Molecular chaperone SES1 positively regulates salt stress resistance by alleviating salt-induced ER stress.
Abstract
Salt stress seriously affects plant growth and development. Through genetic screening, we identified and characterized an Arabidopsis (Arabidopsis thaliana) sensitive to salt1 (ses1) mutant. SES1 was ubiquitously expressed and induced by salt treatment. The salt-sensitive phenotype of ses1 was due neither to the overaccumulation of Na+ nor to the suppression of salt tolerance-associated genes. SES1 encoded an uncharacterized endoplasmic reticulum (ER)-localized protein. Coinciding with its subcellular distribution, ses1 exhibited overactivation of unfolded protein response genes and was largely influenced by severe ER stress. Biochemical evidence revealed that SES1 functions as an important molecular chaperone to alleviate salt-induced ER stress. Furthermore, the ER stress sensor basic leucine zipper factor17 transactivated SES1 by binding directly to its promoter region. These results provide insights into salt stress responses and ER homeostasis and shed light on the mechanism by which SES1 modulates salt resistance.
High salinity in soil, which is a crucial environmental problem, causes severe ion imbalance, osmotic stress, and oxidative damage to glycophytes (Parida et al., 2004; Keyster et al., 2012). Nearly 20% of the world’s cultivated land and half of the irrigated regions are affected by salt stress (Zhu, 2002). Therefore, it is fundamentally important to understand the mechanisms underlying plant responses to salt stress, which is the prerequisite for breeding and genetic engineering strategies to improve salt tolerance in plants.
To minimize the detrimental effects of salt stress, plants have developed various elaborate mechanisms to exclude excess salt from their cells or tolerate salt within cells (Munns and Tester, 2008). The salt overly sensitive (SOS) pathway is a vital and conserved regulatory system in plants to sustain Na+/K+ homeostasis and to tolerate salt stress (Zhu, 2003; Martínez-Atienza et al., 2007; Tang et al., 2010). Other categories of ion channels, including the Na+/H+ antiporter NHX, the Ca2+/H+ antiporter CHX, the voltage-dependent cation channels high-affinity K+ transporter (HKT) and K+ uptake permease, and the proton pumps H+-ATPase and H+-pyrophosphatase, also play important roles in maximizing the compartmentation of Na+ (Gaxiola et al., 2001; Senn et al., 2001; Berthomieu et al., 2003; An et al., 2007; Fuglsang et al., 2007; Yang et al., 2010; Barragán et al., 2012; Jia et al., 2015). In addition to the above-mentioned mechanisms, phytohormones and mitogen-activated protein kinases are closely correlated with salt stress tolerance (Rodriguez et al., 2010; Yu et al., 2010; Colebrook et al., 2014; Ovečka et al., 2014; Kazan, 2015; Ryu and Cho, 2015). Moreover, subcellular organelles such as the mitochondria, chloroplasts, Golgi apparatus, and endoplasmic reticulum (ER) are involved in signal transduction and integration under salt stress (Kang et al., 2008; Zhao et al., 2013; Jin and Daniell, 2014; Rodrigues et al., 2017).
The ER is not only the main site of biosynthesis and processing of all secretory and membrane proteins in eukaryotic cells but also is an indispensable organelle for plant adaptation to diverse environmental stresses (Vitale and Boston, 2008; Liu and Howell, 2010b). Under adverse conditions, the correct folding and assembly of proteins in the ER are disturbed, and a set of genes are transcriptionally induced to cope with such situations, which is referred to as the unfolded protein response (UPR) or the ER stress response (Ron and Walter, 2007; Urade, 2007). High salinity, pathogen attack, drought, and heat stress can easily upset the process of protein folding and trigger ER stress responses (Liu et al., 2007a; Gao et al., 2008; Liu and Howell, 2010b; Deng et al., 2011; Ye et al., 2011; Zhang and Wang, 2012). After the initiation of ER stress, cells triple their efforts to relieve protein overloading in the ER (Schröder and Kaufman, 2005; Wan and Jiang, 2016). First, molecular chaperones are up-regulated and vesicle trafficking is enhanced; second, endoplasmic reticulum-assisted degradation (ERAD) of malfunctional proteins is promoted; and third, the translation of secretory proteins is attenuated (Martínez and Chrispeels, 2003).
The UPR is an important cellular mechanism that controls protein homeostasis in the ER and is triggered by the accumulation of unfolded proteins in the ER lumen (Hollien, 2013). The molecular mechanisms of the UPR have been investigated extensively in yeast and animals (Mori, 2009; Oikawa et al., 2010; Wu et al., 2014). The UPR in yeast is controlled by the inositol-requiring transmembrane kinase/endonuclease1 (IRE1p), which belongs to the type I transmembrane ER proteins (Cox and Walter, 1996; Sidrauski and Walter, 1997). In mammalian cells, the UPR is mediated by two types of ER transmembrane proteins. The type I ER stress sensor consists of IRE1, including two identifiable IRE1 isoforms, IRE1α and IRE1β, and protein kinase RNA-like ER kinase (PERK), whereas the type II ER stress sensor comprises the activating transcription factor6 proteins ATF6α and ATF6β (Hetz et al., 2011). Similar to animals, two UPR pathways have been identified in plants. One is mediated by IRE1 and bZIP60 (Urade, 2007; Vitale and Boston, 2008; Nagashima et al., 2011; Deng et al., 2013). IRE1 is a transmembrane protein with a luminal domain that is capable of sensing the accumulation of unfolded proteins through direct interaction with them or by an indirect manner depending on the availability of binding proteins (BiPs; Oikawa et al., 2009, 2012; Pincus et al., 2010; Gardner and Walter, 2011). In Arabidopsis (Arabidopsis thaliana), the mRNA of bZIP60 can be cleaved at specific sites by IRE1 during the stress-triggered UPR (Deng et al., 2011; Nagashima et al., 2011; Moreno et al., 2012). Then, the truncated version of bZIP60 enters the nucleus and activates the expression of target genes (Iwata and Koizumi, 2005). Other than IRE1, two bZIP transcription factors, bZIP17 and bZIP28, also act as ER stress sensors in Arabidopsis (Liu et al., 2007a, 2007b). Under normal conditions, BiPs bind to the C-terminal tail of bZIP17 and bZIP28, retaining them within the ER membrane (Srivastava et al., 2013, 2014). During ER stress, BiPs bind to the unfolded proteins, resulting in the release of bZIP17 and bZIP28 from the ER membrane (Srivastava et al., 2014). After sequential cleavage by proteases in the Golgi (Liu et al., 2007a, 2007b; Iwata et al., 2017), bZIP17 and bZIP28 enter the nucleus and activate the transcription of ER stress-related genes such as BiP, ER DNAJ (ERDJ), calnexin (CNX), calreticulin, peptidylprolyl isomerases, and protein disulfide isomerases (PDI; Liu et al., 2007a, 2007b; Tajima et al., 2008; Liu and Howell, 2010a; Howell, 2013; Srivastava et al., 2014).
In Arabidopsis, unfolded proteins accumulate rapidly in the ER after salt treatment, triggering ER stress (Liu et al., 2011). Knocking out bZIP17, one of the major ER stress sensors, leads to increased sensitivity to salt stress, whereas transgenic plants overexpressing bZIP17 show enhanced salt tolerance, indicating that ER stress responses are closely correlated with salt stress tolerance (Liu et al., 2007a, 2008; Li et al., 2017). However, the reciprocal regulation between them remains unclear. In this study, we identified and characterized an Arabidopsis mutant termed sensitive to salt1 (ses1), which is hypersensitive to salt stress. SES1, which is induced by salt treatment or ER stress inducers, encodes an ER-localized chaperone. Functional disruption of SES1 markedly aggravates salt-induced ER stress. Moreover, we found that the ER stress sensor bZIP17 acts as the upstream transcriptional activator of SES1.
RESULTS
Identification and Characterization of the Salt-Sensitive Mutant ses1
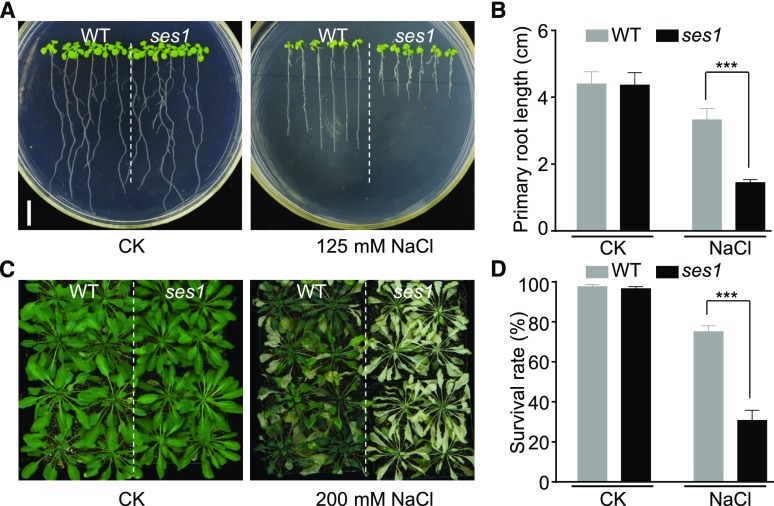
To identify players participating in salt stress tolerance, an ethyl methanesulfonate (EMS) mutagenized M2 population in the Columbia-0 background was carefully screened using the root elongation assay. One mutant designated as ses1 was selected for further analysis. Under normal growth conditions, the ses1 mutant was almost indistinguishable from wild-type plants (Fig. 1, A and B). When seedlings were transferred to NaCl-containing medium, the root growth of ses1 was inhibited more seriously compared with the wild type (Fig. 1, A and B). However, scoring of seed germination based on the emergence of the radicle indicated that the ses1 mutants did not show any obvious difference regardless of salt treatment (Supplemental Fig. S1, A and B). In addition, after treatment with 200 mm NaCl for 2 weeks, the ses1 mutants grown in soil exhibited significantly lower survival rates and more severe leaf chlorosis (Fig. 1, C and D; Supplemental Fig. S1C). These results clearly indicate that ses1 mutants are more vulnerable to salt stress, although no difference in germination rate was observed after salt treatment.
