) was offered as an acceptor substrate. The MXE activity was higher than the XET activity (MXE:XET ratio ∼1.4). Neither water-soluble cellulose acetate (structurally resembling grass xyloglucans; Gibeaut et al., 2005; Fry et al., 2008a) nor insoluble cellulose (filter paper) was utilized as a donor substrate. The pH optima of both XET and MXE activity were determined as 5.3 to 5.4, which falls within the range of the plant apoplast (Fig. 6). BdXTH8 did not yield hydrolysis products separable by thin-layer chromatography when acting on xyloglucan or MLG (Supplemental Fig. S11B). BdXTH8 along with the other three genes in the grass-specific clade are expressed in tissues that accumulate high amounts of MLG, also supporting the idea that they play an important role in metabolism between MLG and xyloglucan in B. distachyon. They may represent novel contributors to MLG synthesis and remodeling pathways in grasses. This result suggests that appreciable MXE activity exists in grasses.
Figure 6.
Activities of Pichia expressed BdXTH8. A, XET, MXE, and CXE (WSCA) activities were measured using soluble donor substrates (xyloglucan, MLG, or water-soluble cellulose acetate; WSCA); CXE (AP) activity was assayed using insoluble alkali-treated filter paper (AP) as a donor. [3H]XXXGol served as acceptor. Statistically significant differences (P < 0.01, ANOVA + Tukey’s post-hoc test) among activities after 24 h or 36 h incubation are indicated by lower- or uppercase letters, respectively. B, pH dependence of XET and MXE activity expressed as % of pH optima. Activities are shown as means ± sd from four (A) or three (B) independent experiments.
BdXTH8 is coexpressed with the transcription factor BdTHX2 in addition to BdTHX1. BdXTH8 and BdTHX2 both have higher expression in elongating internodes and elongating leaf tissue than that in endosperm, while BdTHX1 and BdCSLF6 have nearly equal expression in elongating internodes and endosperm tissues (Supplemental Fig. S1, S6B, and S10B).
To test the DNA binding ability of BdTHX2 to BdCSLF6 and BdXTH8, an EMSA using cell-free wheat germ synthesized BdTHX2 and biotin-labeled probes from BdCSLF6 (F5) and BdXTH8 (X7) were performed and showed that BdTHX2 binds to both BdCSLF6 and BdXTH8 (Supplemental Fig. S12). In order to compare BdTHX2 binding activity to the two genes, both unlabeled F5 and X7 were used to compete for the binding of BdTHX2 to biotin-F5 and biotin-X7. Both competitors can compete for the binding of BdTHX2 to biotin-F5. X7 but not F5 can compete for the binding of BdTHX2 to biotin-X7 (Supplemental Fig. S12). This competition assay indicates that BdTHX2 has higher binding affinity to BdXTH8 than to BdCSLF6. This result is consistent with the fact that BdTHX2 and BdXTH8 are coexpressed and suggests that BdTHX2 may be involved in MLG accumulation in vegetative tissues by regulating the expression of XTH8.
Misregulation of BdTHX1 Is Detrimental to B. distachyon Development
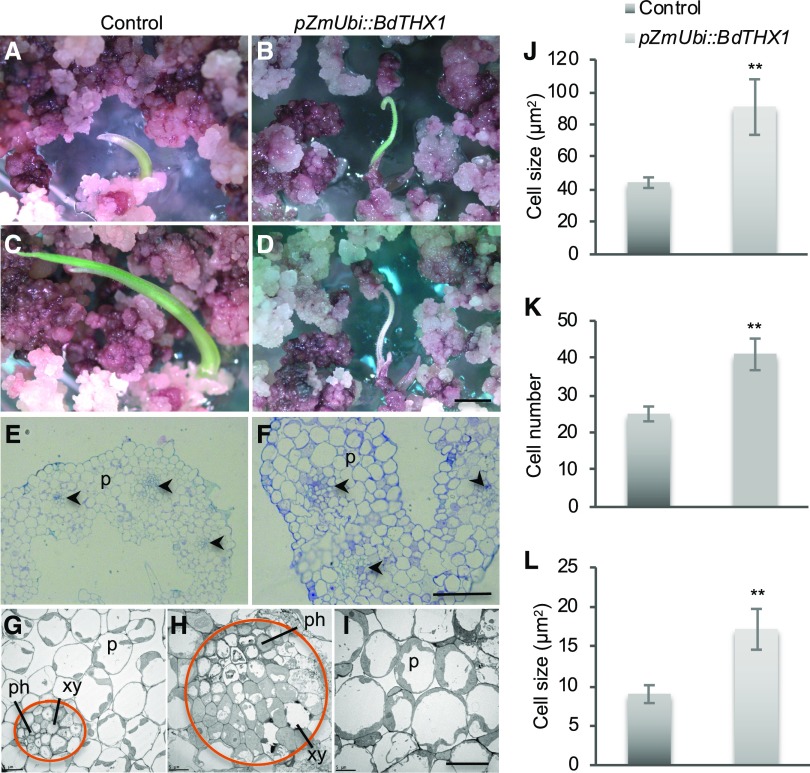
To gain information on the genes regulated by BdTHX1, we transformed B. distachyon with BdTHX1 under the control of the maize ubiquitin promoter (pZmUbi). After selection for transformed calli on hygromycin-containing media for 3 weeks, we transferred the calli onto regeneration medium. After 1 to 2 weeks, green shoots emerged as shown in Figure 7B. Regenerating shoots from pZmUbi::BdTHX1 stopped growing and died, while transgenic shoots from control calli (pZmUbi::GUS) continued growing (Fig. 7, A to D). Fifty T0 shoots (100%) resulting from transformation with pZmUbi::BdTHX1 died when they were less than 0.6 cm tall. This is similar to barley CSLF6 overexpression plants, where most of the p35S::HvCSLF6 transgenic plants cannot survive (Burton et al., 2011). To examine the MLG accumulation in the transgenic shoots, we assayed cross sections of transformed shoots with the MLG monoclonal antibody. Some lines (five out of eight) showed a stronger signal than the control, especially in the vascular bundle (Supplemental Fig. S13). Toluidine blue staining of cross sections showed that the average size of cells in pZmUbi::BdTHX1 transgenic shoots is approximately twice as large as the control (Fig. 7, E, F, and J). Transmission electron microscopy of the regenerated shoots showed that pZmUbi::BdTHX1-transformed plants had larger parenchyma cells and an abnormal arrangement of vascular bundles (Fig. 7, G–I). More cells were found in the vascular bundle of pZmUbi::BdTHX1 transgenic plants, and these cells were larger than those in the control (Fig. 7, K and L). A similar phenotype has been reported in plants overexpressing the barley CSLF6 (Burton et al., 2011). The vascular bundle is an important transport system for water and nutrients. It is possible that the lack of properly organized vascular tissue leads to plant death. This phenotype may be due to the excessive amount of MLG or due to the accumulation of MLG in tissues or developmental stages that normally have low levels of MLG. Additionally, BdTHX1 may regulate genes related to cell wall assembly or restructuring such as BdXTH8. It is reported that XTH members could act as cell wall loosening enzymes to affect cell expansion (Van Sandt et al., 2007).