Figure 1.
The ses1 mutant is sensitive to salt stress. A, Images of wild-type (WT) and ses1 mutant seedlings grown on one-half-strength Murashige and Skoog (1/2 MS) agar plates with or without 125 mm NaCl treatment. Photographs were taken after growing vertically at 22°C for 7 d. Bar = 1 cm. B, Root length of the seedlings in A. Error bars indicate sd (n = 18). ***, P < 0.001 (Student’s t test). C, Images of 3-week-old wild-type and ses1 plants with or without 200 mm NaCl treatment. D, Survival rate of the wild-type and ses1 plants in C. Error bars indicate sd (n = 60). ***, P < 0.001 (Student’s t test).
Positional Cloning of the ses1 Mutant Gene
To investigate the nature of the ses1 mutation, we reciprocally crossed ses1 with wild-type plants. All F1 progeny (97 seedlings) showed a similar salt sensitivity to that of wild-type plants. In the F2 population, the ses1 phenotype segregated at a 1:3 ratio (ses1:wild type = 63:210, χ2 = 0.54, P > 0.05), indicating that ses1 is a recessive mutation at a single nuclear locus.
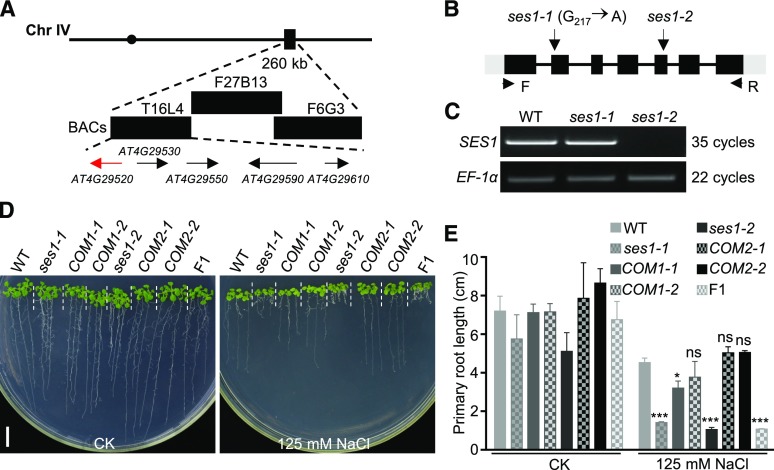
To identify the SES1 gene, the ses1 mutants were crossed with Landsberg erecta, and the F2 progeny exhibiting salt sensitivity were used for genetic mapping. The ses1 mutation was mapped to a 260-kb region on chromosome IV between bacterial artificial chromosome clones T16L4 and F27B13 (Fig. 2A). DNA sequencing of candidate genes in this region revealed a G-to-A transition at nucleotide 217 within the coding region of AT4G29520, which results in the substitution of Glu-73 with a Lys residue (Fig. 2B). AT4G29520 encodes a polypeptide of 306 amino acid residues containing a predicted transmembrane domain and a conserved saposin B domain (Supplemental Fig. S2A). The amino acid substitution in ses1 occurs within the saposin B domain, and the mutated Glu is highly conserved in plants, suggesting that this amino acid is functionally important (Supplemental Fig. S2B). Reverse transcription (RT)-PCR analysis revealed that this point mutation had no obvious effects on the expression level and transcript stability of AT4G29520 (Fig. 2C).
Figure 2.
Map-based cloning of SES1. A, Genetic mapping of SES1. Markers used for the genetic mapping are shown on the top, and the predicted genes are shown at the bottom. The arrows indicate the direction of transcription, and the candidate gene for SES1 is shown in red. BAC, Bacterial artificial chromosome. B, Genome structure of SES1. The black boxes, gray boxes, and lines indicate exons, untranslated regions, and introns, respectively. The positions of the ses1 mutant alleles and the PCR primers for RT-PCR are shown. C, RT-PCR analysis of the SES1 transcripts in wild-type (WT), ses1-1, and ses1-2 mutant plants. EF-1α was used as a loading control. D, Phenotypes of ses1-1, ses1-2, complementary lines (COM), and F1 mutants with or without 125 mm NaCl treatment. COM1-1 and COM1-2 are in the ses1-1 background, while COM2-1 and COM2-2 are in the ses1-2 background. Photographs were taken after growing vertically at 22°C for 10 d. Bar = 1 cm. E, Root length of the seedlings in D. Error bars indicate sd (n = 6). ns indicates no significant difference from the wild type. *, P < 0.05 and ***, P < 0.001 (Student’s t test).
To confirm that AT4G29520 is SES1, we complemented the ses1 mutant with a 3.8-kb genomic DNA fragment containing the native promoter and the coding sequence of AT4G29520. T3 seeds from two independent homozygous single-insertion lines fully restored the salt-sensitive phenotype of ses1 (Fig. 2, D and E). We renamed our original mutant as ses1-1 and obtained an additional T-DNA insertion mutant of AT4G29520 (ses1-2) from the GABI-KAT center (http://www.GABI-Kat.de) in the Columbia-0 background, in which the full-length transcripts of SES1 could not be detected (Fig. 2C). To test possible allelism between ses1-1 and ses1-2, we crossed these two materials and found that all F1 progeny showed a salt-sensitive phenotype (Fig. 2, D and E), demonstrating that ses1-1 and ses1-2 mutants are allelic. The 3.8-kb DNA fragment also completely rescued the salt-sensitive phenotype of ses1-2 (Fig. 2, D and E). Taken together, we conclude that the ses1-1 mutant phenotype is indeed caused by a mutation in AT4G29520 and that SES1 is required for Arabidopsis salt tolerance.
Overexpression of SES1 Does Not Enhance Salt Tolerance
As a loss-of-function mutation involving SES1 results in hypersensitivity to salt, we hypothesized that its overexpression would enhance salt tolerance. To test our hypothesis, we generated transgenic plants overexpressing SES1 under the control of the 35S promoter. The p35S::SES1 transgenic lines all showed substantially higher levels of SES1 expression compared with the wild-type plants (Supplemental Fig. S3A). Three overexpression lines, OE1, OE5, and OE6, were characterized further. No visible enhancement in salt tolerance was observed in the SES1-overexpressing plants (Supplemental Fig. S3, B and C), indicating that excess SES1 is insufficient to improve the salt tolerance of Arabidopsis.
SES1 Is Required for Both Ionic and Osmotic Stress Tolerance
To test whether SES1 is involved specifically in NaCl tolerance, we assayed other kinds of salts, including NaNO3, KCl, and KNO3. ses1-1 and ses1-2 exhibited hypersensitivity to these types of salts (Supplemental Fig. S4, A and B). High salinity causes both ionic and osmotic stresses. To determine in which kind of stress SES1 is involved, we grew 4-d-old wild-type, ses1-1, and ses1-2 seedlings vertically on 1/2 MS agar plates with or without 250 mm mannitol/12 mm LiCl. After treatment for 7 d, both ses1-1 and ses1-2 displayed sensitivity to mannitol, whereas only ses1-2 showed significant differences on the LiCl plates compared with the wild-type seedlings (Supplemental Fig. S4, B and C). When germinated and grown in the presence of 12 mm LiCl, ses1-1 and ses1-2 displayed hypersensitivity (Supplemental Fig. S4D). Moreover, root elongation in the ses1 mutants was suppressed more severely under salt stress conditions compared with the mannitol treatment (Supplemental Fig. S4B). Taken together, these results reveal that SES1 is required for both ionic and osmotic stress tolerance.
SES1 Is Expressed Ubiquitously and Induced by Salt Stress
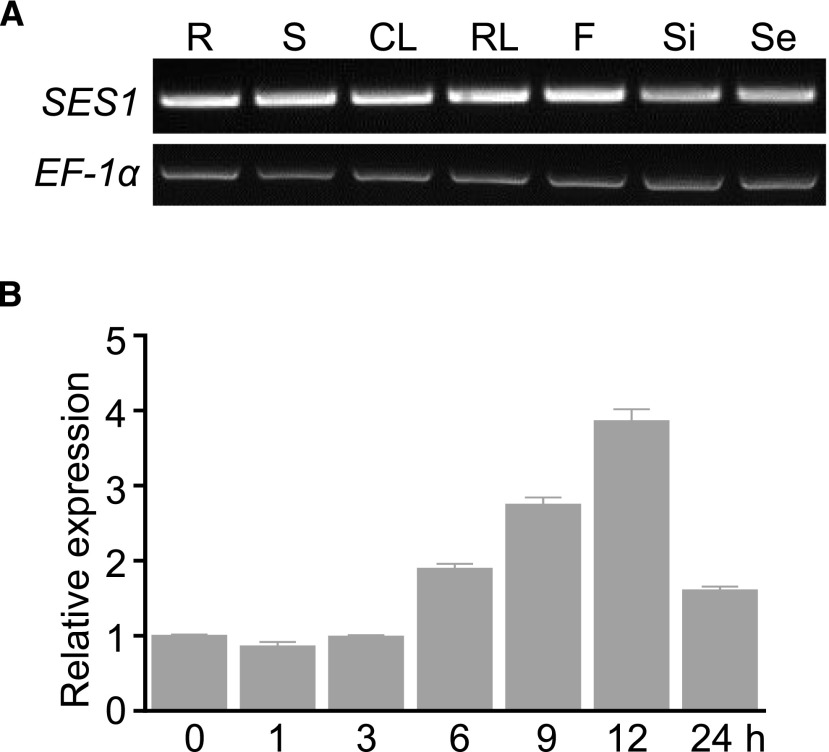
To investigate the expression patterns of SES1, we performed RT-PCR using total RNA extracted from various tissues, including roots, shoots, rosette leaves, cauline leaves, flowers, siliques, and seeds. SES1 expression was detected in all examined tissues (Fig. 3A).
Figure 3.