Figure 7.
Phenotype of pZmUbi::BdTHX1 transgenic shoots. A to D, Transgenic shoots of control (A) and pZmUbi::BdTHX1 (B) regenerated on shoot-regeneration medium. After 4 days (control) or 7 days (pZmUbi::BdTHX1), shoots of control plants continued to grow (C), but the transgenic shoots of pZmUbi::THX1 stopped growing and died (D). Scale bar, 2 mm. E and F, Cross sections of control (E) and pZmUbi::BdTHX1 (F) shoots stained with toluidine blue. The arrowheads indicate the vascular bundle. P, parenchyma cells. Scale bar, 50 μm. G to I, Transmission electron microscopy of control (G) and pZmUbi::BdTHX1 shoots (H and I). The circle in orange indicates the area of vascular bundle. xy, Xylem; ph, phloem; P, parenchyma cells. Scale bar, 10 μm. J, Average cell size of control and pZmUbi::BdTHX1 were shown as means ± sd from different transgenic shoots (n = 3). **P < 0.01 compared with control (unpaired t test). K and L, Cell number (K) and average cell size (L) of cells in vascular bundle of control and pZmUbi::BdTHX1 transgenic shoots were shown as means ± sd (n = 3). **P < 0.01 compared with control (unpaired t test).
Overexpressing BdTHX1 under the control of the Cauliflower mosaic virus 35S promoter resulted in the same phenotype as pZmUbi::BdTHX1 transgenic shoots (Supplemental Table S3). The use of a β-estradiol-inducible system to overexpress BdTHX1 did not produce mature plants. Using the GUS gene as a control to test this inducible system revealed that GUS was expressed in the absence of the inducer 17-β-estradiol indicating that we were also likely still overexpressing the BdTHX1 gene. Transforming with a dominant-negative construct using the BdTHX1-EAR (EAR motif repression domain; Hiratsu et al., 2003) with the Cauliflower mosaic virus 35S promoter produced transgenic shoots, which also did not survive (Supplemental Table S3). To generate RNA interference (RNAi) lines with lower amounts of BdTHX1 transcripts, four different RNAi constructs were made and used to construct transgenic B. distachyon. No visible phenotypes were observed in any of these plants (Supplemental Fig. S14A). More than 120 T0 lines from four different RNAi constructs were analyzed by reverse transcription-quantitative PCR. eight independent lines from the same construct showed reduced expression of BdTHX1. The progeny of four independent lines were analyzed, and the data from the T2 is shown in Supplemental Figure S14B. No segregation was detected in the T2 generation of line #1 and #22. The expression of BdTHX1 and BdXTH8 was down-regulated, and the transcript abundance of BdCSLF6 was not significantly changed. In these four lines, the expression of BdTHX1, BdCSLF6, and BdXTH8 showed the same pattern in the T0, T1, and T2 generation. The lines with the lowest relative expression level still contained 60% of the mRNA as that of control plants suggesting that there is a limit to the reduction of BdTHX1 that is tolerated by the plant. Further exploration and examination of the targets of BdTHX1 will provide more clues on the role of BdTHX1 in plant development.
DISCUSSION
BdTHX1 Binds to Nonpromoter Regions That Likely Function as Enhancers
ChIP-seq is widely used to identify the binding site of DNA-associated proteins. To identify the targets of the transcription factor BdTHX1, we carried out ChIP-seq experiments with the BdTHX1-specific antibody, and BdCSLF6 was one of the highest scoring candidates. BdTHX1 binds to the second intron of BdCSLF6 but not to the promoter, a result supported by the EMSA. The ChIP-seq data also showed that BdTHX1 binds to the 3′ proximal region of BdXTH8. Transcription factors bind to the proximal promoter elements or enhancer regions of the genes they regulate. Unlike proximal promoter elements, enhancers are difficult to identify because they can be located large distances from their target promoters and can be found upstream or downstream of genes and within introns (Deyholos and Sieburth, 2000; Bianchi et al., 2009; Pennacchio et al., 2013). Recently, Zhu et al. (2015) developed an open chromatin signature-based enhancer prediction system in the Arabidopsis genome, and thousands of regions were predicted to be enhancers. As an example, the second intron of AGAMOUS contains an enhancer that controls carpel- and stamen-specific expression of Arabidopsis genes during flower development (Sieburth and Meyerowitz, 1997; Deyholos and Sieburth, 2000; Liu and Liu, 2008). For MLG synthesis, one member of the CSLF gene family, i.e. CSLF6, is likely required for the majority of MLG synthesis (Kim et al., 2015), and this gene is likely regulated in a tissue- and developmental-stage-specific way. It is likely that BdTHX1 is involved in regulating this specific expression of BdCSLF6.
Trihelix Factor BdTHX2 Is Likely Involved in MLG Accumulation by Targeting BdXTH8
The trihelix family is a large transcription factor family that contains 54 members in B. distachyon. One member, BdTHX2, has high protein sequence identity with BdTHX1 and a similar expression pattern as BdTHX1 (Supplemental Fig. S6). Both genes have high expression in young vegetative tissues. BdTHX2 is highly coexpressed with BdXTH8 and has higher expression in elongating internodes than that in endosperm, while BdTHX1 showed high expression in both elongating internodes and endosperm (Supplemental Figs. S1, S6B, and S10B). EMSA showed that BdTHX2 binds to both BdCSLF6 and BdXTH8 and has higher affinity to the latter (Supplemental Fig. S12). These two transcription factors have similar but not identical expression patterns, and their different binding affinity to CSLF6 and XTH8 suggests that THX1 and THX2 may regulate MLG biosynthesis in specific tissues. BdXTH8 might be important for recruiting MLG as a structural cell wall component, but not as a storage compound in endosperm cell walls, which does not show strong BdTHX2 expression. The composition of cell walls in vegetative tissues differs from that in grain. In grasses, cell walls in vegetative tissues contain a high level of cellulose and a low level of MLG (Gibeaut et al., 2005; Burton and Fincher, 2014; Zhang et al., 2014). Studies on trihelix factors have revealed that many members form dimers between trihelix factors or interact with other transcription factors. It is possible that BdTHX1 interacts with BdTHX2 as a heterodimer in vegetative tissues but not endosperm to regulate the expression of target genes. Likely, more genes regulating MLG biosynthesis could be uncovered by performing ChIP-seq experiments using an anti-BdTHX2 antibody and young vegetative tissues, such as elongating internodes or elongating leaves. Examining those genes should lead to a better understanding of MLG deposition in young vegetative tissues and the mechanism of MLG’s effect on plant development. With such information, it might be possible to engineer crops like sorghum with the stem as a storage organ accumulating large amounts of MLG without affecting plant growth.