Expression patterns of SES1. A, SES1 in different tissues detected by RT-PCR (35 cycles). EF-1α was used as a loading control (22 cycles). R, Root; S, stem; CL, cauline leaf; RL, rosette leaf; F, flower; Si, silique; Se, seed. B, Responsiveness of SES1 to NaCl treatment analyzed by RT-qPCR. RNA samples were extracted from 10-d-old seedlings after treatment with 200 mm NaCl for different periods. The data were normalized against the expression of ACTIN7 and UBQ10. The means were calculated from three independent replicates and compared with the no-treatment condition (0 h). Error bars indicate se.
Given that SES1 plays vital roles in salt stress tolerance, we assessed the responsiveness of SES1 to salt stress by reverse transcription quantitative PCR (RT-qPCR). SES1 was up-regulated gradually in Arabidopsis seedlings after treatment with 200 mm NaCl (Fig. 3B). Similar to the observed salt responses in whole seedlings, the expression levels of SES1 also increased in different tissues upon salt treatment, except for the seeds (Supplemental Fig. S5). These data reveal that ubiquitous SES1 is salt inducible.
SES1 Is Localized to the ER
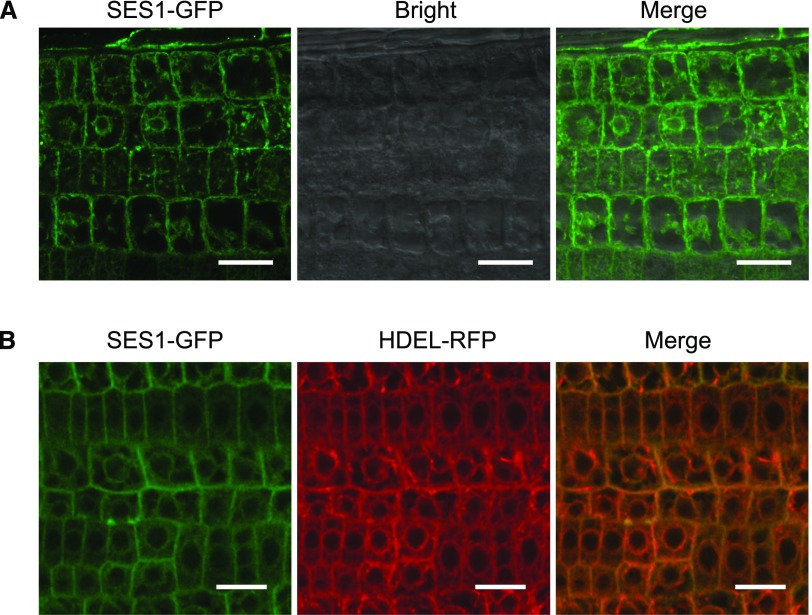
Bioinformatics analysis using the Subcellular Location of Proteins in Arabidopsis Database (http://suba.live), InterPro (http://www.ebi.ac.uk/interpro/), and TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) predicted an N-terminal signal peptide, a transmembrane domain, and a long noncytoplasmic C terminus in SES1, indicating the probable ER location of SES1 and the ER lumen distribution of its C terminus (Supplemental Figs. S2A and S6, A and B). To determine the subcellular localization of SES1, we generated transgenic Arabidopsis plants harboring a SES1-GFP fusion driven by the 35S promoter (p35S::SES1-GFP). To reveal the bona fide subcellular localization of SES1, we first examined whether the biological activity of SES1 is influenced by adding a GFP tag. By stable transformation, we found that the salt-sensitive phenotype of ses1-1 was fully rescued by SES1-GFP (Supplemental Fig. S6C), suggesting that this fusion could report the correct distribution of SES1. Then, SES1 was localized by high-resolution laser confocal microscopy. The GFP fluorescence signals were observed mainly in the perinuclear region, which resembles the ER structure (Fig. 4A). To corroborate this result, we conducted SES1-GFP colocalization experiments with an ER-specific organelle marker, HDEL-RFP. For this assay, we crossed p35S::SES1-GFP transgenic plants with a p35S::HDEL-RFP transgenic line and analyzed the F1 hybrids. Figure 4B shows that the GFP signals almost completely colocalized with HDEL-RFP. In addition, after treatment with 200 mm NaCl, no alteration in the subcellular distribution of SES1 was observed (Supplemental Fig. S6D). Taken together, these findings indicate that SES1 functions in the ER under normal or salt-stressed conditions. SES1 (AT4G29520) is defined as a nucleophosmin in ARAPORT (www.araport.org) and TAIR (www.arabidopsis.org), which is largely a nucleolus-located protein. However, given its ER localization, previous annotations about AT4G29520 seem to be incorrect.
Figure 4.
SES1 is localized to the ER. A, GFP fluorescence of p35S::SES1-GFP transgenic lines was imaged with a high-resolution laser confocal microscope at 488 nm. Bars = 20 µm. B, GFP and RFP fluorescence of transgenic lines containing SES1-GFP and HDEL-RFP was imaged with a high-resolution laser confocal microscope at 488 and 610 nm, respectively. Bars = 20 µm.
Salt Sensitivity of ses1 Is Not Due to the Overaccumulation of Na+
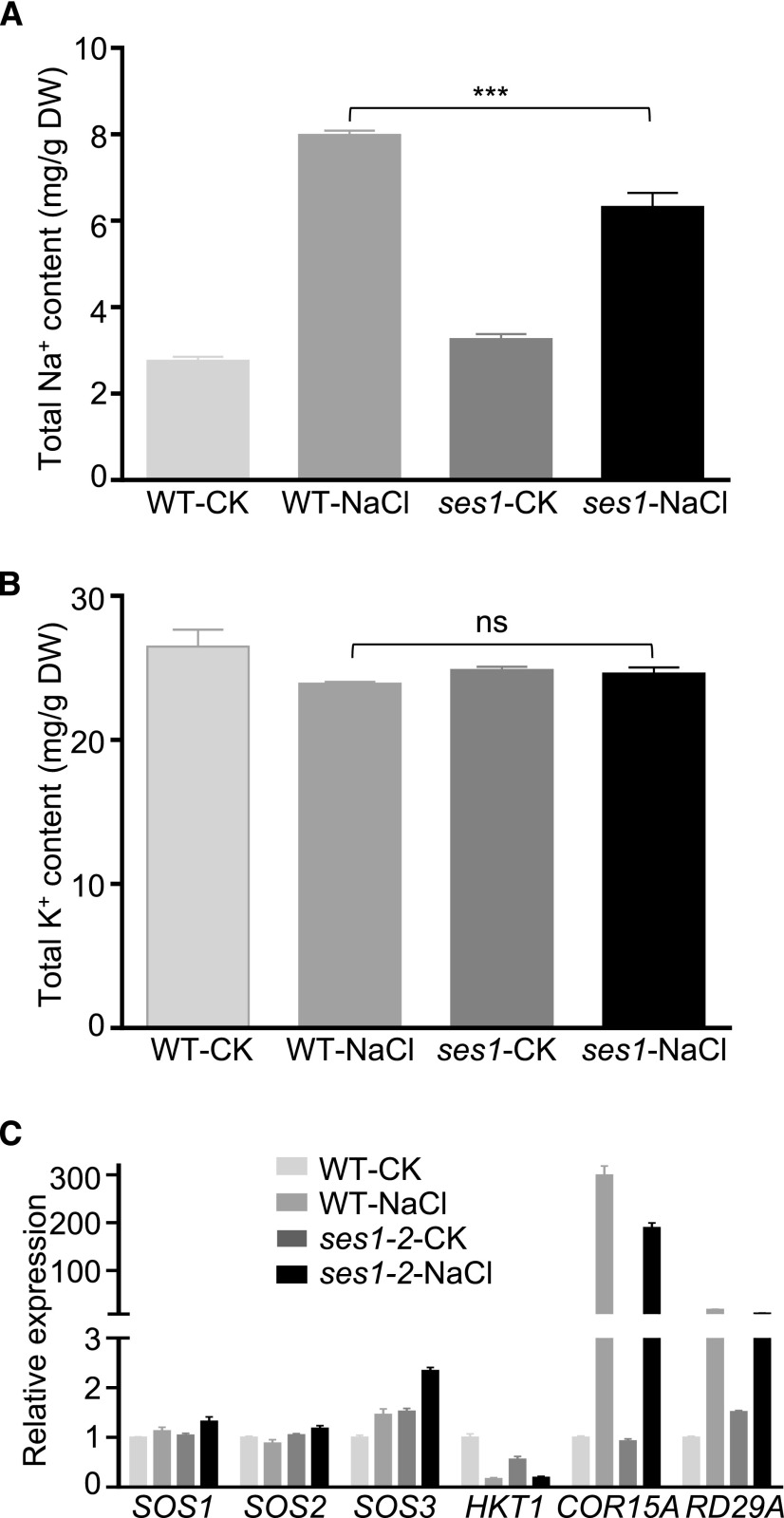
Zhu (2001) reported that more Na+ ions were absorbed under salt stress conditions, which could significantly disrupt normal cellular metabolism. To decipher the underlying salt-sensitive mechanisms of ses1 mutants, we investigated the Na+ and K+ contents in 3-week-old wild-type or ses1-2 plants before and after salt treatment. Under normal conditions, there was no difference in the Na+ or K+ content between ses1-2 and wild-type plants (Fig. 5, A and B). However, after treatment with 200 mm NaCl for 48 h, K+ levels were comparable whereas ses1-2 accumulated less Na+ than the wild-type plants (Fig. 5, A and B). These results suggest that the salt-sensitive phenotype of ses1 is not related to the accumulation of Na+. Additionally, we also detected the expression levels of representative genes involved in salt stress tolerance by RT-qPCR, including SOS1, SOS2, SOS3, HKT1, cold-regulated15A (COR15A), and responsive to desiccation29A (RD29A). None of these marker genes were significantly up-regulated or suppressed in the ses1-2 mutants compared with the wild-type plants (Fig. 5C). These findings reveal that the salt-sensitive trait of ses1 was due neither to the overaccumulation of Na+ nor to the suppression of salt tolerance-associated genes.
Figure 5.