BdTHX1 Is Likely a Key Regulator of MLG Biosynthesis and Remodeling
Using B. distachyon transcriptional profiling data, we identified a transcription factor that is coexpressed with BdCSLF6. Both genes are highly expressed in tissues that accumulate a large amount of MLG. An examination of recent studies of transcriptional profiling data using a variety of tissues in rice and maize revealed that a homolog of BdTHX1 is coexpressed with CSLF6 (Jain et al., 2007; Sekhon et al., 2011; Wang et al., 2014; Stelpflug et al., 2016). These expression data are consistent with BdTHX1 being a transcription factor involved in MLG biosynthesis. Histochemical analysis of the plants transformed with a construct containing the promoter of BdTHX1 driving the expression of GUS showed that expression of BdTHX1 correlates with the MLG accumulation in different tissues of B. distachyon.
Additionally, ChIP-seq assays with a BdTHX1-specific antibody in B. distachyon immature seeds showed that BdTHX1 binds to the second intron of BdCSLF6. The binding activity was confirmed by EMSA. The ChIP-seq data also identified BdXTH8, a member of the large family of GH16 endotransglucosylase/hydrolases. Many GH16 proteins catalyze xyloglucan-to-xyloglucan homo-transglycosylation (i.e. the well-known XET activity). All poalean XTHs studied prior to the present work catalyzed only very slight MLG-to-xyloglucan hetero-transglycosylation (MXE activity) in vitro (Hrmova et al., 2007). It was speculated (Hrmova et al., 2007) that in muro, i.e. in the presence of naturally high MLG concentrations, some poalean XTHs may exert appreciable MXE action; however, direct tests of this hypothesis with radiolabeled substrates in living barley tissues (Fig. 4g of Mohler et al., 2013) showed that they do not. No appreciable MXE action was detectable in leaves, coleoptiles, or roots of young or older barley seedlings and in the second and fifth leaves of 12-week-old plants, whereas XET action was readily detected in all these tissues. The current study found a high expression of BdXTH8 in the youngest internode of 12-week-old B. distachyon plants, elongating leaves of 25-d-old plants and the endosperm and thus did not include tissues tested in barley by Mohler et al. (2013). It is possible that the expression of MLG transglucanases in grasses is restricted to specific tissues during certain life stages (e.g. not in leaves of seedlings but in elongating leaves of older plants and not in established leaves). If true, it is crucial that future studies include various tissues formed during different plant life stages to prevent overlooking the expression and/or action of transglucanases. For example, the endosperm and integument tissues, in which MLG is very abundant and in which two other grass-specific XTHs (Bradi1g09700.1 and Bradi1g09690.1) are particularly strongly expressed, were not tested for transglucanase action (Mohler et al., 2013).
Surprisingly, an enzyme with very high MXE (relative to XET) activity was found, apparently confined to the early-diverging genus of fern allies, Equisetum (Fry et al., 2008a). This enzyme, hetero-transβ-glucanase, also exerts an additional major hetero-transglycosylation reaction — cellulose:xyloglucan endotransglucosylase (CXE) activity. The MXE action of hetero-transβ-glucanase was readily detected in living Equisetum tissues (Mohler et al., 2013). More recently, an Arabidopsis GH16 enzyme, AtXTH3, was also found to exhibit CXE activity (CXE:XET ratio ∼0.45) when using H3PO3-amorphized cellulose as a donor substrate, though its MXE activity was not assayed (Shinohara et al., 2017). With these few known examples of hetero-transglucosylases, and none in the Poales, it was of great interest to discover in this study that BdXTH8 is a poalean XTH that preferentially exhibits MXE, a hetero-transglycosylase activity. With the elucidation of the activity of BdXTH8, enzymes possessing greater MXE than XET activity have therefore been found in both the main groups of land-plants—Equisetales and Poales—that produce the polysaccharide MLG.
BdXTH8 is a grass enzyme that shows (1) appreciable MXE activity and (2) a higher MXE than XET activity. However, it does not exhibit appreciable CXE activity, with either insoluble (paper) cellulose or water-soluble cellulose acetate as the donor substrate. The enzyme’s main role in vivo is likely to be the making and/or breaking of MLG-xyloglucan covalent bonds that stably link two hemicelluloses of the grass cell wall and thus contribute to grass cell wall assembly and/or loosening.
These results support the hypothesis that BdTHX1 regulates genes involved in MLG biosynthesis and remodeling. Examining the function of this transcription factor in B. distachyon likely will allow us to further understand the molecular aspects of MLG synthesis in grasses.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Brachypodium distachyon ecotype Bd21-3 was used in all experiments. Dry seeds of B. distachyon were sown in Jiffy 7 peat pellets (Greenhouse Megastore) and stratified at 4°C for 3 days. Plants were then grown in a controlled growth chamber under long-day conditions with 20-h light/4-h dark cycles at a fluence rate of 150 μmol photons/m2s and relative humidity of 60%. Growth temperatures were 24°C during the day and 18°C at night. Stem and grain tissues used for RNA-seq were from plants grown under stem elongation conditions (16-h-light/8-h-dark cycle) as previously described (Jensen and Wilkerson, 2017).
Plasmid Construction and B. distachyon Transformation
The C-terminal and full-length BdTHX1 coding sequence were amplified and cloned into the Gateway vector pDEST17 (Invitrogen) for expressing 6×HIS-tBdTHX1 and 6×HIS-BdTHX1, respectively. A 3,532-bp genomic DNA fragment of the BdTHX1 promoter upstream of the start codon was amplified by PCR and cloned into p6MoI-asd-GUS for generation of pBdTHX1::GUS. The binary vector p6MoI-asd was ordered from DNA Cloning Service (http://dna-cloning.com). The coding sequences of GUS and BdTHX1 were amplified and cloned into pANIC6B (Mann et al., 2012) to generate pZmUbi::GUS and pZmUbi::BdTHX1, respectively. BdTHX1 cDNA fused with the linker sequence encoding the EAR motif transcriptional repression domain (LDLDLELRLGFA; Hiratsu et al., 2003) was used to generate constructs expressing BdTHX1-EAR. Different regions of the BdTHX1 cDNA fragment were cloned into pANIC12A (Mann et al., 2012) for generation of BdTHX1 RNAi constructs. The coding sequence of BdXTH8 25 to 301 amino acids was amplified and cloned into pPICZα A (Invitrogen) to generate pPICZα A-BdXTH8 for expressing BdXTH8 as a secreted protein with a c-myc epitope and a poly-His tag in P. pastoris. Primers used for cloning are listed in Supplemental Table S4. B. distachyon transformations were performed according to Agrobacterium tumefaciens-mediated infection of Bd21-3 embryonic callus tissue (Vogel and Hill, 2008).