Salt sensitivity of ses1 is not due to the overaccumulation of Na+. A, Na+ content in ses1-2 and wild-type (WT) plants with or without 200 mm NaCl treatment for 48 h. Bars indicate means ± sd of three independent measurements. ***, P < 0.001 (Student’s t test). B, K+ content in ses1-2 and wild-type plants with or without 200 mm NaCl treatment for 48 h. Bars indicate means ± sd of three independent measurements. ns indicates no significant difference (P < 0.05, Student’s t test). DW, Dry weight. C, Expression levels of genes involved in salt stress tolerance in ses1-2 and wild-type plants with or without 200 mm NaCl treatment. The data were normalized against ACTIN7 and UBQ10. The means were calculated from three independent replicates and compared with the no-treatment condition of wild-type plants (WT-CK). Error bars indicate se.
Disruption of SES1 Significantly Aggravates Salt-Induced ER Stress
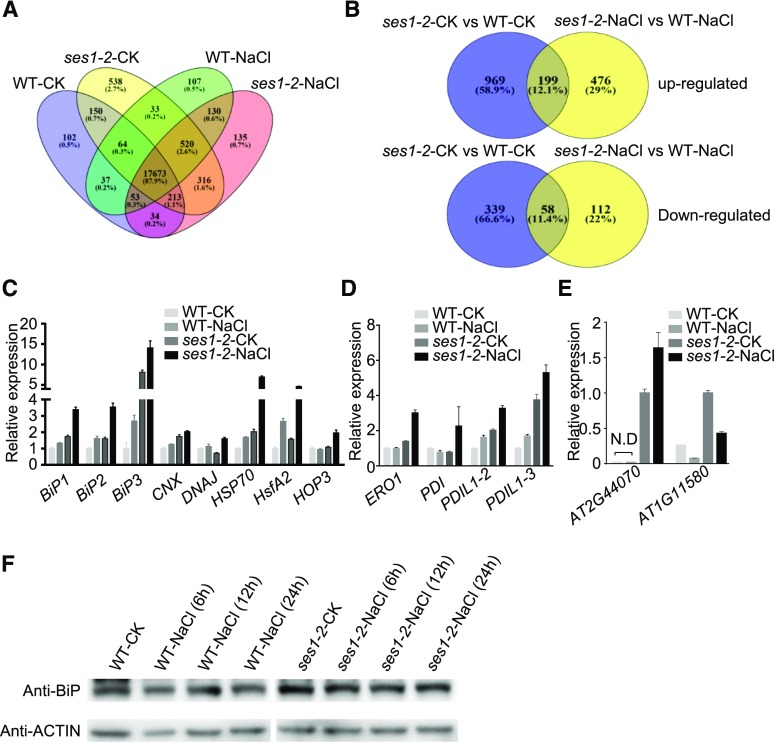
To further explore the mechanisms of SES1 in modulating salt stress tolerance, we performed transcriptome profiling of 10-d-old wild-type and ses1-2 plants before and after salt treatment by RNA sequencing (RNA-seq) with three biological repeats. Out of the total 20,105 transcripts detected by RNA-seq, 17,673 genes were detected in all four materials (Fig. 6A; Supplemental Data Set S1). Under normal conditions, 1,565 genes showed significant differences in expression levels (P < 0.05) between the wild type and ses1-2, including 1,168 up-regulated and 397 down-regulated genes (Fig. 6B; Supplemental Table S1). However, when the plants were treated with 200 mm NaCl for 6 h, 845 genes showed significant differential expression (P < 0.05) between the wild type and ses1-2, including 675 up-regulated and 170 down-regulated genes (Fig. 6B; Supplemental Table S2). Interestingly, the number of up-regulated genes was much higher than that of down-regulated genes, suggesting that SES1 probably plays a negative role in regulating gene expression under salt stress directly or indirectly.
Figure 6.
Altered expression of ER stress-associated genes in ses1-2. A, Venn diagram analysis of genes detected in wild-type (WT) and ses1-2 plants before and after salt treatment. B, Venn diagram analysis of differentially expressed genes in wild-type and ses1-2 plants before and after salt treatment. C, Expression levels of marker genes involved in the UPR pathway. D, Expression levels of marker genes involved in the ERAD pathway. E, Expression levels of marker genes that suppress translation in wild-type and ses1-2 plants before and after salt treatment. The data were normalized against ACTIN7 and UBQ10. The means were calculated from three independent replicates and compared with the no-treatment condition of wild-type (C and D) or ses1-2 (E) plants. Error bars indicate se. N.D, Not detected. F, Analysis of BiP protein levels in wild-type and ses1-2 seedlings before and after salt treatment. Total protein was extract from 10-d-old seedlings.
Gene Ontology (http://www.geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) analyses indicated that genes with increased expression were related closely to ER stress, which include GO0034976 (response to ER stress), GO0006986 (response to UPR), and one Kyoto Encyclopedia of Genes and Genomes term (protein processing in ER). Based on their biological roles, these genes could be classified into four categories: chaperones, ERAD, protein folding, and ER-localized DNAJ proteins (Table 1). The up-regulation of these genes is a canonical characteristic of ER stress responses, suggesting that ses1-2 plants experience more severe ER stress under high-salinity conditions.
Table 1. ER stress-associated genes with higher expression in ses1-2 after salt treatment.
| Gene Ontology Identification | P | Category | Gene List | Description | Log2 Fold Change | Adjusted P |
|---|---|---|---|---|---|---|
| GO:0034976, response to ER stress | 6.12E-08 | AT1G09080 | BiP3 | 3.4198 | 0.0031928 | |
| AT4G12400 | HOP3 | 1.2800 | 0.0023900 | |||
| AT4G25200 | HSP23.6 | 2.8491 | 0.0026638 | |||
| AT5G12020 | HSP17.6 | 2.4297 | 0.00015884 | |||
| Chaperones | AT5G12030 | HSP17.6 | 2.3925 | 9.62E-05 | ||
| AT5G02490 | HSP70 | 2.0938 | 0.010237 | |||
| AT5G51440 | HSP20-like | 1.9370 | 6.49E-08 | |||
| AT1G59860 | HSP20-like | 1.8327 | 0.0042743 | |||
| AT2G29500 | HSP20-like | 1.4372 | 0.0011818 | |||
| AT5G52640 | HSP90 | 1.7769 | 1.33E-07 | |||
| AT1G72280 | ERO1 | 1.2651 | 0.018797 | |||
| ERAD | AT5G38900 | PDI | 1.6651 | 0.0026520 | ||
| AT3G54960 | PDIL1-3 | 1.0417 | 0.0079990 | |||
| AT1G77510 | PDIL1-2 | 0.96635 | 0.021199 | |||
| AT2G26150 | HsfA2 | 2.2342 | 0.042346 | |||
| GO:0006986, response to UPR | 0.000405 | AT4G33050 | EDA39 | 1.5988 | 4.48E-06 | |
| AT1G07000 | EXO70B2 | 1.1158 | 0.0075010 | |||
| AT1G42990 | bZIP60 | 0.91575 | 0.037637 | |||
| AT1G24140 | Unknown | 1.8275 | 4.12E-05 | |||
| Protein folding | AT4G39670 | Unknown | 1.7776 | 9.37E-06 | ||
| AT1G08050 | Unknown | 1.5451 | 0.00071200 | |||
| AT1G19020 | Unknown | 1.5192 | 0.034525 | |||
| AT2G25735 | Unknown | 1.4618 | 0.037124 | |||
| AT5G42010 | Unknown | 1.0913 | 0.042568 | |||
| AT1G16670 | Unknown | 0.99205 | 0.039421 | |||
| DNAJ | AT3G26910 | Unknown | 0.96216 | 0.039261 | ||
| AT3G08970 | ERDJ3A | 1.5309 | 0.00069800 | |||
| AT2G20560 | DNAJ | 1.2315 | 0.0030660 |
When plants undergo ER stress, a series of biological processes are stimulated to alleviate it, such as up-regulating the transcription of UPR genes, accelerating the degradation of misfolded proteins, and suppressing protein biosynthesis. To validate the RNA-seq data and to evaluate the ER stress status of the ses1-2 mutant, we examined the expression levels of ER stress-responsive marker genes by RT-qPCR. Consistent with the RNA-seq data, the genes involved in the UPR and ERAD pathways, including BiP1, BiP2, BiP3, CNX, DNAJ, heat shock protein70 (HSP70), heat shock transcription factor A2 (HsfA2), member 3 of the HSP70-HSP90 organizing protein (HOP3), ER oxidoreductase1 (ERO1), PDI, PDI-like1-2 (PDIL1-2), and PDIL1-3, were up-regulated in the ses1-2 mutants compared with the wild-type plants, particularly under salt stress conditions (Fig. 6, C and D). Similarly, the expression of genes that suppress translation under ER stress conditions, such as AT2G44070 and AT1G11580, also was up-regulated in the ses1-2 mutants (Fig. 6E). In addition, higher protein levels of BiPs were observed in ses1-2 after salt treatment (Fig. 6F). Taken together, we conclude that the disruption of SES1 significantly aggravates salt-induced ER stress.
SES1 Is Involved in ER Stress Responses
RNA-seq and RT-qPCR analyses suggested that SES1 is involved in ER stress responses. To confirm this, we first tested the responses of SES1 to tunicamycin (TM), a potent ER stress chemical inducer. To support our findings, BiP1 and bZIP28 were used as the positive and negative controls, respectively (Koizumi, 1996; Liu et al., 2007b). Figure 7A shows that SES1 was induced remarkably by exogenous TM treatment. We also generated pSES1::GUS transgenic Arabidopsis plants. GUS staining showed that pSES::GUS seedlings were hardly detected under normal conditions, but after TM treatment, GUS activity was expressed strongly (Fig. 7B), indicating that SES1 itself is ER stress responsive.
Figure 7.