GUS Staining Assay and mRNA In Situ Hybridization
The histochemical GUS assay was performed as described previously (Jefferson et al., 1987) using a commercial kit (Sigma-Aldrich) as detailed in the Supplemental Materials and Methods.
Probes and plant samples for mRNA in situ hybridization were prepared as described in the Supplemental Materials and Methods. mRNA in situ hybridization was carried out as previously described (Karlgren et al., 2009), with a minor modification to the hybridization temperature, here using 65°C to hybridize and 70°C for posthybridization washing steps to decrease nonspecific binding. Detection of digoxigenin-labeled nucleic acids was performed using an Anti-Digoxigenin-Alkaline Phosphatase conjugate and color substrate solution, nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate, according to the manufacturer's instructions (Roche). Slides were stored in the dark for 7 d before being washed and fixed with in situ resin (Electron Microscopy Sciences). Images of plant sections were obtained using bright field through a Zeiss Axio Imager.M2 microscope.
Preparation of AIR and MLG Assay
AIR was prepared as previously described (Foster et al., 2010) from different tissues of B. distachyon. MLG content in the AIR was assayed using a commercial Mixed Linkage β-Glucan Assay Kit (Megazyme) with slight modifications. The freeze-dried AIR was weighed, and about 3 mg of AIR from endosperm and 10 mg of AIR from the other tissues were subjected to the MLG assay. One-tenth volume of the buffers and enzymes used in the assay for barley (Hordeum vulgare) and fiber samples were used for each sample in this experiment.
Generation and Purification of Anti-BdTHX1 and Protein Immunoblot Analysis
To obtain the BdTHX1-specific antibody, the protein sequence was compared with other sequences from B. distachyon using the BLAST program. The C-terminal region from 572 to 758 amino acids was found to be specific to BdTHX1. The expressed 6×HIS-tBdTHX1 from Escherichia coli cells was purified by affinity chromatography and sent to a commercial facility (Cocalico Biologicals) to generate an antibody. Final bleeds were taken from two rabbits after 65 d and four consecutive injections. The antiserum was affinity purified using immobilized HIS-tagged antigen columns as previously described (Smith et al., 1992) with details described in the Supplemental Materials and Methods. Protein immunoblot analysis was performed as previously described (Jensen et al., 2014).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed as previously described (Kaufmann et al., 2010; Li et al., 2014) with modifications adapted to B. distachyon immature seeds. Immature seeds (12 to 20 d after fertilization) from 6-week-old plants grown under long-day conditions were harvested and cross linked for 10 min with cross linking buffer (400 mm Suc, 10 mm Tris-HCl, pH 8.0, and 2% (v/v) formaldehyde) under a vacuum and quenched by Gly (Haring et al., 2007). Fixed samples were ground to a fine powder using a Qiagen grind jar set with liquid nitrogen. Nuclei were extracted as described previously (Peterson et al., 2000; Lutz et al., 2011) with modifications and chromatin was sheared as described in the Supplemental Materials and Methods. The sheared chromatin was diluted 10-fold by addition of ChIP dilution buffer (10 mm Tris-HCl, pH 8.0, 1.2 mm EDTA, pH 8.0, 100 mm NaCl, 0.25% (v/v) Triton X-100, 1 mm phenylmethylsulfonyl fluoride and protease inhibitor). The diluted chromatin was precleared by adding preimmune serum and then protein A-agarose beads as described previously (Kaufmann et al., 2010). The precleared chromatin was equally divided into two parts: one, the IP sample, was incubated with purified anti-BdTHX1, and the other, the negative control, was incubated with preimmune serum. Immunoprecipitation was performed, and the ChIP DNA was purified as described previously (Li et al., 2014).
Transcriptome Profiling, ChIP Sequencing, and Data Analysis
Total RNA was prepared using the RNeasy Plant Mini Kit (QIAGEN) according to the manufacturer’s instructions. Total RNA or ChIP DNAs were submitted to the Research Technology Support Facility at MI State University for next-generation sequencing library preparation and sequencing. Libraries were pooled for multiplexed sequencing on an Illumina HiSeq 2500 High Output flow cell (v4) and sequenced in a 1 × 50 bp single-read format using HiSeq SBS reagents.
Sequence reads were aligned to B. distachyon strain Bd21 v1.0 using the Map Reads to Reference tool in CLC Genomics Workbench version 8.5 (similarity fraction, 0.8; length fraction, 0.75). For transcriptome profiling, expression levels were determined as reads per kilobase of transcript per million mapped reads, and gene coexpression was analyzed by using Pearson’s correlation coefficient as the metric. Peak regions of ChIP-seq were identified independently for biological replicates using the callpeaks program in MACS version 2.1.0 (Zhang et al., 2008) to compare positive IP samples versus negative control (q, 0.01). Peaks with overlapping regions in both samples were identified using the batch-consistency analysis Rscript from the irreproducibility discovery rate (IDR) framework (Li et al., 2011). The top 100 peaks by IDR score were selected for motif detection by MEME version 4.11 (Bailey and Elkan, 1994). Three hundred basepairs of sequence centered on each peak summit were analyzed for motifs with zero or one occurrence per sequence (mod, zoops; minw, 6; maxw, 12; nmotifs, 5). The first motif identified by MEME was aligned to the top 500 peaks by IDR score using program Find Individual Motif Occurrences (Grant et al., 2011). Matching peak regions were annotated with nearby gene details, peak summit locations and distance from motif alignment to peak summit using custom scripts.
Cell-free Wheat Germ Protein Synthesis and EMSA
BdTHX1 protein was synthesized using a cell-free wheat germ expression system (Takai et al., 2010) with the Premium PLUS Expression Kit (Cell Free Sciences) according to the manufacturer’s instructions. E. coli DHFR protein synthesized using the same system was used as a control. Labeled DNA probes larger than 100 bp were amplified with biotin-labeled primers, while smaller probes were synthesized by annealing biotin-labeled complementary oligonucleotides. The specific competitors were used at a 200-fold molar excess and had the same sequence as that of labeled probes. Labeled probes were incubated with wheat germ cell-free extract containing the synthesized protein for 20 min on ice in binding buffer (10 mm Tris, 100 mm KCl, 5 mm MgCl2, 50 ng/μL poly(deoxyinosinic-deoxycytidylic), 2.5% [v/v] glycerol, 0.05% [v/v] nonyl phenoxypolyethoxylethanol (NP-40), and 1 mm dithiothreitol, pH 7.5). The reaction mixture was electrophoresed on 5% TBE gels and transferred to a positively charged nylon membrane. The transferred DNA was cross linked to a membrane by UV irradiation, and the biotin-labeled DNA was detected by chemiluminescence.
Secreted Protein Expression in P. pastoris
The pPICZα A-BdXTH8 construct was transformed into yeast P. pastoris strain X-33. To screen clones with multiple inserts, genomic DNA from yeast cells was extracted as previously described (Lõoke et al., 2011), and quantitative genomic PCR, also called Jackpot screening, was performed as described in the Supplemental Materials and Methods. A single clone of P. pastoris cells with multiple copies of BdXTH8 was grown in 10 mL of BMGY (1% [w/v] yeast extract, 2% [w/v] peptone, 100 mm potassium phosphate, pH 6.0, 1.34% [w/v] yeast nitrogen base with ammonium sulfate without amino acids, 1.637 μm biotin, and 0.5% [v/v] methanol) in a 250-mL baffled flask overnight at 28°C. This 10-mL culture was grown in 500 mL of buffered complex glycerol medium (BMGY) in a 4-L flask until the culture reached the log phrase. To induce expression, cells were harvested and resuspended to an optical density at 600 nm (OD600) of1.0 in buffered complex methanol medium (BMMY) (1% [w/v] yeast extract, 2% [w/v] peptone, 100 mm potassium phosphate, pH 6.0, 1.34% [w/v] yeast nitrogen base with ammonium sulfate without amino acids, 1.637 μm biotin, and 0.5% [v/v] methanol). The cells were grown at 18°C, and methanol was added to a final concentration of 0.5% (v/v) every 24 h. The supernatant was obtained after 72 h of induction by centrifugation at 3,000g for 5 min at room tempreture.
Radiochemical Transglucanase Assays
BdXTH8 secreted into the P. pastoris medium was precipitated with ammonium sulfate at 80% saturation and desalted by dialysis. XET, MXE, and CXE activities were assayed essentially as described in Simmons et al. (2015). A 20-µL reaction mixture containing 8 µL concentrated BdXTH8, 0.5% (w/v) tamarind xyloglucan (for XET activity), medium-viscosity barley MLG (for MXE activity) or water-soluble cellulose acetate (WSCA; for CXE activity; Fry et al., 2008a) as the donor substrate, 1 kBq [3H]XXXGol (∼640 MBq µmol−1) as the acceptor substrate, 0.1% (w/v) bovine serum albumin and 0.1 m succinate (Na+, pH 5.5). For testing CXE activity on an insoluble cellulose donor substrate, Whatman filter paper No. 1 (20 mg) pretreated with 6 m NaOH (alkali paper) was soaked with 20 µL reaction mixture, where the soluble donor substrate was replaced by water. NaOH pretreatment was shown to improve paper as a donor substrate for transglycosylation (Simmons et al., 2015). After incubation for up to 36 h, XET and MXE reactions were stopped by addition of formic acid. Reactions with WSCA as a donor substrate were stopped with NH3, converting WSCA to (water-insoluble) cellulose (Fry et al., 2008a), which was then repeatedly washed in water. Soluble transglycosylation products (produced by XET and MXE) were dried on Whatman No. 3 paper, washed in running tap water overnight, and redried, and paper-bound 3H was quantified by scintillation counting in GoldStar ‘O’ scintillation fluid (Meridian, Chesterfield, UK). CXE reactions on alkali paper were stopped with formic acid, and papers were washed in 6 m NaOH at 25°C overnight, followed by 6 m NaOH at 100°C for 0.5 h, running tap water overnight, and 2% acetic acid at 25°C for 1 h; the paper was then dried and assayed for 3H in GoldStar ‘O’ scintillation fluid. To test the effect of pH on XET and MXE activity of BdXTH8, we replaced succinate by citrate buffers (pH 3.2–7.4) in the reaction mixtures. Donor substrate-free assays served as controls.
Immunofluorescence Microscopy
Green regenerating shoots from transformed calli of control and pZmUbi::BdTHX1 were collected and fixed in 0.1 m phosphate buffer with 4% (v/v) paraformaldehyde and 0.5% (v/v) glutaraldehyde, pH 7.2, for at least 4 h. Fixed samples were dehydrated using a series of ethanol and embedded in London Resin-white (Electron Microscopy Sciences). Thin sections were cut on a Power Tome XL ultramicrotome and placed on silane-coated slides (Electron Microscopy Sciences). Sections were blocked in 5% (w/v) nonfat milk in phosphate-buffered saline (PBS) at room temperature for 15 min. After one rinse with PBS, sections were incubated for 15 min at room temperature with MLG monoclonal antibody (diluted 1:25; Biosupplies Australia). After washing three times with PBS, sections were incubated for 15 min at room temperature with anti-mouse immunoglobulin G conjugated to fluorescein isothiocyanate (IgG-FITC) antibody (diluted 1:100, Sigma-Aldrich). Sections were washed three times with PBS and then mounted with CitiFluor. Images were taken with an Olympus FluoView 1000 laser scanning confocal microscope.
Phylogenetic Analysis
Phylogenetic trees were constructed as described by Jensen et al. (2013) with codon-based cDNA alignment and bootstrapping values based on 100 trees. Sequences were obtained from the Phytozome 12 database (Goodstein et al., 2012; http://phytozome.jgi.doe.gov/) from the genome annotations as detailed in the Supplemental Materials and Methods.
Accession Numbers
Sequence data for the genes mentioned in this study can be found in B. distachyon genome as follows: BdTHX1 (Bradi5g17150.1), BdTHX2 (Bradi1g77610.1), BdCSLF6 (Bradi3g16307.1), BdXTH8 (Bradi3g18600.1). The transcriptional profiling data used in this article can be found in the Short Read Archive at NCBI (https://www.ncbi.nlm.nih.gov/sra) and the BioSample accession numbers are as follows: elongating leaf, SAMN10078797; mature leaf, SAMN00727911; node, SAMN10078800; internode 1.1, SAMN10078801; internode 1.2, SAMN10078802; internode 1.3, SAMN10078803; internode 1.4, SAMN10078804; internode 2, SAMN10078805; internode 3, SAMN08097934; internode 4, SAMN08097937; internode 5, SAMN08097940; internode 6, SAMN08097943; endosperm 10 DAF, SAMN10078806; endosperm 12 DAF, SAMN10078807; endosperm 16 DAF, SAMN10078808; integument, SAMN10078809.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Coexpression profile of BdCSLF6 and BdTHX1 from a series of tissues of B. distachyon at different developmental stages.