SES1 is involved in the ER stress response. A, Response of SES1 expression to the ER stress reagent TM (5 μg mL−1) for the indicated durations. The data were normalized against the expression of ACTIN7 and UBQ10. The means were calculated from three independent replicates and compared with the no-treatment condition (0 h). Error bars indicate se. B, Histochemical staining analysis of pSES1::GUS seedlings before and after 5 μg mL−1 TM treatment. Bar = 1 cm. C, Phenotypes of ses1, wild-type (WT), and overexpression (OE) lines with or without TM treatment. Photographs were taken after growing vertically at 22°C for 7 d. Bar = 1 cm. D, Root length of the seedlings in C. Error bars indicate sd (n = 6). Statistical differences from the wild type were determined by Student’s t test: ***, P < 0.001. E, Growth status of ses1, wild-type, and overexpression lines with or without TM treatment. Photographs were taken after growing at 22°C for 7 d. Bar = 1 cm. F, Cotyledon greening rate of ses1, wild-type, and overexpression lines with or without TM treatment. The bars indicate means ± sd of three independent measurements. Statistical differences from the wild type were determined by Student’s t test: ***, P < 0.001.
Then, we transferred 4-d-old wild-type, ses1-1, ses1-2, OE1, and OE5 seedlings grown vertically on 1/2 MS agar plates onto 1/2 MS agar plates with or without 0.08 μm TM. After growing for an additional 7 d, the ses1 mutants showed increased sensitivity to TM, whereas the root length of SES1-overexpressing lines did not exhibit remarkable differences compared with the wild type (Fig. 7, C and D). In addition, we examined the cotyledon greening rate of wild-type, ses1-1, ses1-2, OE1, and OE5 seedlings under control or TM-supplemented conditions. Compared with wild-type seedlings, a significantly lower greening rate was observed in the ses1-1 and ses1-2 mutants (Fig. 7, E and F). These findings show that SES1 is up-regulated by ER stress and plays vital roles in mitigating ER stress.
SES1 Has Chaperone Activity
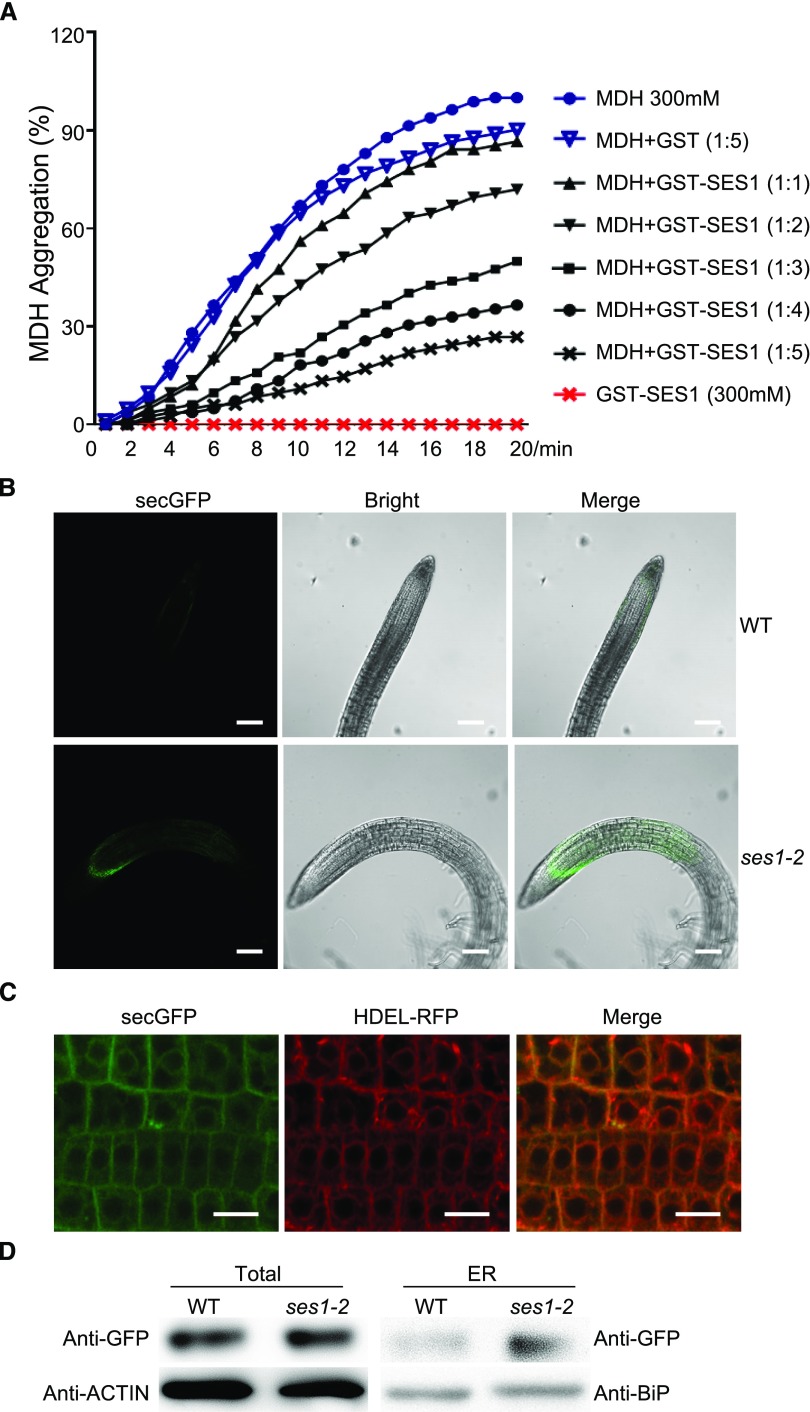
As SES1 was localized to the ER and more severe UPR was detected in the ses1 mutant, we reasoned that SES1 may function in protein folding. To test our hypothesis, we assayed the holdase chaperone activity of SES1 by measuring its capacity to suppress thermal aggregation of the model substrate malate dehydrogenase (MDH; Cha et al., 2009; Li et al., 2015). After codon optimization (Supplemental Fig. S7), SES1 was successfully expressed and purified from Escherichia coli strain BL21. We monitored MDH aggregation by measuring A340 with or without SES1 under thermal denaturing conditions. As shown in Figure 8A, the heat-induced aggregation of MDH was inhibited by GST-SES1 in a dose-dependent manner. Simultaneously, GST-SES1 was heat stable and promoted the stability of MDH under 45°C (Supplemental Fig. S8). Taken together, these results suggest that SES1 acts as a molecular chaperone.
Figure 8.
SES1 has chaperone activity. A, Holdase chaperone activity of SES1 with the model substrate MDH. MDH (300 μm) was incubated without or with recombinant GST-SES1 at 45°C for the indicated times. GST was used as a negative control. The means were calculated from three independent replicates. B, GFP fluorescence of p35S::secGFP in ses1-2 and wild-type (WT) transgenic lines was imaged with a high-resolution laser confocal microscope at 488 nm. Bars = 100 µm. C, GFP and RFP fluorescence of transgenic lines containing secGFP and HDEL-RFP in the ses1-2 background was imaged with a high-resolution laser confocal microscope at 488 and 610 nm, respectively. Bars = 20 µm. D, Analysis of total and ER-localized secGFP in wild-type and ses1-2 seedlings. Protein was extracted from 10-d-old seedlings.
Proteins that do not achieve native conformations can accumulate in the ER (Liu and Howell, 2010b). To further illustrate the chaperone activity of SES1, we employed a marker secretory protein, secGFP. In accordance with previous reports, when secGFP was expressed in the wild-type background, only very dim fluorescence was detected, as most secGFP was secreted to the apoplast (Batoko et al., 2000). On the contrary, much stronger GFP signals were observed in ses1-2 and were emitted mainly from the ER (Fig. 8, B and C). We also examined the protein levels of secGFP from total extracts and the ER. Consistent with the fluorescence data, a larger portion of secGFP was trapped in the ER of ses1-2 (Fig. 8D). These results support that SES1 functions as an important chaperone in the ER and that the protein-folding capacity is weakened significantly in ses1.
Transcription Factor bZIP17 Targets and Activates SES1 Expression
Previous studies have shown that bZIP17, a vital ER stress sensor, also plays important roles in salt stress tolerance (Liu et al., 2007a, 2008; Henriquez-Valencia et al., 2015). It binds to the ER stress-response element (ERSE; CCAATN9CCACG), thereby activating the transcription of target genes (Liu and Howell, 2010a). An ERSEL (ERSE-Like) element (CCAATN9CCACT) is present within the promoter region of SES1. Given that SES1 is correlated strongly with both the ER and salt stress tolerance, we deduced that bZIP17 may be the upstream transcriptional activator of SES1.
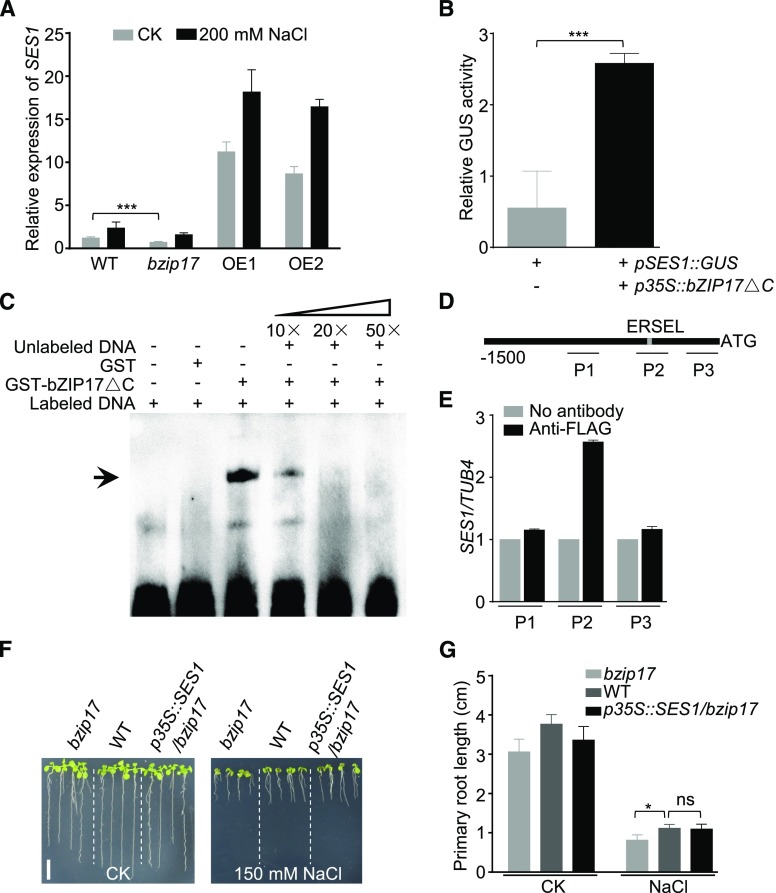
By detecting the expression levels of SES1 in bzip17 knockout mutants and bZIP17 overexpression lines, we found that SES1 expression was significantly lower in bzip17 and much higher in bZIP17 overexpression lines, particularly under salt stress conditions (Fig. 9A). To further investigate whether bZIP17 regulates SES1 directly, we conducted transient expression experiments using Nicotiana benthamiana leaves. When p35S::bZIP17ΔC (the active form of bZIP17 with a C-terminal deletion of 358 amino acids) and pSES1::GUS were coexpressed in N. benthamiana leaves, GUS activity increased remarkably (Fig. 9B). These results demonstrate that bZIP17 positively modulates the expression of SES1.