Supplemental Figure S2. THX1 has a similar expression pattern as CSLF6 in maize and rice.
Supplemental Figure S3. Different stages of tissues used for GUS staining and MLG assay.
Supplemental Figure S4. Histochemical analysis of GUS expression in cross sections of pBdTHX1::GUS transgenic B. distachyon.
Supplemental Figure S5. The specificity of BdTHX1 antibody.
Supplemental Figure S6. Protein sequence alignment and expression profile of BdTHX2.
Supplemental Figure S7. ChIP-seq enrichment and expression profiles of the three highest scoring binding sites identified by the MACS program.
Supplemental Figure S8. Sequences data for BdTHX1 binding regions near BdCSLF6 and BdXTH8.
Supplemental Figure S9. Analysis of the binding motif in BdCSLF6.
Supplemental Figure S10. Phylogenetic analysis and gene expression profiles of four grass-specific BdXTHs.
Supplemental Figure S11. P. pastoris expressed secreted BdXTH8 and lack of hydrolytic activity of BdXTH8.
Supplemental Figure S12. Binding of BdTHX2 to BdCSLF6 and BdXTH8.
Supplemental Figure S13. Immunolabeling of pZmUbi::BdTHX1 transgenic shoots.
Supplemental Figure S14. Morphology and expression of BdTHX1, BdCSLF6 and BdXTH8 in BdTHX1 RNAi lines.
Supplemental Table S1. Top 100 binding sites identified from ChIP-seq experiments.
Supplemental Table S2. Prevalence analysis of GT motifs in introns of grass CSLF6.
Supplemental Table S3. The details of transformation obtaining overexpression plants.
Supplemental Table S4. Primers used in this study.
Acknowledgments
We thank Nick Thrower (Michigan State University) for bioinformatics assistance in analyzing the ChIP-seq data, Dr. Javier Sampedro (Universidad de Santiago, Spain) for generously providing plasmid pERV1, Alicia Withrow (Michigan State University) for help with transmission electron microscopy, Sang-Jin Kim and Linda Danhof (Michigan State University) for sharing Pichia Jackpot screening protocol, and Emily Frankman (Michigan State University) for editing this manuscript.
Footnotes
This work was supported by the US Department of Energy Great Lakes Bioenergy Research Center (grant no. BER DE-FC02-07ER64494 and BER DE-SC0018409) and the UK Biotechnology and Biological Sciences Research Council (BBSRC; grant no. BB/N002458/1).
Articles can be viewed without a subscription.
References
- Bailey TL, Elkan C (1994) Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers. Proc Second Int Conf Intell Syst Mol Biol. 28–36 [PubMed] [Google Scholar]
- Bianchi M, Crinelli R, Giacomini E, Carloni E, Magnani M (2009) A potent enhancer element in the 5′-UTR intron is crucial for transcriptional regulation of the human ubiquitin C gene. Gene 448: 88–101 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Fincher GB (2012) Current challenges in cell wall biology in the cereals and grasses. Front Plant Sci 3: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Fincher GB (2014) Evolution and development of cell walls in cereal grains. Front Plant Sci 5: 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB (2006) Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science 311: 1940–1942 [DOI] [PubMed] [Google Scholar]
- Burton RA, Jobling SA, Harvey AJ, Shirley NJ, Mather DE, Bacic A, Fincher GB (2008) The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol 146: 1821–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Gidley MJ, Fincher GB (2010) Heterogeneity in the chemistry, structure and function of plant cell walls. Nat Chem Biol 6: 724–732 [DOI] [PubMed] [Google Scholar]
- Burton RA, Collins HM, Kibble NAJ, Smith JA, Shirley NJ, Jobling SA, Henderson M, Singh RR, Pettolino F, Wilson SM, et al. (2011) Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-β-d-glucans and alters their fine structure. Plant Biotechnol J 9: 117–135 [DOI] [PubMed] [Google Scholar]
- Carpita NC. (1996) Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol 47: 445–476 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Carpita NC, McCann MC (2010) The maize mixed-linkage (1→3),(1→4)-β-ᴅ-glucan polysaccharide is synthesized at the Golgi membrane. Plant Physiol 153: 1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen U, Alonso-Simon A, Scheller HV, Willats WGT, Harholt J (2010) Characterization of the primary cell walls of seedlings of Brachypodium distachyon—a potential model plant for temperate grasses. Phytochemistry 71: 62–69 [DOI] [PubMed] [Google Scholar]
- Cox JM, Hayward MM, Sanchez JF, Gegnas LD, van der Zee S, Dennis JH, Sigler PB, Schepartz A (1997) Bidirectional binding of the TATA box binding protein to the TATA box. Proc Natl Acad Sci USA 94: 13475–13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehesh K, Hung H, Tepperman JM, Quail PH (1992) GT-2: a transcription factor with twin autonomous DNA-binding domains of closely related but different target sequence specificity. EMBO J 11: 4131–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos MK, Sieburth LE (2000) Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12: 1799–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A (2009) A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci USA 106: 5996–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira SS, Hotta CT, Poelking VG de C, Leite DCC, Buckeridge MS, Loureiro ME, Barbosa MHP, Carneiro MS, Souza GM (2016) Co-expression network analysis reveals transcription factors associated to cell wall biosynthesis in sugarcane. Plant Mol Biol 91: 15–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher GB. (2009) Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol 149: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010) Comprehensive compositional analysis of plant cell walls (Lignocellulosic biomass) part I: lignin. J Vis Exp 3: 5–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franková L, Fry SC (2013) Biochemistry and physiological roles of enzymes that ‘cut and paste’ plant cell-wall polysaccharides. J Exp Bot 64: 3519–3550 [DOI] [PubMed] [Google Scholar]
- Fry SC, Mohler KE, Nesselrode BHWA, Franková L (2008a) Mixed-linkage β-glucan : xyloglucan endotransglucosylase, a novel wall-remodelling enzyme from Equisetum (horsetails) and charophytic algae. Plant J 55: 240–252 [DOI] [PubMed] [Google Scholar]
- Fry SC, Nesselrode BHWA, Miller JG, Mewburn BR (2008b) Mixed-linkage (1→3,1→4)-β-ᴅ-glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytol 179: 104–115 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Carpita NC (1993) Synthesis of (1→3), (1→4)-β-ᴅ-glucan in the Golgi apparatus of maize coleoptiles. Proc Natl Acad Sci USA 90: 3850–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaut DM, Pauly M, Bacic A, Fincher GB (2005) Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 221: 729–738 [DOI] [PubMed] [Google Scholar]
- Gilmartin PM, Sarokin L, Memelink J, Chua NH (1990) Molecular light switches for plant genes. Plant Cell 2: 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon F, Bouchet B, Jamme F, Robert P, Quéméner B, Barron C, Larré C, Dumas P, Saulnier L (2011) Brachypodium distachyon grain: characterization of endosperm cell walls. J Exp Bot 62: 1001–1015 [DOI] [PubMed] [Google Scholar]
- Guillon F, Larré C, Petipas F, Berger A, Moussawi J, Rogniaux H, Santoni A, Saulnier L, Jamme F, Miquel M, et al. (2012) A comprehensive overview of grain development in Brachypodium distachyon variety Bd21. J Exp Bot 63: 739–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, Yokoyama R, Osakabe K, Toki S, Nishitani K (2014) Function of xyloglucan endotransglucosylase/hydrolases in rice. Ann Bot 114: 1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T. (1989) Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol 40: 139–168 [Google Scholar]
- Henriksson H, Denman SE, Campuzano IDG, Ademark P, Master ER, Teeri TT, Brumer H III (2003) N-linked glycosylation of native and recombinant cauliflower xyloglucan endotransglycosylase 16A. Biochem J 375: 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Kondo M, Aya K, Miyao A, Sato Y, Antonio BA, Namiki N, Nagamura Y, Matsuoka M (2013) Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol 54: 1791–1802 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hrmova M, Farkaš V, Lahnstein J, Fincher GB (2007) A Barley xyloglucan xyloglucosyl transferase covalently links xyloglucan, cellulosic substrates, and (1,3;1,4)-β-D-glucans. J Biol Chem 282: 12951–12962 [DOI] [PubMed] [Google Scholar]
- Hrmova M, Farkaš V, Harvey AJ, Lahnstein J, Wischmann B, Kaewthai N, Ezcurra I, Teeri TT, Fincher GB (2009) Substrate specificity and catalytic mechanism of a xyloglucan xyloglucosyl transferase HvXET6 from barley (Hordeum vulgare L.). FEBS J 276: 437–456 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143: 1467–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Wilkerson CG (2017) Brachypodium as an experimental system for the study of stem parenchyma biology in grasses. PLoS One 12: e0173095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Johnson N, Wilkerson CG (2013) Discovery of diversity in xylan biosynthetic genes by transcriptional profiling of a heteroxylan containing mucilaginous tissue. Front Plant Sci 4: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JK, Johnson NR, Wilkerson CG (2014) Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant J 80: 207–215 [DOI] [PubMed] [Google Scholar]
- Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42: D1182–D1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Tian F, Yang D-C, Meng Y-Q, Kong L, Luo J, Gao G (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45(D1): D1040–D1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan-Levy RN, Brewer PB, Quon T, Smyth DR (2012) The trihelix family of transcription factors--light, stress and development. Trends Plant Sci 17: 163–171 [DOI] [PubMed] [Google Scholar]
- Kaplan-Levy RN, Quon T, O’Brien M, Sappl PG, Smyth DR (2014) Functional domains of the PETAL LOSS protein, a trihelix transcription factor that represses regional growth in Arabidopsis thaliana. Plant J 79: 477–491 [DOI] [PubMed] [Google Scholar]
- Karlgren A, Carlsson J, Gyllenstrand N, Lagercrantz U, Sundström JF (2009) Non-radioactive in situ hybridization protocol applicable for Norway spruce and a range of plant species. J Vis Exp 26: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Østerås M, Farinelli L, Krajewski P, Angenent GC (2010) Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat Protoc 5: 457–472 [DOI] [PubMed] [Google Scholar]
- Kaur S, Dhugga KS, Beech R, Singh J (2017) Genome-wide analysis of the cellulose synthase-like (Csl) gene family in bread wheat (Triticum aestivum L.). BMC Plant Biol 17: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Olek AT, Carpita NC (2000) Cell wall and membrane-associated exo-β-D-glucanases from developing maize seedlings. Plant Physiol 123: 471–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-J, Zemelis S, Keegstra K, Brandizzi F (2015) The cytoplasmic localization of the catalytic site of CSLF6 supports a channeling model for the biosynthesis of mixed-linkage glucan. Plant J 81: 537–547 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Zemelis-Durfee S, Jensen JK, Wilkerson CG, Keegstra K, Brandizzi F (2018) In the grass species Brachypodium distachyon, the production of mixed-linkage (1,3;1,4)-β-glucan (MLG) occurs in the Golgi apparatus. Plant J 93: 1062–1075 [DOI] [PubMed] [Google Scholar]
- Lazaridou A, Biliaderis CG (2004) Cryogelation of cereal β-glucans: structure and molecular size effects. Food Hydrocoll 18: 933–947 [Google Scholar]
- Li B, Jiang S, Yu X, Cheng C, Chen S, Cheng Y, Yuan JS, Jiang D, He P, Shan L (2015) Phosphorylation of trihelix transcriptional repressor ASR3 by MAP KINASE4 negatively regulates Arabidopsis immunity. Plant Cell 27: 839–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brown JB, Huang H, Bickel PJ (2011) Measuring reproducibility of high-throughput experiments. Ann Appl Stat 5: 1752–1779 [Google Scholar]
- Li W, Lin Y-C, Li Q, Shi R, Lin C-Y, Chen H, Chuang L, Qu G-Z, Sederoff RR, Chiang VL (2014) A robust chromatin immunoprecipitation protocol for studying transcription factor-DNA interactions and histone modifications in wood-forming tissue. Nat Protoc 9: 2180–2193 [DOI] [PubMed] [Google Scholar]
- Lis M, Walther D (2016) The orientation of transcription factor binding site motifs in gene promoter regions: does it matter? BMC Genomics 17: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little A, Schwerdt JG, Shirley NJ, Khor SF, Neumann K, O’Donovan LA, Lahnstein J, Collins HM, Henderson M, Fincher GB, et al. (2018) Revised phylogeny of the Cellulose Synthase gene superfamily: insights into cell wall evolution. Plant Physiol 177: 1124–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Liu Z (2008) The second intron of AGAMOUS drives carpel- and stamen-specific expression sufficient to induce complete sterility in Arabidopsis. Plant Cell Rep 27: 855–863 [DOI] [PubMed] [Google Scholar]