Figure 9.
Transcription factor bZIP17 targets the promoter of SES1. A, Expression of SES1 in wild-type (WT), bzip17, and two bZIP17ΔC overexpression lines (OE1 and OE2) with or without 200 mm NaCl treatment. Error bars indicate sd (n = 6). Statistical significance was determined by Student’s t test: ***, P < 0.001. B, GUS activity measurement in N. benthamiana leaves after transient expression of p35S::bZIP17ΔC. pSES1::GUS was used as an internal control. Error bars indicate sd (n = 6). Statistical significance was determined by Student’s t test: ***, P < 0.001. C, DNA-binding activity of bZIP17ΔC to the ERSEL element. The positions of bands is indicated with the arrow. D, Diagram of the SES1 promoter region. The gray rectangle represents the position of ERSEL (CCAATN9CCACT). P1, P2, and P3 indicate three fragments used for ChIP-PCR analysis. E, bZIP17ΔC combination with DNA fragments in the ChIP assay. The bars indicate means ± sd of three independent measurements. F, Phenotypes of bzip17, the wild type, and p35S::SES1/bzip17 (overexpression of SES1 in the bzip17 mutant) with or without 150 mm NaCl treatment. Photographs were taken after growing vertically at 22°C for 7 d. Bar = 1 cm. G, Root length of the seedlings in F. Error bars indicate sd (n = 6). Statistical significance was determined by Student’s t test. *, P < 0.05. ns indicates no significant differences (P < 0.05, Student’s t test).
To confirm the direct binding of bZIP17 to the SES1 promoter, an electrophoretic mobility shift assay (EMSA) was performed with the biotin-labeled SES1 promoter DNA fragment containing the ERSEL cis-element. The constitutively active form of bZIP17 (bZIP17ΔC), which was fused to a GST tag, was expressed in E. coli and purified from the soluble fraction. When GST-bZIP17ΔC was incubated with the biotin-labeled DNA probe, a clear band shift was detected, reflecting the formation of the respective complex (Fig. 9C). In addition, the binding activity decreased gradually with the addition of unlabeled competitors (Fig. 9C). As a negative control, GST protein alone did not bind to the probes (Fig. 9C). Furthermore, we performed chromatin immunoprecipitation (ChIP) using a FLAG antibody to assess the DNA-binding activity of bZIP17ΔC-FLAG to the SES1 promoter in vivo. We designed three fragments (P1–P3) within the SES1 promoter region (Fig. 9D). The ChIP-qPCR results indicated that bZIP17 bound specifically to the fragment containing the ERSEL element (Fig. 9E). Moreover, the salt sensitivity of bzip17 was largely rescued by enhancing the expression level of SES1 (Fig. 9, F and G). These findings demonstrate that bZIP17 binds directly to the ERSEL element within the SES1 promoter region and activates its expression.
DISCUSSION
Although remarkable advances have been made during the last decades in understanding the salt tolerance mechanisms in model plants such as Arabidopsis, numerous functional genes that are involved in salt tolerance remain uncharacterized. We aimed at screening and characterizing unknown players involved in salt tolerance. Here, we identified and analyzed a salt tolerance-associated gene, SES1. Our data demonstrate that SES1 encodes an ER-localized chaperone and protects plants from salt stress by alleviating salt-induced ER stress.
Mutation in SES1 caused hypersensitivity to salt stress (Fig. 1). The T-DNA knockout mutant ses1-2 is more vulnerable than the point mutation mutant ses1-1 (Fig. 2), suggesting that the mutated SES1 in ses1-1 remains weakly bioactive. Nevertheless, the salt-sensitive phenotype was not due to the overaccumulation of Na+ in the ses1 mutants (Fig. 5). By RNA-seq and subsequent experimental validation, we found that ER stress responses are more evident in the ses1 mutants compared with the wild-type plants (Fig. 6). When treated with TM, the ses1 mutants also displayed increased sensitivity (Fig. 7), suggesting that ses1 mutants were more largely affected by severe ER stress. Prolonged ER stress responses led to the inhibition of plant growth (Tao et al., 2016). Similarly, ses1-1 and ses1-2 exhibited stunted growth under long-day conditions (Supplemental Fig. S9). However, the exogenous application of tauroursodeoxycholic acid (TUDCA), a compound that relieves ER stress in plants, did not restore the salt-sensitive phenotype of ses1 (Supplemental Fig. S10), revealing that ER stress responses in ses1 are too severe to be rescued by TUDCA.
As a molecular chaperone, the most puzzling and interesting point is which proteins SES1 helps fold. We used yeast two-hybrid and coimmunoprecipitation assays to analyze the targets of SES1. However, no substrate protein was identified, probably due to the transient nature of the interactions between chaperone and target proteins. Given that malformed proteins accumulate in the ER, a comprehensive ER proteomic study comparing ses1-2 and wild-type plants seems a promising way to answer this question. On the other hand, we could not thoroughly exclude the possibility that SES1 also might be involved in biological process other than ER stress responses.
Expression analysis with bzip17 mutants and bZIP17 overexpression plants showed that bZIP17 up-regulates the expression of SES1 (Fig. 9), which was confirmed further by transient activation assays, EMSA, and ChIP analysis (Fig. 9). In addition to bZIP17, two other well-known transcription factors, bZIP28 and bZIP60, also serve as ER stress sensors in Arabidopsis (Iwata and Koizumi, 2005; Liu et al., 2007b). We found that SES1 also was down-regulated in bzip28, bzip60, or the bzip28/60 double mutant after TM treatment (Supplemental Fig. S11), suggesting that the upstream transcription factor of SES1 is not limited to bZIP17. However, no increased salt sensitivity was observed in bzip28, bzip60, or bzip28/60 (Supplemental Fig. S12). The functional redundancy between bZIP17 and bZIP28 in modulating salt stress tolerance is probable, as they show high sequence similarity and strong interactions (Deppmann et al., 2004; Liu and Howell, 2010a). But we could not obtain the bzip17/28 double knockout mutant (SALK_104326 for bzip17 and SALK_132285 for bzip28), suggesting that bZIP17 and bZIP28 are vital for normal growth and development.
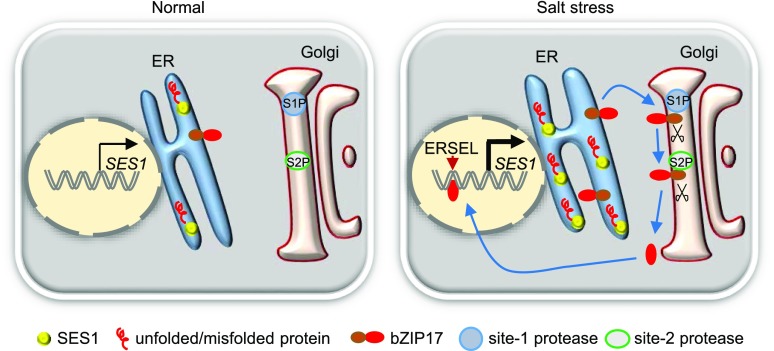
Based on our findings and previous reports, we suggest a preliminary working model for SES1 (Fig. 10). Under normal conditions, protein folding in the ER is accurately balanced. Without ER stress, the ER stress sensor bZIP17 is anchored to the ER membrane and its target gene SES1 is expressed weakly. However, unfolded and/or misfolded proteins accumulate in the ER lumen under salt stress conditions, triggering ER stress and subsequent ER stress responses. To reinforce the protein-folding capability, bZIP17 releases from the ER. After a two-step cleavage, truncated bZIP17 enters the nucleus and then activates the expression of SES1 via binding to the ERSEL cis-element. SES1 then chaperones protein folding and alleviates ER stress under salt stress conditions.
Figure 10.
Simplified working model of SES1 in Arabidopsis.
CONCLUSION
We identified and characterized a salt-sensitive mutant, ses1, in Arabidopsis. SES1 localizes to the ER and can alleviate salt-induced ER stress processes by nursing nascent protein folding. The ER stress sensor bZIP17 activates the expression of SES1 via directly binding to the ERSEL cis-element.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants used in this study were in the Columbia-0 background. Seeds were surface sterilized by soaking in 70% ethanol for 5 min and 10% NaClO for 10 min and then washed six times with sterilized water. The sterile seeds were plated on 1/2 MS medium (Sigma-Aldrich; pH 5.7) containing 1.5% (w/v) Suc. The plates were then kept in the dark at least 48 h at 4°C for stratification before being placed in a growth room for germination. Approximately 7-d-old seedlings were transferred to soil and grown to maturity at 22°C in a greenhouse under long days with a 16-h-light/8-h-dark cycle unless mentioned otherwise. For salt or TM treatment, 10-d-old seedlings were incubated on 1/2 MS agar plates supplemented with 200 mm NaCl or 5 μg mL−1 TM for the indicated periods of time.
The T-DNA insertional mutants ses1-2 (GABI_944F02) and bzip17 (SALK_104326) were obtained from the GABI-KAT center (http://www.GABI-Kat.de) and the Arabidopsis Biological Resource Center (http://www.arabidopsis.org), respectively. Homozygous T-DNA insertion mutants were identified using PCR-based genotyping with a T-DNA-specific primer and gene-specific primers. All primers used in this study are listed in Supplemental Table S3.