- Lõoke M, Kristjuhan K, Kristjuhan A (2011) Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques 50: 325–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz KA, Wang W, Zdepski A, Michael TP (2011) Isolation and analysis of high quality nuclear DNA with reduced organellar DNA for plant genome sequencing and resequencing. BMC Biotechnol 11: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. (2011) Transcriptional activators and activation mechanisms. Protein Cell 2: 879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQuarrie KL, Fong AP, Morse RH, Tapscott SJ (2011) Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet 27: 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DGJ, Lafayette PR, Abercrombie LL, King ZR, Mazarei M, Halter MC, Poovaiah CR, Baxter H, Shen H, Dixon RA, et al. (2012) Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol J 10: 226–236 [DOI] [PubMed] [Google Scholar]
- Mohler KE, Simmons TJ, Fry SC (2013) Mixed-linkage glucan:xyloglucan endotransglucosylase (MXE) re-models hemicelluloses in Equisetum shoots but not in barley shoots or Equisetum callus. New Phytol 197: 111–122 [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G (2013) Enhancers: five essential questions. Nat Rev Genet 14: 288–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DG, Tomkins JP, Frisch DA, Wing RA, Paterson AH (2000) Construction of plant bacterial artificial chromosome (BAC) libraries: An illustrated guide. J Agric Genomics 5: https://wheat.pw.usda.gov/jag/ [Google Scholar]
- Pettolino F, Sasaki I, Turbic A, Wilson SM, Bacic A, Hrmova M, Fincher GB (2009) Hyphal cell walls from the plant pathogen Rhynchosporium secalis contain (1,3/1,6)-β-D-glucans, galacto- and rhamnomannans, (1,3;1,4)-β-D-glucans and chitin. FEBS J 276: 3698–3709 [DOI] [PubMed] [Google Scholar]
- Popper ZA, Fry SC (2003) Primary cell wall composition of bryophytes and charophytes. Ann Bot 91: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Shinohara N, Sunagawa N, Tamura S, Yokoyama R, Ueda M, Igarashi K, Nishitani K (2017) The plant cell-wall enzyme AtXTH3 catalyses covalent cross-linking between cellulose and cello-oligosaccharide. Sci Rep 7: 46099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM (1997) Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons TJ, Fry SC (2017) Bonds broken and formed during the mixed-linkage glucan : xyloglucan endotransglucosylase reaction catalysed by Equisetum hetero-trans-β-glucanase. Biochem J 474: 1055–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons TJ, Mohler KE, Holland C, Goubet F, Franková L, Houston DR, Hudson AD, Meulewaeter F, Fry SC (2015) Hetero-trans-β-glucanase, an enzyme unique to Equisetum plants, functionalizes cellulose. Plant J 83: 753–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MC, Cook JA, Smanik PA, Wakulchik M, Kasher MS (1992) Kinetically Inert Co(III) linkage through an engineered metal binding site: specific orientation of recombinant human papillomavirus type 16 E7 protein on a solid support. Methods 4: 73–78 [Google Scholar]
- Sørensen I, Pettolino FA, Wilson SM, Doblin MS, Johansen B, Bacic A, Willats WGT (2008) Mixed-linkage (1→3,1→4)-β-ᴅ-glucan is not unique to the Poales and is an abundant component of Equisetum arvense cell walls. Plant J 54: 510–521 [DOI] [PubMed] [Google Scholar]
- Stelpflug SC, Sekhon RS, Vaillancourt B, Hirsch CN, Buell CR, de Leon N, Kaeppler SM (2016) An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9: 1–16 [DOI] [PubMed] [Google Scholar]
- Takai K, Sawasaki T, Endo Y (2010) Practical cell-free protein synthesis system using purified wheat embryos. Nat Protoc 5: 227–238 [DOI] [PubMed] [Google Scholar]
- Thompson JE, Fry SC (2001) Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells. Plant J 26: 23–34 [DOI] [PubMed] [Google Scholar]
- Thompson JE, Smith RC, Fry SC (1997) Xyloglucan undergoes interpolymeric transglycosylation during binding to the plant cell wall in vivo: evidence from 13C/3H dual labelling and isopycnic centrifugation in caesium trifluoroacetate. Biochem J 327: 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sandt VST, Suslov D, Verbelen JP, Vissenberg K (2007) Xyloglucan endotransglucosylase activity loosens a plant cell wall. Ann Bot 100: 1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Sánchez ME, Verhertbruggen Y, Christensen U, Chen X, Sharma V, Varanasi P, Jobling SA, Talbot M, White RG, Joo M, et al. (2012) Loss of cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol 159: 56–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Sánchez ME, Verhertbruggen Y, Scheller HV, Ronald PC (2013) Abundance of mixed linkage glucan in mature tissues and secondary cell walls of grasses. Plant Signal Behav 8: e23143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. (2008) Unique aspects of the grass cell wall. Curr Opin Plant Biol 11: 301–307 [DOI] [PubMed] [Google Scholar]
- Vogel J, Hill T (2008) High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep 27: 471–478 [DOI] [PubMed] [Google Scholar]
- Wang L, Czedik-Eysenberg A, Mertz RA, Si Y, Tohge T, Nunes-Nesi A, Arrivault S, Dedow LK, Bryant DW, Zhou W, et al. (2014) Comparative analyses of C4 and C3 photosynthesis in developing leaves of maize and rice. Nat Biotechnol 32: 1158–1165 [DOI] [PubMed] [Google Scholar]
- Wilson SM, Ho YY, Lampugnani ER, Van de Meene AML, Bain MP, Bacic A, Doblin MS (2015) Determining the subcellular location of synthesis and assembly of the cell wall polysaccharide (1,3; 1,4)-β-D-glucan in grasses. Plant Cell 27: 754–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JR, Fincher GB, Stone BA (1983) Water-soluble (1→3),(1→4)-β-ᴅ-glucans from barley (Hordeum vulgate) endosperm. II. Fine structure. Carbohydr Polym 3: 207–225 [Google Scholar]
- Xie ZM, Zou HF, Lei G, Wei W, Zhou QY, Niu CF, Liao Y, Tian AG, Ma B, Zhang WK, et al. (2009) Soybean Trihelix transcription factors GmGT-2A and GmGT-2B improve plant tolerance to abiotic stresses in transgenic Arabidopsis. PLoS One 4: e6898. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang Q, Cheetamun R, Dhugga KS, Rafalski JA, Tingey SV, Shirley NJ, Taylor J, Hayes K, Beatty M, Bacic A, et al. (2014) Spatial gradients in cell wall composition and transcriptional profiles along elongating maize internodes. BMC Plant Biol 14: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou DX. (1999) Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends Plant Sci 4: 210–214 [DOI] [PubMed] [Google Scholar]
- Zhu B, Zhang W, Zhang T, Liu B, Jiang J (2015) Genome-wide prediction and validation of intergenic enhancers in Arabidopsis using open chromatin signatures. Plant Cell 27: 2415–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]