Mutant Isolation
Approximately 63,000 EMS-mutagenized M2 populations were screened for salt-tolerant mutants. After growing 4 d on 1/2 MS agar plates, wild-type and EMS-mutated seedlings were transferred to plates containing 125 mm NaCl and grown vertically for another 7 d. Putative salt-sensitive or -resistant mutants then were transferred to soil. Seeds from the putative mutants were screened again using the same method. For root length assays, ImageJ software was used to measure primary root length, and at least six to eight biological replicates were performed.
Seed Germination and Salt Treatment during the Adult Stage
For the germination assay, at least 100 seeds of each genotype were sterilized and sown on 1/2 MS agar plates supplemented with or without salt. Germination was defined as the first sign of radicle tip emergence, and the germination results were calculated based on at least three independent experiments.
Wild-type and ses1 plants were grown in a greenhouse under short days with an 8-h-light/16-h-dark cycle for 3 weeks and then irrigated with 200 mm NaCl for 2 weeks. One week after recovering, surviving plants were counted.
Chlorophyll Content Measurement
Plant leaves were collected, and their fresh weight was determined. Total chlorophyll was extracted in 80% (v/v) acetone at 25°C in the dark for 24 h, and the concentration of chlorophyll was determined according to Komatsu et al. (2010).
Genetic Mapping and Cloning of the SES1 Gene
To map the ses1 mutation, the ses1 mutant was crossed with Landsberg erecta plants. A total of 1,200 F2 plants exhibiting the salt-sensitive phenotype were selected as a mapping population. Genomic DNA from these F2 plants was extracted and used for PCR-based mapping using simple sequence length polymorphism based on insertions/deletions identified from the Arabidopsis Mapping Platform (http://amp.genomics.org.cn/). Genomic DNA fragments corresponding to candidate genes were PCR amplified from ses1 and used in the DNA sequencing analysis to identify the mutation.
Plasmid Construction and Plant Transformation
For the complementation assay, a 3.8-kb genomic fragment comprising the SES1 promoter, coding region, and 3′ untranslated region was PCR amplified from the genomic DNA of wild-type plants. The PCR product was ligated subsequently into pBI121 to generate the pSES1::SES1 construct. Using primers SES1-OE-F and SES1-OE-R, we amplified the overexpression fragment and ligated it into the binary vector pBI121. To generate the pSES1::GUS fusion, a 1.5-kb genomic fragment upstream of the ATG start codon was PCR amplified using Pro-SES1-F and Pro-SES1-R primers. The amplified fragment was fused with the GUS reporter gene in the binary vector pBI121. The SES1 cDNA fragment lacking a stop codon was PCR amplified using primers SES1-GFP-F and SES1-GFP-R and fused with GFP to generate the expression vector p35S::SES1-GFP. After verification by DNA sequencing, the binary vector was introduced into Agrobacterium tumefaciens strain GV3101. The agrobacteria carrying different constructs were used to transform wild-type or ses1 plants using the floral dip method (Clough and Bent, 1998). The corresponding T2 transgenic seedlings that segregated at a ratio of 3:1 (resistant:sensitive) were selected to propagate T3 individuals.
Na+ and K+ Content Measurement
Three-week-old seedlings were treated with 200 mm NaCl for 48 h and harvested, dried for 48 h at 80°C, and then ground to powder. Then, 100 mg of tissue powder was digested in concentrated nitric acid, hydrochloric acid, and hydrogen peroxide for 30 min in a microwave 3000 digestion system (Anton Paar) for element extraction. Na+ and K+ concentrations were determined using a flame atomic absorption spectrometer (Analytik Jena).
RT-qPCR Analysis
To assay relative expression levels, RT-qPCR analysis was performed with the RNA samples isolated from 10-d-old seedlings before and after treatment with 200 mm NaCl. Total RNA was extracted with TRIzol Reagent (Invitrogen) followed by treatment with RNase-free DNase I (Takara) at 42°C for 2 min. The treated RNA samples (1 μg each) were used as templates for first-strand cDNA synthesis. A SYBR Green real-time PCR master mix (Takara) and a CFX96 real-time system detector (Bio-Rad) were used for real-time PCR. The data are presented after normalizing to the reference genes ACTIN7 (AT5G09810) and UBQ10 (AT4G05320). Three biological replicates under similar conditions were performed for each experiment.
Histochemical GUS Staining, Fluorometric GUS Assay, and Analysis of Subcellular Localization
The histochemical detection of GUS staining and fluorometric GUS assays were performed as described previously (Jefferson et al., 1987). GUS activity was measured with 4-methylumbelliferyl-β-d-glucuronide as a substrate with an F-4500 fluorescence spectrofluorometer (Hitachi). The standard curves were prepared with 4-methylumbelliferone. Average GUS activity was obtained from at least five independent transformants, and each assay was repeated three times.
The roots of 5-d-old transgenic plants expressing p35S::SES1-GFP and p35S::HDEL-RFP were visualized using a high-resolution laser confocal microscope (LSM880; Zeiss).
RNA-seq Analysis
Ten-day-old wild-type and ses1-2 seedlings were treated with 200 mm NaCl for 6 h and then harvested for RNA extraction. The transcriptome analysis was performed by Novogene with three biological repeats. Library construction was performed according to Illumina standard instructions. Reads were aligned to the Arabidopsis genome using TopHat2 (Langmead et al., 2009). Genes with adjusted P < 0.05 were considered to be differentially expressed.
Holdase Chaperone Assay
Recombinant GST-SES1 was expressed in Escherichia coli strain BL21(DE3) and purified using a GST protein purification kit (CWBIO). MDH (EC 1.1.1.37) was bought from Solarbio (M8400). The holdase chaperone activity of GST-SES1 was assayed by measuring its capacity to suppress the thermal aggregation of MDH (Cha et al., 2009; Li et al., 2015). MDH aggregation was monitored in reaction buffer (40 mm HEPES, pH 7.5) by measuring A340 using a UV-visible spectrophotometer attached to a thermostatic cell holder assembly at 45°C. In each set, recombinant GST-SES1 was prepared independently.
EMSA
bZIP17ΔC was cloned using primers pGEX-bZIP17-F and pGEX-bZIP17-R and ligated into the expression vector pGEX4T-3. The recombinant plasmid was transformed into E. coli strain BL21(DE3), and recombinant bZIP17ΔC was purified using a GST protein purification kit (CWBIO). EMSA was conducted using a light shift chemiluminescent EMSA kit (Thermo Fisher Scientific) following the manufacturer’s protocol. The labeled probe was synthesized by Sangon Biotech.
ChIP-qPCR Assay
The ChIP-qPCR assay was performed as described previously (Cheng et al., 2014). Five-day-old transgenic seedlings (p35S::bZIP17ΔC-FLAG) were harvested and used for ChIP assays. Mouse monoclonal FLAG antibody was used.
Accession Numbers
Sequence data for the genes described in this article can be found in the Arabidopsis TAIR database (https://www.arabidopsis.org/index.jsp) under the following accession numbers: SES1 (AT4G29520), bZIP17 (AT2G40950), ERO1 (AT1G72280), PDI (AT5G38900), PDIL1-3 (AT3G549690), PDIL1-2 (AT1G77510), BiP1 (AT5G28540), BiP2 (AT5G42020), BiP3 (AT1G09080), CNX (AT5G61790), DNAJ (AT2G20560), HSP70 (AT5G02490), HsfA2 (AT2G26150), HOP3 (AT4G12400), SOS1 (AT2G01980), SOS2 (AT5G35410), SOS3 (AT5G24270), HKT1 (AT4G10310), RD29A (AT5G52310), COR15A (AT2G42540), ACTIN7 (AT5G09810), UBQ10 (AT4G05320), EF-1α (AT5G60390), and TUB4 (AT5G44340).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Germination rate and chlorophyll content of wild-type and ses1 plants.
Supplemental Figure S2. SES1 has a conserved saposin B domain.
Supplemental Figure S3. Phenotype analysis of SES1-overexpressing lines.
Supplemental Figure S4. SES1 is required for both ionic and osmotic stress tolerance.
Supplemental Figure S5. SES1 was induced by NaCl treatment in different tissues.
Supplemental Figure S6. Subcellular localization analysis of SES1.
Supplemental Figure S7. Coding sequence of SES1 without the N-terminal signal peptide after codon optimization.
Supplemental Figure S8. Chaperone activity of recombinant SES1.
Supplemental Figure S9. ses1 mutants exhibited stunted growth under long-day conditions.
Supplemental Figure S10. TUDCA fails to restore the salt-sensitive phenotype of ses1.
Supplemental Figure S11. SES1 expression levels in bzip17, bzip28, bzip60, and bzip28/60 mutants before and after TM treatment.
Supplemental Figure S12. Responses of bzip28, bzip60, and bzip28/60 to salt treatment.
Supplemental Table S1. Differentially expressed genes between the wild type and ses1-2 before treatment.
Supplemental Table S2. Differentially expressed genes between the wild type and ses1-2 after salt treatment.
Supplemental Table S3. Primer and enzyme site sequences used in this study.
Supplemental Data Set S1. Genes detected in the wild type and ses1-2 before and after salt treatment.
Acknowledgments
We thank Dr. Jianxiang Liu (Institute of Plant Biology, School of Life Sciences, Fudan University) for providing bzip28, bzip60, and bzip28/60 mutant seeds.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31570271 and 31771878) and the Natural Science Foundation of Shandong Province (grant no. ZR2016CM22).
References
- An R, Chen QJ, Chai MF, Lu PL, Su Z, Qin ZX, Chen J, Wang XC (2007) AtNHX8, a member of the monovalent cation:proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li/H antiporter. Plant J 49: 718–728 [DOI] [PubMed] [Google Scholar]
- Barragán V, Leidi EO, Andrés Z, Rubio L, De Luca A, Fernández JA, Cubero B, Pardo JM (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24: 1127–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I (2000) A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthomieu P, Conéjéro G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, et al. (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JY, Jung MH, Ermawati N, Su’udi M, Rho GJ, Han CD, Lee KH, Son D (2009) Functional characterization of orchardgrass endoplasmic reticulum-resident Hsp90 (DgHsp90) as a chaperone and an ATPase. Plant Physiol Biochem 47: 859–866 [DOI] [PubMed] [Google Scholar]
- Cheng ZJ, Zhao XY, Shao XX, Wang F, Zhou C, Liu YG, Zhang Y, Zhang XS (2014) Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26: 1053–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colebrook EH, Thomas SG, Phillips AL, Hedden P (2014) The role of gibberellin signalling in plant responses to abiotic stress. J Exp Biol 217: 67–75 [DOI] [PubMed] [Google Scholar]
- Cox JS, Walter P (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87: 391–404 [DOI] [PubMed] [Google Scholar]
- Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, Howell SH (2011) Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci USA 108: 7247–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Howell SH (2013) Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int J Mol Sci 14: 8188–8212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppmann CD, Acharya A, Rishi V, Wobbes B, Smeekens S, Taparowsky EJ, Vinson C (2004) Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: a comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res 32: 3435–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Guo Y, Cuin TA, Qiu Q, Song C, Kristiansen KA, Bych K, Schulz A, Shabala S, Schumaker KS, et al. (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19: 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Brandizzi F, Benning C, Larkin RM (2008) A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 16398–16403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner BM, Walter P (2011) Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333: 1891–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR (2001) Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci USA 98: 11444–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriquez-Valencia C, Moreno AA, Sandoval-Ibañez O, Mitina I, Blanco-Herrera F, Cifuentes-Esquivel N, Orellana A (2015) bZIP17 and bZIP60 regulate the expression of BiP3 and other salt stress responsive genes in an UPR-independent manner in Arabidopsis thaliana. J Cell Biochem 116: 1638–1645 [DOI] [PubMed] [Google Scholar]
- Hetz C, Martinon F, Rodriguez D, Glimcher LH (2011) The unfolded protein response: integrating stress signals through the stress sensor IRE1α. Physiol Rev 91: 1219–1243 [DOI] [PubMed] [Google Scholar]
- Hollien J. (2013) Evolution of the unfolded protein response. Biochim Biophys Acta 1833: 2458–2463 [DOI] [PubMed] [Google Scholar]
- Howell SH. (2013) Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol 64: 477–499 [DOI] [PubMed] [Google Scholar]
- Iwata Y, Koizumi N (2005) An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc Natl Acad Sci USA 102: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Ashida M, Hasegawa C, Tabara K, Mishiba KI, Koizumi N (2017) Activation of the Arabidopsis membrane-bound transcription factor bZIP28 is mediated by site-2 protease, but not site-1 protease. Plant J 91: 408–415 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Wang C, Huang J, Yang G, Wu C, Zheng C (2015) SCF E3 ligase PP2-B11 plays a positive role in response to salt stress in Arabidopsis. J Exp Bot 66: 4683–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Daniell H (2014) Expression of γ-tocopherol methyltransferase in chloroplasts results in massive proliferation of the inner envelope membrane and decreases susceptibility to salt and metal-induced oxidative stresses by reducing reactive oxygen species. Plant Biotechnol J 12: 1274–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Frank J, Kang CH, Kajiura H, Vikram M, Ueda A, Kim S, Bahk JD, Triplett B, Fujiyama K, et al. (2008) Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci USA 105: 5933–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. (2015) Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci 20: 219–229 [DOI] [PubMed] [Google Scholar]
- Keyster M, Klein A, Ludidi N (2012) Caspase-like enzymatic activity and the ascorbate-glutathione cycle participate in salt stress tolerance of maize conferred by exogenously applied nitric oxide. Plant Signal Behav 7: 349–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N. (1996) Isolation and responses to stress of a gene that encodes a luminal binding protein in Arabidopsis thaliana. Plant Cell Physiol 37: 862–865 [DOI] [PubMed] [Google Scholar]
- Komatsu T, Kawaide H, Saito C, Yamagami A, Shimada S, Nakazawa M, Matsui M, Nakano A, Tsujimoto M, Natsume M, et al. (2010) The chloroplast protein BPG2 functions in brassinosteroid-mediated post-transcriptional accumulation of chloroplast rRNA. Plant J 61: 409–422 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu J, Wang G, Cha JY, Li G, Chen S, Li Z, Guo J, Zhang C, Yang Y, et al. (2015) A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27: 908–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wei H, Liu L, Yang X, Zhang X, Xie Q (2017) Unfolded protein response activation compensates endoplasmic reticulum-associated degradation deficiency in Arabidopsis. J Integr Plant Biol 59: 506–521 [DOI] [PubMed] [Google Scholar]
- Liu JX, Howell SH (2010a) bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH (2010b) Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH (2007a) Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J 51: 897–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH (2007b) An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19: 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Howell SH (2008) Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ 31: 1735–1743 [DOI] [PubMed] [Google Scholar]
- Liu L, Cui F, Li Q, Yin B, Zhang H, Lin B, Wu Y, Xia R, Tang S, Xie Q (2011) The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res 21: 957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez IM, Chrispeels MJ (2003) Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Atienza J, Jiang X, Garciadeblas B, Mendoza I, Zhu JK, Pardo JM, Quintero FJ (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol 143: 1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, Chen Y, Brandizzi F, Dong X, Orellana A, et al. (2012) IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE 7: e31944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K. (2009) Signalling pathways in the unfolded protein response: development from yeast to mammals. J Biochem 146: 743–750 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Nagashima Y, Mishiba K, Suzuki E, Shimada Y, Iwata Y, Koizumi N (2011) Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D, Kimata Y, Kohno K, Iwawaki T (2009) Activation of mammalian IRE1α upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp Cell Res 315: 2496–2504 [DOI] [PubMed] [Google Scholar]
- Oikawa D, Tokuda M, Hosoda A, Iwawaki T (2010) Identification of a consensus element recognized and cleaved by IRE1 α. Nucleic Acids Res 38: 6265–6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D, Kitamura A, Kinjo M, Iwawaki T (2012) Direct association of unfolded proteins with mammalian ER stress sensor, IRE1β. PLoS ONE 7: e51290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovečka M, Takáč T, Komis G, Vadovič P, Bekešová S, Doskočilová A, Šamajová V, Luptovčiak I, Samajová O, Schweighofer A, et al. (2014) Salt-induced subcellular kinase relocation and seedling susceptibility caused by overexpression of Medicago SIMKK in Arabidopsis. J Exp Bot 65: 2335–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parida AK, Das AB, Mittra B (2004) Effects of salt on growth, ion accumulation, photosynthesis and leaf anatomy of the mangrove Bruguiera parviflora. Trees (Berl) 18: 167–174 [Google Scholar]
- Pincus D, Chevalier MW, Aragón T, van Anken E, Vidal SE, El-Samad H, Walter P (2010) BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol 8: e1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues NF, Fonseca GCD, Kulcheski FR, Margis R (2017) Salt stress affects mRNA editing in soybean chloroplasts. Genet Mol Biol (Suppl 1) 40: 200–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Ryu H, Cho YG (2015) Plant hormones in salt stress tolerance. J Plant Biol 58: 147–155 [Google Scholar]
- Schröder M, Kaufman RJ (2005) ER stress and the unfolded protein response. Mutat Res 569: 29–63 [DOI] [PubMed] [Google Scholar]
- Senn ME, Rubio F, Bañuelos MA, Rodríguez-Navarro A (2001) Comparative functional features of plant potassium HvHAK1 and HvHAK2 transporters. J Biol Chem 276: 44563–44569 [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Walter P (1997) The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90: 1031–1039 [DOI] [PubMed] [Google Scholar]
- Srivastava R, Deng Y, Shah S, Rao AG, Howell SH (2013) BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell 25: 1416–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Deng Y, Howell SH (2014) Stress sensing in plants by an ER stress sensor/transducer, bZIP28. Front Plant Sci 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima H, Iwata Y, Iwano M, Takayama S, Koizumi N (2008) Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem Biophys Res Commun 374: 242–247 [DOI] [PubMed] [Google Scholar]
- Tang RJ, Liu H, Bao Y, Lv QD, Yang L, Zhang HX (2010) The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress. Plant Mol Biol 74: 367–380 [DOI] [PubMed] [Google Scholar]
- Tao YK, Yu PL, Bai YP, Yan ST, Zhao SP, Zhang GQ (2016) Role of PERK/eIF2α/CHOP endoplasmic reticulum stress pathway in oxidized low-density lipoprotein mediated induction of endothelial apoptosis. Biomed Environ Sci 29: 868–876 [DOI] [PubMed] [Google Scholar]
- Urade R. (2007) Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J 274: 1152–1171 [DOI] [PubMed] [Google Scholar]
- Vitale A, Boston RS (2008) Endoplasmic reticulum quality control and the unfolded protein response: insights from plants. Traffic 9: 1581–1588 [DOI] [PubMed] [Google Scholar]
- Wan S, Jiang L (2016) Endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) in plants. Protoplasma 253: 753–764 [DOI] [PubMed] [Google Scholar]
- Wu H, Ng BS, Thibault G (2014) Endoplasmic reticulum stress response in yeast and humans. Biosci Rep 34: 321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Qin Y, Xie C, Zhao F, Zhao J, Liu D, Chen S, Fuglsang AT, Palmgren MG, Schumaker KS, et al. (2010) The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22: 1313–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Dickman MB, Whitham SA, Payton M, Verchot J (2011) The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol 156: 741–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, Liu J, Xiao Y, Liang Y, Zhang W (2010) Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol 188: 762–773 [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang A (2012) Virus-induced ER stress and the unfolded protein response. Front Plant Sci 3: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Pan Z, Zhang Y, Qu X, Zhang Y, Yang Y, Jiang X, Huang S, Yuan M, Schumaker KS, et al. (2013) The Actin-Related Protein2/3 complex regulates mitochondrial-associated calcium signaling during salt stress in Arabidopsis. Plant Cell 25: 4544–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2001) Plant salt tolerance. Trends Plant Sci 6: 66–71 [DOI] [PubMed] [Google Scholar]
- Zhu JK. (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6: 441–445 [DOI] [PubMed] [Google Scholar]